Abstract Abstract
Karyotypes and chromosomal characteristics of both minor and major rDNAs in four fish species known popularly as “lambaris”, namely Astyanax abramis (Jenyns, 1842), Astyanax asuncionensis Géry, 1972, Astyanax correntinus (Holmberg, 1891) and Astyanax sp. collected from downstream of the Iguassu Falls (Middle Paraná River basin), preservation area of the Iguassu National Park, were analyzed by conventional and molecular protocols. Astyanax abramis had diploid chromosome number 2n=50 (4m+30sm+8st+8a) and single AgNORs (pair 22), Astyanax asuncionensis had 2n=50 (8m+24sm+6st+12a) and single AgNORs (pair 20), Astyanax sp. had 2n=50 (4m+26sm+8st+12a) and single AgNORs (pair 25), and Astyanax correntinus had 2n=36 (12m+16sm+2st+6a) and multiple AgNORs (pairs 12, 15, 16, 17). FISH with 18S rDNA showed a single site for Astyanax abramis, Astyanax asuncionensis and Astyanax sp. and multiple for Astyanax correntinus (14 sites). FISH with 5S rDNA showed single 5S-bearing loci chromosome pair only for Astyanax asuncionensis and multiple for Astyanax abramis (four sites), Astyanax correntinus (five sites) and Astyanax sp. (four sites). Distinct distribution patterns of heterochromatin were observed for karyotypes of all species, with the exception of the first acrocentric chromosome pair characterized by centromeric, interstitial-proximal and telomeric blocks of heterochromatin on the long arm, which may represent homeology between karyotypes of Astyanax abramis and Astyanax asuncionensis. Our study showed species-specific characteristics which can serve in diagnosis and differentiation between Astyanax abramis and Astyanax asuncionensis, considered cryptic species, as well as strengthening the occurrence of a species of Astyanax not yet described taxonomically. In addition, the data obtained from first cytogenetic studies in Astyanax correntinus suggest a high similarity with Astyanax schubarti Britski, 1964, suggesting that these species may belong to the same morphological group and that can be phylogenetically related.
Keywords: Fish cytogenetics, chromosome banding, rDNA-FISH, karyotype differentiation, rDNA sites multiplication
Introduction
Characiformes are considered one of the most diversified groups in the world freshwater ichthyofauna, comprising 18 families with 270 genera and more than 1,700 species (Nelson 2006). This diversity is recognized mainly in the Neotropical region, which is home to around 1,000 species in Brazilian hydrographic systems alone (Buckup et al. 2007).
Among the families that comprise Characiformes, four are in the African continent (Alestidae, Citharinidae, Distichodontidae and Hepsetidae) and 14 are in Neotropical regions (Acestrorhynchidae, Anostomidae, Characidae, Chilodontidae, Crenuchidae, Ctenolucidae, Curimatidae, Cynodontidae, Erythrinidae, Gasteropelecidae, Hemiodontidae, Lebiasinidae, Parodontidae and Prochilodontidae) (Nelson 2006); however, some authors recognize Serrasalmidae as valid in Characiformes (Jégu et al. 2003, Calcagnotto et al. 2005, Ortí et al. 2008). Recently, Oliveira et al. (2011) proposed a study by rearranging the phylogenetic relationships in the order, suggesting a new definition for Characidae, based on analysis of sequences of 2 mitochondrial genes and 3 nuclear genes, obtained from 166 genera distributed in 18 acknowledged families, and another 56 genera were considered as incertae sedis. This study raises the subfamilies Bryconinae, Iguanodectinae and Triportheinae to families Bryconidae, Iguanodectidae and Thiportheidae, respectively. Albert et al. (2011) added Chalceidae (comprised only by Chalceus), resulting in a new classification for Characiformes composed of 23 families (including Serrasalmidae, above mentioned).
Astyanax Baird & Girard, 1854 known popularly as “lambaris”, includes around 140 valid species and probably many not yet discovered and/or awaiting formal description (Froese and Pauly 2014). Being a genus with the highest species count in Characidae distributed in Central and South America, in Brazilian basins Astyanax comprises around 50 valid species (Buckup et al. 2007). The major inconsistencies shown in the family occur in incertae sedis, in which the taxonomic construction is a catch-all assemblage and includes several distinct lineages with absence of proven monophyly (Lima et al. 2003, Mirande 2010). Belonging to this group, Astyanax was first allocated in Tetragonopterinae; however, systematic reviews in the subfamily have listed all genera (except for Tetragonopterus) as incertae sedis (Lima et al. 2003). Recently, Mirande (2009) proposed, with phylogenetic contributions, a new relationship among genera incertae sedis, resulting in a phylogenetic lineage within the Characidae named “clade Astyanax”.
Ichthyofaunal researches in river systems of southern Brazil were carried out particularly in systems that comprise the Upper Paraná River basin (Langeani et al. 2007, Graça and Pavanelli 2007) and the Iguassu River basin (Ingenito et al. 2004, Bifi et al. 2006, Baumgartner et al. 2012). In the hydrographic system of Paraná-Paraguay basin there were 110 species identified, eight being represented by Astyanax (Neris et al. 2010). Astyanax altiparanae Garutti & Britski, 2000 was described for the Upper Paraná River basin and Astyanax asuncionensis Géry, 1972 for the Middle-Lower Paraná River and Paraguay River basins (Lima et al. 2003). In this same region, Astyanax abramis (Jenyns, 1842) is a sister-group of Astyanax asuncionensis, and may be considered highly cryptic by presenting similar morphological characteristics (Britski et al. 2007). Astyanax correntinus (Holmberg, 1891) and Astyanax pellegrini Eigenmann, 1907 are grouped in “clade Astyanax” next to Astyanax asuncionensis and Astyanax abramis (Mirande 2009). Despite the proximity in the clade, Astyanax correntinus does not morphologically fit in any of the artificial groups recognized in Astyanax, otherwise occurring in complexes Astyanax bimaculatus (Jenyns, 1842), Astyanax fasciatus (Cuvier, 1819) and Astyanax scabripinnis (Jenyns, 1842).
