Methot et al. identify a mechanism for cytoplasmic retention of activation-induced deaminase (AID) in cells. Interactions of AID with Hsp90 and eEF1A proteins, both of which stabilize AID, promote sequential folding and retention of functional AID in the cytoplasm. Inhibition of the translation elongation factor eEF1A blocks its interaction with AID, which then accumulates in the nucleus, increasing class switch recombination and the generation of chromosomal translocation byproducts.
Abstract
Activation-induced deaminase (AID) initiates mutagenic pathways to diversify the antibody genes during immune responses. The access of AID to the nucleus is limited by CRM1-mediated nuclear export and by an uncharacterized mechanism of cytoplasmic retention. Here, we define a conformational motif in AID that dictates its cytoplasmic retention and demonstrate that the translation elongation factor eukaryotic elongation factor 1 α (eEF1A) is necessary for AID cytoplasmic sequestering. The mechanism is independent of protein synthesis but dependent on a tRNA-free form of eEF1A. Inhibiting eEF1A prevents the interaction with AID, which accumulates in the nucleus and increases class switch recombination as well as chromosomal translocation byproducts. Most AID is associated to unspecified cytoplasmic complexes. We find that the interactions of AID with eEF1A and heat-shock protein 90 kD (HSP90) are inversely correlated. Despite both interactions stabilizing AID, the nature of the AID fractions associated with HSP90 or eEF1A are different, defining two complexes that sequentially produce and store functional AID in the cytoplasm. In addition, nuclear export and cytoplasmic retention cooperate to exclude AID from the nucleus but might not be functionally equivalent. Our results elucidate the molecular basis of AID cytoplasmic retention, define its functional relevance and distinguish it from other mechanisms regulating AID.
During immune responses, B cells producing high-affinity antibodies of the IgG, IgA, and IgE classes are generated from the low-affinity, IgM+ B cells that initially recognize the invading antigens. The molecular mechanisms underpinning the affinity maturation and change of class of the antibody response are somatic hypermutation (SHM) and class switch recombination (CSR), respectively; mutagenic processes that modify the antibody genes. The enzyme activation-induced deaminase (AID) initiates SHM and CSR by converting deoxycytidine to deoxyuridine in DNA. Repair enzymes that recognize uracil in DNA trigger further mutagenic processing to generate the full spectrum of SHM or the DNA breaks that are necessary for CSR. Human patients lacking AID have no antibody affinity maturation or class switching and are immunodeficient (Revy et al., 2000). On the other hand, excessive or deregulated AID activity can be cytotoxic (Zahn et al., 2014), contribute to autoimmunity (Diaz, 2013), or predispose to B cell lymphomas (Robbiani et al., 2009). The delicate balance between the physiological and pathological effects of AID is enforced by multiple levels of AID regulation (Vuong and Chaudhuri, 2012; Keim et al., 2013).
Subcellular localization and protein stability are major points of AID regulation (Orthwein and Di Noia, 2012; Vuong and Chaudhuri, 2012). AID is a nuclear-cytoplasmic shuttling protein (Brar et al., 2004; Ito et al., 2004; McBride et al., 2004) and its stability is intricately related to its compartmentalization. AID is stabilized in the cytoplasm by a heat-shock protein 90 kD (HSP90) molecular chaperoning pathway that requires the DnaJa1 HSP40 (Orthwein et al., 2010; 2012) and destabilized in the nucleus by ubiquitin-dependent and -independent pathways (Aoufouchi et al., 2008; Uchimura et al., 2011). The small size of AID (24 kD) should allow it to diffuse through the nuclear pores; however, it requires active import to enter the nucleus (Patenaude et al., 2009) and 90% of AID is localized to the cytoplasm under steady-state conditions (Rada et al., 2002; Pasqualucci et al., 2004). Two mechanisms that exclude AID from the nucleus have been identified. AID is exported from the nucleus by CRM1, which recognizes a Leucine-rich nuclear export signal (NES) within positions 188–198 of AID (McBride et al., 2004). AID is also retained in the cytoplasm by a still ill-defined mechanism that requires residues Asp 187 and 188 in human AID, which overlap with the NES (Patenaude et al., 2009). The relative contribution of CRM1-mediated nuclear export and cytoplasmic retention to nuclear exclusion and functional regulation of endogenous AID is also unknown because of the lack of reagents to block each mechanism without resorting to AID mutants that might affect both processes.
HSP90 and DnaJa1 bind cytoplasmic AID but do not mediate its retention (Orthwein et al., 2012; 2010). Cytoplasmic AID also interacts with the translation elongation factor eukaryotic elongation factor 1 α (eEF1A) in human, mouse and chicken B cells (Häsler et al., 2011). This factor delivers aminoacyl-tRNA to the elongating ribosomes (Andersen et al., 2003) but it has other functions that are unrelated to protein synthesis (Mateyak and Kinzy, 2010). Because mutations in AID residues Asp187 and Asp188 disrupt cytoplasmic retention (Patenaude et al., 2009), as well as the interaction with eEF1A (Häsler et al., 2011), it is possible that eEF1A is part of a complex retaining AID in the cytoplasm (Häsler et al., 2012). However, mutating Asp187/188 could also disrupt the interaction of AID with other factors. Moreover, the N-terminal region of AID is required for the interaction with eEF1A (Häsler et al., 2011) but not for cytoplasmic retention (Patenaude et al., 2009). In addition, eEF1A and AID form part of an uncharacterized ∼350 kD cytoplasmic complex (Häsler et al., 2011), which could contain redundant or additional factors. Thus, the mechanism of cytoplasmic retention and the involvement of eEF1A remain to be elucidated. In addition, whether AID forms distinct complexes with HSP90 and eEF1A and the possibility that AID shuttles between these two factors, as well as the comparative functional relevance of each of these interactions also remain to be determined.
Here, we demonstrate that the mechanism of AID cytoplasmic retention depends on a specific AID conformation and on eEF1A and that it limits AID function. We also identify differences between eEF1A-, HSP90-, and CRM1-mediated AID regulation that suggest a model to explain the transit and functional maturation of AID in the cytoplasm, which impacts on the efficiency of antibody gene diversification and its collateral damage.
RESULTS
AID cytoplasmic retention is evolutionarily conserved
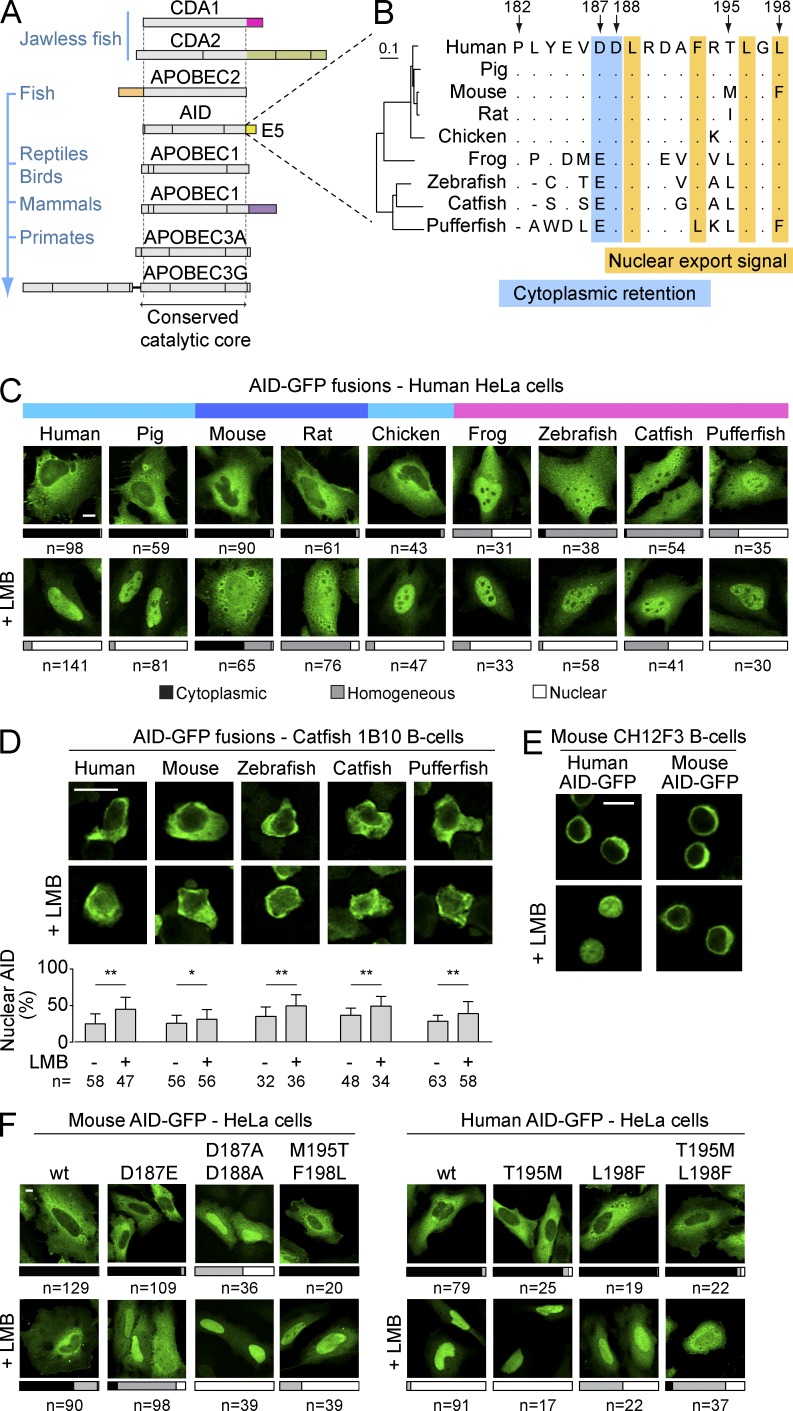
The structure of the AID/apolipoprotein B mRNA editing enzyme catalytic polypeptide-like (APOBEC) family proteins along evolution shows that many of these deaminases have acquired unique extensions at either end of the common catalytic domain, as epitomized by the C-terminal tail that APOBEC1 acquired in mammals (Severi et al., 2011; Fig. 1 A). These extensions are often encoded by a single exon of the corresponding gene, such as exon 1 of APOBEC2 or exon 5 of mammalian APOBEC1 (Fig. 1 A). Similarly, AID has a specific C–terminal extension of 17 residues encoded by exon 5 (henceforth E5). E5 is critical for AID nuclear exclusion, as it harbors both the NES and a poorly defined cytoplasmic retention motif (Brar et al., 2004; Ito et al., 2004; McBride et al., 2004; Patenaude et al., 2009). The NES is conserved between AID homologues (Fig. 1 B). Two acidic residues important for cytoplasmic retention are also conserved, albeit in poikilotherms Glu replaces Asp187 and the region preceding it is very different (Fig. 1 B). To determine whether both nuclear exclusion mechanisms of AID were functional throughout evolution, we analyzed the subcellular localization of multiple AID orthologs. AID-GFP fusions from all tested homeotherms were excluded from the nucleus of HeLa cells, whereas AID-GFP from poikilotherms was not (Fig. 1 C). These differences in localization did not reflect proteolytic generation of free GFP, as controlled for by Western blot, and were also observed in HEK293 and mouse NIH3T3 cells (unpublished data). Treatment with the CRM1 inhibitor leptomycin B (LMB) led to increased nuclear abundance of every AID ortholog in all cell lines tested (Fig. 1 C and not depicted), indicating that the NES of all the orthologs was functional. Thus, the homogenous distribution of AID from poikilotherms in mammalian cells could reflect defective cytoplasmic retention. However, the sequence divergence of the E5 domain during evolution could prevent the interaction of fish and frog AID with the human/mouse retention factors. We therefore expressed AID from poikilotherms species in the catfish B cell line 1B10. We found that all fish AID were excluded from the nucleus in 1B10 cells and that this exclusion was partially relieved by LMB treatment (Fig. 1 D). These results indicate that nuclear export was functional in 1B10 cells and that fish AID were retained in the cytoplasm in the proper cellular context (Fig. 1 D).
