Zhou et al. demonstrate a requirement for the Let-7–Lin28b axis regulating a shift in development between fetal liver and bone marrow B lymphocyte progenitors in the generation of B1 versus B2 B cells. Specifically, the transcription factor Arid3a, induced by Lin28b and a target of Let-7 miRNA, is sufficient to recapitulate fetal B cell development from bone marrow progenitors.
Abstract
Mouse B cell precursors from fetal liver and adult bone marrow (BM) generate distinctive B cell progeny when transplanted into immunodeficient recipients, supporting a two-pathway model for B lymphopoiesis, fetal “B-1” and adult “B-2.” Recently, Lin28b was shown to be important for the switch between fetal and adult pathways; however, neither the mechanism of Lin28b action nor the importance of B cell antigen receptor (BCR) signaling in this process was addressed. Here, we report key advances in our understanding of the regulation of B-1/B-2 development. First, modulation of Let-7 in fetal pro-B cells is sufficient to alter fetal B-1 development to produce B cells resembling the progeny of adult B-2 development. Second, intact BCR signaling is required for the generation of B1a B cells from Lin28b-transduced BM progenitors, supporting a requirement for ligand-dependent selection, as is the case for normal B1a B cells. Third, the VH repertoire of Lin28b-induced BM B1a B cells differs from that of normal B1a, suggesting persisting differences from fetal progenitors. Finally, we identify the Arid3a transcription factor as a key target of Let-7, whose ectopic expression is sufficient to induce B-1 development in adult pro-B cells and whose silencing by knockdown blocks B-1 development in fetal pro-B cells.
B cells, a key arm of the immune system responsible for humoral immunity, are generated through a tightly regulated sequence of developmental stages, in the liver before birth and in the BM of adults. During B cell development, Ig heavy and light chains are rearranged and selected, yielding a diverse antigen receptor repertoire that is largely purged of high-affinity pathogenic self-reactivity (Nemazee, 2006; Goodnow, 2007). Importantly, mature B cells in mice are not completely homogenous across anatomical sites. In particular, certain functionally distinct subsets, such as the CD5+ B cell (“B1a”) subset, show some degree of self-reactivity (Hayakawa et al., 1984). A key unresolved issue is how these self-reactive cell types diverge from the primary B cell development pathway that generates follicular B cells. Study of the Ig heavy and light chains rearranged in these cells has shown that they constitute a biased set of B cell antigen receptors (BCRs; Förster et al., 1988; Pennell et al., 1989; Carmack et al., 1990), some of which have been shown to be selected by interaction with self-determinants (Hayakawa et al., 1999), suggesting an instructive antigen-dependent model for CD5+ B cell generation.
Early BM transfer experiments revealed poor generation of CD5+ B cells (B1a) in adult hosts (Hayakawa et al., 1985). Later experiments, using more defined populations of B cell progenitors from fetal and adult sources, showed that fetal precursors supported efficient production of B1a B cells, but repopulation of typical follicular B cells was inefficient (Hardy and Hayakawa, 1991). These results led us to propose a switch in B cell lymphopoiesis during ontogeny, similar to the well-known switch from fetal to adult hemoglobin in erythropoiesis (Groudine et al., 1983). Specifically, we suggested that the fetal pathway of development (termed “B-1”) is responsible for generating most of the CD5+ B cell pool, whereas an adult pathway (termed “B-2”) generates most of the CD5− B cells that populate the adult (Hardy and Hayakawa, 2001). The latter cells are often identified as follicular or “B2” B cells. We hypothesized that a distinctive gene program operating in B cell progenitors is responsible for the fetal-biased B-1 generation of CD5+ B cells.
Therefore we analyzed fetal- and adult-origin B cell precursors for mRNA and microRNA (miRNA) expression differences by microarray to rigorously identify potential regulators that might play a role in the B-1/B-2 developmental switch. Cell fractions where initial Ig heavy chain rearrangement takes place, pre-pro-B (Fr. A) and pro-B (Fr. BC), were analyzed because these populations span the stage at which B lineage commitment occurs (Rumfelt et al., 2006). Based on this idea of distinctive fetal and adult lymphopoiesis, Yuan et al. (2012) performed and published a similar analysis of pro-B stage cells, comparing gene expression and miRNA expression in such cells purified from fetal liver (FL) and adult BM of mice. They found that retroviral expression of Lin28b in BM stem cells generated innate-type B and T cells in transfer recipients (Yuan et al., 2012). Considering the differential expression of Lin28b and Let-7 that they identified, they hypothesized that this “axis” functions to promote fetal development of B1a B cells and innate-type T cells.
Here we have asked whether perturbation of this regulatory axis can reprogram cells later than stem cells, at the committed pro-B cell stage. We have also asked whether reprogrammed cells still depend on BCR signaling for B1a B cell generation and identified genes altered by this axis to understand the mechanism of reprogramming. We found that ectopic provision of Lin28b as late as the pro-B stage in adults can redirect cells to the B-1 developmental pathway. Conversely, expression of Let-7b miRNA in fetal pro-B cells can switch fetal development to B-2. Transduction of Btk-deficient precursor cells from Xid mouse BM (Hayakawa et al., 1983; Rawlings et al., 1993; Khan et al., 1995) reveals that normal BCR signaling is required for accumulation of CD5+ B1 B cells, even when they are generated from Lin28b-redirected BM pro-B cells in the adult. Finally, we report a critical new role for the Arid3a (Bright, E2fbp1) transcription factor in promoting B-1 development. We found this factor in a gene expression screen designed to identify genes whose expression was reciprocally regulated by the Lin28b–Let-7 axis. Significantly, the Arid3a mRNA contained multiple Let-7 target motifs, consistent with regulation by this miRNA. Strikingly, retroviral transduction of adult BM pro-B cells with Arid3a induced generation of B1a B cells, and conversely, Arid3a shRNA inhibited B1a generation from FL pro-B cells, revealing its key role in the switch from B-2 to B-1 development.
RESULTS
Fetal and adult B cell progenitors: Distinctive progeny and differential Lin28b–Let-7 expression
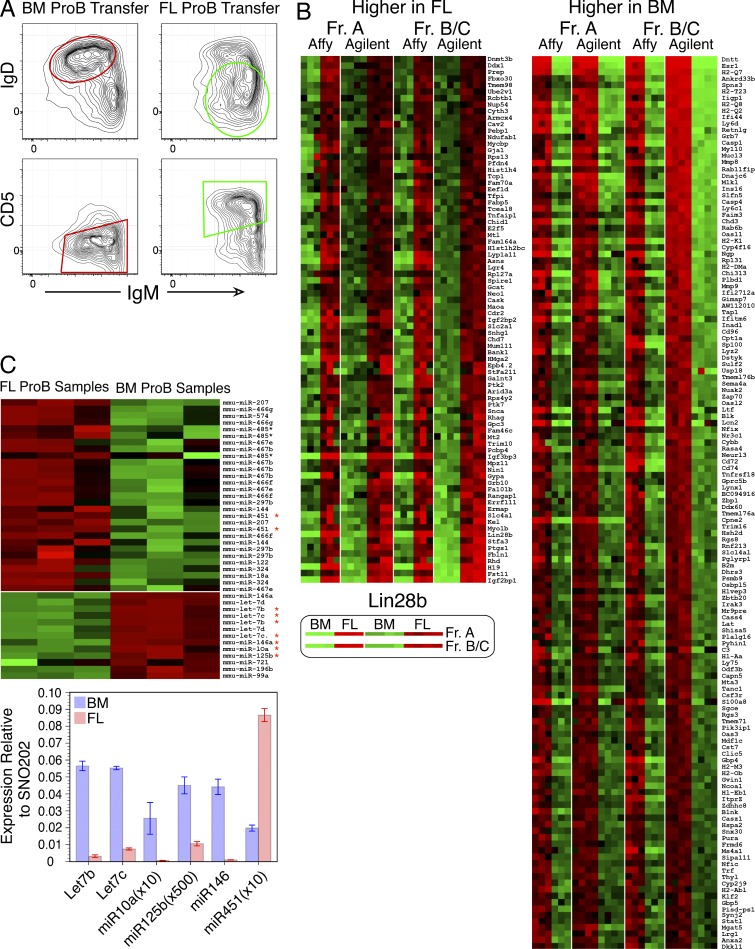
Pro-B cells transferred into SCID mice generate B cells in the spleen and peritoneal cavity: most B cells arising from adult BM transfers are IgM+IgD2+CD23+CD5−, in contrast to FL transfers which yield IgM2+IgD+/−CD23+/− B cells that are predominantly CD5+ (Fig. 1 A). Thus, B cells with distinctive cell surface phenotypes are generated in adult SCID mice from fetal and adult CD19+ B cell precursors. Microarray analyses were performed to identify genes whose expression distinguished B-1 and B-2 development at pre-pro-B and pro-B stage cells, using two different platforms, selecting genes that showed concordant results in both stages and platforms. As shown previously by RT-PCR analysis, expression of several genes key for B cell development, including Rag1, Rag2, Lambda5/Igll1, and VpreB1, does not differ significantly between these two cell types, whereas terminal deoxynucleotidyl transferase (TdT; encoded by the Dntt gene) is strikingly abundant in adult precursors but absent from those in FL (Li et al., 1993). These criteria filtered 567 Fr. A genes and 592 Fr. BC genes to a final set of 219, shown in the heat map in Fig. 1 B. Gene lists and measurements used to derive this set are provided in Datasets S1–S7.