Astyanax comprises interesting species for cytogenetic studies, with different evolutionary models that show from maintenance of a preserved chromosomal condition to derived karyotype characteristics, used as important tools in the differentiation and identification of species (Moreira-Filho and Bertollo 1991, Vicari et al. 2008, Ferreira-Neto et al. 2009, Peres et al. 2008). Available cytogenetic data reveal diploid chromosome numbers within Astyanax that vary from 2n = 36 in Astyanax schubarti Britski, 1964 (Morelli et al. 1983) to 2n = 50, as observed Astyanax scabripinnis, Astyanax fasciatus, Astyanax altiparanae and Astyanax jacuhiensis (Cope, 1894) (Souza and Moreira-Filho 1995, Artoni et al. 2006, Ferreira-Neto et al. 2009, Pacheco et al. 2010). Martinez et al. (2012) carried out a review of twenty populations of Astyanax altiparanae, observing intraspecific differences in the karyotypes, and the number and position of the nucleolus organizing regions (NORs). Likewise, for chromosomes of Astyanax fasciatus (Pazza et al. 2006, Medrado et al. 2008) and Astyanax scabripinnis (Mantovani et al. 2004, Santos and Morelli 2006), interpopulation differences mainly associated with heterochromatin distribution patterns were observed.
The physical mapping of genes 5S rDNA and 18S rDNA has also been used to characterize different populations in the species of Astyanax scabripinnis complex (Souza et al. 2001, Mantovani et al. 2005, Fernandes and Martins-Santos 2006, Peres et al. 2008), Astyanax altiparanae complex (Almeida-Toledo et al. 2002, Fernandes and Martins-Santos 2006), Astyanax fasciatus complex (Ferreira-Neto et al. 2012) and Astyanax jacuhiensis (Pacheco et al. 2010). These data documented highly variable NORs phenotype diversity in the representatives of the genus (Almeida-Toledo et al. 2002, Mantovani et al. 2005, Fernandes and Martins-Santos 2006, Peres et al. 2008).
The aim of the present study was to characterize using the conventional and molecular cytogenetic techniques, the karyotypes and chromosomal characteristics of rDNA in the species Astyanax abramis, Astyanax asuncionensis, Astyanax sp. and Astyanax correntinus, collected downstream from the Iguassu River Falls (middle Parana River), to contribute to the taxonomy of one of the major component of Neotropical Characidae fish diversity.
Methods
The specimens analyzed were deposited in Coleção Ictiológica do Núcleo de Pesquisas em Limnologia, Ictiologia e Aquicultura - (NUP), Universidade Estadual de Maringá: nine specimens of Astyanax abramis (four males and five females, NUP 14581), 25 specimens of Astyanax asuncionensis (13 males and 12 females, NUP 14584), 25 specimens of Astyanax correntinus (11 males and 14 females, NUP 14582) and one specimen of Astyanax sp. (female, NUP 14583), in the Iguassu River, in the stretch with around 25 km between downstream of the Iguassu Falls and its mouth on the Paraná River, Middle Parana River basin, located in the preservation area of the Iguassu National Park (25°38'18.72"S; 54°28'4.74"W).
All the specimens were anesthetized and sacrificed by an overdose of clove oil (Griffiths 2000). The chromosome preparations were obtained from anterior kidney cells by means of the techniques by Bertollo et al. (1978) and Foresti et al. (1993) using 0.02% colchicine treatment for 40 or 30 minutes, respectively. Thirty metaphases spreads from each fish were analyzed and ten of the best mitotic metaphases were used to measure karyotypes.
The AgNORs were revealed by silver impregnation according to Howell and Black (1980) and C-banding followed Sumner (1972), with modifications suggested by Lui et al. (2012).
The physical mapping of 5S rDNA and 18S rDNA loci was carried out by fluorescence in situ hybridization (FISH) according to Pinkel et al. (1986) and modifications suggested by Margarido and Moreira-Filho (2008), using probes obtained from Leporinus elongatus (Martins and Galetti 1999) and from Prochilodus argenteus (Hatanaka and Galetti 2004) DNAs, respectively. The hybridization was performed under high stringency condition (77%). Probes were labeled by nick translation with digoxigenin-11-dUTP (5S rDNA) and biotin-16-dUTP (18S rDNA) (Roche®). The detection of signals was performed with antidigoxigenin-rhodamine (Roche®) for probe of 5S rDNA and amplified avidin-FITC with biotinylated anti-avidin (Sigma-Aldrich) for probe of 18S rDNA, the chromosomes were counterstained with 4’,6-diamidino-2-phenylindole (DAPI, 50 μg/mL).
The metaphases were photographed using a BX 61 epifluorescence microscope, coupled with Olympus DP 71 digital camera with the Olympus DP Controller software 3.2.1.276. The chromosomes were classified and organized in accordance with Levan et al. (1964) in metacentric (m), submetacentric (sm), subtelocentric (st) and acrocentric (a). The fundamental number (FN) was calculated considering m, sm and st chromosomes as having two arms, and a chromosomes as having only one chromosome arm.