Figure 1.
Cytoplasmic retention of AID is conserved. (A) In-scale representation of AID/APOBEC family members, indicating protein regions encoded by different exons and their phylogenetic distribution. Domains unique to specific members of the family are colored. (B) Amino acid alignment of the E5 region from AID homologues. Arrows indicate positions according to, and dots denote identities with, human AID. The NES and a motif required for cytoplasmic retention are highlighted. An N-J tree built on the full-length proteins’ alignment (left) illustrates the phylogenetic distances between homologues. (C) Representative confocal microscopy images of HeLa cells transiently expressing AID-GFP homologues under steady-state conditions or after inhibition of nuclear export with LMB (50 ng/ml, 2 h). Bars represent the proportion of n cells from ≥3 experiments showing the indicated types of subcellular distribution (cytoplasmic, black; homogenous, gray; nuclear, white). The color bars at the top identify AID-GFP homologues that show similar subcellular distributions in human cells (human-like, teal; stronger retention, blue; weaker retention, pink). (D) Representative confocal microscopy images of AID-GFP homologues transiently expressed in catfish 1B10 B cells and treated as in (C). Nuclear GFP signals were measured by line intensity analysis for n cells from ≥2 experiments and the mean + SEM, plotted (graph below). Differences were assessed by Student’s t test (*, P < 0.05; **, P < 0.001). (E) Representative confocal microscopy images of mouse CH12F3 B cells overexpressing human or mouse AID-GFP and stimulated with CIT (1 µg/ml anti-CD40, 1 ng/ml TGF-β, and 10 ng/ml IL-4) for 24 h before treatment with either vehicle or 10 ng/ml LMB for 2 h. Images are representative of two independent experiments. (F) Representative confocal microscopy images of HeLa cells transiently expressing mouse and human AID-GFP and indicated mutants. Cells were treated as in C and subcellular localization scored from three independent, pooled experiments. Bars, 10 µm.
We discovered the mechanism of cytoplasmic retention using human AID (Patenaude et al., 2009). However, for reasons described in this study, the C-terminal GFP fusion weakens the retention of human AID-GFP so that this mechanism is not readily apparent in HeLa cells (Fig. 1 C). This is probably also the case for pig and chicken AID-GFP that have E5 regions identical to the human’s (Fig. 1 B). In contrast, the mouse and rat orthologs accumulated only partially in the nuclei of HeLa cells after LMB treatment (Fig. 1 C). Even in mouse CH12F3 B cells stimulated for CSR, mouse AID-GFP remained largely cytoplasmic after LMB treatment, whereas human AID-GFP accumulated in the nucleus (Fig. 1 E). Given that the mouse AID NES is functional (McBride et al., 2004; Fig. 1, B and C), our results suggested that there are amino acid differences between mouse and human AID that could modulate the strength of cytoplasmic retention in the context of AID-GFP fusions. We confirmed that cytoplasmic retention was preventing mouse AID-GFP nuclear accumulation in LMB-treated cells by mutating Asp187 and 188. Substituting Asp187 with the Glu residue present in fish was sufficient to render mouse AID-GFP responsive to LMB (Fig. 1 F), which could partially explain the lack of retention of fish AID-GFP in HeLa cells. Mutating both positions to Ala eliminated its nuclear exclusion (Fig. 1 F). We then focused on the only two positions within E5 that are different between human and mouse AID (Fig. 1 B). Mutating mouse AID Met195 and Phe198 to the human residues (Thr and Leu, respectively) allowed nuclear accumulation of mouse AID-GFP when nuclear export was inhibited (Fig. 1 F), suggesting reduced cytoplasmic retention. Conversely, mutations T195M and L198F in human AID-GFP synergistically reduced its response to LMB (Fig. 1 F), thus suggesting increased retention.
We conclude that AID cytoplasmic retention was acquired early on during evolution and therefore likely plays an important function in AID biology. We also find that amino acid sequence differences between AID homologues could dictate cytoplasmic retention efficiency, at least in the context of AID-GFP fusions.
Cytoplasmic retention requires a specific AID conformation
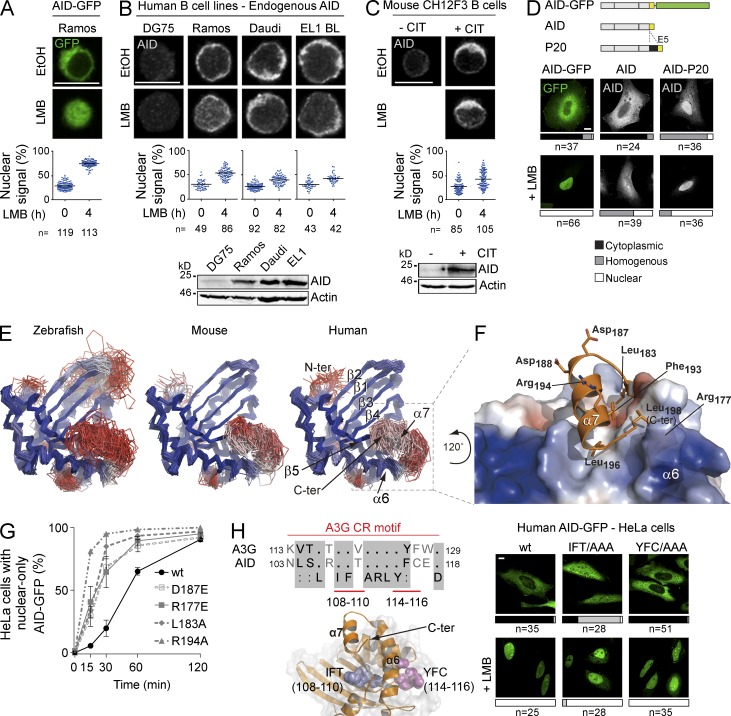
We then asked whether the difference in cytoplasmic retention efficiency between human and mouse AID-GFP held true for the endogenous enzymes in B cells. In the human Ramos B cell line, in which transfected human AID-GFP completely relocalized to the nucleus after LMB treatment (Fig. 2 A), endogenous AID detected by immunofluorescence (IF) responded significantly less (Fig. 2 B). The same modest response to LMB was observed for endogenous AID in two other human B cell lines (Fig. 2 B), as well as for the endogenous mouse AID in stimulated CH12F3 cells (Fig. 2 C). Thus, in contrast to their C-terminal GFP fusion counterparts, endogenous human and mouse AID show similar cytoplasmic retention, which plays a large role in their nuclear exclusion (Fig. 2, B and C).
Figure 2.
A specific conformation of AID is necessary for cytoplasmic retention. (A) Representative confocal microscopy images of AID-GFP in human Ramos B cell stable transfectants treated for 4 h with either vehicle (EtOH) or 10 ng/ml LMB. The proportion of nuclear AID signal was calculated as the ratio between the AID signal overlapping with propidium iodide signal to the total AID signal, and was plotted for n individual cells (dots). The mean for each group is indicated by a horizontal line (graph below). (B) Representative confocal microscopy images of endogenous AID detected by IF in the indicated human B cell lymphoma lines treated, and quantified as in A (graph below). DG75 cells were used as negative control. A Western blot of AID and actin expression in total cell extracts for each cell line is shown (bottom). (C) Representative confocal microscopy images of endogenous AID detected by IF in mouse CH12F3 B cells stimulated with CIT for 24 h before LMB treatment and quantification as in (A; graph below). Nonstimulated cells were used as negative control. A Western blot as in B is shown (bottom). (A–C) Representative data from one out of two independent experiments. (D) Representative confocal microscopy images of HeLa cells transiently expressing human AID-GFP or untagged AID WT or variant P20 with 34 amino acids inserted upstream of E5. Localization was assessed by detecting GFP, or by IF for overexpressed untagged AID. Cells were treated with either vehicle or 50 ng/ml LMB for 2 h. The aligned schemes of the proteins are shown on the top panel and quantification of subcellular localization in n cells from 2 independent, pooled experiments is shown below each image (cytoplasmic, black; homogenous, gray; nuclear, white). (E) Ensemble of top scoring models from the best represented cluster were derived from the conformational sampling of the E5 C-terminal tail for zebrafish, mouse, and human AID in which the catalytic core was modeled on the APOBEC3C structure. Models were colored according to the root-mean-square deviation (rmsd) differences for each position with a ramp from blue (low) to red (high). The lower the rmsd, the better the coincidence between multiple models at any given position. (F) Top scoring model of human AID from the best represented cluster. The electrostatic potential (negative in red, positive in blue) was mapped onto the molecular surface of the core domain, while the C-terminal tail is represented as a cartoon. Selected residues are represented as sticks. This model is rotated 120° counter-clockwise relative to the models in E. (G) Nuclear localization kinetics of the indicated human AID-GFP variants were assessed in HeLa cells. Cells were treated with 50 ng/ml LMB and fixed at the indicated times. The mean ± SEM proportion of cells with full nuclear localization were plotted for each time point from 2 independent experiments, after scoring >40 cells per time point, per experiment. (H) Sequence alignment of an APOBEC3G (A3G) cytoplasmic retention motif with its homologous AID region. The structural model of AID shows the relative position of residues 108–110 (blue spheres) and 114–116 (magenta spheres; left). Representative confocal images of HeLa cells transiently expressing human AID-GFP variants in which either one of these residue stretches were mutated to Ala. Cells were treated with either vehicle or 50 ng/ml LMB for 2 h, and scored as in D, from 2 independent, pooled experiments (right panels). Bars, 10 µm.
The effect of the GFP fusion on the human AID C terminus was still informative. The different behavior of human AID-GFP and endogenous AID was not an artifact of overexpression. Untagged human AID also accumulated much less than AID-GFP in the nucleus of HeLa cells after LMB treatment (Fig. 2 D), suggesting that the C-terminal GFP fusion weakened cytoplasmic retention. We also observed that a natural human AID variant bearing an insertion of 34 amino acids between exons 4 and 5 (Ito et al., 2004) was not excluded from the nucleus despite having an intact NES (Fig. 2 D). Together, our results suggested that cytoplasmic retention requires that the E5 domains adopt a certain conformation with respect to the enzyme core. This conformation would be variably affected by a GFP fusion depending on the sequence of E5 (Fig. 1 F).
Because AID has not been crystallized, we resorted to molecular modeling to ask whether an interaction between the E5 region and the catalytic core of AID was thermodynamically possible. We modeled the AID catalytic core using as template APOBEC3C (Kitamura et al., 2012), which among the AID paralogs of known 3D structure displays the highest sequence similarity (42%). The C-terminal tail of AID is absent in the APOBECs (Fig. 1 A) and cannot be modeled by comparison. However, secondary structure predictions for AID of all species consistently showed an amphipathic helix within the E5 (helix α7), in which the hydrophobic residues of the NES and the Asp187 and 188 are on opposite sides (not depicted). We used the Rosetta suite (Das and Baker, 2008) to perform a conformational sampling of the AID E5 from human, mouse and zebrafish, all of which yielded a similar result (Fig. 2 E). In all the best energy clusters (see Materials and methods), E5 sat within the same circumscribed region interacting with the helix α6 and the strand β5 from the catalytic core. This convergence of all three models despite significant sequence differences (zebrafish and human AID are 59% similar) strongly suggests that E5 spontaneously adopts this conformation.
