Tesio at al. identify a novel pathway controlled by the tumor suppressor and deubiquitinase cylindromatosis (CYLD), which is involved in the regulation of hematopoietic stem cell quiescence and repopulation potential.
Abstract
The status of long-term quiescence and dormancy guarantees the integrity of hematopoietic stem cells (HSCs) during adult homeostasis. However the molecular mechanisms regulating HSC dormancy remain poorly understood. Here we show that cylindromatosis (CYLD), a tumor suppressor gene and negative regulator of NF-κB signaling with deubiquitinase activity, is highly expressed in label-retaining dormant HSCs (dHSCs). Moreover, Cre-mediated conditional elimination of the catalytic domain of CYLD induced dHSCs to exit quiescence and abrogated their repopulation and self-renewal potential. This phenotype is dependent on the interactions between CYLD and its substrate TRAF2 (tumor necrosis factor–associated factor 2). HSCs expressing a mutant CYLD with an intact catalytic domain, but unable to bind TRAF2, showed the same HSC phenotype. Unexpectedly, the robust cycling of HSCs lacking functional CYLD–TRAF2 interactions was not elicited by increased NF-κB signaling, but instead by increased activation of the p38MAPK pathway. Pharmacological inhibition of p38MAPK rescued the phenotype of CYLD loss, identifying the CYLD–TRAF2–p38MAPK pathway as a novel important regulator of HSC function restricting HSC cycling and promoting dormancy.
Hematopoietic stem cells (HSCs) are defined by their ability to both life-long self-renew and give rise to all mature blood cell lineages. A tight balance between self-renewal and differentiation is crucial to maintain the integrity of the entire hematopoietic tissue, preventing exhaustion of the stem cell pool or development of hematopoietic malignancies such as leukemia. In the healthy murine BM, the highest self-renewal capacity has been attributed to dormant HSCs (dHSCs; Wilson et al., 2008; Foudi et al., 2009; Takizawa et al., 2011). These cells are long-term label retaining and are characterized by a deep long-term quiescent state, as in the absence of stress they divide only five times per lifetime. Although during homeostasis dHSCs constitute a silent stem cell reservoir, during stress situations such as infection or chemotherapy, they enter the cell cycle and start to proliferate, thereby replenishing the hematopoietic system of the cells that have been damaged or lost during injury (Wilson et al., 2008). Despite their important role at the helm of the hematopoietic hierarchy, very limited knowledge is available with respect to the molecular mechanism of the complex function of dHSCs (Trumpp et al., 2010).
Ubiquitination is a posttranslational process whereby the highly conserved protein ubiquitin is covalently attached to target proteins through a multistep process involving ubiquitin-activating or -conjugating enzymes and ubiquitin ligases. The ubiquitin coupling to substrate proteins occurs on seven different lysine residues (K6, K11, K27, K29, K33, K48, or K63) and may involve a single ubiquitin molecule or a chain of them (Peng et al., 2003). Among the seven linkage types, K48, K11, and K63 are the most abundant ones. Lys11-linked polyubiquitin chains play important roles in the control of the cell cycle (Bremm and Komander, 2011), whereas lysine-48–linked polyubiquitin chains affect the stability of the substrate proteins, marking them for proteasomal degradation. Lysine-63–linked polyubiquitin chains have signaling functions instead, and they have been implicated in the control of DNA repair (Hofmann and Pickart, 1999), activation of the IκB kinase complex IKK (Deng et al., 2000), the IL-1/Toll-like receptor, and the NF-κB pathways (Chen, 2005; Conze et al., 2008).
Ubiquitination is a reversible process and is antagonized by deubiquitinases (DUBs), enzymes hydrolyzing polyubiquitin chains. One the most studied DUBs, both in human patients and in mouse models, is cylindromatosis (CYLD; Bignell et al., 2000). The C-terminal catalytic domain of this protein possesses unique structural features that confer the enzyme specificity for Lys63-linked ubiquitin chains (Komander et al., 2008). This specific DUB activity is strictly linked to a tumor suppressor function. Mutations inactivating the C-terminal deubiquitination domain have been originally identified in patients affected by familial cylindromatosis, an autosomal-dominant disease which predisposes for the development of tumors of skin appendages (Bignell et al., 2000). Recently, the loss of CYLD expression and/or deubiquitination function has been described in multiple human tumors such as melanoma (Massoumi et al., 2006), hepatocellular carcinoma (Pannem et al., 2014), breast (Hutti et al., 2009), and adenoid cystic carcinoma (Stephens et al., 2013).
CYLD inhibits tumor development mostly by preventing the activation of the NF-κB pathway. By removing lysine-63–linked polyubiquitin chains from Bcl-3, NF-κB essential modulator (NEMO), and TNF receptor–associated factors (TRAFs) such as TRAF2, CYLD interferes with TNF-induced activation of the classical NF-κB signaling cascade, thereby inhibiting cell proliferation and survival (Brummelkamp et al., 2003; Kovalenko et al., 2003; Trompouki et al., 2003; Massoumi et al., 2006). However, the biological role of CLYD is not limited to its tumor-suppressive function. By negatively regulating NF-κB activation, CYLD limits the inflammatory response during infections, thus minimizing tissue damage (Zhang et al., 2011). Furthermore, in vivo studies demonstrated that CYLD plays multiple roles during immune cell development and homeostasis (Sun, 2008).
In this study, we use genetics to demonstrate that HSC dormancy is controlled by the DUB CYLD at the posttranslational level. The conditional inactivation of the CYLD DUB domain abolishes HSC quiescence, dormancy, and repopulation potential. Mechanistically, our data show that the CYLD–TRAF2 interaction is crucial to maintain dHSCs as it precludes p38MAPK activation, down-regulation of dormancy-associated genes, and entry of dHSCs into the cell cycle, ultimately preventing HSC exhaustion.
RESULTS
CYLD is crucial for the long-term repopulating capacity of HSCs
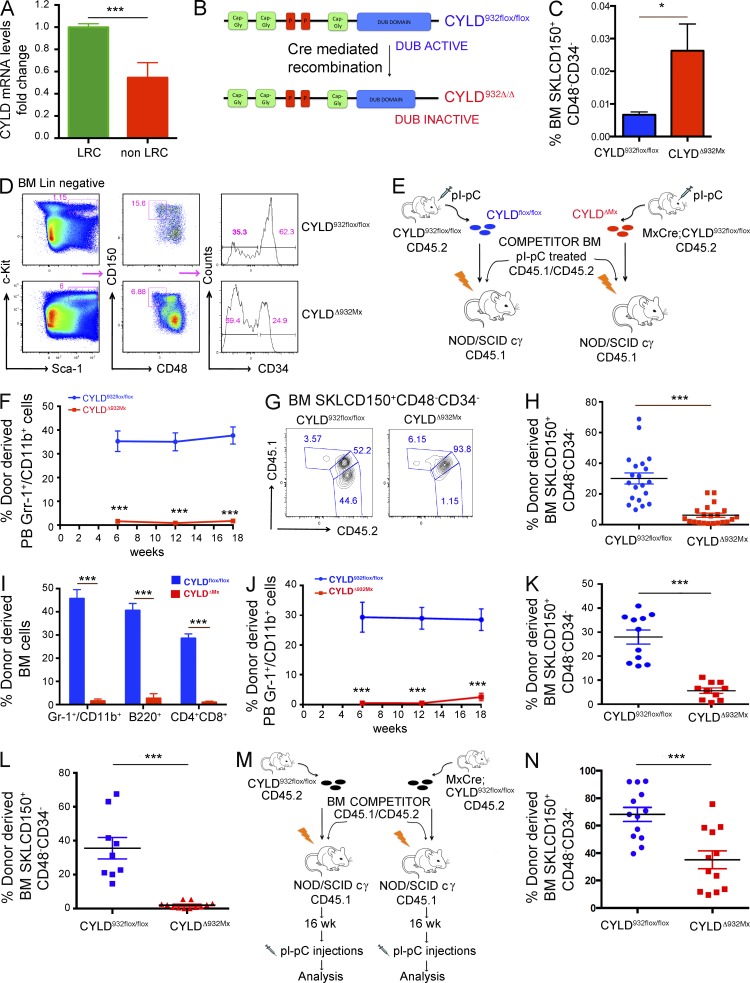
Extending our previous data suggesting that CYLD is differentially expressed between dHSCs (label-retaining cells [LRCs]) and active HSCs (non-LRCs; not depicted; Wilson et al., 2008), we now show that dHSCs express about twofold higher levels of CYLD transcripts compared with active HSCs (Fig. 1 A). During differentiation toward multipotent progenitor cells, CYLD expression remains rather constant both at the mRNA and protein level (Cabezas-Wallscheid et al., 2014), suggesting that CYLD might play a predominant role in dHSCs. Because CYLD encodes a DUB, these results prompted us to examine whether the catalytic activity of this molecule is important to preserve HSC dormancy. To genetically address this issue, we used a conditional knockout model recapitulating the human mutations that inactivate CYLD in patients suffering from cylindromatosis. In this gene-targeted mouse strain (CYLDflox/flox), Cre-mediated recombination replaces the last exon of the gene by a mutated copy, inducing the expression of a C-terminal truncated form of CYLD (CYLDΔ932) that lacks its catalytic DUB activity (Fig. 1 B; Welz et al., 2011). This inactivating mutation mimics the c.2806C>T mutation identified in numerous patients in distinct studies (Bignell et al., 2000; Bowen et al., 2005; Young et al., 2006; Saggar et al., 2008; Kazakov et al., 2009). To inactivate CYLD in HSCs, we crossed CYLDflox/flox mice with a transgenic line carrying the Mx1-Cre allele in which Cre recombinase is induced by type I interferons or polyinosine-polycytidine (pI-pC; Kühn et al., 1995). As expected, after two pI-pC injections, CYLD alleles were efficiently recombined in BM cells of mutant mice, hereafter referred to as CYLDΔ932Mx (not depicted). By 12 d after the last pI-pC injection, we already observed a significant expansion of (Sca-1+cKit+Lin− [SKL] CD150+CD48−CD34− cells highly enriched in functional HSCs in the BM of mutant mice (Fig. 1, C and D). To investigate the repopulation potential of these mutant phenotypic HSCs, we performed competitive transplantation assays using immune-compromised recipients as the mice have been bred on a mixed genetic background. BM cells were harvested from pI-pC–treated CYLDflox/flox or MxCre;CYLDflox/flox (CD45.2) mice and i.v. transplanted into sublethally irradiated NOD/SCID cg (NSG) recipients (CD45.1) together with BM competitor cells collected from pI-pC–treated C57BL/6 mice (CD45.1/CD45.2; Fig. 1 E). To exclude that a difference in the repopulation potential of control and mutant BM cells could arise from the higher HSC numbers observed in CYLDΔ932Mx mice, we transplanted total BM cells containing equivalent ratios of phenotypically defined mutant SKLCD150+CD48−CD34− donor and normal competitor cells of the same phenotype (1:3.4 mutant/competitor and 1:1 control/competitor ratios). Strikingly, by 6 wk after transplant (earliest time point analyzed), CYLDΔ932Mx cells had failed to reconstitute the peripheral blood compartments of recipient mice (Fig. 1 F and not depicted). Furthermore, flow cytometric analysis of the BM chimerism at 18 wk after transplant revealed that mutant cells not only failed to reconstitute the HSC compartment (Fig. 1, G and H) in the BM, but also failed to give rise to mature lineages (Fig. 1 I). To exclude that the failure of reconstitution of mutant HSCs was caused by the different total BM cell ratio transplanted, we performed additional competitive transplantation assays whereby control and mutant BM cells were both transplanted in a 1:1 ratio with competitor cells. Also using these conditions, CYLDΔ932Mx cells failed to reconstitute the peripheral blood and BM HSC compartment of recipient mice (Fig. 1, J and K). Importantly, mutant cells were unable to reconstitute the HSC pool of lethally irradiated recipients even when transplanted intrafemorally, excluding a homing defect of mutant HSCs (Fig. 1 L). To further confirm these results and determine whether the impaired reconstitution of mutant cells is caused by a defective self-renewal capacity, we transplanted 106 BM cells harvested from CYLDflox/flox or unrecombined MxCre;CYLDflox/flox mice together with the same number of competitor cells into sublethally irradiated recipients. After confirming the establishment of stable hematopoiesis (16 wk), the reconstituted recipients were injected with pI-pC to delete CYLD in a homeostatic situation, followed by subsequent evaluation of donor-derived chimerism (Fig. 1 M). This experimental setting enabled us to evaluate the impact of CYLD deletion on HSC function specifically during homeostatic conditions, excluding inflammatory effects or putative roles of CYLD in homing or niche engraftment. As shown in Fig. 1 N, pI-pC–mediated induction of the CYLD mutation in homeostatic HSCs resulted in a significant depletion of HSCs in the BM of the recipient mice, demonstrating that mutant HSCs are not maintained in the absence of functional CYLD activity (Fig. 1 N). Hence, CYLD catalytic activity is not only essential for the self-renewal capacity of HSCs in a reconstitution setting after conditioning (irradiation), but also during steady-state homeostasis.