Results
Astyanax abramis
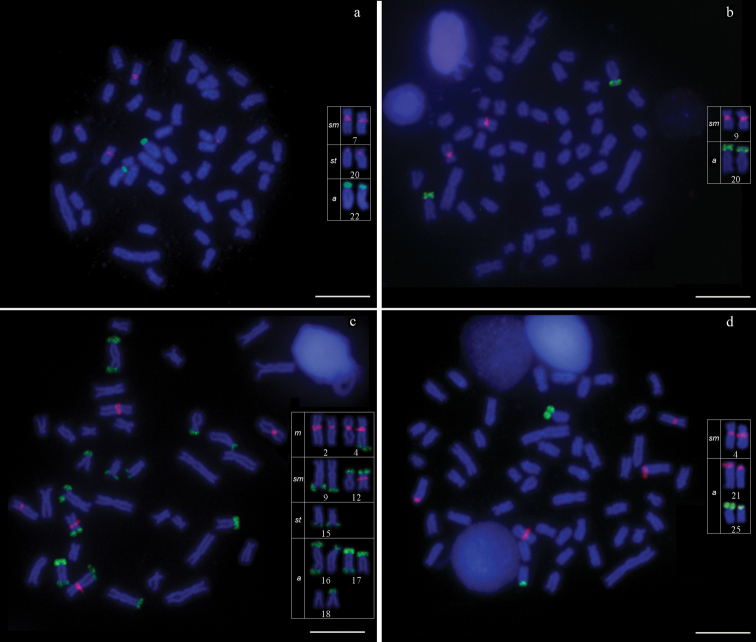
The 2n was 50 chromosomes (4m+30sm+8st+8a, FN=92) for males and females (Fig. 1a). A single pair of NORs was located in terminal position on the p arm of chromosome pair 22 (Fig. 1a, in box). C-banding showed centromeric heterochromatin blocks in pairs 7, 14 and 21, pericentromeric on the q arm of pairs 22 and 24, telomerics on the p and q arms in pair 22, and coincident with the NORs (Fig. 1b). The FISH revealed multiple sites of 5S rDNA in centromeric position in the sm pair 7 and the sm pair 20, and a single site of 18S rDNA in terminal position on the p arm of the a pair 22 (Fig. 2a).
Figure 1.
Karyotypes arranged from Giemsa-stained chromosomes of: a Astyanax abramis c Astyanax asuncionensis e Astyanax correntinus g Astyanax sp.; and from C-banded chromosomes of: b Astyanax abramis d Astyanax asuncionensis f Astyanax correntinus h Astyanax sp. The AgNORs bearing chromosomes are framed. Bar = 10 µm.
Figure 2.
Metaphases chromosomes spreads after FISH with 5S rDNA probe (red) and 18S rDNA probe (green) of: a Astyanax abramis b Astyanax asuncionensis c Astyanax correntinus d Astyanax sp. The 5S rDNA and 18S rDNA bearing chromosomes are framed. Bar = 10 µm.
Astyanax asuncionensis
The 2n was 50 chromosomes (8m+24sm+6st+12a, FN=88) for males and females (Fig. 1c). A single pair of NORs was located in a terminal position on the p arm of chromosome pair 20 (Fig. 1c, in box). C-banding showed centromeric heterochromatin blocks in pairs 2, 3 and 20, pericentromeric on the p arm of pair 8, on the q arm of pairs 9, 13 and 14, telomerics on the q arm of pair 8, on the p and q arms in pair 20, and coincident with the NORs (Fig. 1d). FISH revealed a single site of 5S rDNA in centromeric position in the sm pair 9, and a single site of 18S rDNA in terminal position on the p arm of the a pair 20 (Fig. 2b).
Astyanax correntinus
The 2n was 36 chromosomes (4m+26sm+8st+12a, FN=66) for males and females (Fig. 1e). Multiple AgNORs bearing pairs were located in terminal position on the p arm of chromosome pair 12, on the p and q arms in pair 17, on the q arm of one chromosome from pair 15 and on the q arm of one chromosome from pair 16 (Fig. 1e, in box). C-banding showed centromeric heterochromatin blocks in pairs 1, 2, 4, 7, 9, 10, 12 and 14, telomeric on the q arm of pair 17, on the p arm of pair 18, on the p and q arms in pair 16, and coincident with the NORs (Fig. 1f). FISH revealed multiple sites of 5S rDNA in centromeric position in the m pairs 2 and 4, and in one chromosome from the sm pair 12. Multiple sites of 18S rDNA were observed in terminal position on the p arm of the sm pair 12, the a pairs 16, 17 and 18, and on the q arm of the sm pair 9 and st pair 15, and on the q arm of one chromosome from the m pair 4 and the a pairs 16 and 17 (Fig. 2c).
Astyanax sp.
The 2n was 50 chromosomes (12m+16sm+2st+6a, FN=88) (Fig. 1g). A single pair of AgNORs was located in terminal position on the p arm of chromosome pair 25 (Fig. 1g, in box). C-banding showed pericentromeric heterochromatin blocks on the q arm of pairs 4, 16, 21, 22 and 24, telomeric on the q arm of pair 5, and coincident with the NORs (Fig. 1h). FISH revealed multiple sites of 5S rDNA in centromeric position in the sm pair 4 and the a pair 21, and single site of 18S rDNA in terminal position on the p arm of the a pair 25 (Fig. 2d).