The predicted binding of the helix α7 buries the hydrophobic residues of the NES (Phe193, Leu196, and Leu 198 in human AID) in a groove on the catalytic core conformed by residues Leu126, Ala137, Ile138, Leu176, Leu180, and Leu181, thus avoiding their exposure to the solvent (Fig. 2 F). The model shows several contacts between NES residues and the AID core, such as a salt bridge between the side chain of Arg177 and the C-terminal carboxylate in Leu198 and multiple van der Waals contacts (Leu183:Arg127, Leu183:His130, Arg177:Phe193, Phe109:Leu196, etc.). Such a conformation predicts that mutating the NES would not only eliminate nuclear export but also compromise cytoplasmic retention by affecting the position of E5. Indeed, we have shown that mutating the NES caused more efficient AID nuclear accumulation than inhibiting CRM1 (Patenaude et al., 2009). Moreover, the reduced retention of AID-GFP (compare Fig. 2 A to Fig. 2 B) and of the P20 variant (Fig. 2 D), were also consistent with our model, as the GFP fusion or the insertion separating E5 from the core would interfere with this conformation. To further validate the model, we mutated other residues, outside the NES, that would participate in the interaction between E5 and the enzyme core. Mutations L183A and R177E compromised cytoplasmic retention of AID, as judged by faster nuclear import kinetics (Fig. 2 G). Charged residues Asp187, Asp 188, and Arg194, were predicted to be on the solvent-exposed face of helix α7 (Fig. 2 F) and could mediate the interaction with retention factors. Consistently, mutants R194A and D187E had faster nuclear localization kinetics than WT AID (Fig. 2 G).
The AID paralogs APOBEC1, APOBEC3A, and APOBEC3G have mechanisms for nuclear exclusion (Chester et al., 2003; Bennett et al., 2006; Stenglein et al., 2008; Land et al., 2013), which might have evolved from those of AID. Interestingly, the AID model predicted interactions between E5 and an internal region corresponding to residues 103–118, which is homologous to a cytoplasmic retention determinant identified in APOBEC3G (Stenglein et al., 2008; Bennett et al., 2006; Fig. 2 H). Mutating AID Ile108-Phe109-Thr110 to Ala, in which Phe109 would form van der Waals interactions with Leu196 in E5, abrogated AID nuclear exclusion (Fig. 2 H). On the other hand, mutating residues 114 to 116, which do not contact E5 in the 3D model, did not affect AID localization (Fig. 2 H).
We conclude that cytoplasmic retention of AID relies on a specific conformation in which E5 interacts with the enzymatic core through the NES, leaving residues around Asp187 poised to interact with the retention factors. The measurable effect of the conservative D187E substitution in human and mouse AID (Figs. 2 G and 1 F) suggested that even subtle changes on the surface could affect this interaction.
Mutually exclusive interaction of AID with eEF1A and HSP90
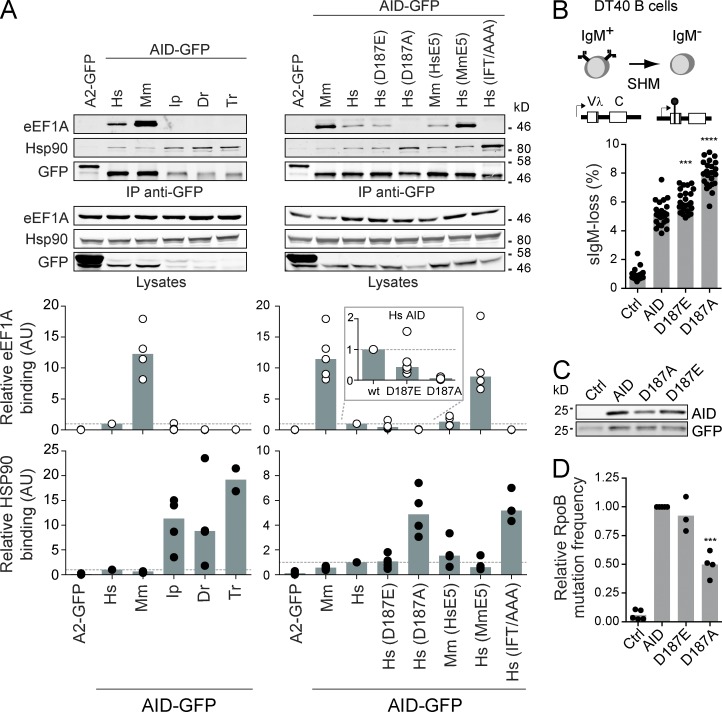
The factors mediating AID retention in the cytoplasm are unknown. eEF1A is a likely candidate because its interaction with cytoplasmic AID is prevented by AID mutations D187A/D188A (Häsler et al., 2011), but whether it is necessary for AID retention is unknown. Moreover, eEF1A is in a large cytoplasmic complex with AID and whether HSP90 is part of the same or a distinct AID complex is unknown (Häsler et al., 2011). The graded cytoplasmic retention efficiency of AID-GFP proteins from different species and of human AID-GFP with amino acid substitutions in E5 provided us with unique tools to probe the interaction of AID with eEF1A and HSP90. Co-immunoprecipitation (coIP) experiments showed that the binding efficiency of each tested AID homologue or point mutant to eEF1A in human cells directly correlated with its relative cytoplasmic retention efficiency and was inversely correlated to its interaction with HSP90 (Fig. 3 A). Thus, the fish AID-GFP homologues, which were not retained, barely interacted with eEF1A, whereas mouse AID-GFP, of stronger retention, bound more efficiently than human AID-GFP. The reciprocal replacement of the two amino acid differences between human and mouse AID E5 affected their eEF1A binding, exactly mirroring the effect of each replacement on cytoplasmic retention. Human AID-GFP with the Ile108-Phe109-Thr110 motif mutated to Ala failed to interact with eEF1A, in line with its loss of nuclear exclusion. Finally, substituting human AID Asp187 with Glu or Ala showed a progressive decrease in their interaction with eEF1A that was proportional to their relative effect on AID localization. As mentioned, the relative interaction of each AID variant with HSP90 was inversely correlated to their eEF1A binding. Because the effect of mutating Asp187 on cytoplasmic retention of human AID does not depend on the GFP tag (Häsler et al., 2011), we compared the relative ability of untagged human AID WT, D187E, and D187A to perform SHM. We used an engineered AID-deficient DT40 B cell line, in which the SHM capacity of transduced AID variants is directly proportional to the fraction of cells losing surface IgM expression over time (Arakawa et al., 2004; Zahn et al., 2014; Fig. 3 B). In line with their relative effect on retention, AID D187E and D187A displayed increasingly higher SHM capacity (Fig. 3 B) despite the fact that their expression levels and catalytic activity were similar to or lower than WT AID (Fig. 3, C and D). These results strongly suggest that eEF1A is part of the AID cytoplasmic retention complex and that this interaction limits SHM, with the caveat that mutations in AID E5 could affect other AID interactions that we cannot control for. We also conclude that eEF1A and HSP90 form distinct complexes with AID.
Figure 3.
Cytoplasmic AID is present in at least two mutually exclusive complexes. (A) Total lysates from HEK293 cells transfected with GFP-tagged APOBEC2 (A2), AID orthologs, or AID mutants were immunoprecipitated with anti-GFP microbeads and analyzed by Western blot as indicated (top). One of ≥3 independent experiments is shown. Hs, human; Mm, mouse; Ip, catfish; Dr, zebrafish; Tr, pufferfish. Quantification of Western blot signals by densitometry from ≥3 coIP for each AID variant were plotted to compare their binding to eEF1A (white circles) and HSP90 (black circles; bottom). Values were normalized to that of human AID in each experiment, indicated with a dotted line across the plot. Gray columns indicate medians. Inset shows an expanded view of eEF1A binding for human AID and D187 point mutants that are used in B–D. (B) SHM assay in IgM+ DT40 ΔψVL Aicda−/− B cells complemented with AID variants in pMXs-(AID)-ires-GFP or empty vector (Ctrl). SHM frequency is proportional to the median accumulation of surface IgM− cells in multiple populations over time (top). Medians IgM-loss (gray bars) for 24 individual populations (black dots) per variant were plotted (bottom). One representative out of 2 experiments is shown. (C) Western blots for AID and GFP expression in total cell lysates from (C) are shown. One representative out of 2 repeats. (D) Relative enzymatic activity of AID variants was estimated from the frequency of rpoB mutation they induce in E. coli. Means (gray bars) of the relative medians (black dots) from ≥3 independent experiments normalized to AID are shown. (B and D) Differences relative to wt AID were tested by Anova with Dunnett’s post-test (***, P < 0.0004; ****, P < 0.0001).