Figure 1.
CYLD catalytic activity is required to preserve HSC activity. (A) Expression of CYLD in label-retaining dormant and non–label-retaining homeostatic HSCs. (B) Cre-mediated recombination of the CYLD932flox/flox allele generates a truncated and catalytically inactive protein. (C and D) Analysis of BM SKLCD150+CD48−CD34− in CYLD932flox/flox and CYLDΔ932Mx mice by flow cytometry. (E) Experimental scheme to generate and analyze competitive BM chimeras (i.v. injection) normalized to equal numbers of phenotypic HSCs shown in F–I. (F) Donor-derived myeloid chimerism in the peripheral blood of recipient mice. (G–I) BM of recipient mice (18 wk after transplant) analyzed for donor-derived HSCs (G and H) and myeloid, B, and T cells (I). (J and K) Peripheral blood myeloid (J) and BM HSC chimerism (K) of mice transplanted with an equivalent ratio of test/competitor total BM cells. (L) Similar analysis as shown in E, but transplantation was performed by intrafemoral injection. Donor-derived HSC chimerism in the BM of recipients is shown 18 wk after transplant. (M) Experimental scheme to generate competitive BM chimeras in which Cre-mediated recombination was induced by four pI-pC injections after stable engraftment (16 wk) has been achieved in the chimeras. (N) Analysis of donor-derived HSCs chimerism 18 wk after pI-pC induction. Results are shown of two (F–I: 20/21; J and K: 11/12; L: 9/12; N: 13/12) or three (A: 6/6; C and D: 14/7) independent experiments, with the numbers of analyzed control/mutant mice indicated in parentheses. Error bars indicate SEM. *, P < 0.05; ***, P < 0.001.
CYLD preserves HSC dormancy
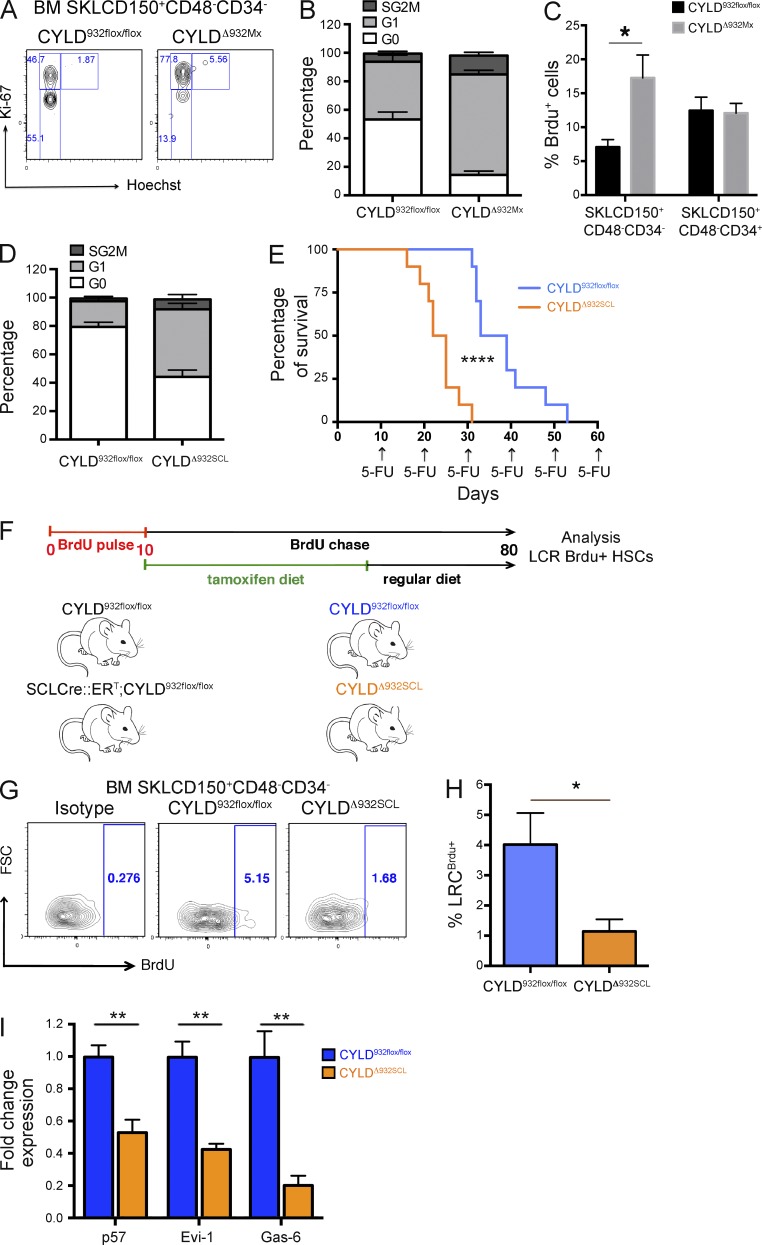
Having observed that HSCs dramatically lose their functional activity upon CYLD inactivation, we next investigated whether CYLD controls HSC dormancy. To this end, we first examined the cell cycle distribution of control and mutant HSCs. As shown in Fig. 2 (A and B), 53 ± 5.2% of CYLDflox/flox control HSCs were in the G0 phase, as expected; in contrast, only 14 ± 2.7% of CYLDΔ932Mx HSCs were in G0. The higher proliferation rate was further confirmed by BrdU assays, demonstrating a threefold increased BrdU incorporation in CYLD mutant SKLCD150+CD48−CD34− cells compared with control cells. Importantly, whereas the absence of a functional DUB activity strongly impacted the proliferation of primitive HSCs, no differences in the proliferation rate were observed in SKLCD15+CD48−CD34+ progenitor cells (Fig. 2 C). To exclude the possibility that these changes are indirectly related by pI-pC–induced activation of the interferon cascade (Essers et al., 2009), we also used mice in which Cre was driven by a tamoxifen (Tx)-inducible Cre (SCL-Cre::ERT; Göthert et al., 2005; Tesio et al., 2013). After 42 d of Tx diet, the CYLD flox alleles were recombined in BM cells, albeit to a lesser extent than what we observe using the Mx-Cre system (not depicted). Nevertheless, also in the SCL-CreERT model, the percentage of quiescent HSCs was twofold decreased in the BM of mutant mice (referred to as CYLDΔ932SCL), confirming the results obtained in CYLDΔ932Mx animals (Fig. 2 D). Because HSC dormancy mediates their resistance to antiproliferative agents (Essers et al., 2009), we serially administered the chemotherapeutic drug 5-fluouracil (5-FU) to CYLDΔ932SCL mice and monitored their survival. As shown in Fig. 2 E, mutant mice succumbed significantly earlier than CYLDflox/flox animals to serial 5-FU injections, indicating that the increased HSC proliferation in CYLD mutants correlates with a faster exhaustion of HSCs in response to an antiproliferative drug. Next, we determined the number of LRC-HSCs in control and CYLDΔ932SCL mice by a pulse chase experiment (Wilson et al., 2008; Tesio et al., 2013). To do this, SCLCre::ERT;CYLDflox/flox mice and their control counterparts were pulsed for 10 d with BrdU and then chased for 70 d. During the first 40 d of the chase period, mice were fed a Tx diet to induce CYLD inactivation and subsequently chased for an additional 30 d with a regular diet (Fig. 2 F). As shown in Fig. 2 (G and H), at the end of the chase period a fourfold decrease of SKLCD150+CD48−CD34−BrdU+ LRCs was observed in the BM of CYLD mutants. These data demonstrate that CYLD DUB activity is essential for HSC dormancy and that its loss induces HSCs to proliferate. In agreement with these functional data, dormancy-associated genes such as the CDK inhibitor p57 (Cdkn1c), the transcriptional factor Evi-1, and Gas-6 (Goyama et al., 2008; Matsumoto et al., 2011; Zou et al., 2011; unpublished data) were strongly down-regulated in CYLD mutant HSCs (Fig. 2 I). Altogether, these results demonstrate that CYLD activity is essential to preserve dHSCs in vivo.