Table 1 presents a comparison of the cytogenetical data (2n, karyotype formula, AgNORs, C-banding, 18S rDNA and 5S rDNA) obtained for the Astyanax species analyzed in the present study.
Table 1.
Summary of the cytogenetical data for the Astyanax species analyzed in the present study.
| Species | Astyanax abramis | Astyanax asuncionensis | Astyanax correntinus | Astyanax sp. |
|---|---|---|---|---|
| 2n | 50 | 50 | 36 | 50 |
| Karyotype formula | 4m+30sm+8st+8a | 8m+24sm+6st+12a | 4m+26sm+8st+12a | 12m+16sm+2st+6a |
| AgNORs | Single: - pair 22, a, tel, p arm |
Single: - pair 20, a, tel, p arm |
Multiple: - pair 12, sm, tel, p arm - pair 15, st, tel, q arm - pair 16, a, tel, q arm - pair 17, a, bitel |
Single: - pair 25, a, tel, p arm |
|
Heterochromatin (C-banding) |
Centromeric, pericentromeric and telomeric | Centromeric, pericentromeric and telomeric | Centromeric and telomeric | Pericentromeric and telomeric |
| 18S rDNA | Single: - pair 22, a, tel, p arm |
Single: - pair 20, a, tel, p arm |
Multiple: - pair 4, m, tel, q arm - pair 9, sm, tel, q arm - pair 12, sm, tel, p arm - pair 15, st, tel, q arm - pair 16, a, tel/bitel - pair 17, a, tel/bitel - pair 18, a, tel, p arm |
Single: - pair 25, a, tel, p arm |
| 5S rDNA | Single: - pair 7, sm, cent |
Single: - pair 9, sm, cent |
Multiple: - pair 2, m, cent - pair 4, m, cent - pair 12, sm, cent |
Multiple: - pair 4, sm, cent - pair 21, a, tel |
m: metacentric; sm: submetacentric; st: subtelocentric; a: acrocentric; cent: centromeric; tel: telomeric; bitel: bitelomeric; p: short arm; q: long arm.
Discussion
Diploid numbers and karyotype formulae
Although the present study revealed the same diploid chromosome number (2n= 50) for Astyanax abramis, Astyanax asuncionensis and Astyanax sp., with karyotypes dominated by bi-armed chromosomes, karyotypes differed among these three species, and can be used as a species-specific cytogenetic profile (Fig. 1a, c, g). Similar results were also found in other species of Astyanax (Oliveira et al. 1988, Daniel-Silva and Almeida-Toledo 2001, Kavalco et al. 2003), including the presence of the first large-sized m chromosome pair, these characteristics being assigned to an ancestral condition in genus (e.g. Portella et al. 1988, Ferreira-Neto et al. 2009, Kavalco et al. 2009, among others). Different from the species that possess these ancestral conditions in the genus, Astyanax correntinus has 2n = 36 and a karyotype containing eight large m-sm chromosome pairs (Fig. 1e). These findings are similar to that found in Astyanax schubarti by Morelli et al. (1983) and Almeida-Toledo et al. (2002) that shares with Astyanax correntinus a low 2n originating from chromosome fusions, presence of large meta-submetacentric chromosomal pairs, in addition to low number of subtelo-acrocentric chromosomes. In addition, the external appearance of Astyanax correntinus and Astyanax schubarti shows similarly high body, a horizontal silver band on the side of the body and a large amount of non-branching rays in anal fin in relation to other Astyanax species. Based on the cytogenetic results and morphological similarities it is possible to hypothesize that the two species may be phylogenetically closely related.
Nucleolus organizing regions and 18S rDNA
The number and position of NORs (Ag-impregnation and 18S rDNA-FISH), i.e. NOR phenotypes, observed in karyotypes of Astyanax species under study were conserved for the three species with 2n=50, with presence of a single site always located on the p arm in terminal position of a chromosome pair in Astyanax abramis, Astyanax asuncionensis and Astyanax sp. (Fig. 2a, b, d). Astyanax abramis and Astyanax asuncionensis are part of the Astyanax bimaculatus complex, which is diagnosed by showing an oval humeral spot and caudal peduncle blotch, extending to the end of middle caudal rays. Similar results were observed in populations of Astyanax altiparanae, which is also part of the Astyanax bimaculatus complex (Domingues et al. 2007, Peres et al. 2008, Pacheco et al. 2011), although for Astyanax altiparanae intraspecific variations were also observed when different populations were compared (Fernandes and Martins-Santos 2006, Ferreira-Neto et al. 2009). However, in chromosomes of Astyanax correntinus multiple NORs were observed both by Ag- impregnation (Fig. 1e, seven sites) and by 18S rDNA-FISH (Fig. 2c, 14 sites), with some pairs presenting these sites in only one of the homologous chromosomes. Multiple NORs were also observed in karyotype of Astyanax schubarti with of four ribosomal sites observed through 18 rDNA-FISH (Almeida-Toledo et al. 2002). In Astyanax scabripinnis, up to 16 chromosomes bearing these ribosomal genes were observed (Ferro et al. 2001, Mantovani et al. 2005), showing the high degree of number variability observed for the genus.