eEF1A is necessary for AID cytoplasmic retention
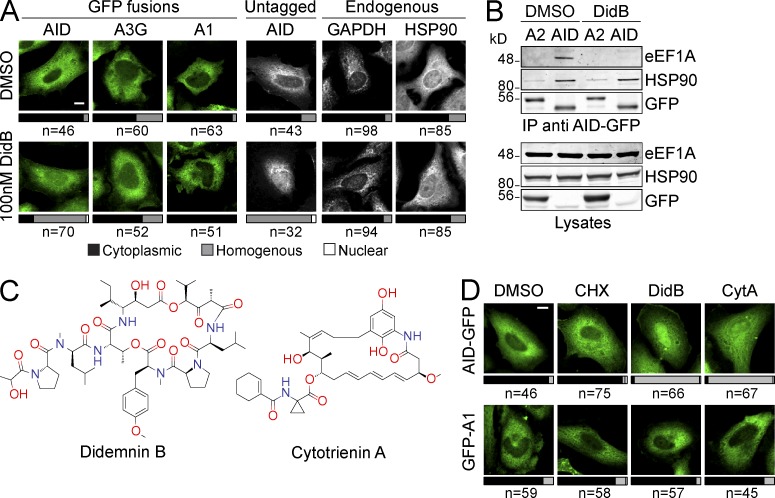
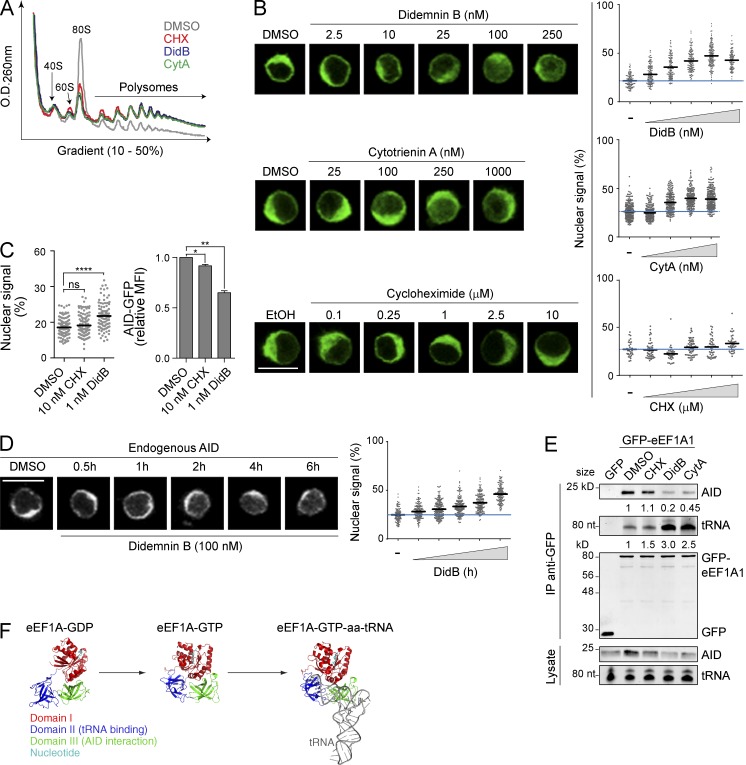
The fact that eEF1A is extremely abundant and an essential cellular factor (Trachsel, 1996) precludes genetic ablation approaches to test whether it is necessary to retain AID in the cytoplasm. To circumvent this limitation, we resorted to didemnin B (DidB), a natural cyclic depsipeptide that binds eEF1A to inhibit its function and block protein synthesis (Crews et al., 1996; Marco et al., 2004). DidB treatment rapidly increased the nuclear abundance of both AID-GFP and untagged AID in HeLa cells (Fig. 4 A). This effect was specific; APOBEC3G-GFP and GFP-APOBEC1, which do not interact with eEF1A (Häsler et al., 2011), as well as endogenous GAPDH and HSP90, remained largely cytoplasmic after treatment (Fig. 4 A). We also found that DidB prevented the coIP of eEF1A with AID (Fig. 4 B). To exclude the possibility that these effects were a result of off-target consequences of DidB, we used two other inhibitors of translation elongation. These were cytotrienin A (CytA), an ansamycin compound chemically distinct from DidB (Fig. 4 C) that we previously showed inhibits translation elongation specifically by interfering with eEF1A function (Lindqvist et al., 2010) and cycloheximide (CHX), an elongation inhibitor that targets the ribosome (Robert et al., 2009). CytA had similar effects to DidB in inducing nuclear accumulation of AID-GFP, but not GFP-APOBEC1, in HeLa cells, whereas CHX did not affect either protein (Fig. 4 D).
Figure 4.
eEF1A inhibition causes nuclear accumulation of AID. (A) Representative confocal microscopy images of HeLa cells transiently expressing GFP-tagged AID, APOBEC3G (A3G), or APOBEC1 (A1) or untagged AID are shown. Localization was assessed by detecting GFP, or by IF for overexpressed untagged AID and endogenous GAPDH and HSP90. Cells were treated with either vehicle or 100 nM Didemnin B (DidB) for 2 h before fixation. Bars represent the proportion of n cells from two independent, pooled experiments showing each subcellular distribution (cytoplasmic, black; homogenous, gray; nuclear, white). (B) Total cell extracts from HEK293 cells transiently transfected with GFP-tagged APOBEC2 (A2) or AID and treated for 4 h with either vehicle or 100 nM DidB were immunoprecipitated with anti-GFP microbeads and analyzed by Western blot as indicated. One representative out of three independent experiments is shown. (C) Chemical structure of Didemnin B and Cytotrienin A. (D) Representative confocal microscopy images of HeLa cells transiently transfected with GFP-A1 or AID-GFP are shown. Cells were treated with DMSO, 2.5 µM CHX, 100 nM DidB or 1 µM CytA for 2 h and scored as in (A) from 2 independent, pooled experiments. Bars, 10 µm.
To confirm these results in B cells, we first used Ramos B cells stably expressing human AID-GFP (twofold over endogenous AID, not depicted). As expected, the three drugs inhibited translation elongation in Ramos to similar extents, as inferred from polysome stabilization profiles (Fig. 5 A). None of the treatments changed the relative distribution of AID, HSP90, or eEF1A in sucrose gradients, according to Western blots of the fractions (unpublished data). However, DidB and CytA significantly increased nuclear AID accumulation in a dose-dependent and saturable manner, whereas CHX had little effect (Fig. 5 B). Anticipating toxicity issues (see below), we tested very low doses of DidB that were tolerated for extended periods of time. Even at 1 nM DidB, we could detect a significant increase in nuclear AID-GFP in Ramos cells by 24 h (Fig. 5 C). Because nuclear AID is actively destabilized (Aoufouchi et al., 2008), the reduction in cellular AID-GFP levels in DidB-treated cells quantified by flow cytometry (Fig. 5 C), was also consistent with an increased proportion of nuclear AID. CHX did not increase nuclear AID-GFP and had a much smaller effect on its levels (Fig. 5 C), indicating that inhibiting translation per se was not at the root of our observations with DidB and CytA. Finally, we confirmed that endogenous AID in Ramos B cells also relocalized to the nucleus in response to DidB treatment, although with slower kinetics than AID-GFP (Fig. 5 D), in keeping with the weaker cytoplasmic retention of AID-GFP compared with untagged AID (Fig. 2 D).
Figure 5.
eEF1A is necessary for cytoplasmic retention of AID. (A) UV absorbance profile of total extracts from Ramos B cells treated for 2 h with vehicle, 2.5 µM CHX, 100 nM DidB, or 1 µM CytA, and fractionated by sucrose gradient. One representative out of three independent experiments is shown. (B) Representative confocal microscopy images (left) and nuclear signal quantitation (right) of Ramos B cells stably expressing AID-GFP treated for 2 h with either vehicle or increasing concentrations of DidB (top), CytA (middle), or CHX (bottom), as indicated. Plots (right graphs) indicate the quantitated nuclear signal in >40 individual cells per point, from 1 out of 2 independent experiments. Mean values are shown as black horizontal bars, and the mean value of untreated cells is shown as a blue line across the plot. (C) Ramos B cells stably expressing AID-GFP were treated for 24 h with DMSO, 10 nM CHX, or 1 nM DidB, and the nuclear GFP signal was quantified as in (B, left). One out of two independent experiments is shown. The mean fluorescence intensity (MFI) of AID-GFP measured by flow cytometry and normalized to DMSO + SD was plotted (right) for three independent experiments. Differences relative to DMSO were analyzed by ANOVA with Dunnett’s post-test (*, P < 0.05; **, P < 0.005; ****, P < 0.0001). (D) Representative confocal microscopy images are shown (left) of endogenous AID (detected by IF) in Ramos B cells after DMSO or 100 nM DidB treatment for the indicated times. Plots show nuclear signal quantified as in B from one out of two independent experiments (right). (E) Total cell extracts from HeLa cells transiently expressing AID and GFP or GFP-eEF1A1 and treated for 4 h with either vehicle, 2.5 µM CHX, 100 nM DidB, or 1 µM CytA, were immunoprecipitated with anti-GFP microbeads and analyzed by Western blot or by Urea-PAGE, stained with SYBR Gold to detect tRNA. One representative out of three independent experiments is shown. The mean binding of GFP-eEF1A1 to AID or tRNA normalized to DMSO (measured by densitometry) is indicated under the relevant blots. (F) Structure of the eEF1A homologue EF-Tu (33% identical to eEF1A1) in the GDP- (PDB:1TUI) and GTP-bound forms (PDB:1TTT, drawn with and without the tRNA molecule (gray)). The domains of EF-Tu (I, red; II, blue; III, green), and bound nucleotide (teal) are indicated. Bars, 10 µm.
Mechanistically, DidB and CytA, but not CHX, impaired the interaction of eEF1A with AID (Fig. 5 E). There are two distinct conformers of eEF1A depending on whether it is bound to GTP or GDP (Andersen et al., 2003; Fig. 5 F). It is unknown if AID binds specifically to one form or the other, but the GTP-bound form of eEF1A binds to the aminoacyl-tRNA, which largely overlaps with the eEF1A domain III that is necessary for the interaction with AID (Häsler et al., 2011; Fig. 5 F). It is thought that DidB freezes eEF1A in the GTP-bound conformation (Marco et al., 2004). Accordingly, we found that in cells treated with DidB or CytA, the association of eEF1A with tRNA was increased at the same time that the interaction with AID was decreased; neither interaction was affected by CHX (Fig. 5 E). This suggests that the interaction of AID and tRNA with eEF1A is mutually exclusive. We conclude that cytoplasmic retention of AID is mediated by a mechanism dependent on the tRNA-free conformer of eEF1A but independent of eEF1A’s role in translation.
eEF1A limits isotype switching and chromosomal translocations
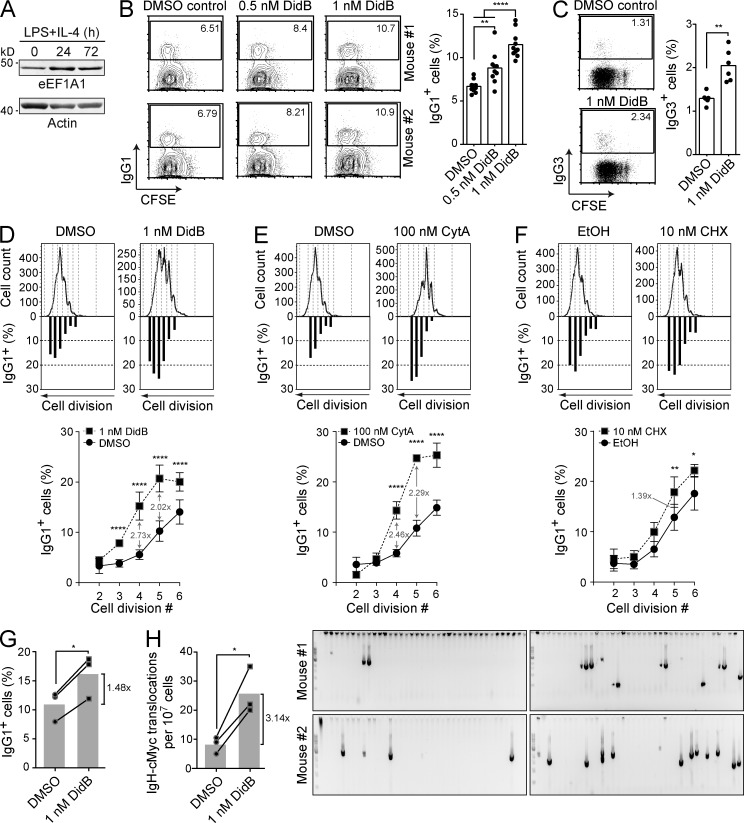
eEF1A could be expected to limit AID activity through cytoplasmic retention, and/or promote it by stabilizing AID (Fig. 5 C). To measure the biological relevance of the AID cytoplasmic retention mechanism by eEF1A, we induced isotype switching in mouse splenic B cells, which also induces the expression of eEF1A (Fig. 6 A), and 24 h later treated with DidB. We used DidB at around 1 nM, which had low toxicity over several days and still caused a measurable accumulation of AID in the nucleus (Fig. 5 C). We observed a dose-dependent increase in overall CSR efficiency to IgG1 in B cells treated with DidB, with the 1 nM dose increasing CSR by ∼70% and to IgG3 by ∼50% (Fig. 6, B and C). Even at these low concentrations, DidB still reduced B cell proliferation, as measured by CFSE dilution (Fig. 6, B and C), so the results likely underestimate the positive effect of DidB on CSR. Indeed, when we compared cells that had undergone the same number of cell divisions, DidB increased CSR to IgG1 by >100% (Fig. 6 D). CytA treatment caused an increase in CSR per cell division similar to that caused by DidB (Fig. 6 E), whereas CHX caused only a marginal increase in CSR (Fig. 6 F). Next, we asked whether cytoplasmic retention would limit the potential of AID to induce the chromosomal translocations that occur as side effects of CSR (Ramiro et al., 2004). We used B cells from TP53−/− mice because they facilitate the detection of cells with translocations (Ramiro et al., 2006). As observed for WT B cells, 1 nM DidB increased CSR by ∼50% in activated TP53−/− B cells (Fig. 6 G) and concomitantly increased the frequency of translocations by ∼3-fold (Fig. 6 H). We conclude that cytoplasmic retention of AID mediated by eEF1A limits CSR and oncogenic translocations.