Figure 2.
The catalytic activity of CYLD maintains HSC dormancy. (A and B) Cell cycle analysis of HSCs isolated from CYLD932flox/flox (control) and MxCre;CYLD932flox/flox (experimental) mice 12 d after the last pI-pC injection (P < 0.001). (C) BrdU incorporation in SKLCD150+CD48−CD34− or SKLCD150+CD48−CD34+ HSCs from control or CYLDΔ932Mx mutant mice (*, P < 0.01). (D) Same analysis as in B, but using CYLD932flox/flox and SCLCre::ERT;CYLD932flox/flox mice fed for 42 d with a Tx diet (P < 0.001). (E) Kaplan–Meier survival curve of CYLD932flox/flox and CYLDΔ932SCL mice treated with 5-FU every 10 d (****, P < 0.0001). (F) Experimental scheme for the long-term label-retaining assay used in G and H. (G and H) Percentage of BM BrdU label–retaining HSCs determined by flow cytometry (*, P < 0.05). (I) mRNA expression levels of dormancy-associated genes determined by qRT-PCR of FACS-sorted HSCs from CYLD932flox/flox and CYLDΔ932SCL mice (**, P < 0.01). Results are shown of two (C: 4/4; E: 10/10; F: 7/11; I: 4/6) or three (A,B: 6/8; D: 7/6) independent experiments, with the numbers of analyzed control/mutant mice indicated in parentheses. Error bars indicate SEM.
The maintenance of HSC dormancy by CYLD requires interaction with TRAF2
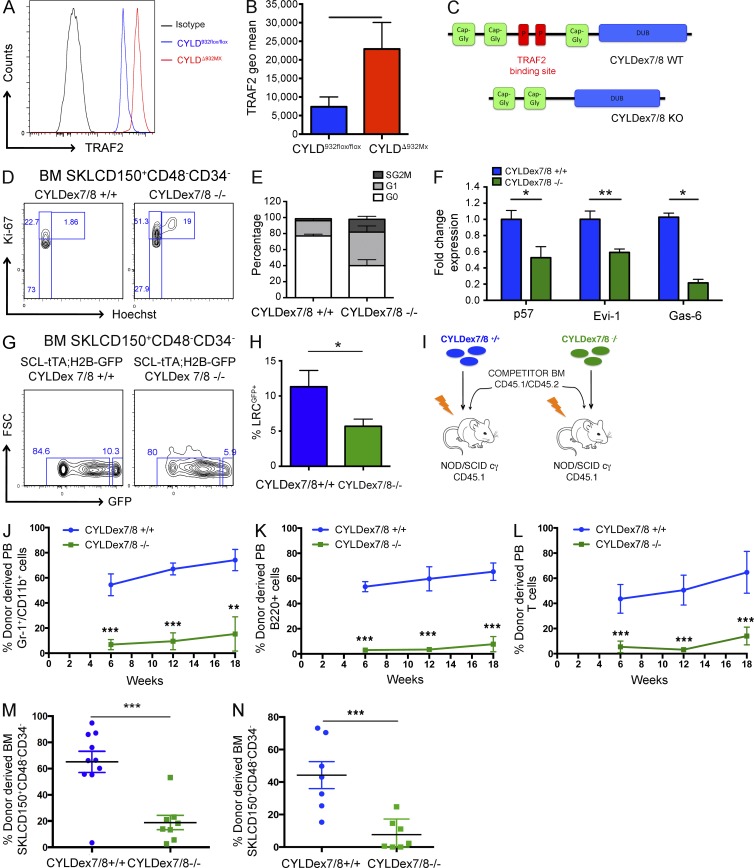
We next determined the signaling cascade downstream of CYLD and found that CYLD-deficient HSCs show a threefold up-regulation of TRAF2 expression (Fig. 3, A and B). Because TRAF2 is a critical CYLD substrate (Brummelkamp et al., 2003; Kovalenko et al., 2003; Trompouki et al., 2003), these results raise the possibility that CYLD-mediated deubiquitination of TRAF2 may be a critical step to maintain HSC dormancy. To verify this hypothesis, we examined CYLDex7/8−/− mice, which lack the full CYLD molecule but overexpress a shorter CYLD isoform (sCYLD) that is unable to bind to and thus deubiquitinate TRAF2 (Fig. 3 C; Hövelmeyer et al., 2007). As a consequence, mutant cells show an accumulation of hyperubiquitinated TRAF2 (Hövelmeyer et al., 2007).
Figure 3.
CYLD interaction with TRAF2 is crucial to preserve HSC dormancy. (A and B) TRAF2 expression determined by flow cytometry in BM HSCs isolated from CYLD932flox/flox and CYLDΔ932Mx mice. (C) Scheme showing normal CYLD and the CYLDex7/8 mutant protein. (D and E) Cell cycle analysis by flow cytometry of BM HSCs isolated from control and CYLDex7/8−/− mice. (F) mRNA expression levels of dormancy-associated genes determined by qRT-PCR of FACS-sorted HSCs from control and CYLDex7/8−/− mice. (G and H) Label-retaining assays using SCL-tTA;H2B-GFP;CYLDex7/8+/+ (control) and SCL-tTA;H2B-GFP;CYLDex7/8−/− (experimental) mice. Percentage of LRCs was determined by flow cytometry. (I) Experimental scheme for the generation of BM chimeras (i.v.) used for the analysis shown in J–M. (J–M) Donor-derived level of myeloid (J), B cell (K), and T cell (L) chimerism in the peripheral blood (6, 12, and 18 wk after transplant), as well as BM of recipient mice (18 wk after transplant) analyzed for HSCs (M). (L) Same analysis as shown in E, but transplantation was performed by intrafemoral injection. Donor-derived HSC chimerism in the BM of recipients was analyzed 18 wk after transplant. (N) Same analysis as shown in I, but transplantation was performed by intrafemoral injection. Donor-derived HSC chimerism in the BM of recipients is shown 18 wk after transplant. Results are shown of two (F: 6/4; J–M: 10/8; N: 7/7) or three (A and B: 7/8; D and E: 10/8; G and H: 5/7) independent experiments, with the numbers of analyzed control/mutant mice indicated in parentheses. Error bars indicate SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Strikingly, similarly to what we observed in CYLDΔ932Mx and in CYLDΔ932SCL mice, CYLDex7/8−/− HSCs lost their quiescent status. Only 36.5 ± 7.4% of mutant HSCs were in G0 phase, in contrast to control cells (78 ± 2% in G0; Fig. 3, D and E). Moreover, mutant HSCs also expressed lower levels of the dormancy-associated genes p57 (Cdkn1c), Evi-1, and Gas-6 (Fig. 3 F). To investigate whether the interactions between CYLD and TRAF2 are important to preserve the pool of dHSCs, we crossed CYLDex7/8 mutant animals with SCL-tTA;H2B-GFP mice and again performed label-retaining assays (Wilson et al., 2008). Although after 100 d of chase (+doxycycline) 11.0 ± 2.3% of CYLDex7/8 WT SKLCD150+CD48−CD34− cells retained the H2B-GFP label, only 5.6 ± 1.02% of mutant cells remained GFP positive (Fig. 3, G and H), demonstrating that CYLD binding to TRAF2 is a crucial step to maintain HSC dormancy.
To confirm these data on a functional level, we transplanted into sublethally irradiated NSG recipients total BM cells containing almost equivalent ratios of phenotypically defined mutant SKLCD150+CD48−CD34− donor and normal competitor cells (equivalent ratio at the HSC level corresponded to a total BM ratio of 1:1 for control/competitor cells and a ratio of 1:1.5 for mutant/competitor cells; Fig. 3 I and not depicted). As shown in Fig. 3 (J–L), 6 wk after transplant CYLDex7/8−/− cells were outcompeted by WT cells and failed to give rise to circulating myeloid and lymphoid cells. Most importantly, mutant cells failed to reconstitute the BM stem cell compartment when injected into the recipient mice either i.v. (Fig. 3 M) or intrafemorally (Fig. 3 N), thus demonstrating that the self-renewal potential of CYLDex7/8−/− cells is severely impaired.