5S rDNA
As to the 5S rDNA-FISH, simple sites were observed in karyotype of Astyanax asuncionensis located in centromeric position (Fig. 2b), similar to that found in different populations of Astyanax altiparanae, although they differ in location (interstitial-proximal position, Ferreira-Neto et al. 2009, Pacheco et al. 2011). Multiple sites were observed in karyotypes of Astyanax abramis (Fig. 2a, four sites), Astyanax correntinus (Fig. 2c, five sites) and Astyanax sp. (Fig. 2d, four sites), in centromeric position for all 5S rDNA-bearing chromosomes. Our findings indicate thus interspecific differences, which can be used as a diagnostic tool for their differentiation, because they are morphologically diagnosed only by differences in the number of perforated scales on the lateral line – up to 40 in Astyanax asuncionensis and 42 or more in Astyanax abramis (Britski et al. 2007). Likewise, Astyanax correntinus shows a greater number of chromosomes bearing sites of 5S rDNA compared to that observed in Astyanax schubarti (four sites) (Almeida-Toledo et al. 2002). In addition to Astyanax correntinus showing a high number of chromosomes bearing sites of 5S rDNA, it was observed synteny of 5S and 18S sites in one of the chromosomes from pairs 4 and 12 (Fig. 2c). Mantovani et al. (2005) also observed synteny in a population of Astyanax scabripinnis for these ribosomal genes, being a characteristic considered derived for Astyanax in terms of genomic organization and chromosomal evolution. Therefore, despite the distribution of 5S rDNA sites being considered conserved for some groups of fish, the results observed show variation regarding the number and location of these ribosomal genes in Astyanax.
Distribution pattern of heterochromatin
With regards to the distribution pattern of heterochromatin, although low amount in Astyanax abramis, Astyanax asuncionensis and Astyanax sp., it was found mainly in centromeric and interstitial-proximal position, in addition to NORs associated (Fig. 1b, d, h). These results were also observed in other phylogenetically close species of the genus, as in Astyanax altiparanae (Domingues et al. 2007, Ferreira-Neto et al. 2009) and Astyanax jacuhiensis (Pacheco et al. 2010). Still, in karyotypes Astyanax abramis and Astyanax asuncionensis, the first pair of a chromosomes, with the NORs on the p arm, both share the same pattern of bands: centromeric heterochromatin, interstitial-proximal heterochromatin on the q arm, and telomeric heterochromatin on the q arm, and this pair may represent homeological chromosomes (Fig. 1b, d). In Astyanax correntinus, centromeric heterochromatins were observed in most m-sm chromosome pairs (Fig. 1f), being this pattern similar to that observed in Astyanax schubarti (Daniel-Silva and Almeida-Toledo 2001). Differently from Astyanax abramis, Astyanax asuncionensis and Astyanax correntinus, Astyanax sp. showed a particular pattern, with the presence of five st-a chromosome pairs carrying heterochromatin in interstitial-proximal position on the q arm, in addition to some sm chromosome pairs bearing heterochromatin both in centromeric and telomeric positions on the q arm (Fig. 1h). According to our morphological observation, Astyanax sp. is part of the Astyanax scabripinnis complex defined by Bertaco and Malabarba (2001), but does not fit into any taxonomically described species in this complex. The cytogenetic data corroborate this hypothesis; therefore, we believe that this is a new species, and that a greater number of specimens are required to confirm it.
Conclusions
The present study shows species-specific cytogenetic markers which can serve in diagnosis and differentiation between Astyanax abramis and Astyanax asuncionensis, considered cryptic species (deep body; presence of a well defined, black, horizontal humeral spot; absence of maxillary tooth; and the presence of circuli in posterior field of scales), as well as strengthening the occurrence of a species of Astyanax not yet described taxonomically (elongated body; absence of a well defined, black, horizontal humeral spot; presence of one maxillary tooth; and the absence of circuli in posterior field of scales). In addition, the data obtained from first cytogenetic studies in Astyanax correntinus suggest a high similarity with Astyanax schubarti, suggesting that these species may belong to the same morphological group (deep body; absence of a well defined, black, horizontal humeral spot; presence of one maxillary tooth; and the absence of circuli in posterior field of scales; broad silvery lateral band) and that can be phylogenetically related. Further studies, including another species of the “clade Astyanax” and molecular analyses of mitochondrial genes sequences, may confirm these hypotheses.
Acknowledgements
The authors are grateful to Instituto Chico Mendes de Conservação da Biodiversidade (ICMBio) for authorizing the capture of the fishes (License number: SISBIO 10522-1). The authors thank to Universidade Estadual do Oeste do Paraná (UNIOESTE), Parque Nacional do Iguaçu, Macuco Safari, Grupo de Pesquisa em Tecnologia de Produção e Conservação de Recursos Pesqueiros e Hídricos (GETECH) and to the Núcleo de Pesquisas em Limnologia, Ictiologia e Aquicultura (NUPÉLIA) for logistical support. This study was financed by CAPES (Coordenadoria de Aperfeiçoamento de Ensino Superior), Fundação Araucária (Fundação Araucária de Apoio e Desenvolvimento Científico e Tecnológico do Estado do Paraná) and CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico).