Figure 1.
Fetal and adult pro-B cells generate B cells with distinct phenotype and show differential expression of mRNAs and miRNAs. (A) Pro-B cells were sorted from FL and BM and then transferred i.v. into immunodeficient SCID mice. Peritoneal cavity washout cells (PerC) were analyzed 3 wk later for the indicated surface proteins by flow cytometry. Data are representative of five independent transfer experiments with two mice per group. (B) Heat map displays of mRNA fold change comparing fetal and adult pre-pro-B (Fr. A) and pro-B (Fr. BC) stage cell samples by microarray of genes higher in FL or in BM using both the Affymetrix (C57BL/6J) and Agilent Technologies (BALB/c ICR) platforms. This analysis identifies 219 differentially expressed genes. (C) Heat map display of microarray data (top) and Q-PCR analysis (bottom) of miRNA species differentially expressed between adult BM and FL pro-B cells (Fr. BC). miRNAs in the heat map that are also analyzed by Q-PCR are indicated by a red asterisk. Q-PCR data were normalized to SNO202 expression. All expression analyses were performed in duplicate or triplicate, with each sample representing RNA or miRNA from at least four mice. Error bars represent ±SEM.
miRNA expression was analyzed using the Agilent Technologies miRNA microarray platform, identifying a set of miRNAs that were differentially expressed, including many Let-7 family miRs that were up-regulated in BM and miR-451 that was up-regulated in FL (Fig. 1 C, top; and all data in Dataset S8). This pattern of expression was confirmed using miRNA TaqMan assays, showing the most significant differences among those tested to be miR-10a, miR-125b, miR-146, and Let-7 members in BM and miR-451 in FL (Fig. 1 C, bottom). Importantly, Lin28b and Let-7 can function as a molecular switch, in that Let-7 targets Lin28b mRNA for degradation and Lin28b functions to sequester pre–Let-7 from processing to functional miRNA (Mayr et al., 2012).
Redirecting fetal and adult B cell development at the B cell–committed pro-B stage
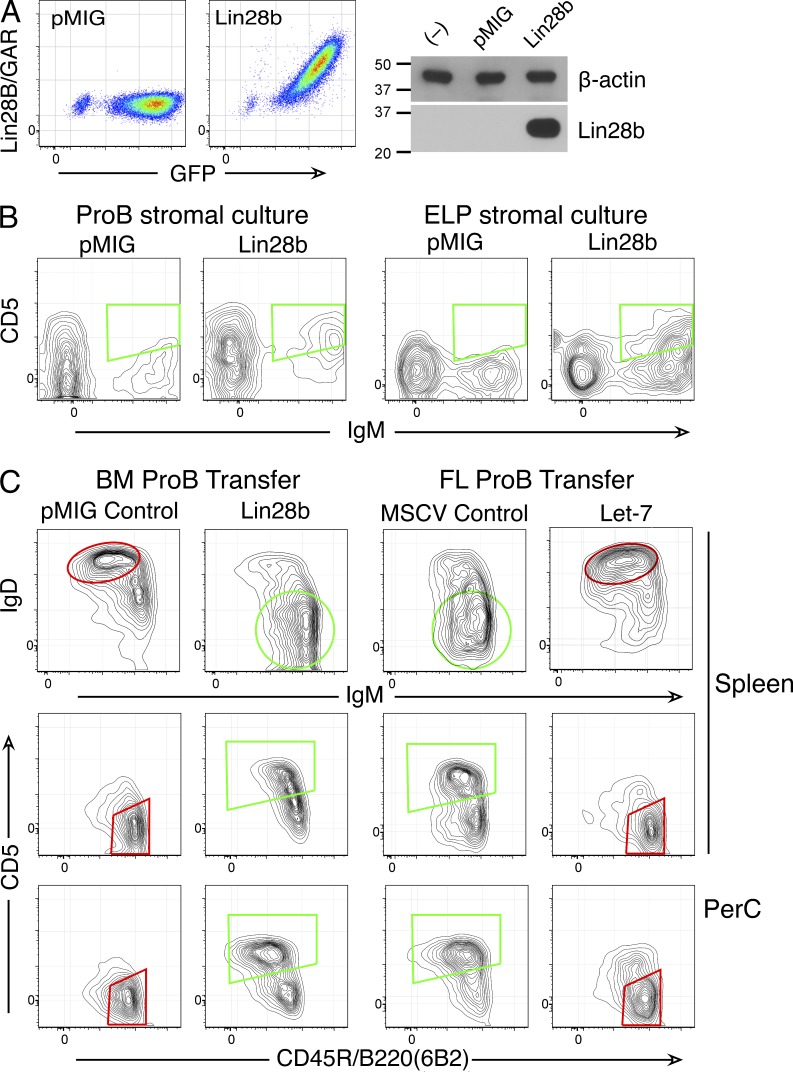
To assess the developmental stage at which these regulators determine development, fetal and adult cells were retrovirally transduced to express Let-7b and Lin28b. Quantitative PCR (Q-PCR) validated expression of Let-7b in FL pro-B (not depicted); cytoplasmic staining and Western blotting verified Lin28b expression in a pro-B cell line (Fig. 2 A). The developmental consequences of Lin28b expression in BM pro-B or early lymphoid progenitors were assessed by culturing transduced cells for an extended period. Many of the IgM+ cells produced in these cultures were CD5+, whereas cells transduced with empty vector control retrovirus were not (Fig. 2 B). Mature B cells are not generated efficiently in vitro, so we next performed cell transfer experiments with pro-B cells transduced to inappropriately express these regulators, i.e., Let-7 in FL and Lin28b in BM. Results, shown in Fig. 2 C, were very clear. SCID mice receiving empty vector BM pro-B cells generated GFP+ B cells that were IgM+IgD2+CD5−CD23+CD21med (follicular type); in contrast, Lin28b BM pro-B cells generated GFP+ B cells that were IgM2+IgD−CD23−CD21med and many CD5+. In contrast, SCID mice receiving empty vector FL pro-B cells generated GFP+ B cells that were IgM2+IgD+/−CD23lo and many CD5+; in contrast, animals receiving Let-7b FL pro-B cells generated GFP+ B cells that were IgM+IgD2+CD5−CD23+. These findings indicate that alterations in the Lin28–Let-7 axis are both necessary and sufficient to specify fetal and adult lymphopoietic programs even after progenitors have adopted the B cell fate. These differential B cell phenotypes are shown for spleen B cells in Fig. 3 and for peritoneal cavity–resident B cells in Fig. 4. Quantitation of cell numbers engrafted in SCID mice 3 wk after transfer, shown in Table 1, reveals somewhat increased numbers in Lin28b-transduced recipients and reduced numbers in Let-7–transduced recipients. These differences likely arise from changes in cell proliferation at the pre-B stage resulting from expression of these regulators.
Figure 2.