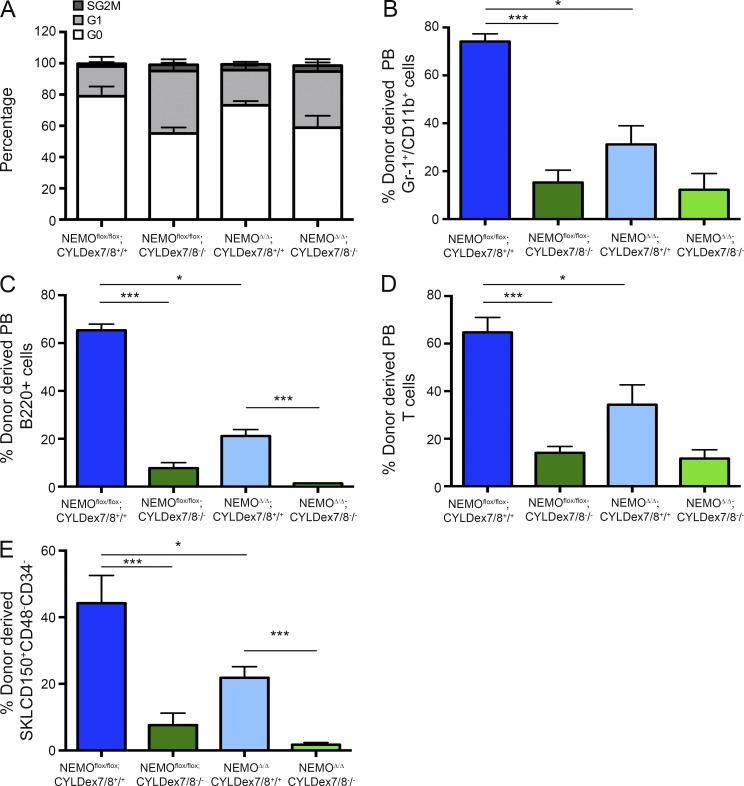
Although the short CYLD isoform expressed in CYLDex7/8−/− mice causes an accumulation of hyperubiquitinated TRAF2 without affecting NEMO levels (Hövelmeyer et al., 2007), we cannot exclude that the loss of dormancy observed in mutant HSCs is exclusively caused by impaired TRAF2 deubiquitination. In fact, although the inability of sCYLD to bind and deubiquitinate NEMO remains to be demonstrated, sCYLD has been shown to be devoid of the putative NEMO-binding site. Thus, to exclude a contribution of NEMO hyperubiquitination to the phenotype observed in CYLDex7/8−/− mice, we crossed these animals to a NEMO conditional knockout line in which NEMO deletion is controlled by SCL-Cre::ERT (SCL-Cre::ERT;NEMOflox/flox;CYLDex7/8−/− mice). Cell cycle analysis of SKLCD150+CD48−CD34− cells from single and double mutant animals revealed that NEMO deletion could not restore quiescence in CYLDex7/8−/− HSCs (Fig. 4 A). Furthermore, competitive repopulation assays demonstrated that NEMOΔSCL;CYLDex7/8−/− BM cells lack reconstitution activity, similar to NEMOflox/flox;CYLDex7/8−/− BM cells (Fig. 4, B–E). Altogether, these data demonstrate that lack of NEMO does not rescue the HSC defects observed in mutant CYLDex7/8 mice. Thus, the loss of HSC dormancy that occurs in these mice is most likely solely caused by an impaired CYLD–TRAF2 interaction.
Figure 4.
The loss of dormancy in CYLDex7/8 mutant HSCs is not caused by impaired NEMO binding. (A) Cell cycle analysis of BM SKLCD150+CD48−CD34− cells harvested from NEMOflox/flox;CYLDex7/8+/+ or NEMOflox/flox;CYLDex7/8−/− or SCLCre;NEMOflox/flox;CYLDex7/8+/+ or SCLCre;NEMOflox/flox;CYLDex7/8−/− mice fed for 15 d with a Tx diet. (B–D) Mice from all four genotypes (CD45.2) and WT (CD45.1/CD45.2) animals were fed for 15 d with a Tx diet. Donor BM cells were harvested, combined with BM competitor cells, and injected into sublethally irradiated (CD45.1) NOD/SCID cγ recipients. The level of reconstitution of myeloid (B), B (C), and T (D) cells in peripheral blood 6, 12, and 18 wk after transplant was determined by flow cytometry. (E) Donor-derived chimerism of BM HSCs 18 wk after transplant as determined by flow cytometry. Results are shown of two (A: 6/6; B–E: 7/7) independent experiments, with the numbers of analyzed control/mutant mice indicated in parentheses. Error bars indicate SEM. *, P < 0.05; ***, P < 0.001.
CYLDex7/8 mutant cells do not activate NF-κB signaling
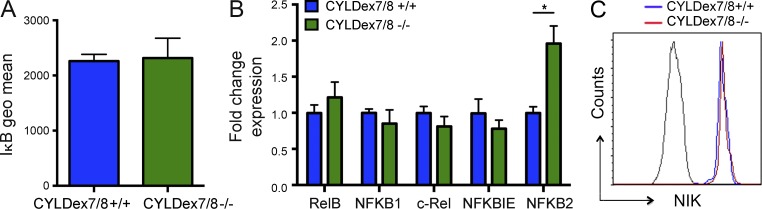
To further investigate the signaling cascade downstream of CYLD–TRAF2 interactions, we first determined whether mutant CYLDex7/8−/− HSCs exhibit increased canonical NF-κB signaling. To this purpose, we analyzed the degradation of IκBα, an inhibitory kinase which sequesters NF-κB dimers in the cytosol (Baeuerle and Baltimore, 1988). Surprisingly, not only were IκBα levels the same in mutant and WT HSCs (Fig. 5 A), they were not decreased after in vitro stimulation with TNF (not depicted). In line with these results, expression of the major NF-κB signaling effectors was not increased in CYLDex7/8−/− SKLCD150+CD48−CD34− cells, with the notable exception of NFKB2 (Fig. 5 B).
Figure 5.
CYLD–TRAF2 interaction has no significant effect on NF-κB signaling. (A) IκBα levels in BM HSCs from control and CYLDex7/8−/− mice. (B) Expression analysis using qRT-PCR of NF-κB target genes in BM HSCs sorted from control and CYLDex7/8−/− mice. (C) NIK expression analyzed by flow cytometry in BM HSCs from control and CYLDex7/8−/− mice. Results are shown of two (A: 4/4; B: 5/7) or three (C: 4/6) independent experiments, with the numbers of analyzed control/mutant mice indicated in parentheses. Error bars indicate SEM. *, P < 0.05.
By regulating the turnover of the NIK kinase, TRAF2 also contributes to activation of the alternative NF-κB signaling pathway (Vallabhapurapu et al., 2008). We thus examined NIK levels in CYLDex7/8 mutant and control HSCs. Again, we did not detect any changes in NIK expression in knockout cells (Fig. 5 C). Moreover, NIK levels did not change after in vitro stimulation with TNF (not depicted). Altogether, these data suggest that neither the canonical nor the noncanonical NF-κB signaling pathways are activated in CYLDex7/8 mutant HSCs, suggesting that alternative pathways must contribute to the loss of HSC dormancy in these mice.
By interacting with TRAF2, CYLD prevents p38MAPK activation
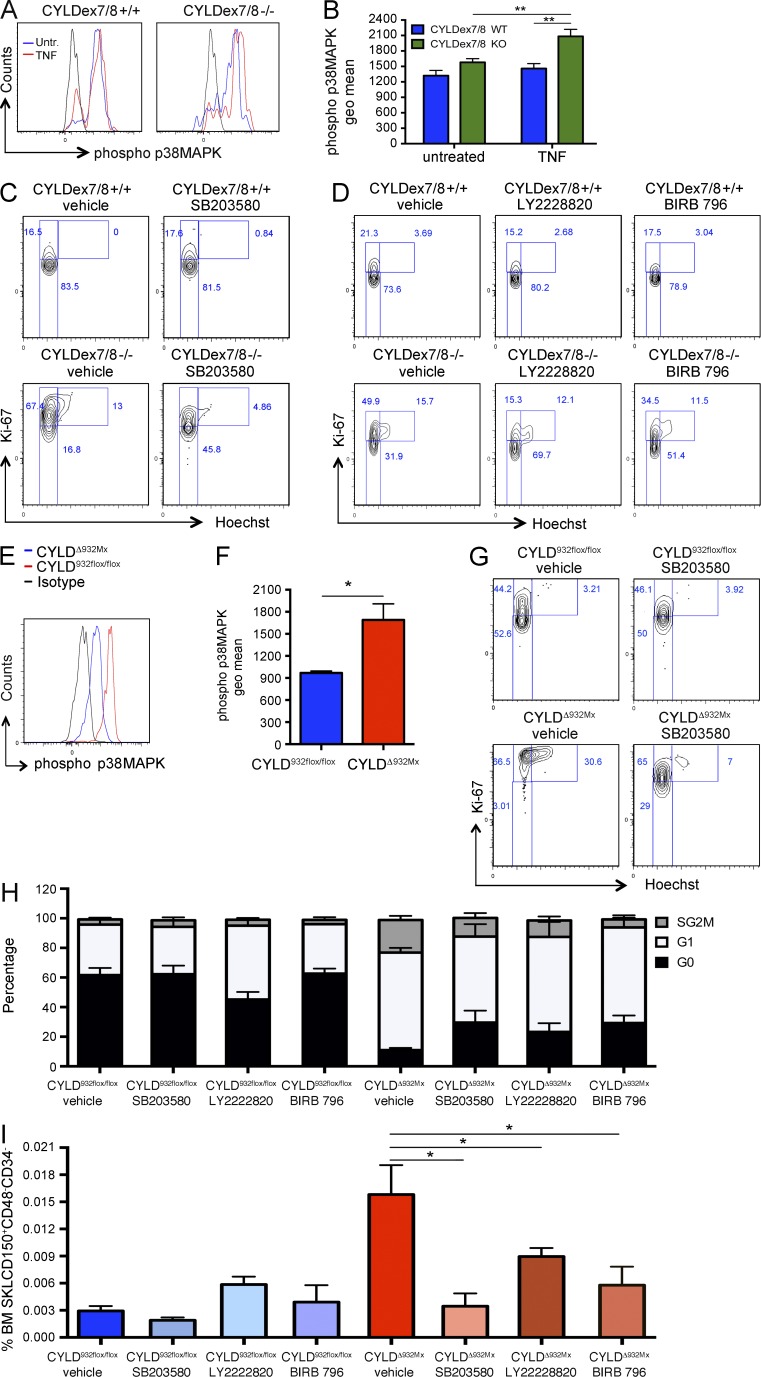
In addition to activating NF-κB, TRAF2 controls the activity of members of the MAPK kinase family, such as p38MAPK (Carpentier et al., 1998). This kinase was shown to be involved in reactive oxygen species (ROS)–induced loss of HSC quiescence (Ito et al., 2006). We thus investigated whether the impaired CYLD–TRAF2 interactions may alter p38MAPK activity. We then evaluated the levels of phospho-p38MAPK after exposure to TNF. As shown in Fig. 6 (A and B), 5-min stimulation with 100 ng/ml TNF was sufficient to up-regulate phospho-p38MAPK levels in mutant CYLDex7/8 HSCs but not in their WT counterparts. We next blocked p38MAPK activation in vivo by administering the selective p38MAPK inhibitor SB203580 (Ito et al., 2006). Although SB203580 did not alter the cell cycle distribution in WT HSCs, it restored quiescence in CYLDex7/8-deficient HSCs (Fig. 6 C). To exclude putative unspecific activities of this inhibitor, we administered to two additional compounds: LY2228820 and BIRB 796, known to selectively and potently block p38MAPK signaling (Pargellis et al., 2002; Campbell et al., 2014; Thalheimer et al., 2014). As shown in Fig. 6 D, both compounds at least partially restored HSC quiescence in CYLDex7/8 mutant HSCs but did not alter the cell cycle distribution of control cells, thus confirming a specific role for p38MAPK signaling downstream of CYLD–TRAF2. Importantly, increased p38MAPK activation also was detected in CYLDΔ932Mx mutant even in the absence of additional stimulation with TNF (Fig. 6, E and F). In line with these results, all three tested p38MAPK inhibitors rescued the loss of quiescence in CYLDΔ932Mx HSCs (Fig. 6, G and H). Moreover, prevention of the extensive proliferation of mutant HSCs by SB203580, LY2228820, and BIRB 796 resulted in the normalization of HSC numbers in the BM of CYLDΔ932Mx mice (Fig. 6 I). In summary, these results suggest that the CYLD–TRAF2 pathway mediates its effect on dHSCs not by repressing NF-κB, but by negatively regulating p38MAPK activity.