Citation
Paiz LM, Baumgärtner L, da Graça WJ, Margarido VP (2015) Basic cytogenetics and physical mapping of ribosomal genes in four Astyanax species (Characiformes, Characidae) collected in Middle Paraná River, Iguassu National Park: considerations on taxonomy and systematics of the genus. Comparative Cytogenetics 9(1): 51–65. doi: 10.3897/CompCytogen.v9i1.9002
References
- Albert JS, Bart Jr. HL, Reis RE. (2011) Species richness and cladal diversity. In: Albert JS, Reis RE. (Eds) Historical Biogeography of Neotropical freshwater fishes, First Edition University of California Press, California, 89–104. [Google Scholar]
- Almeida-Toledo LF, Ozouf-Costaz C, Foresti F, Bonillo C, Porto-Foresti F, Daniel-Silva MFZ. (2002) Conservation of the 5S-bearing chromosome pair and co-localization with major rDNA clusters in five species of Astyanax (Pisces, Characidae). Cytogenetic and Genome Research 97: 229–233. doi: 10.1159/000066609 [DOI] [PubMed] [Google Scholar]
- Artoni RF, Shibatta AO, Gross MC, Schneider CH, De Almeida MC, Vicari MR, Bertollo LAC. (2006) Astyanax aff. fasciatus Cuvier, 1819 (Teleostei; Characidae): evidences of a species complex in the upper rio Tibagi basin (Paraná, Brazil). Neotropical Ichthyology 4: 197–202. doi: 10.1590/S1679-62252006000200005 [Google Scholar]
- Baumgartner G, Pavanelli CS, Baumgartner D, Bifi AG, Debona T, Frana VA. (2012) Peixes do baixo rio Iguaçu. First Edition Eduem, Maringá, 203 pp. doi: 10.7476/9788576285861 [Google Scholar]
- Bertaco VA, Malabarba LR. (2001) Description of new species of Astyanax (Teleostei: Characidae) from headwater streams of Southern Brazil, with comments on the ‘‘A. scabripinnis species complex’’. Ichthyological Exploration of Freshwaters 12: 221–234. [Google Scholar]
- Bertollo LAC, Takahashi CS, Moreira-Filho O. (1978) Cytotaxonomic considerations on Hoplias lacerdae (Pisces, Erythrinidae). Brazilian Journal of Genetics 1: 103–120. [Google Scholar]
- Bifi AG, Baumgartner D, Baumgartner G, Frana VA, Debona T. (2006) Composição específica e abundância da ictiofauna do rio dos Padres, bacia do rio Iguaçu, Brasil. Acta Scientiarum 28: 203–211. doi: 10.4025/actascibiolsci.v28i3.193 [Google Scholar]
- Britski HA, Silimon KZS, Lopes BS. (2007) Peixes do Pantanal: Manual de identificação. Brasília, 227 pp. [Google Scholar]
- Buckup PA, Menezes NA, Guazzi MS. (2007) Catálogo das espécies de peixes de água doce do Brasil. Museu Nacional, Universidade Federal do Rio de Janeiro, Rio de Janeiro, 195 pp. [Google Scholar]
- Calcagnotto D, Schaefer SA, Desalle R. (2005) Relationships among characiform fishes inferred from analysis of nuclear and mitochondrial gene sequences. Molecular Phylogenetics and Evolution 36: 135–153. doi: 10.1590/S1415-47572008000200030 [DOI] [PubMed] [Google Scholar]
- Daniel-Silva MFZ, Almeida-Toledo LF. (2001) Chromosome R-banding pattern and conservation of a marker chromosome in four species, genus Astyanax (Characidae, Tetragonopterinae). Caryologia 54: 209–215. doi: 10.1080/00087114.2001.10589228 [Google Scholar]
- Domingues MS, Vicari MR, Abilhoa V, Wamser JP, Cestari MM, Bertollo LAC, Almeida MC, Artoni RF. (2007) Cytogenetic and comparative morphology of two allopatric populations of Astyanax altiparanae Garutti and Britski, 2000 (Teleostei: Characidae) from upper rio Parana basin. Neotropical Ichthyology 5: 37–44. doi: 10.1590/S1679-62252007000100005 [Google Scholar]
- Fernandes CA, Martins-Santos IC. (2006) Mapping of the 18S and 5S ribosomal RNA genes in Astyanax altiparanae Garutti and Britski 2000 (Teleostei, Characidae) from the upper Paraná river basin, Brazil. Genetics and Molecular Biology 29: 464–468. doi: 10.1590/S1415-47572006000300011 [Google Scholar]
- Ferreira-Neto M, Vicari MR, Camargo EF, Artoni RF, Moreira-Filho O. (2009) Comparative cytogenetics among populations of Astyanax altiparanae (Characiformes, Characidae, Incertae Sedis). Genetics and Molecular Biology 32: 792–796. doi: 10.1590/S1415-47572009005000078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira-Neto M, Artoni RF, Vicari MR, Moreira-Filho O, Camacho JPM, Bakkali M, Oliveira C, Foresti F. (2012) Three sympatric karyomorphs in the fish Astyanax fasciatus (Teleostei, Characidae) do not seem to hybridize in natural populations. Comparative Cytogenetics 6: 29–40. doi: 10.3897/CompCytogen.v6i1.2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro DAM, Moreira-Filho O, Bertollo LAC. (2001) Nucleolar organizing regions, 18S and 5S rDNA in Astyanax scabripinnis (Pisces, Characidae): Populations distribution and functional diversity. Genetica 110: 55–62. doi: 10.1023/A:1017963217795 [DOI] [PubMed] [Google Scholar]