Lin28b and Let-7 miRNA reciprocally redirect adult and fetal B cell development. (A) The Ret02 pro-B stage cell line, which lacks expression of Lin28b, was transduced with a Lin28b-containing retrovirus or with empty vector (pMIG). 1 d after transduction, cells were examined for Lin28b expression by cytoplasmic staining using flow cytometry (left) and Western blotting (right). Molecular mass is indicated in kilodaltons. (B) BM pro-B and ELP stage cells, 1 d after initiation of stromal cell culture, were transduced with empty vector or a Lin28b-containing retrovirus. After 11 (pro-B) or 15 d (ELP), lymphoid cells were recovered from cultures and analyzed by flow cytometry. CD19+GFP+ cells (pro-B) or CD19+Cherry+ cells (for ELP) are shown in the plots. 20–40% of IgM+ cells were CD5+ in cultures of Lin28b-transduced BM, whereas <5% were positive with pMIG. Representative data from three independent experiments are shown in flow cytometry plots. (C) Analysis of B cells generated in SCID mice, 3 wk after transfer of Lin28b (or pMIG empty vector)-transduced BM pro-B or Let-7 (or MSCV empty vector)–transduced FL pro-B cells. Lin28b-induced fetal-type B cells from BM (green regions of interest; IgM2+IgD+/−CD45R+ CD5+) and empty vector B cells (follicular/B2 phenotype; red regions of interest; IgM+IgD2+CD45R2+CD5−) are shown. Plots are gated for CD19+GFP+ cells. Data are representative of five independent experiments, with two mice per group. PerC, peritoneal cavity washout cells.
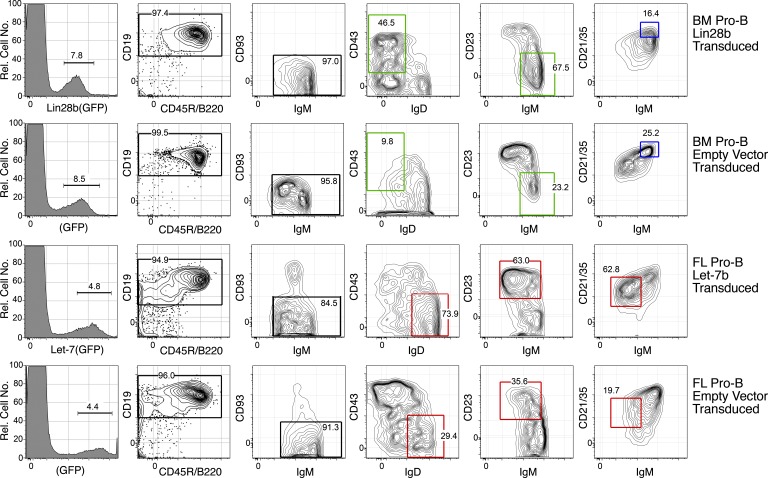
Figure 3.
Phenotype of Lin28b-transduced BM and Let-7–transduced FL generates B cells in recipient spleen that resemble B1 and FO/B2. CD19+GFP+ mature (CD93−) B cells in spleen were generated from transfer of the indicated pro-B–transduced cells, 3 wk after transfer in SCID mice, and analyzed by flow cytometry using the indicated markers. The red boxes indicate an adult-type and the green boxes indicate a fetal-type phenotype. The blue gates represent IgMhiCD21hi cells, a marginal zone B cell type. Data are representative of five independent transfer experiments with two mice per group.
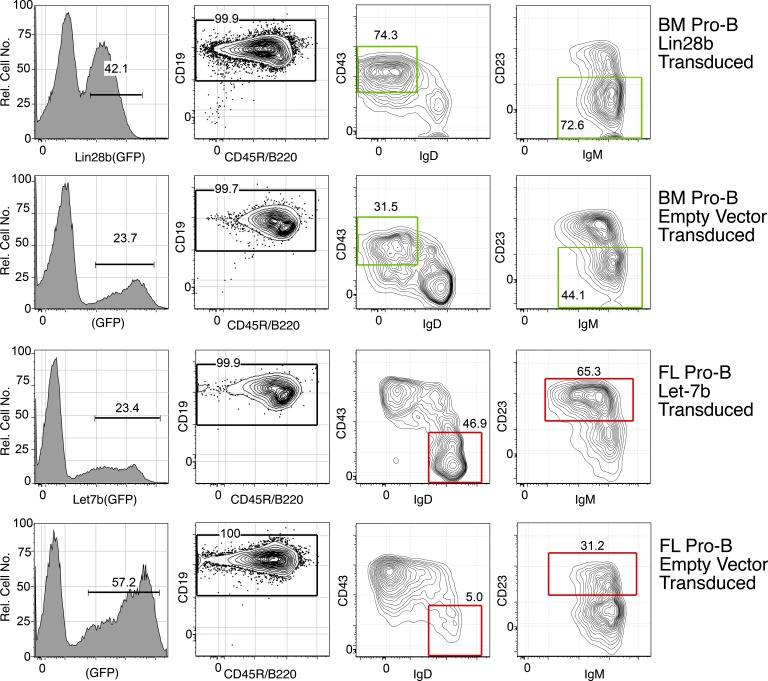
Figure 4.
Phenotype of Lin28b-transduced BM and Let-7–transduced FL generates B cells in recipient peritoneal cavity that resemble B1 and B2. CD19+GFP+ cells in the peritoneal cavity were generated from transfer of the indicated pro-B–transduced cells, 3 wk after transfer in SCID mice, and analyzed by flow cytometry using the indicated markers. The red boxes indicate an adult-type and the green boxes indicate a fetal-type phenotype. Data are representative of five independent transfer experiments with two mice per group.
Table 1.
Cell numbers engrafted in SCID mice by transfer of transduced pro-B cells
| Cell type (×104) | BM pro-B | FL pro-B | ||||||
| pMIG | SE | Lin28b | SE | MSCV | SE | Let-7b | SE | |
| Spleen B1a | 4.74 | 1.59 | 34.81 | 8.44 | 10.56 | 0.74 | 1.08 | 0.25 |
| Spleen Fo | 32.81 | 8.71 | 10.78 | 2.45 | 4.99 | 0.96 | 6.12 | 1.25 |
| Spleen MZ | 29.57 | 8.01 | 12.22 | 3.11 | 12.83 | 1.12 | 4.09 | 0.47 |
| PerC B1a | 7.02 | 2.94 | 51.04 | 10.67 | 40.12 | 10.78 | 3.14 | 0.71 |
MZ, marginal zone. CD19+GFP+ cells of the indicated cell type were engrafted in SCID mice 3 wk after cell transfer of transduced pro-B cells. Mean and standard error (SE) are shown; for each transfer sample type, n = 4. pMIG and MSCV are control empty vectors.
Lin28b-induced BM B1a B cells: Requirement for intact BCR signaling
The generation of normal CD5+ B cells depends on the presence of antigen (Hayakawa et al., 1999). Furthermore, mouse mutants that weaken BCR signaling have a deficit in CD5+ B cells (Tarakhovsky et al., 1995; Inaoki et al., 1997; Suzuki et al., 1999). For example, the Btk-deficient Xid mouse has fewer B cells in spleen, accumulates B cells with a distinctive phenotype, has very low levels of serum IgM, and completely lacks CD5+ B cells (Hayakawa et al., 1983; Khan et al., 1995). However, the ability of ectopic expression of Lin28B to divert cells to the B-1 fate in progenitors with defective BCR signaling has never been directly tested. Therefore, we asked whether BCR signaling is required for production of Lin28b-induced BM-derived B1a B cells. For this purpose, transduced pro-B cell isolated from BM of Xid mice were transferred into SCID mice, and then engrafted cells were examined for generation of CD5+ B cells. As shown in Fig. 5 A, even provision of Lin28b did not induce production of CD5+ B cells from Btk-deficient pro-B cells, demonstrating a continuing dependence on intact (Btk dependent) BCR signaling.
Figure 5.