Figure 6.
AID cytoplasmic retention limits CSR and chromosomal translocations. (A) Western blot analysis was performed with anti-eEF1A and anti-actin antibodies on 40 µg of total lysate from mouse splenic B cells at 0, 24, and 72 h after stimulation with 5 µg/ml LPS and 5 ng/ml IL-4. One representative experiment out of two is shown (n = 1 mouse/experiment). (B–F) CSR assays were performed on purified mouse naive splenic B cells loaded with CFSE and stimulated with 5 µg/ml LPS and 5 ng/ml IL-4 to induce switching to IgG1 or with 15 µg/ml LPS for IgG3. The indicated inhibitors were added to the medium 24 h after stimulation and CSR was assayed 3 d after stimulation by flow cytometry using biotinylated anti-IgG1 or anti-IgG3 antibodies, followed by anti-biotin APC. (B) Representative flow cytometry plots (left) of B cells induced for class switching to IgG1, and treated with vehicle or DidB as indicated. The graph (right) compiles the proportion of IgG1+ B cells from 10 mice in 6 independent experiments. Means were compared by one-way ANOVA (**, P = 0.0029; ****, P < 0.0001). (C) Representative flow cytometry (left) and compilation plots (right) as in (B) for class switching assays to IgG3 of 6 mice in 3 independent experiments (n = 2 mice/experiment). Means were compared by Student’s t test (**, P = 0.0047). (D–F) Representative graphs of the proportion of IgG1+ splenic B cells per cell division were plotted below their corresponding CFSE plots as shown (top). Graphs in the bottom panels show compiled data from 6 mice in 4 independent experiments in which B cells were treated with DidB (D) or CytA (E), and from B cells which were treated with CHX (n = 4 mice; 2 experiments; F). Differences for each point were analyzed by two-way Anova (*, P < 0.05; **, P < 0.005; ****, P < 0.0001). (G) CSR to IgG1 in B cells from three TP53−/− mice was performed as in (B) in 2 independent experiments, compared by Student’s t test (*, P = 0.0151). (H) Igh-cMyc chr12 derivative translocations were measured in B cells from (G; left graph). Each dot indicates the frequency of translocations for an individual mouse and bars show the means, compared by Student’s t test (*, P = 0.0387). The gels show representative ethidium bromide staining of PCR bands for Igh-cMyc fusions from 2 of the mice (right). Translocations were confirmed by sequencing (not depicted).
Functional integration of mechanisms regulating cytoplasmic AID
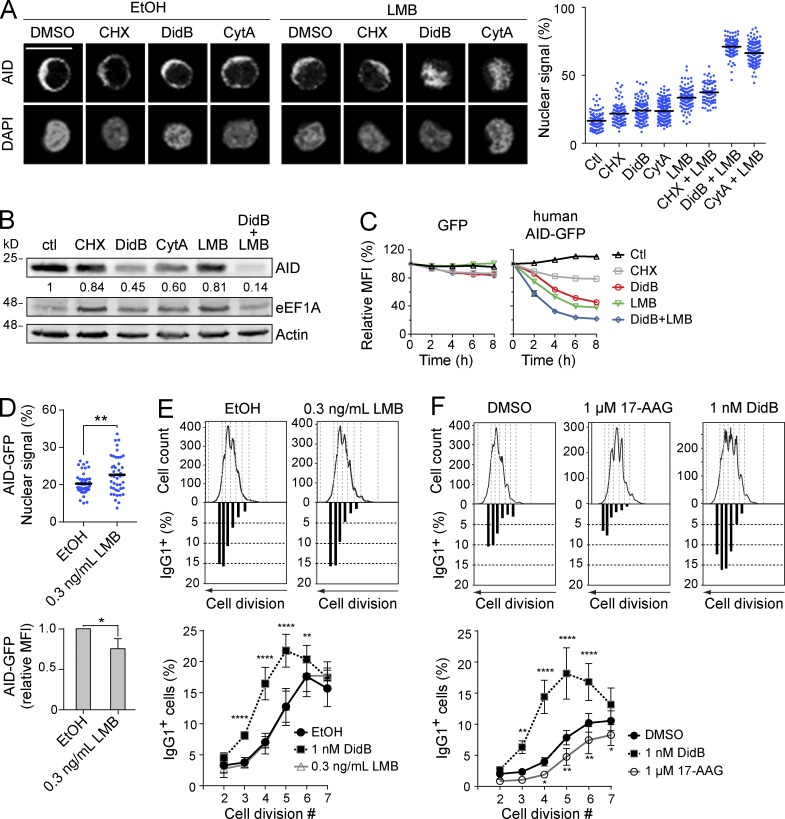
The existence of two mechanisms to exclude AID from the nucleus (i.e., nuclear export and cytoplasmic retention) begs the question as to their relative roles in regulating AID. We analyzed the localization of endogenous AID in Ramos B cells treated with the eEF1A inhibitors and/or LMB. Incubation with 100 nM DidB, 1 µM CytA or 10 ng/ml LMB each modestly increased nuclear signal of AID after 2 h (Fig. 7 A). However, the combination of DidB or CytA with LMB had a synergistic effect, resulting in full nuclear accumulation of AID (Fig. 7 A). CHX showed little effect alone and no interaction with LMB (Fig. 7 A). An increase in the proportion of nuclear AID would be reflected in reduced AID stability (Aoufouchi et al., 2008). Indeed, DidB and CytA reduced AID stability in Ramos B cells, as shown by Western blot of endogenous AID, and by the kinetics of AID-GFP decay (Fig. 7, B and C). CHX had a very small effect on AID stability, indicating that the effect of DidB and CytA was not caused by inhibiting translation. Note that DidB and CHX have equal effects on translation at the doses used (Robert et al., 2009). DidB and CytA synergized with LMB to destabilize AID, whereas CHX did not (Fig. 7, B and C). However, we noted that LMB alone did not destabilize endogenous AID to the same extent as DidB or CytA (Fig. 7 B). We then asked whether nuclear export and cytoplasmic retention were also complementary in limiting AID function. Treating activated mouse splenic B cells with LMB was toxic, but low doses that significantly increased the proportion of AID-GFP in the nucleus of Ramos B cells over 24 h (Fig. 7 D) were tolerated, although they reduced cell proliferation (Fig. 7 E). Nonetheless, LMB failed to increase CSR, whereas DidB increased CSR in the same cells (Fig. 7 E). Together, these results show that cytoplasmic retention is complementary to nuclear export in excluding AID from the nucleus. However, the data suggest that these two mechanisms might not be functionally analogous.
Figure 7.
eEF1A, CRM1, and Hsp90 associate with functionally distinct AID fractions. (A) Representative confocal microscopy images are shown of endogenous AID (detected by IF) in Ramos B cells (left). Cells were treated for 2 h with DMSO, 2.5 µM CHX, 100 nM DidB, or 1 µM CytA in combination with either vehicle or 10 ng/ml LMB. Scale bar, 10 µm. The proportion of nuclear AID signal for ≥100 cells per point was assessed as the ratio of AID signal overlapping DAPI signal to total AID signal × 100. The nuclear AID signal of each cell (dots), and the mean for each group (black bars) are shown for one out of 2 independent experiments, (right graph). (B) Ramos B cells were treated as in (A) for 6 h and expression of AID, actin and eEF1A were analyzed by Western blot. One representative out of 2, independent experiments is shown. The mean ratio of AID to actin measured by densitometry, normalized to DMSO is indicated under the relevant blots. (C) Ramos B cells stably expressing either GFP or human AID-GFP were treated in duplicate as in (A). The MFI ± SEM of GFP was measured by flow cytometry at different times post-treatment and normalized to t0 = 100%, and was plotted over time for one out of ≥2 representative, independent experiments. (D) Ramos B cells stably expressing AID-GFP were treated for 24 h, as indicated, and the nuclear GFP signal was quantified as in A (top). The MFI + SD of AID-GFP was measured by flow cytometry and normalized to vehicle, and was plotted from 4 independent experiments (bottom). Means were compared by Student’s t test (*, P < 0.05; **, P < 0.005). (E and F) Representative plots of the proportion of IgG1+ cells per division are shown below the corresponding CFSE profiles in wt mouse naive splenic B cells stimulated with 5 µg/ml LPS and 5 ng/ml IL-4 in the presence of the indicated inhibitors 24 h later. The proportions of IgG1+ cells were determined by flow cytometry 3 d after stimulation (shown in bottom graphs). Data were compiled from 4 mice from 2 experiments, (n = 2 mice/experiment). Differences relative to vehicle were analyzed by two-way ANOVA (*, P < 0.05; **, P < 0.005; ****, P < 0.0001).
We then compared the functional roles of the interaction of AID with HSP90 and eEF1A. Despite the HSP90 inhibitor 17-AAG affecting cell proliferation and AID stability (Orthwein et al., 2010) similarly to DidB, testing both drugs in parallel showed a clear difference, with DidB increasing and 17-AAG decreasing CSR per cell division (Fig. 7 F). We conclude that, unlike HSP90-associated AID, the eEF1A complex stores functionally competent AID in the cytoplasm. Altogether our results suggest a model in which shuttling between different complexes underpins the production and storage of CSR-competent AID in the cytoplasm.
DISCUSSION
We identify a mechanism that retains AID in the cytoplasm through eEF1A, thereby excluding functional AID from the nucleus of B cells and limiting CSR per cell division and oncogenic chromosomal translocations. We also compare cytoplasmic retention to nuclear export and cytoplasmic stabilization of endogenous AID by HSP90 in B cells to propose a minimal model for the transit of AID through distinct cytoplasmic complexes to acquire functional competence.