- Foresti F, Oliveira C, Almeida-Toledo LF. (1993) A method for chromosome preparations from large fish specimens using in vitro short-term treatment with colchicine. Experientia 49: 810–813. doi: 10.1007/BF01923555 [Google Scholar]
- Froese R, Pauly D. (2014) FishBase. http://www.fishbase.org [accessed 01 November 2014]
- Graça WJ, Pavanelli CS. (2007) Peixes da planície de inundação do alto rio Paraná e áreas adjacentes. First Edition. Eduem, Maringá, 241 pp. [Google Scholar]
- Griffiths SP. (2000) The use of clove oil as an anaesthetic and method for sampling intertidal rockpool fishes. Journal of Fish Biology 57: 1453–1464. doi: 10.1006/jfbi.2000.1406 [Google Scholar]
- Hatanaka T, Galetti Jr PM. (2004) Mapping of the 18S and 5S ribosomal RNA genes in the fish Prochilodus argenteus Agassiz, 1829 (Characiformes, Prochilodontidae). Genetica 122: 239–244. doi: 10.1007/s10709-004-2039-y [DOI] [PubMed] [Google Scholar]
- Howell WM, Black DA. (1980) Controlled silver-staining of nucleolus organizer regions with a protective colloidal developer: A 1-step method. Experientia 36: 1014–1015. doi: 10.1007/BF01953855 [DOI] [PubMed] [Google Scholar]
- Ingenito LFS, Duboc LF, Abilhoa V. (2004) Contribuição ao conhecimento da ictiofauna da bacia do alto rio Iguaçu, Paraná, Brasil. Arquivos de Ciência e Veterinária 7: 23–26. [Google Scholar]
- Jégu M, Keith P, Bail PY. (2003) Myloplus planquettei n. sp. (Teleostei, Characidae, Serrasalminae), une nouvelle espèce de grand Serrasalminae phytophage du bouclier guyanais (Guyane française). Revue Suisse de Zoologie 110: 823–853. [Google Scholar]
- Kavalco KF, Moreira-Filho O. (2003) Cytogenetical analyses in four species of the genus Astyanax (Pisces, Characidae) from Paraiba do Sul river basin. Caryologia 56: 453–461. doi: 10.1080/00087114.2003.10589358 [Google Scholar]
- Kavalco KF, Brandao KO, Pazza R, Almeida-Toledo LF. (2009) Astyanax hastatus Myers, 1928 (Teleostei, Characidae): a new species complex within the genus Astyanax. Genetics and Molecular Biology 32: 477–483. doi: 10.1590/S1415-47572009005000055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langeani F, Castro RMC, Oyakawa OT, Shibatta OA, Pavanelli CS, Casatti L. (2007) Diversidade da ictiofauna do alto rio Paraná: composição atual e perspectivas futuras. Biota Neotropica 7: 181–197. [Google Scholar]
- Levan A, Fredga K, Sandberg AA. (1964) Nomenclature for centromeric position on chromosomes. Heredity 52: 201–220. [Google Scholar]
- Lima FCT, Malabarba LR, Buckup PA, Silva JF, Vari RPP, Harold A, Benine R, Oyakawa OT, Pavanelli CS, Menezes NA, Lucena CAS, Malabarba MCSL, Lucena ZMS, Reis RE, Langeani F, Casatti L, Bertaco VA, Moreira C, Lucinda PHF. (2003) Genera Incertae Sedis in Characidae. In: Reis RE, Kullander SO, Ferraris CJ. (Eds) Check List of the Freshwater Fishes of South and Central America. Rio Grande do Sul, 134–141. [Google Scholar]
- Lui RL, Blanco DR, Moreira-Filho O, Margarido VP. (2012) Propidium iodide for making heterochromatin more evident in the C-banding technique. Biotechnic and Histochemistry 87: 433–438. doi: 10.3109/10520295.2012.696700 [DOI] [PubMed] [Google Scholar]
- Mantovani M, Abel LDS, Mestriner CA, Moreira-Filho O. (2004) Evidence of the differentiated structural arrangement of constitutive heterochromatin between two populations of Astyanax scabripinnis (Pisces, Characidae). Genetics and Molecular Biology 27: 536–542. doi: 10.1590/S1415-47572004000400012 [Google Scholar]
- Mantovani M, Abel LDS, Moreira-Filho O. (2005) Conserved 5S and variable 45S rDNA chromosomal localization revealed by FISH in Astyanax scabripinnis (Pisces, Characidae). Genetica 123: 211–216. doi: 10.1007/s10709-004-2281-3 [DOI] [PubMed] [Google Scholar]
- Margarido VP, Moreira-Filho O. (2008) Karyotypic differentiation through chromosome fusion and number reduction in Imparfinis hollandi (Ostariophysi Heptapteridae). Genetics and Molecular Biology 31: 235–238. doi: 10.1590/S1415-47572008000200012 [Google Scholar]
- Martinez ERM, Alves AL, Silveira SM, Foresti F, Oliveira C. (2012) Cytogenetic analysis in the incertae sedis species Astyanax altiparanae Garutti and Britzki, 2000 Hyphessobrycon eques Steindachner, 1882 (Characiformes, Characidae) from the upper Paraná river basin. Comparative Cytogenetics 6: 41–51. doi: 10.3897/CompCytogen.v6i1.1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins C, Galetti Jr PM. (1999) Chromosomal localization of 5S rDNA genes in Leporinus fish (Anostomidae, Characiformes). Chromosome Research 7: 363–367. doi: 10.1023/A:1009216030316 [DOI] [PubMed] [Google Scholar]
- Medrado AS, Figueiredo AVA, Waldschmidt AM, Affonso PRAM, Carneiro PLS. (2008) Cytogenetic and morphological diversity in populations of Astyanax fasciatus (Teleostei, Characidae) from Brazilian northeastern river basins. Genetics and Molecular Biology 31: 208–214. doi: 10.1590/S1415-47572008000200007 [Google Scholar]