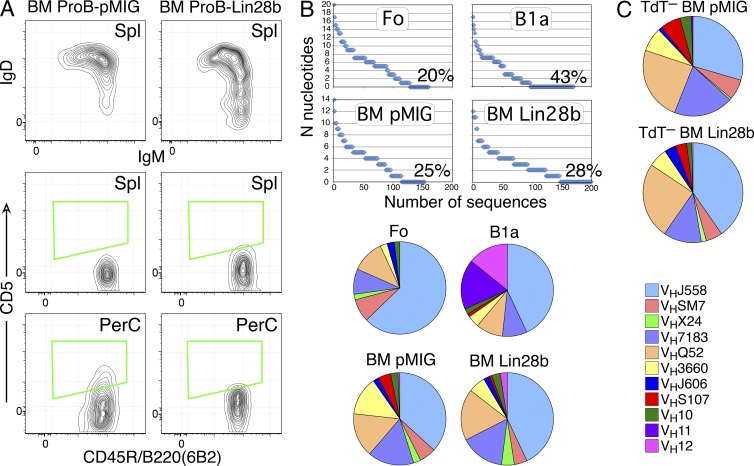
Lin28b BM–generated CD5+ B cells require intact BCR signaling but do not express the usual B1a repertoire, regardless of TdT expression. (A) Btk-deficient BM pro-B cells, transduced with Lin28b or empty (pMIG) retrovirus, were transferred to SCID mice, and spleen cells were analyzed 3 wk after transfer by flow cytometry using the indicated markers. Plots are gated for CD19+GFP+ cells. Data are representative of two independent transfer experiments with two mice per group. (B) The level of N-addition and distribution of VH gene usage was analyzed in individual sorted follicular/B2 (Fo) and CD5+ B1 (B1a) B cells from intact mice and in B cells from SCID mice repopulated with BM pro-B cells (transduced with empty vector [pMIG] or Lin28b-containing retrovirus). Data were combined from three separate experiments for each cell type, each experiment using two mice for each sample. (C) The VH gene distribution in B cells from SCID mice repopulated with BM pro-B cells from TdT− mice (transduced as in B) is illustrated in pie charts. Data were combined from three separate experiments for each cell type, each experiment using two mice for each sample.
Lin28b-induced BM B1a B cells: VH repertoire different from wild-type B1a B cells
CD5+ B cells (B1a) generated during fetal development have been shown to have a biased BCR repertoire (Förster et al., 1988; Pennell et al., 1989; Carmack et al., 1990), enriched for self-reactivity and production of natural autoantibodies (Hayakawa et al., 1983, 1984). Therefore, single cell sequence analysis of VDJs expressed in normal B cell subsets and in transduced B cells generated in SCID mice was performed to compare the VH segments used in B cells generated from Lin28b- and empty vector–transduced BM pro-B cells (Fig. 5 B, including data from normal CD5+ B cells and follicular/B2 B cells, sequences in Dataset S9). There were some repeated sequences in Lin28b B1a cells, but fewer than in the normal CD5+ B cell population (not depicted), and segments lacking N-addition were less frequent. The most striking difference observed was in VH usage, where few cells used VH11 or VH12 genes, in contrast with normal CD5+ B cells, where such rearrangements are abundant.
We then asked whether the failure to generate a typical B1a fetal-type repertoire was the result of relatively weak down-regulation of Dntt, the gene which codes for TdT, by Lin28b. Because increased N-addition in Ig heavy chains sequenced from B1a B cells generated by Lin28b-transduced BM pro-B cells was observed, similar transfer experiments were performed using pro-B cells from TdT− mice. Results from single cell sequence analysis still showed a repertoire quite distinct from that of normal B1a B cells (Fig. 5 C), indicating that N-addition in Lin28b-transduced pro-B cell development is not the reason for the different VH gene distribution.
Genes regulated by the Lin28b–Let-7 axis: Role of Arid3a
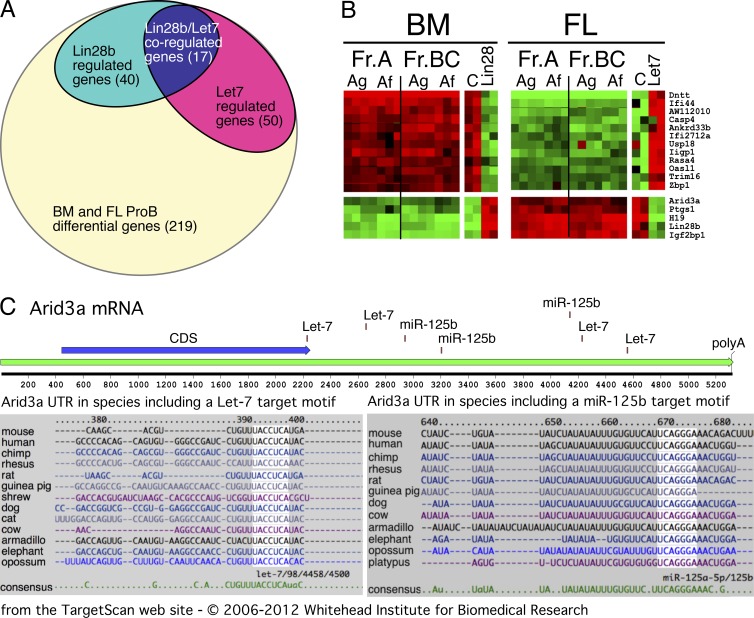
To gain insight into genes that may be influencing the development of fetal versus adult B cells as a consequence of expression of Lin28b or Let-7, fetal and adult pro-B cells were transduced as above, and then, 4 d after transduction, RNA was prepared from sorted GFP+ cells. Microarray analysis identified 219 genes as differentially expressed between normal fetal and adult pro-B cells (data in Dataset S7). Of these, 40 genes showed altered expression in Lin28b-transduced BM pro-B cells and 50 genes in Let-7–transduced FL pro-B cells (Fig. 6 A and Dataset S10). The reciprocal overlap (Fig. 6 B) identified 16 genes, in addition to Lin28b, showing an expression pattern that might be relevant to the fetal–adult developmental switch. One particularly interesting gene in the list is a transcription factor, Arid3a, also known as Bright, that has been previously implicated in enhancing Ig heavy chain expression and altering B cell responses (Schmidt et al., 2009). Importantly, the 3′ untranslated region (UTR) of the Arid3a mRNA contains several Let-7 target sites (shown in Fig. 6 C), indicating that this transcription factor could be regulated by the miR Let-7. Another miRNA differentially expressed in fetal and adult pro-B stage cells is miR-125b, previously shown to regulate Arid3a function in human and mouse B cell progenitors (Puissegur et al., 2012). Target sites for this miRNA are also shown in Fig. 6 C.
Figure 6.
Analysis of mRNAs regulated by the Lin28b–Let-7 axis in pro-B cells identified Arid3a as a key regulator of B cell fate. (A) Venn diagram illustrating the identification of a set of 17 genes whose expression is perturbed reciprocally by alteration of Lin28b–Let-7 in pro-B stage cells. BM pro-B cells sorted from BALB/c mice were transduced after 1 d in culture with either Lin28b or empty vector retrovirus and then cultured an additional 4 d and finally sorted as CD19+GFP+ cells. FL pro-B cells from the same strain were treated similarly, except that transduction was with Let-7b or empty retrovirus and cells were sorted as CD19+GFP+ (results are incorporated from B). (B) Heat map of mRNA expression fold change perturbations (relative to control-transduced samples) induced by retroviral transduction of Lin28b–Let-7b (as diagrammed in A). Two samples representative of expression changes in each cell type are shown. For BM cells, genes normally high (red) switch to green, whereas those normally low (green) switch to red (left). A reciprocal change is seen with FL cells (right). Data from unperturbed pre-pro-B (Fr. A) and pro-B (Fr. BC) microarray analysis of FL and BM, using both the Agilent (Ag) and Affymetrix (Af) platforms, are included for comparison. Results are representative of two independent experiments. (C) ARID3a, one of the genes whose expression was increased by Lin28b and decreased by Let-7 miRNA, has Let-7 and miR-125b target seed sites in its 3′-UTR. The Arid3a diagram was produced in MacVector, and species alignments were obtained from the TargetScan website, Whitehead Institute for Biomedical Research.