Figure 6.
CYLD–TRAF2 interaction controls HSC quiescence by inhibiting p38MAPK activation. (A and B) Expression levels of phospho-p38MAPK as determined by flow cytometry in BM HSCs (SKLCD150+CD48−CD34−) from control and CYLDex7/8−/− mice after 5-min stimulation with TNF. (C) Cell cycle analysis of BM HSCs isolated from control and CYLDex7/8−/− mice receiving the p38MAPK inhibitor SB203580 or vehicle (DMSO). (D) Cell cycle analysis of BM HSCs isolated from control and CYLDex7/8−/− mice receiving the p38MAPK inhibitors LY2228820 and BIRB 796 or vehicle (DMSO). (E and F) Phospho-p38MAPK levels in BM HSCs (SKLCD150+CD48−CD34−) from control and CYLDΔ932Mx mice. (G) Cell cycle analysis as in C, but analyzing CYLD932flox/flox (control) and CYLDΔ932Mx (mutant) mice. (H) Cell cycle analysis of BM HSCs isolated from control and CYLDΔ932Mx mice receiving the p38MAPK inhibitors LY2228820 and BIRB 796 or vehicle (DMSO). (I) HSC frequency as determined by flow cytometry in the BM of CYLD932flox/flox and CYLDΔ932Mx mice receiving SB203580, LY2228820, BIRB 796, or vehicle (DMSO). Results are shown of two (A–D: 4/4; E and F: 3/5; G and H: 5/5; I: 5/5) independent experiments, with the numbers of analyzed control/mutant mice or control/treated groups indicated in parentheses. Error bars indicate SEM. *, P < 0.05; **, P < 0.01.
DISCUSSION
Our study identified a novel CYLD–TRAF2–p38MAPK signaling cascade that is crucial to maintain HSC dormancy and function and thus the integrity of the hematopoietic compartment. Whereas our data demonstrated that the conditional inactivation of the CYLD DUB domain dramatically impacts HSCs function, no defects had been reported in straight knockout mice so far. Similarly, the absence of the full protein is compatible with life, whereas the lack of its catalytical domain induces the formation of a hyperplastic lung mesenchyme that prevents the proper development of the alveolar–capillary barrier. As a result, mice lacking CYLD DUB activity encounter respiratory dysfunctions, which lead to their perinatal death (Trompouki et al., 2009). The lack of CYLD in straight knockout animals may trigger compensatory mechanisms by, e.g., expression of other DUBs, which cannot be installed after acute deletion. It is noteworthy that a functional redundancy between CYLD and A20 (another K-63–specific DUB) has been suggested as both molecules are critical for HSC function and negatively regulate the NF-κB pathway by targeting the same substrates (Reiley et al., 2004; Wertz et al., 2004; Nakagawa et al., 2015). The conditional models thus allowed us to identify a previously unknown and essential role for CYLD in the control of stem cell dormancy and function. However, CYLD may not be exclusively controlling dormant cells but also their immediate progeny where it remains expressed, albeit at lower levels as compared with dHSCs.
Recently, a few studies have revealed that protein ubiquitylation plays an important role in HSC homeostasis. The E3 ubiquitin ligases c-Cbl and Itch were shown to negatively regulate hematopoietic long-term repopulation potential (Rathinam et al., 2008, 2011). Conversely, the genetic depletion of other E3 ubiquitin ligases such as Cul4A or Fbxw7 severely impaired HSC self-renewal (Li et al., 2007; Matsuoka et al., 2008; Rathinam et al., 2008, 2011; Thompson et al., 2008). Moreover, Abbas et al. (2010) also demonstrated that HSC survival relies on an ubiquitin ligase, Mdm2, which is essential to prevent HSC death in response to stress. The role of these E3 ubiquitin ligases in controlling HSC homeostasis has been mostly ascribed to their role in ubiquitin-mediated protein degradation (Thompson et al., 2008; Abbas et al., 2010; Saur et al., 2010; Rathinam et al., 2011). However, in addition to mediating protein degradation, ubiquitination regulates numerous other cellular functions, including protein trafficking and signal transduction (Emmerich et al., 2011; Haglund and Dikic, 2012), suggesting that posttranslational ubiquitin modifications may regulate HSC biology through more intricate mechanisms. Furthermore, the covalent attachment of ubiquitin chains to target proteins is a reversible, dynamic process, being antagonized by the action of DUBs. Whereas the role of ubiquitin ligases in HSC self-renewal starts to be clarified, the importance of deubiquitination processes in HSCs remains largely elusive. Our study adds a novel level of complexity to this scenario, revealing that the interaction between the DUB CYLD and the adaptor protein, E3 ubiquitin ligase TRAF2, elicits a signaling pathway that promotes dormancy and thus prevents HSCs from unscheduled proliferation and exhaustion. Interestingly, TRAF2 is involved in the signal transduction elicited by members of the TNF receptor superfamily such as TNFR1 and TNFR2 (Wajant and Scheurich, 2001), which have been shown to suppress HSC activity (Pronk et al., 2011). In addition, TRAF2 is part of the signaling cascade downstream of members of the Toll-like receptors (Sasai et al., 2010), a family of proteins whose stimulation activates HSCs upon inflammatory stimuli (Baldridge et al., 2011). Although our CYLDex7/8 mouse model indicates that the TRAF2-binding domain is crucial to control HSC function by promoting the interaction between CYLD and TRAF2, we cannot formally exclude that unknown substrates bind to this domain as well. However, this scenario seems remote as CYLD has a restricted substrate specificity and the TRAF2-binding domain was shown to specifically interact with TRAF2 and not with other TRAF family members, nor with other proteins undergoing a K-63 deubiquitination such as RIP, NIK, and cIAP1 (Kovalenko et al., 2003). However, future investigations should further characterize the TRAF2-binding domain and explore whether other putative interaction partners in hematopoietic/stem progenitor cells do exist.
Studies conducted in cancer cell lines demonstrated that TRAF2 deubiquitylation is required for CYLD-mediated inhibition of the NF-κB pathway (Brummelkamp et al., 2003; Kovalenko et al., 2003; Trompouki et al., 2003). Furthermore, in immune cells TRAF2 was shown to control the activation of the canonical and noncanonical NF-κB signaling pathways in response to different stimuli (Grech et al., 2004; Gardam et al., 2008; Vallabhapurapu et al., 2008). In line with this evidence, CYLDex7/8 mutant B cells and BM dendritic cells showed increased NF-κB activation (Hövelmeyer et al., 2007; Srokowski et al., 2009). Surprisingly, however, CYLDex7/8 mutant HSCs did not significantly activate the NF-κB pathway, but instead p38MAPK signaling, indicating that CYLD–TRAF2 interactions elicit a signaling cascade that is HSC specific. Recent studies also point to a negative role for p38MAPK on HSC maintenance. Activation of the p38MAPK pathway in response to ROS has been reported to cause HSC exhaustion (Ito et al., 2006), and p38MAPK has been placed downstream of GADD45G, which induces lineage-specific HSC differentiation at the expense of self-renewal (Thalheimer et al., 2014). Pharmacological inhibition of p38MAPK maintained stemness of cultured mouse and human HSCs (Baudet et al., 2012; Zou et al., 2012). Additional in vitro studies showed that p38MAPK regulates p57 (Cdkn1c) activity (Joaquin et al., 2012), a known critical regulator of HSC quiescence (Matsumoto et al., 2011; Zou et al., 2011) and which we now show is strongly down-regulated in CYLDΔ932 and CYLDex7/8 mutant HSCs. The data strongly suggest that p38MAPK has potent inhibitory activity on HSCs downstream of several pathways, including ROS, generated by mitochondrial oxidative phosphorylation, GADD45G, and CYLD. As our study is based on a genetic mouse model recapitulating mutations present in CYLD mutant patients, our data on HSCs may suggest examining the BM of cylindromatosis patients.
In conclusion, our genetic data demonstrate that the DUB CYLD is crucial for maintaining dHSCs by preventing unscheduled HSC proliferation through a TRAF2–p38MAPK cascade and thus identify a novel signaling pathway involved in controlling the balance between stem cell dormancy and self-renewal.
MATERIALS AND METHODS
Mice.
CYLD932flox/flox (control) mice, generated as previously described (Welz et al., 2011), were crossed with the Mx1-Cre transgenic mice (Kühn et al., 1995) to obtain MxCre;CYLD932flox/flox (mutant) animals. IFN-α–mediated deletion was induced by two i.p. injections of 10 mg/kg pI-pC (InvivoGen) every 2 d, and unless otherwise indicated, mice were sacrificed after 12 d from the last pI-pC injection. CYLD932flox/flox mice were also crossed with SCL-CreERT transgenic animals (Göthert et al., 2005) to obtain SCL-Cre::ERT;CYLD932flox/flox (mutant) mice. The recombination of CYLD alleles in this model was achieved by a 42-d Tx diet (1 g/kg food; Sigma-Aldrich). CYLDex7/8−/− mice (Hövelmeyer et al., 2007) were provided by A. Waismann (Institute for Molecular Medicine, University Medical Center of the Johannes Gutenberg University of Mainz, Mainz, Germany). These mice were crossed with the transgenic line SCL-tTA;H2B-GFP (Wilson et al., 2008) to obtain SCL-tTA;H2B-GFP;CYLDex7/8−/− and SCL-tTA;H2B-GFP;CYLDex7/8+/+ animals.