The stoichiometric interaction between AID and eEF1A in the cytoplasm, which required AID Asp187 and 188, suggested a role for eEF1A in cytoplasmic retention of AID (Häsler et al., 2011). This observation was not conclusive because the same mutations prevent the interaction of AID with other factors, at least with PABPC1 and hnRNPA1 (unpublished data). Moreover, it is unknown whether AID and eEF1A interact directly or not (Häsler et al., 2012). Thus, those observations were insufficient to tell whether eEF1A was necessary for AID cytoplasmic retention or just part of a larger complex containing both proteins (Häsler et al., 2011). We demonstrate that eEF1A is a necessary component of the complex that retains AID in the cytoplasm. First, the cytoplasmic retention efficiency of different AID-GFP variants correlates perfectly with their relative ability to interact with eEF1A. Second, two translation elongation inhibitors that target eEF1A, DidB, and CytA, disrupt its interaction with AID, increase the AID nuclear accumulation, and boost CSR and Igh-cMyc fusions. The very different chemical nature and structures of DidB and CytA make it extremely unlikely that nonspecific (i.e., eEF1A-independent) effects can explain their identical effects on AID biology at so many levels. In addition, eEF1A is by far the most abundant target of DidB (Crews et al., 1994, 1996), which has a ∼20 nM IC50 for translation inhibition (Robert et al., 2009). The only other known target of DidB, a lysosomal palmitoyl thioesterase, is inhibited at 5 µM IC50 (Meng et al., 1998). Thus, at the ∼1 nM doses we used in functional assays, eEF1A is in all likelihood the main target of DidB. The mechanism whereby eEF1A retains AID in the cytoplasm is neither related to its role in mRNA translation, as shown by the lack of effect of CHX, nor to the transcription-dependent nuclear export pathway in which eEF1A participates. The latter pathway requires a sequence signal that is absent in AID and is inhibited by Actinomycin D (Khacho et al., 2008), which does not affect AID localization (unpublished data). Rather, our data indicates a mechanism in which DidB and CytA favor a specific conformer of eEF1A that is unable to interact, directly or indirectly, with AID. Although DidB does not inhibit the GTPase activity of eEF1A, it stabilizes a GTP-bound-like conformer by bridging domains I and II of eEF1A and preventing the conformational change after GTP hydrolysis (Marco et al., 2004). Our results are consistent with this mechanism, as DidB and CytA both favor the association of eEF1A to tRNA (Fig. 5 E). The tRNA binds to eEF1A domain II but it is large enough to sterically interfere with AID (or an adaptor) binding to domain III (Fig. 5 F). Thus, AID is retained in the cytoplasm in a complex with the eEF1A-GDP conformer. The complex does not contain polyribosomes or ribosomal units, as judged from sucrose gradient fractionations, either under basal conditions or after DidB treatment (unpublished data), and is distinct from the complex that AID forms with HSP90. Future work will determine the composition of this complex.
Optimal binding of AID to eEF1A, and thus cytoplasmic retention, is conformation dependent. We present a structural model in which the C-terminal E5 domain is folded onto the catalytic core of AID, interacting largely through the hydrophobic NES residues. We obtained the same model of AID from phylogenetically distant species simply based on thermodynamic restrictions. Additionally, the model is consistent with several experimental results: (1) mutations in the NES are more efficient than LMB at disrupting nuclear exclusion of AID (Ito et al., 2004; McBride et al., 2004; Geisberger et al., 2009; Patenaude et al., 2009); (2) the C-terminal GFP fusion, or an insertion separating the E5 from the core, interferes with AID retention (Fig. 2 D); (3) several model-guided mutation of non-NES residues have the predicted effect on nuclear import kinetics (Fig. 2, G and H); (4) Asp187 and 188, which mediate cytoplasmic retention and the eEF1A interaction (Patenaude et al., 2009; Häsler et al., 2011), are exposed on the surface.
The need for a specific AID conformation for interacting with eEF1A is consistent with AID forming a distinct complex with HSP90 (Fig. 4). Because HSP90 does not assist the initial folding of proteins but stabilizes metastable conformations (Young et al., 2004), one straightforward interpretation of our data are that this chaperoning pathway facilitates an eEF1A-compatible AID conformation (Orthwein et al., 2010, 2012). Accordingly, inhibiting HSP90 releases mostly unstable AID that is degraded in the cytoplasm (Orthwein et al., 2010), thus placing the interaction of AID with HSP90 before its association to eEF1A during the cellular life cycle of AID. This would also explain the slow kinetics of degradation of AID after HSP90 inhibition compared with other HSP90 client proteins in B cells (Orthwein et al., 2010). Indeed, the pool of AID in complex with eEF1A is insensitive to HSP90 inhibitors and stable in the cytoplasm, thereby delaying the decay of cellular AID levels observed after HSP90 inhibition.
Our results definitely demonstrate that nuclear export and cytoplasmic retention of AID are different mechanisms, as they are mediated by different factors and inhibited by different drugs. The AID 3D model suggests how this is possible despite being mediated by the same very small region of AID. The NES is hidden to expose the cytoplasmic retention motif. In turn, the E5 region must at some stage adopt an extended conformation for interacting with CRM1 (Sun et al., 2013). The combined use of DidB or CytA and LMB indicates that cytoplasmic retention and nuclear export are complementary in excluding endogenous AID from the nucleus. However, the data suggest that they are not functionally equivalent because the eEF1A inhibitors increase CSR whereas the CRM1 inhibitor does not, despite both similarly augmenting the nuclear abundance of AID (Fig. 7 E, and compare Fig. 5 C to (Fig. 7 D). Thus AID in complex with eEF1A is competent for CSR, as indicated by the increased CSR upon its release by DidB. It has been reported that DidB can increase the titer of antigen-specific antibodies after immunization (Montgomery et al., 1987), suggesting it could work similarly in vivo. It is also conceivable that boosting the activity of AID through DidB or CytA could be used to sensitize cells expressing AID to DNA repair inhibitors (Lamont et al., 2013). In contrast, the AID pool that is exported by CRM1 seems unable to perform CSR. We cannot rule out that LMB prevents CSR by some other means, but it is intriguing that LMB has a smaller effect on endogenous AID stability than DidB or CytA (Fig. 7 B). We speculate that AID may enter the nucleus in either CSR-proficient or not-proficient states. The transit of AID through the eEF1A complex may promote the association of AID with some factors that masks the NES and/or are necessary for its function. AID imported without those associated factors would be recognized by CRM1 and exported, rather than degraded in the nucleus. Other possibilities exist, but our results provide a working model to test in future work.
Finally, our findings have implications for the well-established observation that the probability of activated B cells to undergo CSR increases with the number of cell divisions (Hodgkin et al., 1996). Because AID levels also increase with subsequent B cell divisions, AID expression probably requires reaching a certain threshold, contributing to division-linked CSR (Rush et al., 2005). Cytoplasmic retention of AID is another mechanism limiting CSR per cell division. Releasing AID from eEF1A increases the probability of CSR by increasing the effective concentration of functional AID in the nucleus, thereby reducing the threshold level of total AID required. This release comes associated to a large increase in chromosomal translocations, indicating that cytoplasmic retention is important in moderating the oncogenic potential of AID.
MATERIALS AND METHODS
Mice.
C57BL6/J WT and TP53−/− mice (JAX) were kept under specific pathogen-free conditions at the IRCM animal house. All animal experimentation was approved by the IRCM Animal Protection Committee according to the guidelines from the Canadian Council of Animal Care.
DNA constructs.
GFP fusions of APOBEC2, rat APOBEC1, and APOBEC3G have been described previously (Orthwein et al., 2010). Xenopus tropicalis AID, a gift from A. Bottaro (University of Rochester, Rochester, NY), was PCR amplified using oligonucleotides OJ492 and OJ493 and cloned into pEGFP-N3 as a BamHI–EcoRI fragment. All other AID orthologs were a gift from R.S. Harris (University of Minnesota, Minneapolis, MN) and were cloned in pEGFP-N3 as HindIII–SalI fragments. Retroviral expression vectors were made by subcloning HindIII–NotI fragments from pEGFP-N3 into pMXs. Point mutations were introduced by QuikChange (Stratagene). Oligonucleotide sequences are available upon request. Human eEF1A1 was PCR amplified from cDNA using oligonucleotides OJ641 and OJ642 and cloned into pEGFP-C1 as an EcoRI–SalI fragment.
Reagents and antibodies.
Stock aliquots of 50 µg/ml LMB (LC Laboratories) and 100 mM CHX (Sigma-Aldrich) were made in Ethanol. 17-AAG (LC Laboratories) and DidB (NSC 325319; provided by the Natural Products Branch, National Cancer Institute, Bethesda, MD), were prepared as 20 mM stock solutions in DMSO. CytA (Cayman Chemical) was prepared as a 1-nM stock solution in DMSO. Drugs were kept in the dark at −20°C, and diluted fresh before each use. Antibodies (dilutions) for IF were as follows: mAb anti-GAPDH (1:500; SC Biotechnology), mAb anti-HSP90 (1:500; BD), mouse mAb anti-AID (to detect human AID; 1:500; Invitrogen), rat mAb anti-AID (1:500; eBioscience or Cell Signaling Technology), mAID-2 (to detect mouse AID; 1:250; eBioscience). Secondary antibodies (all 1:500; Invitrogen) were anti–mouse Alexa Fluor 488, anti–rat Alexa Fluor 680, anti–rabbit 546, anti–rabbit 680. For Western blot, we used rat mAb anti-AID (1:1,000; Cell Signaling Technology), rabbit anti-GFP (1:2,000; Invitrogen), mouse mAb anti-PCNA (1:3,000; Abcam), rabbit anti-eEF1A (1:2,000; Abcam), mouse mAb anti-HSP90 (1:3,000; BD), and rabbit mAb anti-Actin (1:3,000; Sigma-Aldrich).
Microscopy and immunofluorescence.
HeLa, HEK293, and NIH3T3 cells were plated on coverslips and transfected using TransIT-LT1 (Mirus) according to manufacturer’s instructions. Cells were treated as indicated 36–48 h after transfection and before fixation. Human Ramos B cells stably expressing AID-GFP were described previously (Patenaude et al., 2009). Mouse CH12F3 B cells were retrovirally transduced using culture medium of HEK293 cells co-transfected with pMXs vectors encoding the protein of interest and vectors encoding VSV-G and GAG-Pol. Virus was attached to 6-well plates coated with 0.2 µg/ml Retronectin (Takara) by centrifugation (32°C, 30 min, 2000 g), cells were layered on top by centrifugation (32°C, 30 min, 600 g) and cultured for 2 d. Infected cells were sorted for similar levels of GFP expression. For GFP visualization, cells were fixed with 3.7% (wt/vol) formaldehyde for 10 min, washed three times with PBS, and nuclei stained with propidium iodide (PI). Catfish 1B10 B cells (Miller et al., 1994) were cultured at 28°C, cells were electroporated as described (Ellestad and Magor, 2005), treated for 2 h with EtOH or 50 ng/ml LMB 48 h later, and then fixed as above. Fixed cells were cytospun onto microscopy slides and nuclei were stained with PI before mounting for imaging. EL1 BL, Daudi, and DG75 B cell lymphoma lines were a gift from M. Neuberger (Medical Research Council, London, UK). B cells were kept in suspension at 106 cells/ml during treatments, after which they were attached to poly-L lysine (Sigma-Aldrich)–coated coverslips and processed for IF. Cells were fixed in either 3.7% formaldehyde (for AID) or −20°C methanol (for GAPDH and HSP90), before permeabilization and incubation for 30 min in blocking solution (PBS, 0.5% [vol/vol] Triton-X100, 1% [wt/vol] BSA, 5% [vol/vol] goat serum). Cells were then stained overnight at 4°C in blocking solution containing primary antibodies followed by 3 washes with 0.01% Triton X-100 PBS (PBS-T) and a 1-h incubation with secondary antibody in blocking solution and 3 PBS-T washes.