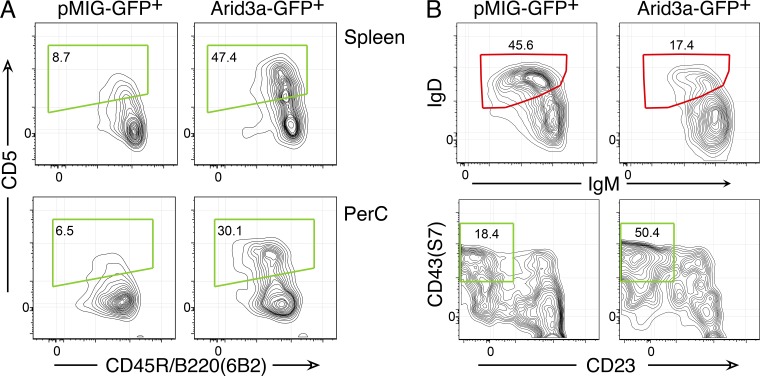
Transduction of BM pro-B cells with Arid3a-expressing retrovirus showed that this transcription factor was sufficient to support the generation of B1a B cells (Fig. 7 A). Thus, Arid3a is a key transcription factor induced by Lin28b and targeted by Let-7b for reprogramming adult B cell development to resemble that ongoing in FL. The phenotype of peritoneal cavity B cells induced by retroviral provision of Arid3a is shown in more detail in Fig. 7 B.
Figure 7.
Arid3a induces fetal-type B cell generation from BM pro-B cells. (A) BM pro-B cells were transduced with an Arid3a retrovirus or pMIG control and then transferred to SCID recipients. 3 wk later, spleen or PerC B cells, gated as CD19+GFP+, were analyzed by flow cytometry. The green regions indicate a fetal phenotype. Data are representative of three independent experiments using two mice per group. (B) The expression of other markers on PerC B cells generated from Arid3a-reprogrammed BM pro-B cells was analyzed by flow cytometry. Cells were gated as CD19+GFP+. The red regions indicate an adult-type and the green regions indicate a fetal-type phenotype. Data are representative of three independent experiments using two mice per group.
Retroviral knockdown of Arid3a in fetal pro-B cells reprograms development
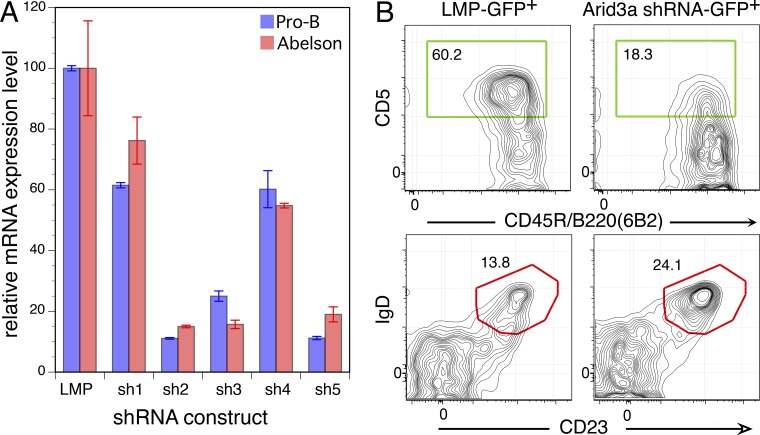
We performed a loss-of-function experiment, asking how B cell development would be affected if we reduced the amount of Arid3a in fetal pro-B cells by retroviral transduction of shRNA specific for this gene. We screened five shRNA hairpins specific for Arid3a from the Broad Institute Public TRC portal, expressing them in the Open Biosystems LMP retroviral vector, and found three that decreased the level of Arid3a 5–10-fold, as tested by Q-PCR (Fig. 8 A). We isolated pro-B stage cells from FL, cultured cells overnight, transduced them the next day with a pool of the shRNAs, and then transferred cells to SCID mice the following day. 3 wk later, recipients were sacrificed and analyzed for engraftment of GFP+ B cells. As is clear from Fig. 8 B, knockdown of Arid3a by this procedure reduced the frequency of CD5+ B cells generated from FL pro-B cells, reprogramming development to generate cells with a B2 B cell phenotype.
Figure 8.
shRNA knockdown of Arid3a inhibits FL pro-B generation of B1a B cells. (A) FL pro-B cells or the N38 Abelson mouse line was transduced with empty vector (LMP) or vector containing each of five different shRNAs. 3 d after transduction, GFP+ cells were sorted for RNA. ARID expression was quantified by Q-PCR. Arid3a expression was first normalized to β-actin, and then the level in control vector (LMP) was set to 100. Error bars represent ±SEM. (B) Phenotype of CD19+GFP+ B cells generated in SCID mice 3 wk after transfer of fetal pro-B cells, transduced with empty vector or a pool of three shRNA constructs. The red regions indicate an adult-type and the green boxes indicate a fetal-type phenotype. Data are representative of three independent experiments using two mice per group.
DISCUSSION
Yuan et al. (2012) recently showed that retroviral transduction of BM hematopoietic progenitors and stem cells could generate large numbers of fetal-type T and B lymphocytes. Based on differential expression of Lin28b and Let-7 in FL and BM B cell progenitors, they proposed that the Lin28b–Let-7 axis functions to regulate the switch from fetal-type to adult-type lymphopoiesis, generating innate-like T and B cells. However, based on this work, it remained unclear whether altering Lin28B–Let-7 could reprogram development after commitment to the B cell fate had occurred. Here we show that B cell development can be efficiently reprogrammed as late as the CD19+ pro-B stage. We also show that retroviral provision of Let-7 is sufficient to reprogram fetal pro-B cells to generate adult-type (B2) B cell progeny. Thus, we confirm and extend the earlier study, showing that the Lin28b–Let-7 axis mediates the B cell developmental switch that we proposed many years ago (Hardy and Hayakawa, 1991, 2001).
In contrast, although we find that generation of CD5+ B cells from Lin28b-transduced adult pro-B cells requires Btk signaling, as with fetal B1a generation, it does not fully recapitulate the fetal B1a BCR repertoire. Specifically, we observe a deficit of anti-PtC B cells that use VH11 and VH12 BCRs. One possible reason for lack of VH11 B cells, where the prototypic VDJs lack TdT-mediated N-addition at the junctions, is relatively modest regulation of TdT by Lin28b. However, even in BM pro-B cells lacking TdT, transduction of Lin28b did not recapitulate the normal fetal B1a repertoire. Considering that the typical B1a fetal-type repertoire requires appropriate initial BCR repertoire generation, selection, and accumulation, the failure of Lin28b-induced adult pro-B cells to generate a normal B1a repertoire may indicate further requirements for the generation of an appropriate initial BCR repertoire and/or fetal antigens lacking in the adult microenvironment.
Global analysis of genes reciprocally perturbed by misexpression of these regulators generated a relatively short list of candidate fetal/adult regulators. We considered the Arid3a transcription factor to be particularly interesting, based on its known functions of altering Ig heavy chain expression and inducing autoantibody production when overexpressed as a transgene (Oldham et al., 2011). Moreover, analysis of Arid3a on the TargetScan website revealed conserved Let-7 and miR-125b miRNA target sites in its 3′-UTR, both miRs which show relatively elevated expression in BM. Transduction of Arid3a into BM pro-B cells induced CD5+ B1 B cells in SCID recipients, similar to results obtained by Lin28b transduction. Finally, loss-of-function shRNA knockdown showed that diminishing Arid3a mRNA levels in fetal pro-B cells could reprogram B-1 development, generating B cells with an adult-type phenotype. Thus, we conclude that a key consequence of altering the Lin28b–Let-7 axis is changing expression levels of Arid3a.
How might Arid3a be inducing the generation of B1a B cells from BM pro-B cells? This transcription factor was originally identified by its capacity to bind to IgH V segments, increasing their expression level in a cell line treated with antigen and IL-5 (Herrscher et al., 1995). Later work showed that Arid3a was not restricted to activated B cells, but was also expressed in B cell progenitors (Webb et al., 1998). Further analysis showed that Arid3a-binding site motifs were common in the promoter regions of both human and mouse VH genes (Goebel et al., 2002). We speculate that Arid3a binding could alter the accessibility of VH genes to rearrangement, biasing the expression of these genes, selecting for B1a “preferred” BCRs. In addition, Arid3a overexpression has been shown to perturb B cell subset generation (Oldham et al., 2011), and a dominant-negative transgene diminishes B1 B cell function (Nixon et al., 2008). Thus, there is clear precedent for BRIGHT/Arid3a altering B cell development.