NEMOflox/flox mice were crossed with the SCL-CreERT transgenic lines to obtain SCL-CreERT;NEMOflox/flox animals. Subsequently, SCL-CreERT;NEMOflox/flox mice were crossed to CYLDex7/8−/− mice to obtain SCL-CreERT;NEMOflox/flox;CYLDex7/8−/− and SCL-CreERT;NEMOflox/flox;CYLDex7/8+/+ mice. To induce NEMO deletion, these animals were subjected to a 15-d Tx diet (1 g/kg food). ROSA26EYFPflox/flox mice (Srinivas et al., 2001) were crossed with MxCre;CYLD932flox/flox and SCL-CreERT;CYLD932flox/flox mice to monitor the recombination of CYLD alleles in BM HSCs upon pI-pC injections or treatment with Tx food.
All animal procedures were performed according to protocols approved by the German authorities no. G-93/11 and no. G-232-12. Mice were maintained in the DKFZ animal facility under specific pathogen–free (SPF) conditions and housed in individually ventilated cages (HIVC).
Long-term label-retaining assays.
To evaluate the number of LRCBrdU+ cells, CYLD932flox/flox and SCL-CreERT; CYLD932flox/flox mice were BrdU labeled for 10 d using 0.8 mg/ml BrdU water (glucose), followed by 70 d of chase. During the chase period, mice were fed for the first 40 d with Tx (1 g/kg food) and for the remaining 30 d with a regular diet. To evaluate the number of LRCBrdU+ cells by FACS, BM cells were lineage depleted, stained for surface markers, and then fixed and permeabilized with Cytofix/Cytoperm buffer (BD). This step was followed by incubation with Cytoperm Plus buffer (BD) and a second round of fixation/permeabilization with Cytofix/Cytoperm buffer. After DNase I digestion, intracellular staining for BrdU was performed by incubating the cells with anti–BrdU-APC or -FITC antibody in Perm/Wash buffer.
To evaluate the number of LRCGFP+ cells, CYLDex7/8−/− and control counterparts were crossed to SCL-tTA;H2B-GFP mice (Wilson et al., 2008) to obtain SCL-tTA;H2B-GFP;CYLDex7/8−/− and SCL-tTA;H2B-GFP;CYLDex7/8+/+ animals. These mice were administered with doxycycline-containing water for 100–150 d, according to a previously described protocol (Wilson et al., 2008).
5-FU survival assay.
SCL-CreERT;CYLD932flox/flox mice and control counterparts were fed for 42 d with a Tx diet (1 g/kg food) to induce CYLD inactivation. Mice were then i.p. injected with 5-FU (150 mg/kg; Sigma-Aldrich) every 10 d.
In vivo p38MAPK inhibition.
After being injected twice with pI-pC, CYLD932flox/flox and MxCre;CYLD932flox/flox mice were administered the selective p38MAPK inhibitors SB203580 (50 mg/kg; Selleckchem), LY2228820 (1.5 mg/kg; Selleckchem), BIRB 796 (15 mg/kg; Selleckchem), or vehicle (DMSO). These compounds were injected i.p. every second day for a week and then daily for an additional 4 d. CYLDex7/8−/− and WT counterparts received four consecutive injections with SB203580 (50 mg/kg), LY2228820 (1.5 mg/kg), BIRB 796 (15 mg/kg), or vehicle (DMSO).
Competitive repopulation assays.
BM test cells (CD45.2) and competitor cells (C57BL/6 mice, CD45.1/CD45.2) were T cell depleted and mixed together to obtain a 1:1 test/competitor ratio of SKLCD150+CD48−CD34− cells, which were than i.v. injected into sublethally irradiated (250 Rads) NOD/SCID cg (CD45.1) recipients. All chimeric mice were maintained on antibiotic-containing water (Bactrim; Roche) for 3 wk after irradiation, and long-term reconstitution of peripheral blood and BM was analyzed 6–18 wk later.
Isolation of BM, spleen, and blood cells.
To collect BM cells, mouse legs and spinal cords were dissected and flesh removed, bones were crushed using a mortar and pestle, and cell suspensions were filtered before further use. Mouse spleens were isolated and crushed using the bottom of a syringe punch and filtered to obtain cell suspensions. Blood cells were collected from cheek vein bleedings into tubes containing 10,0000 U/ml heparin (Sigma-Aldrich), and peripheral red blood cells were lysed using ACK Lysing Buffer (Life Technologies).
Flow cytometric analysis and sorting.
Flow cytometric analyses were performed using an LSRII or Fortessa (BD) flow cytometer equipped with 488-nm and UV (305 nm) lasers. Data were analyzed using FlowJo software.
To sort HSCs, lineage-negative cells were enriched through a lineage magnetic depletion. To this end, BM cells were incubated with a cocktail of antibodies (CD4, CD8, CD11b, Gr1, B220, and Ter119), and lineage-positive cells were removed using magnetic selection with sheep anti–rat IgG-coated M450 Dynabeads (Invitrogen). Lineage-negative cells were subsequently stained for lineage markers (CD4, CD8, CD11b, Gr1, B220, and Ter119) and Sca-1, c-Kit, CD150, CD48, and CD34 antibodies. SKLCD150+CD48−CD34− cells were than sorted using a FACSAria (BD).
Cell cycle analysis.
BM cells were stained using a cocktail of anti-lineage markers (CD4, CD8, CD11b, Gr1, B220, and Ter119) and Sca-1, c-Kit, CD150, CD48, and CD34 antigens. Subsequently, the cells were fixed and permeabilized with Cytofix/Cytoperm solution. Intracellular Ki-67 staining was performed by next incubating the cells with anti–Ki-67 Alexa Fluor 647 or FITC antibody in Perm/Wash buffer. Before FACS analysis, Hoechst 33342 (Molecular Probes) was added for 5–10 min at 20 µg/ml.
Intracellular staining.
BM cells were stained using lineage antibodies (CD4, CD8, CD11b, Gr1, B220, and Ter119) and Sca-1, c-Kit, CD150, CD48, and CD34 antibodies. Cells were then fixed and permeabilized using Cytofix/Cytoperm solution. Subsequently, the cells were stained overnight with a polyclonal rabbit anti-TRAF2 antibody, an anti-NIK antibody, or a matched isotype control (Abcam). Staining with a secondary anti–rabbit antibody PE conjugate followed.
Antibodies.
To stain for lineage makers, the following antibodies were used: Gr-1 (Ly-6G, RB6-8C5)–PECy7, Ter 119–PECy7, B220 (RA3-6B2)-PECy7, CD11b-PECy7, CD4 (clone GK1.5)-PECy7, and CD8α (53.6.7)-PECy7 (eBioscience). CD34 (RAM34)-Al700, CD117 (2B8)-PE and APCeFluor780, Sca1 (D7)-APC, CD150-PECy5, and CD48-PE or –Pacific blue (all from eBioscience) were used to stain HSCs. Primary anti-TRAF2 and NIK antibodies were obtained from Abcam.
Real-time RT-PCR.
Total RNA was isolated from sorted BM SKLCD150+CD48−CD34− cells using the PicoPure RNA Isolation kit (Arcturus) according to the manufacturer’s instructions. RNA samples were then reverse transcribed using the SuperScript VILO cDNA synthesis kit (Invitrogen). Real-time PCR was performed with an ABI 7000 machine (Applied Biosystems) with the SYBR Green PCR Master Mix (Applied Biosystems) and the following primers: p57 (Cdkn1c) sense, 5′-GAAGGACCAGCCTCTCTCG-3′; p57 (Cdkn1c) antisense, 5′-GTTCTCCTGCGCAGTTCTCT-3′; Evi-1 sense, 5′-AACCATGTGTTTGGGGAAAA-3′; Evi-1 antisense, 5′-AGCTTCAAGCGGGTCAGTTA-3′; Gas-6 sense, 5′-GACCCCGAGACGGAGTATTTC-3′; and Gas-6 antisense, 5′-TGCACTGGTCAGGCAAGTTC-3′.
The expression of target genes was normalized using the following housekeeping genes: OAZ sense, 5′-TTTCAGCTAGCATCCTTGTACTCC-3′; OAZ antisense, 5′-GACCCTGGTCTTGTCGTTAGA-3′; SDHA sense, 5′-AAGTTGAGATTTGCCGATGG-3′; and SDHA antisense, 5′-TGGTTCTGCATCGACTTCTG-3′. The data shown in this study refer to data normalized to OAZ. Similar results were obtained when the same data were normalized to SDHA.
Statistical analysis.
Significance levels of data were determined by Student’s t test for the differences in mean values. Asterisks refer to the following p-values: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Acknowledgments
We thank Dr. Reinhard Fässler for providing help with mutant mice at the beginning of the project. We are grateful to Dr. Michael Milsom and Dr. Anne Wilson for critically reading the manuscript and useful discussions. We also wish to thank the members of the DKFZ FACS Core facility and particularly Steffen Schmitt, Gelo de la Cruz, and Ann Atzberger for excellent technical support and the animal caretakers in the DKFZ animal facility for excellent animal husbandry.