- Mirande JM. (2009) Weighted parsimony phylogeny of the family Characidae (Teleostei: Characiformes). Cladistics 25: 574–613. doi: 10.1111/j.1096-0031.2009.00262.x [DOI] [PubMed] [Google Scholar]
- Mirande JM. (2010) Phylogeny of the family Characidae (Teleostei: Characiformes): from characters to taxonomy. Neotropical Ichthyology 8: 385–568. doi: 10.1590/S1679-62252010000300001 [Google Scholar]
- Moreira-Filho O, Bertollo LAC. (1991) Astyanax scabripinnis (Pisces, Characidae): A species complex. Brazilian Journal of Genetics 14: 331–357. [Google Scholar]
- Morelli S, Bertollo LAC, Foresti F, Moreira-Filho O, Toledo-Filho AS. (1983) Cytogenetic considerations on the genus Astyanax (Pisces, Characidae). I. Kariotypic Variability. Caryologia 36: 235–244. [Google Scholar]
- Nelson JS. (2006) Fishes of the World. John Wiley and Sons, New Jersey, 601 pp. [Google Scholar]
- Neris N, Villalba F, Kamada D, Viré S. (2010) Guide to the Fish of Paraguay. First Edition Itaipú Binacional, Asunción, 299 pp. [Google Scholar]
- Oliveira C, Almeida-Toledo LF, Foresti F, Britsky HÁ, Toledo-Filho AS. (1988) Chromosome formulae of neotropical freshwater fishes. Brazilian Journal of Genetics 11: 577–624. [Google Scholar]
- Oliveira C, Avelino GS, Abe KT, Mariguela TC, Benine RC, Ortí G, Vari RP, Corrêa e Castro RM. (2011) Phylogenetic relationships within the speciose family Characidae (Teleostei: Ostariophysi: Characiformes) based on multilocus analysis and extensive in group sampling. BMC Evolutionary Biology 11: . doi: 10.1186/1471-2148-11-275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortí G, Sivasundar A, Dietz K, Jégu M. (2008) Phylogeny of the Serrasalmidae (Characiformes) based on mitochondrial DNA sequences. Genetics and Molecular Biology 31: 343–351. doi: 10.1590/S1415-47572008000200030 [Google Scholar]
- Pacheco RB, Giuliano-Caetano L, Júlio Jr HF, Dias AL. (2010) Cytogenetic data on Astyanax jacuhiensis (Characidae) in the lago Guaíba and tributaries, Brazil. Neotropical Ichthyology 8: 667–671. doi: 10.1590/S1679-62252010000300013 [Google Scholar]
- Pacheco RB, Rosa R, Giuliano-Caetano L, Júlio Jr HF, Dias AL. (2011) Cytogenetic comparison between two allopatric populations of Astyanax altiparanae Garutti et Britski, 2000 (Teleostei, Characidae), with emphasis on the localization of 18S and 5S rDNA. Comparative Cytogenetics 5: 237–246. doi: 10.3897/compcytogen.v5i3.1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazza R, Kavalco KF, Bertollo LAC. (2006) Chromosome polymorphism in Astyanax fasciatus (Teleostei, Characidae). Karyotype analysis, Ag-NORs and mapping of the 18S and 5S ribosomal genes in sympatric karyotypes and their possible hybrid forms. Cytogenetic and Genome Research 112: 313–319. doi: 10.1159/000089886 [DOI] [PubMed] [Google Scholar]
- Peres WAM, Bertollo LAC, Moreira-Filho O. (2008) Physical mapping of the 18S and 5S ribosomal genes in nine Characidae species (Teleostei, Characiformes). Genetics and Molecular Biology 31: 222–226. doi: 10.1590/S1415-47572008000200009 [Google Scholar]
- Pinkel D, Straume T, Gray JW. (1986) Cytogenetic analysis using quantitative, high sensitivity, fluorescence hybridization. Proceedings of National Academy of Sciences 83: 2934–2938. doi: 10.1073/pnas.83.9.2934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portela ALBS, Galetti Jr PM, Bertollo LAC. (1988) Considerations on the chromosome evolution of Tetragonopterinae (Pisces, Characidae). Brazilian Journal of Genetics 11: 307–316. [Google Scholar]
- Santos AC, Morelli S. (2006) Comparação citogenética de duas populações de Astyanax scabripinnis (Pisces, Characidae) da região do triangulo Mineiro. Bioscience Journal 22: 145–150. [Google Scholar]
- Souza IL, Moreira-Filho O. (1995) Cytogenetic diversity in the Astyanax scabripinnis species complex (Pisces, Characidae). I. Allopatric distribution in a small stream. Cytologia 60: 1–11. doi: 10.1508/cytologia.60.1 [Google Scholar]
- Souza IL, Galián J, De La Rúa P, Bertollo LAC, Moreira-Filho O. (2001) Non-random distribution of the GC-rich heterochromatin and nucleolar rDNA sites on Astyanax scabripinnis chromosomes. Cytologia 66: 85–91. doi: 10.1508/cytologia.66.85 [Google Scholar]
- Sumner AT. (1972) A simple technique for demonstrating centromeric heterochromatin. Experimental Cell Research 74: 304–306. doi: 10.1016/0014-4827(72)90558-7 [DOI] [PubMed] [Google Scholar]
- Vicari MR, Noleto RB, Artoni RF, Moreira-Filho O, Bertollo LAC. (2008) Comparative cytogenetic among species of the Astyanax scabripinnis complex. Evolutionary and biogeographical inferences. Genetics and Molecular Biology 31: 173–179. doi: 10.1590/S1415-47572009005000078 [Google Scholar]