Specific AID signal in IF was confirmed using DG75 and unstimulated CH12F3 cells. The ability of anti-AID antibodies to detect nuclear AID was confirmed using LMB-treated AID-GFP Ramos B cells. The results were identical with the three different anti-AID antibodies listed above. After nuclear staining with either PI or Dapi (300 nM in PBS), coverslips were washed with ddH2O and mounted on slides using Lerner Aqua-Mount (Thermo Fisher Scientific). Images were acquired at room temperature using ZEN 2010 on either a Zeiss LSM 510, or LSM 700 confocal microscope with excitation lasers at 405 nM (DAPI), 488 nM (EGFP and Alexa Fluor 488), 543 nM (PI and Alexa Fluor 546), and 633 nM (Alexa Fluor 633, Alexa Fluor 680), using either 40×/1.3 or 63×/1.4 oil immersion objectives, and collected with a Hamamatsu PMT. Subcellular localization of fluorescent signal in HeLa, 293T, and NIH3T3 cells was scored using ImageJ (National Institutes of Health, Bethesda, MD). In ImageJ, we quantified the proportion of nuclear signal intensity using the profile tool to calculate the mean GFP fluorescence signal across the nucleus (i.e., overlapping with PI signal) over the total mean fluorescence signal along the line for multiple cells. In Volocity (Perkin Elmer), for each individual cell, we made masks for nuclear and total GFP or IF signal. The proportion of nuclear signal was calculated as the ratio of nuclear signal/total signal × 100. For each experiment, multiple fields were analyzed for subcellular localization. Cells showing saturated signal, abnormal DNA structure, or mitotic figures were excluded. For making figures, images were transferred to Photoshop for adjusting contrast throughout the whole image when necessary to enhance visibility and for cropping.
Structural modeling.
We generated a structural model of the core domain of AID using the Rosetta suite (version: rosetta_2013wk51) and APOBEC3C (PDB ID 3VOW) as template by comparative homology modeling. To systematically sample suitable positions of the C-terminal tail of AID, we generated 20,000 models using the core domain as a rigid seed and the E5 was sampled with fragments of 9, 3, and 1 residues. All models were evaluated by the Rosetta energy score and the top-ranking ones were clustered using VMD (Humphrey et al., 1996), eventually selecting the most populated ones. Whereas human and mouse models resulted in one major cluster, zebrafish displayed three isoenergetic groups, one of them being equivalent to the major cluster found in the other two species.
coIP.
For AID-GFP coIP, HEK293 cells were transiently transfected with APOBEC/AID-GFP constructs using CaPO4 precipitation 48 h before lysis. Cells were lysed for 30 min on ice in lysis buffer (20 mM Hepes pH 7.5, 150 mM NaCl, 10% glycerol, 0.2% Triton X-100, 1× Complete protease inhibitor cocktail (Roche) and 2x HALT protease and phosphatase inhibitor cocktail (Thermo)]. For GFP-eEF1A1 coIP, HeLa cells were transiently transfected with AID and GFP or GFP-eEF1A1 using TransIT-LT1 48 h before lysis. Cells were lysed for 10 min on ice in lysis buffer (5 mM Tris, pH 7.5, 1.5 mM KCl, 2.5 mM MgCl2, 0.1% Triton X-100, 0.1% Na deoxycholate, 1 mM DTT, 40 U/ml RNaseIN [Promega], and 2× Complete). After lysate clarification, GFP immunoprecipitation was performed using the μMACS GFP isolation kit according to manufacturer’s instructions (Miltenyi Biotec), with the exception of washing. For AID-GFP coIP, washing was done 3× with 200 µl of wash buffer (20 mM Hepes, pH 7.5, 150 mM NaCl, 0.1% Triton X-100, and 1× Complete). For GFP-eEF1A1 coIP, washing was done 5× with 200 µl of wash buffer (50 mM Tris, pH 7.5, 300 mM KCl, 12 mM MgCl2, 1% Triton X-100, 1 mM DTT, and 1X Complete). When indicated, cells were treated with DMSO, 2.5 µM CHX, 100 nM DidB or 1 µM CytA for 4 h before lysis. Elutions and total lysates were analyzed by Western blot using the Li-Cor Odyssey infrared imaging system (Li-COR Biosciences).
Protein stability assays and biochemistry.
For endogenous protein stability, Ramos cells were plated at 0.5 × 106 cells/ml density and treated with indicated drugs for 6 h. After drug incubation, cells were counted, and 2 × 106 cells were harvested, washed with PBS, and lysed directly in SDS-PAGE loading buffer. Protein levels were analyzed by Western blot. For GFP-fused protein stability assays, the GFP signal in stably transfected Ramos B cells was measured by flow cytometry at specific times after the indicated drug treatments, as done previously (Orthwein et al., 2010). Protein expression in mouse splenic B cells was measured by Western blot. Protein lysates were collected from either resting B cells, or B cells stimulated for 24 or 72 h with 5 µg/ml LPS and 5 ng/ml IL-4. At least 5 × 106 cells were pelleted, resuspended in PBS + 5X complete protease inhibitor cocktail and lysed in NP-40 lysis buffer [20 mM Tris-HCl pH.8, 137 mM NaCl, 10% glycerol, 2 mM EDTA and 1% NP-40] for 30 min on ice. Protein was quantified in clarified lysates using a BCA protein assay kit (Thermo Fisher Scientific), and 40 µg total protein loaded for each Western blot. Sucrose gradient fractionation was done as previously described (Robert et al., 2009) using 20 × 106 Ramos B cells. Cells were treated for 2 h with DMSO, 2.5 µM CHX, 100 nM DidB, or 1 µM CytA, washed with PBS, then lysed in hypotonic lysis buffer (5 mM Tris, pH 7.5, 2.5 mM MgCl2, 1.5 mM KCl, 2 mM DTT, 1% Triton X-100 and 0.5% sodium deoxycholate) in the presence of the appropriate inhibitor. Cleared lysates were loaded onto a continuous 10–50% sucrose gradient column (10% to 50% sucrose, 20 mM Hepes pH 7.6, 100 mM KCl, 5 mM MgCl2, and 1 mM DTT), and centrifuged at 35,000 rpm for 2 h at 4°C in a SW41 rotor. Polysomes were visualized and fractionated using a fraction collector connected to a UV254 nm detector (ISCO) and 10-drop fractions were collected. Proteins from each fraction were then concentrated using StrataClean resin (Agilent Technologies) and separated on SDS-PAGE gels for analysis by Western blot.
Monitoring AID activity.
E. coli rpoB mutation assays were performed using the Δung BW310 strain expressing 6XHis-AID fusions, as previously described (Zahn et al., 2014). SHM activity was assessed in IgM+ DT40 ΔψVL Aicda−/− cells (Arakawa et al., 2004) retrovirally complemented with AID variants. Fluctuation analysis of IgM phenotype was performed as previously described (Zahn et al., 2014) using initial populations of 1,000 GFP+ IgM+ cells FACS-sorted into 96-well plates and expanded for 15 d in 24-well plates before staining with anti–chicken IgM-RPE (Southern Biotech) and measuring the proportion of GFP+ IgM− cells by flow cytometry. CSR was measured in resting B cells purified from spleens of C57BL6/J mice using MACS anti-CD43 microbeads depletion (Miltenyi Biotech), as described (Orthwein et al., 2010). Cells were stained with CFSE (Invitrogen) according to manufacturer’s instructions, plated at 106 cells/well in 24-well plates, and stimulated with 5 µg/ml LPS and 5 ng/ml IL-4 for IgG1 or 15 µg/ml LPS for IgG3. 24 h after stimulation, cells were treated as indicated and analyzed 48 h later. B cells were stained with biotinylated rat anti–mouse IgG1 or IgG3 antibody (BD), followed by APC-conjugated anti-biotin antibody (Miltenyi Biotec). IgG1 or IgG3, as well as CFSE dilution, were measured by flow cytometry on a FACSCalibur (BD), and analyzed on FlowJo. Chromosomal translocations were detected by nested PCR amplification of the Igh-cMyc fusion chr12 derivative as previously described (Ramiro et al., 2004; Zahn et al., 2014). PCR products were purified and sequenced at Macrogen using oligonucleotide OJ317.
Acknowledgments
We thank Andrea Bottaro, Reuben S. Harris, and Michael S. Neuberger for reagents. We are indebted to the Natural Products branch of the National Cancer Institute for providing Didemnin B. We thank A.K. Eranki, Dr. A. Orthwein and Dr. J.F. Côté for discussions, and Dr. C. Rada for critical reading of the manuscript. We are thankful for the assistance of D. Fillion with microscopy, E. Massicotte with flow cytometry, and Dr. J. Fraszcsak for the TP53−/− mice.