This work was supported by the Sonderforschungsbereich (SFB) 873 funded by the Deutsche Forschungsgemeinschaft (DFG) and the Dietmar Hopp Foundation (to A. Trumpp).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- 5-FU
- 5-fluouracil
- dHSC
- dormant HSC
- DUB
- deubiquitinase
- HSC
- hematopoietic stem cell
- LRC
- label-retaining cell
- NEMO
- NF-κB essential modulator
- pI-pC
- polyinosine-polycytidine
- ROS
- reactive oxygen species
- Tx
- tamoxifen
References
- Abbas H.A., Maccio D.R., Coskun S., Jackson J.G., Hazen A.L., Sills T.M., You M.J., Hirschi K.K., and Lozano G.. 2010. Mdm2 is required for survival of hematopoietic stem cells/progenitors via dampening of ROS-induced p53 activity. Cell Stem Cell. 7:606–617 10.1016/j.stem.2010.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeuerle P.A., and Baltimore D.. 1988. I kappa B: a specific inhibitor of the NF-kappa B transcription factor. Science. 242:540–546 10.1126/science.3140380 [DOI] [PubMed] [Google Scholar]
- Baldridge M.T., King K.Y., and Goodell M.A.. 2011. Inflammatory signals regulate hematopoietic stem cells. Trends Immunol. 32:57–65 10.1016/j.it.2010.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudet A., Karlsson C., Safaee Talkhoncheh M., Galeev R., Magnusson M., and Larsson J.. 2012. RNAi screen identifies MAPK14 as a druggable suppressor of human hematopoietic stem cell expansion. Blood. 119:6255–6258 10.1182/blood-2012-01-403949 [DOI] [PubMed] [Google Scholar]
- Bignell G.R., Warren W., Seal S., Takahashi M., Rapley E., Barfoot R., Green H., Brown C., Biggs P.J., Lakhani S.R., et al. . 2000. Identification of the familial cylindromatosis tumour-suppressor gene. Nat. Genet. 25:160–165. [DOI] [PubMed] [Google Scholar]
- Bowen S., Gill M., Lee D.A., Fisher G., Geronemus R.G., Vazquez M.E., and Celebi J.T.. 2005. Mutations in the CYLD gene in Brooke–Spiegler syndrome, familial cylindromatosis, and multiple familial trichoepithelioma: Lack of genotype–phenotype correlation. J. Invest. Dermatol. 124:919–920 10.1111/j.0022-202X.2005.23688.x [DOI] [PubMed] [Google Scholar]
- Bremm A., and Komander D.. 2011. Emerging roles for Lys11-linked polyubiquitin in cellular regulation. Trends Biochem. Sci. 36:355–363 10.1016/j.tibs.2011.04.004 [DOI] [PubMed] [Google Scholar]
- Brummelkamp T.R., Nijman S.M., Dirac A.M., and Bernards R.. 2003. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-κB. Nature. 424:797–801 10.1038/nature01811 [DOI] [PubMed] [Google Scholar]
- Cabezas-Wallscheid N., Klimmeck D., Hansson J., Lipka D.B., Reyes A., Wang Q., Weichenhan D., Lier A., von Paleske L., Renders S., et al. . 2014. Identification of regulatory networks in HSCs and their immediate progeny via integrated proteome, transcriptome, and DNA Methylome analysis. Cell Stem Cell. 15:507–522 10.1016/j.stem.2014.07.005 [DOI] [PubMed] [Google Scholar]
- Campbell R.M., Anderson B.D., Brooks N.A., Brooks H.B., Chan E.M., De Dios A., Gilmour R., Graff J.R., Jambrina E., Mader M., et al. . 2014. Characterization of LY2228820 dimesylate, a potent and selective inhibitor of p38 MAPK with antitumor activity. Mol. Cancer Ther. 13:364–374 10.1158/1535-7163.MCT-13-0513 [DOI] [PubMed] [Google Scholar]
- Carpentier I., Declercq W., Malinin N.L., Wallach D., Fiers W., and Beyaert R.. 1998. TRAF2 plays a dual role in NF-κB-dependent gene activation by mediating the TNF-induced activation of p38 MAPK and IκB kinase pathways. FEBS Lett. 425:195–198 10.1016/S0014-5793(98)00226-9 [DOI] [PubMed] [Google Scholar]
- Chen Z.J.2005. Ubiquitin signalling in the NF-κB pathway. Nat. Cell Biol. 7:758–765 10.1038/ncb0805-758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conze D.B., Wu C.J., Thomas J.A., Landstrom A., and Ashwell J.D.. 2008. Lys63-linked polyubiquitination of IRAK-1 is required for interleukin-1 receptor- and toll-like receptor-mediated NF-κB activation. Mol. Cell. Biol. 28:3538–3547 10.1128/MCB.02098-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L., Wang C., Spencer E., Yang L., Braun A., You J., Slaughter C., Pickart C., and Chen Z.J.. 2000. Activation of the IκB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 103:351–361 10.1016/S0092-8674(00)00126-4 [DOI] [PubMed] [Google Scholar]
- Emmerich C.H., Schmukle A.C., and Walczak H.. 2011. The emerging role of linear ubiquitination in cell signaling. Sci. Signal. 4:re5 10.1126/scisignal.2002187 [DOI] [PubMed] [Google Scholar]
- Essers M.A., Offner S., Blanco-Bose W.E., Waibler Z., Kalinke U., Duchosal M.A., and Trumpp A.. 2009. IFNα activates dormant haematopoietic stem cells in vivo. Nature. 458:904–908 10.1038/nature07815 [DOI] [PubMed] [Google Scholar]
- Foudi A., Hochedlinger K., Van Buren D., Schindler J.W., Jaenisch R., Carey V., and Hock H.. 2009. Analysis of histone 2B-GFP retention reveals slowly cycling hematopoietic stem cells. Nat. Biotechnol. 27:84–90 10.1038/nbt.1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardam S., Sierro F., Basten A., Mackay F., and Brink R.. 2008. TRAF2 and TRAF3 signal adapters act cooperatively to control the maturation and survival signals delivered to B cells by the BAFF receptor. Immunity. 28:391–401 10.1016/j.immuni.2008.01.009 [DOI] [PubMed] [Google Scholar]
- Göthert J.R., Gustin S.E., Hall M.A., Green A.R., Göttgens B., Izon D.J., and Begley C.G.. 2005. In vivo fate-tracing studies using the Scl stem cell enhancer: embryonic hematopoietic stem cells significantly contribute to adult hematopoiesis. Blood. 105:2724–2732 10.1182/blood-2004-08-3037 [DOI] [PubMed] [Google Scholar]
- Goyama S., Yamamoto G., Shimabe M., Sato T., Ichikawa M., Ogawa S., Chiba S., and Kurokawa M.. 2008. Evi-1 is a critical regulator for hematopoietic stem cells and transformed leukemic cells. Cell Stem Cell. 3:207–220 10.1016/j.stem.2008.06.002 [DOI] [PubMed] [Google Scholar]
- Grech A.P., Amesbury M., Chan T., Gardam S., Basten A., and Brink R.. 2004. TRAF2 differentially regulates the canonical and noncanonical pathways of NF-κB activation in mature B cells. Immunity. 21:629–642 10.1016/j.immuni.2004.09.011 [DOI] [PubMed] [Google Scholar]
- Haglund K., and Dikic I.. 2012. The role of ubiquitylation in receptor endocytosis and endosomal sorting. J. Cell Sci. 125:265–275 10.1242/jcs.091280 [DOI] [PubMed] [Google Scholar]
- Hofmann R.M., and Pickart C.M.. 1999. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell. 96:645–653 10.1016/S0092-8674(00)80575-9 [DOI] [PubMed] [Google Scholar]
- Hövelmeyer N., Wunderlich F.T., Massoumi R., Jakobsen C.G., Song J., Wörns M.A., Merkwirth C., Kovalenko A., Aumailley M., Strand D., et al. . 2007. Regulation of B cell homeostasis and activation by the tumor suppressor gene CYLD. J. Exp. Med. 204:2615–2627 10.1084/jem.20070318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutti J.E., Shen R.R., Abbott D.W., Zhou A.Y., Sprott K.M., Asara J.M., Hahn W.C., and Cantley L.C.. 2009. Phosphorylation of the tumor suppressor CYLD by the breast cancer oncogene IKKε promotes cell transformation. Mol. Cell. 34:461–472 10.1016/j.molcel.2009.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Hirao A., Arai F., Takubo K., Matsuoka S., Miyamoto K., Ohmura M., Naka K., Hosokawa K., Ikeda Y., and Suda T.. 2006. Reactive oxygen species act through p38 MAPK to limit the lifespan of hematopoietic stem cells. Nat. Med. 12:446–451 10.1038/nm1388 [DOI] [PubMed] [Google Scholar]
- Joaquin M., Gubern A., González-Nuñez D., Josué Ruiz E., Ferreiro I., de Nadal E., Nebreda A.R., and Posas F.. 2012. The p57 CDKi integrates stress signals into cell-cycle progression to promote cell survival upon stress. EMBO J. 31:2952–2964 10.1038/emboj.2012.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazakov D.V., Zelger B., Rütten A., Vazmitel M., Spagnolo D.V., Kacerovska D., Vanecek T., Grossmann P., Sima R., Grayson W., et al. . 2009. Morphologic diversity of malignant neoplasms arising in preexisting spiradenoma, cylindroma, and spiradenocylindroma based on the study of 24 cases, sporadic or occurring in the setting of Brooke-Spiegler syndrome. Am. J. Surg. Pathol. 33:705–719 10.1097/PAS.0b013e3181966762 [DOI] [PubMed] [Google Scholar]
- Komander D., Lord C.J., Scheel H., Swift S., Hofmann K., Ashworth A., and Barford D.. 2008. The structure of the CYLD USP domain explains its specificity for Lys63-linked polyubiquitin and reveals a B box module. Mol. Cell. 29:451–464 10.1016/j.molcel.2007.12.018 [DOI] [PubMed] [Google Scholar]
- Kovalenko A., Chable-Bessia C., Cantarella G., Israël A., Wallach D., and Courtois G.. 2003. The tumour suppressor CYLD negatively regulates NF-κB signalling by deubiquitination. Nature. 424:801–805 10.1038/nature01802 [DOI] [PubMed] [Google Scholar]
- Kühn R., Schwenk F., Aguet M., and Rajewsky K.. 1995. Inducible gene targeting in mice. Science. 269:1427–1429 10.1126/science.7660125 [DOI] [PubMed] [Google Scholar]
- Li B., Jia N., Waning D.L., Yang F.C., Haneline L.S., and Chun K.T.. 2007. Cul4A is required for hematopoietic stem-cell engraftment and self-renewal. Blood. 110:2704–2707 10.1182/blood-2006-12-064154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massoumi R., Chmielarska K., Hennecke K., Pfeifer A., and Fässler R.. 2006. Cyld inhibits tumor cell proliferation by blocking Bcl-3-dependent NF-κB signaling. Cell. 125:665–677 10.1016/j.cell.2006.03.041 [DOI] [PubMed] [Google Scholar]