When expression of the Arid3a gene was eliminated in a gene-targeted mouse, the result was that >99% of embryos died at midgestation from a failure of hematopoiesis (Webb et al., 2011). Analysis of hematopoietic precursors and common lymphoid progenitors present in embryonic day 12.5 (E12.5) FL showed reduced numbers of both cell types, and colony-forming assays revealed reduced function of hematopoietic progenitor cells. Rare surviving progeny (<1%) were significantly smaller in size and had perturbed B lineage development, with fewer pro-B, pre-B, and immature B cells in BM and decreased numbers of transitional B cells in spleen. The follicular compartment was nearly normal, likely because of homeostatic compensation, but marginal zones were decreased and B1a B cells were essentially lacking. Thus, expression of Arid3a is required for normal early hematopoiesis and B lineage development. The difference that we describe between fetal and adult pro-B is therefore not all or none, but a difference in expression level, higher in fetal and lower in adult, that may serve to tune the response of the BCR, as described below, promoting B1 generation from FL and B2 production from adult BM.
Arid3a consists of an acidic N-terminal domain, an A-T–rich ARID DNA-binding domain, a transactivation domain, a helix-loop-helix (HLH) protein interaction domain, and a short C-terminal domain. Interestingly, Arid3a has been found to shuttle between the cytoplasm and the nucleus, under the control of its HLH domain (Kim and Tucker, 2006). The transcriptional activation function in mature B cells was found to be dependent on this domain, and specific residues, mapped by mutational analysis, were shown to be critical for nuclear or cytoplasmic localization. Arid3a has been shown to interact with the Tec kinase Btk (Webb et al., 2000), and in fact, functional Btk is required for Arid3a activity (Rajaiya et al., 2005), by phosphorylation of TFII-I which then forms a tripartite complex with Arid3a and Btk. Thus, Arid3a is not functional in B lineage cells from Btk-deficient Xid mice, known to lack B1a B cells (Hayakawa et al., 1983).
Importantly, Arid3a has also been found to alter BCR signaling because of its association with BCR-containing lipid rafts (Schmidt et al., 2009). Arid3a can be palmitoylated, redirecting its localization to lipid rafts where it associates with the BCR signalosome and interacts with sumoylation enzymes, blocking calcium flux and phosphorylation of Btk and TFII-I. Thus, higher levels of Arid3a decrease B cell responsiveness to BCR cross-linking and so function to tune BCR responsiveness. We speculate that the threshold for selection into the B1a B cell pool is altered in Lin28b/Arid3a-expressing early B lineage cells, facilitating selection of cells with BCRs that normally would be eliminated in BM (B-2) development because of their recognition of self-ligands. The requirement for selection by self-reactivity (Hayakawa et al., 1999) accounts for the observation that many B cells developing from pro-B that express Lin28b (or Arid3a) do not enter the CD5+ B cell pool. Manipulating Arid3a will be key in advancing our understanding of differences in BCR selection during fetal and adult B cell development.
MATERIALS AND METHODS
mRNA expression analysis using Agilent Technologies whole genome arrays.
Total RNA, isolated as described previously (Rumfelt et al., 2006), was used for production of fluorescent-labeled probe and then hybridized to the array. RNA purity and integrity were evaluated using the 2100 Bioanalyzer (Agilent Technologies) and NanoDrop 1000 (Thermo Fisher Scientific) before probe generation. For mouse BM and FL gene expression microarray analysis, a two-color array was used. Experimental samples, BM and FL cells, were labeled with Cy3, and a common reference RNA, a pool of total RNAs from several mouse organs generated in the laboratory, was Cy5 labeled. A single color (Cy3) array format was used for the Lin28b and Let-7 transduction experiment. Hybridized slides were scanned on an Agilent Technologies scanner, and fluorescent intensities of hybridization signals were extracted using Agilent Technologies Feature Extraction software. Statistical analysis was performed in the Fox Chase Biostatistics Facility.
miRNA expression analysis using Agilent Technologies miRNA arrays.
Total RNA was isolated using TRIzol reagent (Invitrogen) and washed with 80% ethanol to better retain small RNAs. The RNA quality and integrity were evaluated using the 2100 Bioanalyzer and NanoDrop 1000. 100 ng total RNA was dephosphorylated with calf intestinal phosphatase and then end-labeled with Cyanine-3-pCp using T4 RNA ligase. Labeled RNA was hybridized to the Agilent Technologies mouse miRNA microarray at 55°C for 20 h. Slides were washed according to the Agilent Technologies miRNA protocol and scanned as above, and signals were extracted as above.
Real-time PCR.
miRNAs were analyzed by TaqMan miRNA assays (Life Technologies) according to the manufacturer’s protocol. In brief, cDNA was reverse transcribed from total RNA samples using specific target miRNA primers from the TaqMan MicroRNA Assay and reagents from the TaqMan MicroRNA Reverse Transcription kit. PCR was then performed using the TaqMan assay together with the TaqMan Universal PCR Master Mix using an ABI 7500 real-time PCR system (Applied Biosystems). Data were normalized using SNO202 expression. Q-PCR Applied Biosystems IDs are as follows: miR-10a, 000387; miR-125b, 002508; miR-146, 000468; Let-7b, 000378; Let-7c, 000379; miR-451, 001141; and Sno202, 001232.
Production of Lin28B pMIG-eGFP/mCherry and Let-7b pMSCV-CFP constructs.
The mouse full-length Lin28b cDNA flanked by NotI and EcoRI sites in pQCXIP retroviral vector was obtained from D. Wiest (Fox Chase Cancer Center). Lin28b cDNA was subcloned into pMIG-EGFP retroviral vector by digesting the original construct with NotI, followed by filling in the cohesive end with T4 polymerase; this construct was then digested with EcoRI, releasing the Lin28b cDNA fragment. The pMIG-EGFP vector was first digested with BglII and filled in to generate a blunt end, then followed by digestion with EcoRI. The Lin28b fragment (EcoRI– blunt) was then ligated into pMIG-EGFP (EcoRI– blunt) yielding the Lin28b retrovirus. A similar procedure was used to generate an mCherry retrovirus containing Lin28b. For the miRNA expression construct, mouse Let-7b miRNA stem loop sequence identified from miRBase, along with its flanking genomic sequence of ∼200 bp to preserve the putative hairpin structure and proper endogenous processing, was amplified by PCR with primers that incorporated BglII and XhoI sites. The PCR fragment was then cloned into the retroviral vector pMSCV-CFP at BglII and XhoI sites. Both retroviral constructs were verified by DNA sequencing and then used to generate retroviral supernatant by calcium phosphate–mediated transfection of Phoenix-E cells.
shRNA constructs for knockdown of Arid3a.
The retroviral vector LMP from GE Healthcare was used to express the shRNAmir construct from RNA Polymerase II (Pol II) promoters (Dickins et al., 2005, 2007). These vectors produce highly efficient knockdown even when present at single copy. Hairpin sequences specific for Arid3a were from the Broad Institute Public TRC portal. Oligos for hairpins were synthesized and cloned into LMP according to a standard protocol, selected for appropriate size, and then verified by sequence analysis. The five hairpin vectors were compared with empty vector to assess the extent of knockdown, and the three most effective were selected for use as a pool in transduction of fetal pro-B stage cells, used in cell transfer experiments.
Animals.
BALB/c, BALB/c XID, and BALB.TdT− female mice, 6–12 wk old, bred in our animal facility were used as sources of BM cells for retroviral transduction. Embryos generated from timed mating of BALB/c were used as sources of day 16 FL. BM from BALB/c Rag1-GFP reporter mice was used for isolation of early lymphoid progenitors. C.B-17 SCID female mice, 6–12 wk old, were used as recipients in transfer experiments. All experiments were conducted under an animal protocol approved by the Fox Chase Institutional Animal Care and Use Committee.
Retroviral transduction of pro-B cells for cell transfer and microarray analysis.
OP9 cells were grown in a 37°C humidified, 10% CO2 gassed incubator in 16% FBS/αMEM (supplemented with 1× glutamine and sodium pyruvate, 10 mM Hepes, 5 × 10−5 M 2-ME, and 0.5 mg/ml gentamicin). Cells were passaged into 24-well plates in the same medium 1–2 d before use, so as to avoid confluence. Immediately before precursor cell deposition, medium was replaced with fresh medium containing cytokine (100 U/ml IL-7; R&D Systems). Pro-B cells were purified from BALB/c BM and FL by flow cytometry, directly depositing cells onto a previously prepared layer of OP9-R cells. Cells were cultured overnight and then transduced with retroviral supernatant by spinfection in the presence of polybrene; medium was replaced after 2–3 h. For transfer experiments, cells were cultured for an additional 24 h, and then 105 cells/recipient were injected i.v. into CB17 SCID mice, irradiated (3 Gy) 24 h before injection. For microarray experiments, cells were cultured for a further 2 d, and then GFP+ cells were sorted into RNA lysis buffer (“Solution D”).