This work was funded by the Canadian Institutes of Health Research operating grant MOP125991 (to J.M. Di Noia) and the National Science and Engineering Research Council of Canada (to B.G. Magor). S.P. Methot was supported by fellowships from the Canadian Institutes of Health Research, Cole Foundation and Fonds de Recherche du Québec – Santé. J.M. Di Noia holds a Canadian Research Chair tier 2.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- AID
- activation-induced deaminase
- APOBEC
- apolipoprotein B mRNA editing enzyme catalytic polypeptide-like
- CHX
- cycloheximide
- CSR
- class switch recombination
- CytA
- cytotrienin A
- DidB
- didemnin B
- eEF1A
- eukaryotic elongation factor 1 α
- HSP90
- heat-shock protein 90 kD
- LMB
- leptomycin B
- NES
- nuclear export signal
- SHM
- somatic hypermutation
References
- Andersen G.R., Nissen P., and Nyborg J.. 2003. Elongation factors in protein biosynthesis. Trends Biochem. Sci. 28:434–441 10.1016/S0968-0004(03)00162-2 [DOI] [PubMed] [Google Scholar]
- Aoufouchi S., Faili A., Zober C., D’Orlando O., Weller S., Weill J.-C., and Reynaud C.-A.. 2008. Proteasomal degradation restricts the nuclear lifespan of AID. J. Exp. Med. 205:1357–1368 10.1084/jem.20070950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakawa H., Saribasak H., and Buerstedde J.-M.. 2004. Activation-induced cytidine deaminase initiates immunoglobulin gene conversion and hypermutation by a common intermediate. PLoS Biol. 2:E179 10.1371/journal,pbio.0020179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett R.P., Diner E., Sowden M.P., Lees J.A., Wedekind J.E., and Smith H.C.. 2006. APOBEC-1 and AID are nucleo-cytoplasmic trafficking proteins but APOBEC3G cannot traffic. Biochem. Biophys. Res. Commun. 350:214–219 10.1016/j.bbrc.2006.09.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar S.S., Watson M., and Diaz M.. 2004. Activation-induced cytosine deaminase (AID) is actively exported out of the nucleus but retained by the induction of DNA breaks. J. Biol. Chem. 279:26395–26401 10.1074/jbc.M403503200 [DOI] [PubMed] [Google Scholar]
- Chester A., Somasekaram A., Tzimina M., Jarmuz A., Gisbourne J., O’Keefe R., Scott J., and Navaratnam N.. 2003. The apolipoprotein B mRNA editing complex performs a multifunctional cycle and suppresses nonsense-mediated decay. EMBO J. 22:3971–3982 10.1093/emboj/cdg369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crews C.M., Collins J.L., Lane W.S., Snapper M.L., and Schreiber S.L.. 1994. GTP-dependent binding of the antiproliferative agent didemnin to elongation factor 1 alpha. J. Biol. Chem. 269:15411–15414. [PubMed] [Google Scholar]
- Crews C.M., Lane W.S., and Schreiber S.L.. 1996. Didemnin binds to the protein palmitoyl thioesterase responsible for infantile neuronal ceroid lipofuscinosis. Proc. Natl. Acad. Sci. USA. 93:4316–4319 10.1073/pnas.93.9.4316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R., and Baker D.. 2008. Macromolecular modeling with rosetta. Annu. Rev. Biochem. 77:363–382 10.1146/annurev.biochem.77.062906.171838 [DOI] [PubMed] [Google Scholar]
- Diaz M.2013. The role of activation-induced deaminase in lupus nephritis. Autoimmunity. 46:115–120 10.3109/08916934.2012.750303 [DOI] [PubMed] [Google Scholar]
- Ellestad K.K., and Magor B.G.. 2005. Evolution of transcriptional enhancers in the immunoglobulin heavy-chain gene: functional characteristics of the zebrafish Emu3′ enhancer. Immunogenetics. 57:129–139 10.1007/s00251-005-0785-3 [DOI] [PubMed] [Google Scholar]
- Geisberger R., Rada C., and Neuberger M.S.. 2009. The stability of AID and its function in class-switching are critically sensitive to the identity of its nuclear-export sequence. Proc. Natl. Acad. Sci. USA. 106:6736–6741 10.1073/pnas.0810808106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häsler J., Rada C., and Neuberger M.S.. 2011. Cytoplasmic activation-induced cytidine deaminase (AID) exists in stoichiometric complex with translation elongation factor 1α (eEF1A). Proc. Natl. Acad. Sci. USA. 108:18366–18371 10.1073/pnas.1106729108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häsler J., Rada C., and Neuberger M.S.. 2012. The cytoplasmic AID complex. Semin. Immunol. 24:273–280 10.1016/j.smim.2012.05.004 [DOI] [PubMed] [Google Scholar]
- Hodgkin P.D., Lee J.H., and Lyons A.B.. 1996. B cell differentiation and isotype switching is related to division cycle number. J. Exp. Med. 184:277–281 10.1084/jem.184.1.277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W., Dalke A., and Schulten K.. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:33–38: 27–28 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- Ito S., Nagaoka H., Shinkura R., Begum N., Muramatsu M., Nakata M., and Honjo T.. 2004. Activation-induced cytidine deaminase shuttles between nucleus and cytoplasm like apolipoprotein B mRNA editing catalytic polypeptide 1. Proc. Natl. Acad. Sci. USA. 101:1975–1980 10.1073/pnas.0307335101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keim C., Kazadi D., Rothschild G., and Basu U.. 2013. Regulation of AID, the B-cell genome mutator. Genes Dev. 27:1–17 10.1101/gad.200014.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khacho M., Mekhail K., Pilon-Larose K., Pause A., Côté J., and Lee S.. 2008. eEF1A is a novel component of the mammalian nuclear protein export machinery. Mol. Biol. Cell. 19:5296–5308 10.1091/mbc.E08-06-0562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura S., Ode H., Nakashima M., Imahashi M., Naganawa Y., Kurosawa T., Yokomaku Y., Yamane T., Watanabe N., Suzuki A., et al. 2012. The APOBEC3C crystal structure and the interface for HIV-1 Vif binding. Nat. Struct. Mol. Biol. 19:1005–1010 10.1038/nsmb.2378 [DOI] [PubMed] [Google Scholar]
- Lamont K.R., Hasham M.G., Donghia N.M., Branca J., Chavaree M., Chase B., Breggia A., Hedlund J., Emery I., Cavallo F., et al. 2013. Attenuating homologous recombination stimulates an AID-induced antileukemic effect. J. Exp. Med. 210:1021–1033 10.1084/jem.20121258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land A.M., Law E.K., Carpenter M.A., Lackey L., Brown W.L., and Harris R.S.. 2013. Endogenous APOBEC3A DNA cytosine deaminase is cytoplasmic and nongenotoxic. J. Biol. Chem. 288:17253–17260 10.1074/jbc.M113.458661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist L., Robert F., Merrick W., Kakeya H., Fraser C., Osada H., and Pelletier J.. 2010. Inhibition of translation by cytotrienin A—a member of the ansamycin family. RNA. 16:2404–2413 10.1261/rna.2307710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco E., Martín-Santamaría S., Cuevas C., and Gago F.. 2004. Structural basis for the binding of didemnins to human elongation factor eEF1A and rationale for the potent antitumor activity of these marine natural products. J. Med. Chem. 47:4439–4452 10.1021/jm0306428 [DOI] [PubMed] [Google Scholar]
- Mateyak M.K., and Kinzy T.G.. 2010. eEF1A: thinking outside the ribosome. J. Biol. Chem. 285:21209–21213 10.1074/jbc.R110.113795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride K.M., Barreto V., Ramiro A.R., Stavropoulos P., and Nussenzweig M.C.. 2004. Somatic hypermutation is limited by CRM1-dependent nuclear export of activation-induced deaminase. J. Exp. Med. 199:1235–1244 10.1084/jem.20040373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L., Sin N., and Crews C.M.. 1998. The antiproliferative agent didemnin B uncompetitively inhibits palmitoyl protein thioesterase. Biochemistry. 37:10488–10492 10.1021/bi9804479 [DOI] [PubMed] [Google Scholar]
- Miller N.W., Rycyzyn M.A., Wilson M.R., Warr G.W., Naftel J.P., and Clem L.W.. 1994. Development and characterization of channel catfish long term B cell lines. J. Immunol. 152:2180–2189. [PubMed] [Google Scholar]
- Montgomery D.W., Celniker A., and Zukoski C.F.. 1987. Didemnin B—an immunosuppressive cyclic peptide that stimulates murine hemagglutinating antibody responses and induces leukocytosis in vivo. Transplantation. 43:133–139 10.1097/00007890-198701000-00029 [DOI] [PubMed] [Google Scholar]
- Orthwein A., and Di Noia J.M.. 2012. Activation induced deaminase: how much and where? Semin. Immunol. 24:246–254 10.1016/j.smim.2012.05.001 [DOI] [PubMed] [Google Scholar]
- Orthwein A., Patenaude A.-M., Affar B., Lamarre A., Young J.C., and Di Noia J.M.. 2010. Regulation of activation-induced deaminase stability and antibody gene diversification by Hsp90. J. Exp. Med. 207:2751–2765 10.1084/jem.20101321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orthwein A., Zahn A., Methot S.P., Godin D., Conticello S.G., Terada K., and Di Noia J.M.. 2012. Optimal functional levels of activation-induced deaminase specifically require the Hsp40 DnaJa1. EMBO J. 31:679–691 10.1038/emboj.2011.417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L., Guglielmino R., Houldsworth J., Mohr J., Aoufouchi S., Polakiewicz R., Chaganti R.S.K., and Dalla-Favera R.. 2004. Expression of the AID protein in normal and neoplastic B cells. Blood. 104:3318–3325 10.1182/blood-2004-04-1558 [DOI] [PubMed] [Google Scholar]
- Patenaude A.-M., Orthwein A., Hu Y., Campo V.A., Kavli B., Buschiazzo A., and Di Noia J.M.. 2009. Active nuclear import and cytoplasmic retention of activation-induced deaminase. Nat. Struct. Mol. Biol. 16:517–527 10.1038/nsmb.1598 [DOI] [PubMed] [Google Scholar]
- Rada C., Jarvis J.M., and Milstein C.. 2002. AID-GFP chimeric protein increases hypermutation of Ig genes with no evidence of nuclear localization. Proc. Natl. Acad. Sci. USA. 99:7003–7008 10.1073/pnas.092160999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramiro A.R., Jankovic M., Eisenreich T., Difilippantonio S., Chen-Kiang S., Muramatsu M., Honjo T., Nussenzweig A., and Nussenzweig M.C.. 2004. AID is required for c-myc/IgH chromosome translocations in vivo. Cell. 118:431–438 10.1016/j.cell.2004.08.006 [DOI] [PubMed] [Google Scholar]
- Ramiro A.R., Jankovic M., Callen E., Difilippantonio S., Chen H.T., McBride K.M., Eisenreich T.R., Chen J., Dickins R.A., Lowe S.W., et al. 2006. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature. 440:105–109 10.1038/nature04495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revy P., Muto T., Levy Y., Geissmann F., Plebani A., Sanal O., Catalan N., Forveille M., Dufourcq-Labelouse R., Gennery A., et al. 2000. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2). Cell. 102:565–575 10.1016/S0092-8674(00)00079-9 [DOI] [PubMed] [Google Scholar]
- Robbiani D.F., Bunting S., Feldhahn N., Bothmer A., Camps J., Deroubaix S., McBride K.M., Klein I.A., Stone G., Eisenreich T.R., et al. 2009. AID produces DNA double-strand breaks in non-Ig genes and mature B cell lymphomas with reciprocal chromosome translocations. Mol. Cell. 36:631–641 10.1016/j.molcel.2009.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert F., Carrier M., Rawe S., Chen S., Lowe S., and Pelletier J.. 2009. Altering chemosensitivity by modulating translation elongation. PLoS ONE. 4:e5428 10.1371/journal.pone.0005428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rush J.S., Liu M., Odegard V.H., Unniraman S., and Schatz D.G.. 2005. Expression of activation-induced cytidine deaminase is regulated by cell division, providing a mechanistic basis for division-linked class switch recombination. Proc. Natl. Acad. Sci. USA. 102:13242–13247 10.1073/pnas.0502779102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severi F., Chicca A., and Conticello S.G.. 2011. Analysis of reptilian APOBEC1 suggests that RNA editing may not be its ancestral function. Mol. Biol. Evol. 28:1125–1129 10.1093/molbev/msq338 [DOI] [PubMed] [Google Scholar]
- Stenglein M.D., Matsuo H., and Harris R.S.. 2008. Two regions within the amino-terminal half of APOBEC3G cooperate to determine cytoplasmic localization. J. Virol. 82:9591–9599 10.1128/JVI.02471-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., Carrasco Y.P., Hu Y., Guo X., Mirzaei H., Macmillan J., and Chook Y.M.. 2013. Nuclear export inhibition through covalent conjugation and hydrolysis of Leptomycin B by CRM1. Proc. Natl. Acad. Sci. USA. 110:1303–1308 10.1073/pnas.1217203110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachsel H.1996. Isolation and characterization of eukaryotic translation initiation, elongation and termination factors. InPost-transcriptional Control of Gene Expression. Resnekov O., and Gabain A., Springer; Berlin Heidelberg: pp. 165–176 10.1007/978-3-642-60929-9_14 [DOI] [Google Scholar]
- Uchimura Y., Barton L.F., Rada C., and Neuberger M.S.. 2011. REG-γ associates with and modulates the abundance of nuclear activation-induced deaminase. J. Exp. Med. 208:2385–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong B.Q., and Chaudhuri J.. 2012. Combinatorial mechanisms regulating AID-dependent DNA deamination: interacting proteins and post-translational modifications. Semin. Immunol. 24:264–272 10.1016/j.smim.2012.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J.C., Agashe V.R., Siegers K., and Hartl F.U.. 2004. Pathways of chaperone-mediated protein folding in the cytosol. Nat. Rev. Mol. Cell Biol. 5:781–791 10.1038/nrm1492 [DOI] [PubMed] [Google Scholar]
- Zahn A., Eranki A.K., Patenaude A.-M., Methot S.P., Fifield H., Cortizas E.M., Foster P., Imai K., Durandy A., Larijani M., et al. 2014. Activation induced deaminase C-terminal domain links DNA breaks to end protection and repair during class switch recombination. Proc. Natl. Acad. Sci. USA. 111:E988–E997 10.1073/pnas.1320486111 [DOI] [PMC free article] [PubMed] [Google Scholar]