- Matsumoto A., Takeishi S., Kanie T., Susaki E., Onoyama I., Tateishi Y., Nakayama K., and Nakayama K.I.. 2011. p57 is required for quiescence and maintenance of adult hematopoietic stem cells. Cell Stem Cell. 9:262–271 10.1016/j.stem.2011.06.014 [DOI] [PubMed] [Google Scholar]
- Matsuoka S., Oike Y., Onoyama I., Iwama A., Arai F., Takubo K., Mashimo Y., Oguro H., Nitta E., Ito K., et al. . 2008. Fbxw7 acts as a critical fail-safe against premature loss of hematopoietic stem cells and development of T-ALL. Genes Dev. 22:986–991 10.1101/gad.1621808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M.M., Thummar K., Mandelbaum J., Pasqualucci L., and Rathinam C.V.. 2015. Lack of the ubiquitin-editing enzyme A20 results in loss of hematopoietic stem cell quiescence. J. Exp. Med. 212:203–216 10.1084/jem.20132544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannem R.R., Dorn C., Ahlqvist K., Bosserhoff A.K., Hellerbrand C., and Massoumi R.. 2014. CYLD controls c-MYC expression through the JNK-dependent signaling pathway in hepatocellular carcinoma. Carcinogenesis. 35:461–468 10.1093/carcin/bgt335 [DOI] [PubMed] [Google Scholar]
- Pargellis C., Tong L., Churchill L., Cirillo P.F., Gilmore T., Graham A.G., Grob P.M., Hickey E.R., Moss N., Pav S., and Regan J.. 2002. Inhibition of p38 MAP kinase by utilizing a novel allosteric binding site. Nat. Struct. Biol. 9:268–272 10.1038/nsb770 [DOI] [PubMed] [Google Scholar]
- Peng J., Schwartz D., Elias J.E., Thoreen C.C., Cheng D., Marsischky G., Roelofs J., Finley D., and Gygi S.P.. 2003. A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 21:921–926 10.1038/nbt849 [DOI] [PubMed] [Google Scholar]
- Pronk C.J., Veiby O.P., Bryder D., and Jacobsen S.E.. 2011. Tumor necrosis factor restricts hematopoietic stem cell activity in mice: involvement of two distinct receptors. J. Exp. Med. 208:1563–1570 10.1084/jem.20110752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam C., Thien C.B., Langdon W.Y., Gu H., and Flavell R.A.. 2008. The E3 ubiquitin ligase c-Cbl restricts development and functions of hematopoietic stem cells. Genes Dev. 22:992–997 10.1101/gad.1651408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam C., Matesic L.E., and Flavell R.A.. 2011. The E3 ligase Itch is a negative regulator of the homeostasis and function of hematopoietic stem cells. Nat. Immunol. 12:399–407 10.1038/ni.2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiley W., Zhang M., and Sun S.C.. 2004. Negative regulation of JNK signaling by the tumor suppressor CYLD. J. Biol. Chem. 279:55161–55167 10.1074/jbc.M411049200 [DOI] [PubMed] [Google Scholar]
- Saggar S., Chernoff K.A., Lodha S., Horev L., Kohl S., Honjo R.S., Brandt H.R., Hartmann K., and Celebi J.T.. 2008. CYLD mutations in familial skin appendage tumours. J. Med. Genet. 45:298–302 10.1136/jmg.2007.056127 [DOI] [PubMed] [Google Scholar]
- Sasai M., Tatematsu M., Oshiumi H., Funami K., Matsumoto M., Hatakeyama S., and Seya T.. 2010. Direct binding of TRAF2 and TRAF6 to TICAM-1/TRIF adaptor participates in activation of the Toll-like receptor 3/4 pathway. Mol. Immunol. 47:1283–1291 10.1016/j.molimm.2009.12.002 [DOI] [PubMed] [Google Scholar]
- Saur S.J., Sangkhae V., Geddis A.E., Kaushansky K., and Hitchcock I.S.. 2010. Ubiquitination and degradation of the thrombopoietin receptor c-Mpl. Blood. 115:1254–1263 10.1182/blood-2009-06-227033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S., Watanabe T., Lin C.S., William C.M., Tanabe Y., Jessell T.M., and Costantini F.. 2001. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1:4 10.1186/1471-213X-1-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srokowski C.C., Masri J., Hövelmeyer N., Krembel A.K., Tertilt C., Strand D., Mahnke K., Massoumi R., Waisman A., and Schild H.. 2009. Naturally occurring short splice variant of CYLD positively regulates dendritic cell function. Blood. 113:5891–5895 10.1182/blood-2008-08-175489 [DOI] [PubMed] [Google Scholar]
- Stephens P.J., Davies H.R., Mitani Y., Van Loo P., Shlien A., Tarpey P.S., Papaemmanuil E., Cheverton A., Bignell G.R., Butler A.P., et al. . 2013. Whole exome sequencing of adenoid cystic carcinoma. J. Clin. Invest. 123:2965–2968 10.1172/JCI67201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S.C.2008. Deubiquitylation and regulation of the immune response. Nat. Rev. Immunol. 8:501–511 10.1038/nri2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa H., Regoes R.R., Boddupalli C.S., Bonhoeffer S., and Manz M.G.. 2011. Dynamic variation in cycling of hematopoietic stem cells in steady state and inflammation. J. Exp. Med. 208:273–284 10.1084/jem.20101643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesio M., Oser G.M., Baccelli I., Blanco-Bose W., Wu H., Göthert J.R., Kogan S.C., and Trumpp A.. 2013. Pten loss in the bone marrow leads to G-CSF–mediated HSC mobilization. J. Exp. Med. 210:2337–2349 10.1084/jem.20122768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalheimer F.B., Wingert S., De Giacomo P., Haetscher N., Rehage M., Brill B., Theis F.J., Hennighausen L., Schroeder T., and Rieger M.A.. 2014. Cytokine-regulated GADD45G induces differentiation and lineage selection in hematopoietic stem cells. Stem Cell Rev. 3:34–43 10.1016/j.stemcr.2014.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson B.J., Jankovic V., Gao J., Buonamici S., Vest A., Lee J.M., Zavadil J., Nimer S.D., and Aifantis I.. 2008. Control of hematopoietic stem cell quiescence by the E3 ubiquitin ligase Fbw7. J. Exp. Med. 205:1395–1408 10.1084/jem.20080277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trompouki E., Hatzivassiliou E., Tsichritzis T., Farmer H., Ashworth A., and Mosialos G.. 2003. CYLD is a deubiquitinating enzyme that negatively regulates NF-κB activation by TNFR family members. Nature. 424:793–796 10.1038/nature01803 [DOI] [PubMed] [Google Scholar]
- Trompouki E., Tsagaratou A., Kosmidis S.K., Dollé P., Qian J., Kontoyiannis D.L., Cardoso W.V., and Mosialos G.. 2009. Truncation of the catalytic domain of the cylindromatosis tumor suppressor impairs lung maturation. Neoplasia. 11:469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trumpp A., Essers M., and Wilson A.. 2010. Awakening dormant haematopoietic stem cells. Nat. Rev. Immunol. 10:201–209 10.1038/nri2726 [DOI] [PubMed] [Google Scholar]
- Vallabhapurapu S., Matsuzawa A., Zhang W., Tseng P.H., Keats J.J., Wang H., Vignali D.A., Bergsagel P.L., and Karin M.. 2008. Nonredundant and complementary functions of TRAF2 and TRAF3 in a ubiquitination cascade that activates NIK-dependent alternative NF-κB signaling. Nat. Immunol. 9:1364–1370 10.1038/ni.1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wajant H., and Scheurich P.. 2001. Tumor necrosis factor receptor-associated factor (TRAF) 2 and its role in TNF signaling. Int. J. Biochem. Cell Biol. 33:19–32 10.1016/S1357-2725(00)00064-9 [DOI] [PubMed] [Google Scholar]
- Welz P.S., Wullaert A., Vlantis K., Kondylis V., Fernández-Majada V., Ermolaeva M., Kirsch P., Sterner-Kock A., van Loo G., and Pasparakis M.. 2011. FADD prevents RIP3-mediated epithelial cell necrosis and chronic intestinal inflammation. Nature. 477:330–334 10.1038/nature10273 [DOI] [PubMed] [Google Scholar]
- Wertz I.E., O’Rourke K.M., Zhou H., Eby M., Aravind L., Seshagiri S., Wu P., Wiesmann C., Baker R., Boone D.L., et al. . 2004. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signalling. Nature. 430:694–699 10.1038/nature02794 [DOI] [PubMed] [Google Scholar]
- Wilson A., Laurenti E., Oser G., van der Wath R.C., Blanco-Bose W., Jaworski M., Offner S., Dunant C.F., Eshkind L., Bockamp E., et al. . 2008. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 135:1118–1129 10.1016/j.cell.2008.10.048 [DOI] [PubMed] [Google Scholar]
- Young A.L., Kellermayer R., Szigeti R., Tészás A., Azmi S., and Celebi J.T.. 2006. CYLD mutations underlie Brooke-Spiegler, familial cylindromatosis, and multiple familial trichoepithelioma syndromes. Clin. Genet. 70:246–249 10.1111/j.1399-0004.2006.00667.x [DOI] [PubMed] [Google Scholar]
- Zhang M., Lee A.J., Wu X., and Sun S.C.. 2011. Regulation of antiviral innate immunity by deubiquitinase CYLD. Cell. Mol. Immunol. 8:502–504 10.1038/cmi.2011.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Qin H., He Y., Huang H., Lu Y., and Chu X.. 2012. Inhibiting p38 mitogen-activated protein kinase attenuates cerebral ischemic injury in Swedish mutant amyloid precursor protein transgenic mice. Neural Regen Res. 7:1088–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou P., Yoshihara H., Hosokawa K., Tai I., Shinmyozu K., Tsukahara F., Maru Y., Nakayama K., Nakayama K.I., and Suda T.. 2011. p57(Kip2) and p27(Kip1) cooperate to maintain hematopoietic stem cell quiescence through interactions with Hsc70. Cell Stem Cell. 9:247–261 10.1016/j.stem.2011.07.003 [DOI] [PubMed] [Google Scholar]