Culture of retrovirally transduced B cell precursors for B cell differentiation.
Early B lineage precursors were purified as “early lymphocyte progenitors” (ELPs) from BALB/c Rag1-GFP transgenic BM by flow cytometry, depositing them onto OP9 cells prepared as described above. Medium was supplemented with 10 ng/ml stem cell factor (SCF), 10 ng/ml Flt3L, and 100 U/ml IL-7, all obtained from R&D Systems. Retroviral transduction was performed as above, and cells were cultured for an additional 3 d. Then cells were passaged onto FLST2 stromal cells in 24-well plates, growing in 5% FBS/RPMI1640 medium (supplemented with 1× glutamine, 10 mM Hepes, 5 × 10−5 M 2-ME, 0.5 mg/ml gentamicin, and 100 U/ml IL-7). After a further 4 d, cells were again passaged into RPMI medium, but with 100-fold reduced IL-7 to promote B cell differentiation. Cells were split once more into low–IL-7 medium and then analyzed 3 d later by flow cytometry (day 15). In other experiments, pro-B cells from BALB/c BM were deposited onto FLST2 stromal cells in 24-well plates (100 U/ml IL-7), transduced the following day, and then replated onto FLST2 cells in 100-fold reduced IL-7. Cells were split 3 d later, split again after 3 d, and finally analyzed by flow cytometry after 4-d further culture (day 11).
Flow cytometry sorting and analysis.
Cells were sorted using a BD FACSVantage SE/DiVa equipped with lasers enabling 12-color detection or a FACSAria equipped with lasers enabling 15-color detection. For microarray analysis, pre-pro-B cells and pro-B cells were isolated: for pre-pro-B as (Ter119,Ly6c,CD3,CD11b,GR1,IgM)−CD93+CD43+CD117+IL7Ra+B220+CD24loCD19− and for pro-B as (Ter119,Ly6c,CD3,CD11b,GR1,IgM)−CD93+CD43+B220+CD24medCD19+. For cell culture and transfer experiments, yield was maximized by sorting pro-B cells from BM and FL as (Ter119,Ly6c,CD3,CD11b,GR1,IgM)−CD43+B220+CD24medCD19+. ELPs were purified from BM of Rag1-GFP reporter mice as (Ter119,Ly6c,CD3,CD11b,GR1,IgM,IL7Ra,CD19)−CD117+GFP+. Mice were sacrificed 3 wk after transfer, and tissues (BM, thymus, spleen, and peritoneal cavity) were analyzed by flow cytometry for generation of lymphoid cells. Analysis was performed using a BD LSR-II flow cytometer, equipped with four lasers (violet, blue, green, and red) for 12-color detection. Staining reagents used in the analyses are described in the figure legends.
Single cell VH sequence analysis.
Ig VH sequences were determined in individual cells using the Advalytix single cell PCR system. In brief, single cells were deposited onto AmpliGrid slides, and then RNA was reverse transcribed and cDNA was amplified for 35 cycles. Samples were transferred to a 96-well plate containing PCR master mix and semi-nested primers. After 40 cycles of PCR using an ABI 9600 plate cycler (Applied Biosystems), 1/10 volume of each sample was subject to agarose electrophoresis to verify appropriate-sized fragments. DNA from the remaining sample was purified using a Nucleofast (MACHEREY-NAGEL) manifold plate system. 1/10 of each sample showing a band of appropriate size was then subject to Sanger sequence analysis in the Fox Chase DNA Sequencing Facility.
Bioinformatics analysis.
Raw expression data obtained from Agilent Technologies two-channel microarrays were background corrected and normalized (Lowess). Raw expression data obtained from single channel Agilent Technologies and Affymetrix arrays were background corrected and normalized (Quantile) across experimental conditions. The LIMMA (linear models for microarray data) methodology was applied to the log2-transformed expression data to identify differentially expressed genes in each comparison. The LIMMA module in the Open Source R/Bioconductor package was used in the computations. Differentially expressed genes were identified based on statistical significance as well as biological significance. Statistical significance was measured by the false discovery rate (FDR) to account for multiple testing. For Agilent Technologies two-channel microarrays, a cutoff of FDR ≤5% and fold change ≥2 were used; for Affymetrix microarrays, a cutoff of FDR ≤20% and fold change ≥1.5 were used. Gene expression heat maps were generated using Java TreeView (version 1.1.5r2). The gene interaction networks of significant genes were generated through the use of IPA (Ingenuity Systems). All edges are supported by at least one reference from the literature, from a textbook, or from canonical information stored in the Ingenuity Knowledge Base. The intensity of the node color indicates the degree of up (red)- or down (green)-regulation. Nodes are displayed using various shapes that represent the functional class of the gene product. Raw microarray data are available under GEO accession no. GSE65536.
Online supplemental material.
Datasets S1–S7 present microarray expression data. Dataset S8 shows miRNA expression microarray data. Dataset S9 shows heavy chain sequence data used to produce Fig. 5. Dataset S10 contains gene expression perturbed by the Lin28b–Let-7 axis for 219 genes differentially expressed in fetal and adult B cell progenitors. All datasets are included in separate Excel files. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20141510/DC1.
Supplementary Material
Acknowledgments
This work utilized key facilities, DNA Sequencing, Flow Cytometry, Laboratory Animal, Genomics, and Biostatistics and Bioinformatics, supported by the Fox Chase Cancer Center Core Grant (P30CA006927). R.R. Hardy was supported by a grant from the National Institutes of Health (NIH; R01AI026782) and PA Department of Health Tobacco Settlement funds. (This project is funded, in part, under a grant with the Pennsylvania Department of Health. The Department specifically disclaims responsibility for any analyses, interpretations, or conclusions.) K. Hayakawa was supported by a grant from the NIH (R01AI49335). All Affymetrix expression analyses were generated by the Immunological Genome Project Consortium.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- BCR
- B cell antigen receptor
- ELP
- early lymphocyte progenitor
- FDR
- false discovery rate
- FL
- fetal liver
- miRNA
- microRNA
- Q-PCR
- quantitative PCR
- TdT
- terminal deoxynucleotidyl transferase
- UTR
- untranslated region
References
- Carmack C.E., Shinton S.A., Hayakawa K., and Hardy R.R.. 1990. Rearrangement and selection of VH11 in the Ly-1 B cell lineage. J. Exp. Med. 172:371–374 10.1084/jem.172.1.371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickins R.A., Hemann M.T., Zilfou J.T., Simpson D.R., Ibarra I., Hannon G.J., and Lowe S.W.. 2005. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat. Genet. 37:1289–1295 10.1038/ng1651 [DOI] [PubMed] [Google Scholar]
- Dickins R.A., McJunkin K., Hernando E., Premsrirut P.K., Krizhanovsky V., Burgess D.J., Kim S.Y., Cordon-Cardo C., Zender L., Hannon G.J., and Lowe S.W.. 2007. Tissue-specific and reversible RNA interference in transgenic mice. Nat. Genet. 39:914–921 10.1038/ng2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förster I., Gu H., and Rajewsky K.. 1988. Germline antibody V regions as determinants of clonal persistence and malignant growth in the B cell compartment. EMBO J. 7:3693–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel P., Montalbano A., Ayers N., Kompfner E., Dickinson L., Webb C.F., and Feeney A.J.. 2002. High frequency of matrix attachment regions and cut-like protein x/CCAAT-displacement protein and B cell regulator of IgH transcription binding sites flanking Ig V region genes. J. Immunol. 169:2477–2487 10.4049/jimmunol.169.5.2477 [DOI] [PubMed] [Google Scholar]
- Goodnow C.C.2007. Multistep pathogenesis of autoimmune disease. Cell. 130:25–35 10.1016/j.cell.2007.06.033 [DOI] [PubMed] [Google Scholar]
- Groudine M., Kohwi-Shigematsu T., Gelinas R., Stamatoyannopoulos G., and Papayannopoulou T.. 1983. Human fetal to adult hemoglobin switching: changes in chromatin structure of the beta-globin gene locus. Proc. Natl. Acad. Sci. USA. 80:7551–7555 10.1073/pnas.80.24.7551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy R.R., and Hayakawa K.. 1991. A developmental switch in B lymphopoiesis. Proc. Natl. Acad. Sci. USA. 88:11550–11554 10.1073/pnas.88.24.11550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy R.R., and Hayakawa K.. 2001. B cell development pathways. Annu. Rev. Immunol. 19:595–621 10.1146/annurev.immunol.19.1.595 [DOI] [PubMed] [Google Scholar]
- Hayakawa K., Hardy R.R., Parks D.R., and Herzenberg L.A.. 1983. The “Ly-1 B” cell subpopulation in normal immunodefective, and autoimmune mice. J. Exp. Med. 157:202–218 10.1084/jem.157.1.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa K., Hardy R.R., Honda M., Herzenberg L.A., Steinberg A.D., and Herzenberg L.A.. 1984. Ly-1 B cells: functionally distinct lymphocytes that secrete IgM autoantibodies. Proc. Natl. Acad. Sci. USA. 81:2494–2498 10.1073/pnas.81.8.2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa K., Hardy R.R., Herzenberg L.A., and Herzenberg L.A.. 1985. Progenitors for Ly-1 B cells are distinct from progenitors for other B cells. J. Exp. Med. 161:1554–1568 10.1084/jem.161.6.1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa K., Asano M., Shinton S.A., Gui M., Allman D., Stewart C.L., Silver J., and Hardy R.R.. 1999. Positive selection of natural autoreactive B cells. Science. 285:113–116 10.1126/science.285.5424.113 [DOI] [PubMed] [Google Scholar]
- Herrscher R.F., Kaplan M.H., Lelsz D.L., Das C., Scheuermann R., and Tucker P.W.. 1995. The immunoglobulin heavy-chain matrix-associating regions are bound by Bright: a B cell-specific trans-activator that describes a new DNA-binding protein family. Genes Dev. 9:3067–3082 10.1101/gad.9.24.3067 [DOI] [PubMed] [Google Scholar]
- Inaoki M., Sato S., Weintraub B.C., Goodnow C.C., and Tedder T.F.. 1997. CD19-regulated signaling thresholds control peripheral tolerance and autoantibody production in B lymphocytes. J. Exp. Med. 186:1923–1931 10.1084/jem.186.11.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan W.N., Alt F.W., Gerstein R.M., Malynn B.A., Larsson I., Rathbun G., Davidson L., Müller S., Kantor A.B., Herzenberg L.A., et al. 1995. Defective B cell development and function in Btk-deficient mice. Immunity. 3:283–299 10.1016/1074-7613(95)90114-0 [DOI] [PubMed] [Google Scholar]
- Kim D., and Tucker P.W.. 2006. A regulated nucleocytoplasmic shuttle contributes to Bright’s function as a transcriptional activator of immunoglobulin genes. Mol. Cell. Biol. 26:2187–2201 10.1128/MCB.26.6.2187-2201.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.S., Hayakawa K., and Hardy R.R.. 1993. The regulated expression of B lineage associated genes during B cell differentiation in bone marrow and fetal liver. J. Exp. Med. 178:951–960 10.1084/jem.178.3.951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr F., Schütz A., Döge N., and Heinemann U.. 2012. The Lin28 cold-shock domain remodels pre-let-7 microRNA. Nucleic Acids Res. 40:7492–7506 10.1093/nar/gks355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemazee D.2006. Receptor editing in lymphocyte development and central tolerance. Nat. Rev. Immunol. 6:728–740 10.1038/nri1939 [DOI] [PubMed] [Google Scholar]
- Nixon J.C., Ferrell S., Miner C., Oldham A.L., Hochgeschwender U., and Webb C.F.. 2008. Transgenic mice expressing dominant-negative bright exhibit defects in B1 B cells. J. Immunol. 181:6913–6922 10.4049/jimmunol.181.10.6913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham A.L., Miner C.A., Wang H.C., and Webb C.F.. 2011. The transcription factor Bright plays a role in marginal zone B lymphocyte development and autoantibody production. Mol. Immunol. 49:367–379 10.1016/j.molimm.2011.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennell C.A., Mercolino T.J., Grdina T.A., Arnold L.W., Haughton G., and Clarke S.H.. 1989. Biased immunoglobulin variable region gene expression by Ly-1 B cells due to clonal selection. Eur. J. Immunol. 19:1289–1295 10.1002/eji.1830190721 [DOI] [PubMed] [Google Scholar]
- Puissegur M.P., Eichner R., Quelen C., Coyaud E., Mari B., Lebrigand K., Broccardo C., Nguyen-Khac F., Bousquet M., and Brousset P.. 2012. B-cell regulator of immunoglobulin heavy-chain transcription (Bright)/ARID3a is a direct target of the oncomir microRNA-125b in progenitor B-cells. Leukemia. 26:2224–2232 10.1038/leu.2012.95 [DOI] [PubMed] [Google Scholar]
- Rajaiya J., Hatfield M., Nixon J.C., Rawlings D.J., and Webb C.F.. 2005. Bruton’s tyrosine kinase regulates immunoglobulin promoter activation in association with the transcription factor Bright. Mol. Cell. Biol. 25:2073–2084 10.1128/MCB.25.6.2073-2084.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings D.J., Saffran D.C., Tsukada S., Largaespada D.A., Grimaldi J.C., Cohen L., Mohr R.N., Bazan J.F., Howard M., Copeland N.G., et al. 1993. Mutation of unique region of Bruton’s tyrosine kinase in immunodeficient XID mice. Science. 261:358–361 10.1126/science.8332901 [DOI] [PubMed] [Google Scholar]
- Rumfelt L.L., Zhou Y., Rowley B.M., Shinton S.A., and Hardy R.R.. 2006. Lineage specification and plasticity in CD19- early B cell precursors. J. Exp. Med. 203:675–687 10.1084/jem.20052444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt C., Kim D., Ippolito G.C., Naqvi H.R., Probst L., Mathur S., Rosas-Acosta G., Wilson V.G., Oldham A.L., Poenie M., et al. 2009. Signalling of the BCR is regulated by a lipid rafts-localised transcription factor, Bright. EMBO J. 28:711–724 10.1038/emboj.2009.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Terauchi Y., Fujiwara M., Aizawa S., Yazaki Y., Kadowaki T., and Koyasu S.. 1999. Xid-like immunodeficiency in mice with disruption of the p85alpha subunit of phosphoinositide 3-kinase. Science. 283:390–392 10.1126/science.283.5400.390 [DOI] [PubMed] [Google Scholar]
- Tarakhovsky A., Turner M., Schaal S., Mee P.J., Duddy L.P., Rajewsky K., and Tybulewicz V.L.. 1995. Defective antigen receptor-mediated proliferation of B and T cells in the absence of Vav. Nature. 374:467–470 10.1038/374467a0 [DOI] [PubMed] [Google Scholar]
- Webb C.F., Smith E.A., Medina K.L., Buchanan K.L., Smithson G., and Dou S.. 1998. Expression of bright at two distinct stages of B lymphocyte development. J. Immunol. 160:4747–4754. [PubMed] [Google Scholar]
- Webb C.F., Yamashita Y., Ayers N., Evetts S., Paulin Y., Conley M.E., and Smith E.A.. 2000. The transcription factor Bright associates with Bruton’s tyrosine kinase, the defective protein in immunodeficiency disease. J. Immunol. 165:6956–6965 10.4049/jimmunol.165.12.6956 [DOI] [PubMed] [Google Scholar]
- Webb C.F., Bryant J., Popowski M., Allred L., Kim D., Harriss J., Schmidt C., Miner C.A., Rose K., Cheng H.L., et al. 2011. The ARID family transcription factor bright is required for both hematopoietic stem cell and B lineage development. Mol. Cell. Biol. 31:1041–1053 10.1128/MCB.01448-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J., Nguyen C.K., Liu X., Kanellopoulou C., and Muljo S.A.. 2012. Lin28b reprograms adult bone marrow hematopoietic progenitors to mediate fetal-like lymphopoiesis. Science. 335:1195–1200 10.1126/science.1216557 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.