Abstract
Cerebral cartography and connectomics pursue similar goals in attempting to create maps that can inform our understanding of the structural and functional organization of the cortex. Connectome maps explicitly aim at representing the brain as a complex network, a collection of nodes and their interconnecting edges. This article reflects on some of the challenges that currently arise in the intersection of cerebral cartography and connectomics. Principal challenges concern the temporal dynamics of functional brain connectivity, the definition of areal parcellations and their hierarchical organization into large-scale networks, the extension of whole-brain connectivity to cellular-scale networks, and the mapping of structure/function relations in empirical recordings and computational models. Successfully addressing these challenges will require extensions of methods and tools from network science to the mapping and analysis of human brain connectivity data. The emerging view that the brain is more than a collection of areas, but is fundamentally operating as a complex networked system, will continue to drive the creation of ever more detailed and multi-modal network maps as tools for on-going exploration and discovery in human connectomics.
Keywords: brain mapping, connectome, fMRI, brain connectivity
1. Introduction
Cartography, or map-making, is the conception and creation of a graphical representation of a system or object. In most cases creating maps means projecting a selected subset of traits or features onto a low-dimensional space for purposes of visualization. For example, geographical maps are typically flat (two-dimensional) representations of morphological features of the Earth's surface. Geographical maps distill vast amounts of knowledge, give a comprehensive picture of spatial relations among places, and enable search and navigation. Quite analogously, brain mapping involves building representations of the brain's major constituent regions, recording their topography (e.g. their extent, location and geometric/spatial relations), and displaying their physiological and anatomical characteristics. ‘Cerebral cartography’ in particular has occupied an important role in the study of the human cortex, where brain maps have been indispensable tools in the on-going quest of correlating brain anatomy with psychological and cognitive function [1].
In the modern era map-making is increasingly tied to ‘sense-making’, the visualization of complex data in order to reveal patterns of organization that enable data management and discovery [2,3]. Particularly important are efforts to map the networked interactions that pervade complex socio-technological and biological systems [4]. Prominent examples of such network maps chart protein–protein interactions in cells [5], genetic associations among common human diseases [6], the global spreading of epidemics [7] and interactions among species in an ecosystem [8]. Building network maps is an integral part of an emerging scientific discipline called ‘network science’ [9,10], the cross-disciplinary study of the structure and function of complex interconnected systems. Network science capitalizes on the fact that many complex systems can be decomposed into sets of elements (nodes) and their mutual relations (edges or connections). Jointly, the arrangement of these nodes and edges defines the network's topology. The topology summarizes how nodes and edges link together, essentially corresponding to a map of ‘what is connected to what’ within the system of interest. While many network maps emphasize non-spatial aspects of topology over geometric or spatial relations, these two aspects of mapping complex systems are often strongly related, since the presence or absence of connections may depend on the arrangement of nodes in physical space.
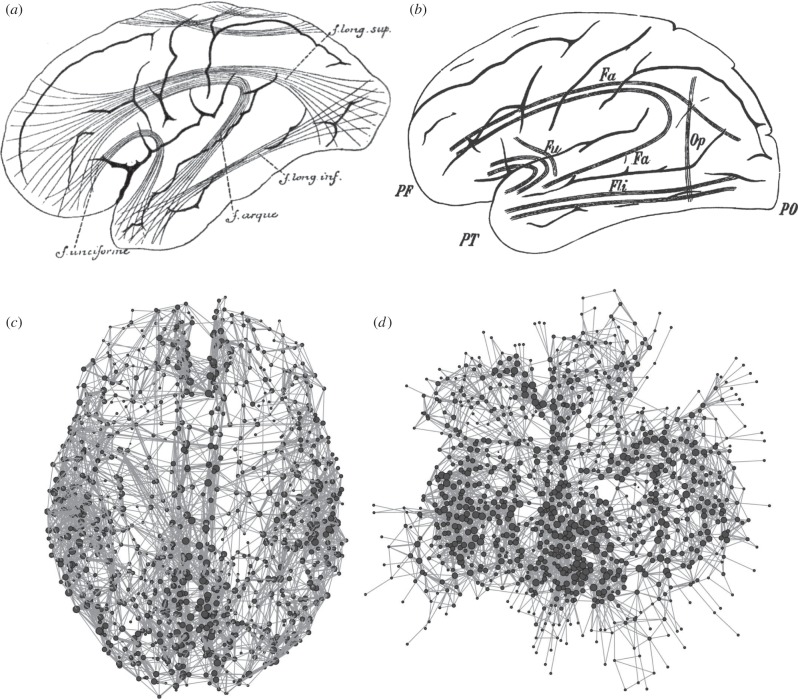
While the informal use of the term ‘network’ has a long history in the neurosciences (often interchangeably used with the term ‘circuit’), the direct application of quantitative tools for mapping and analysing networks to making maps of the brain is a relatively recent development. Starting with seminal efforts to create connectivity maps of anatomical brain systems, especially the primate cortex [11–13], the goal of building such maps for the human brain gave rise to ‘human connectomics’, the project of creating comprehensive network maps of the human brain [14–17] (figure 1). Connectomics has become a broad field of inquiry—it comprises studies of anatomical networks of individual neurons, neuronal populations and large-scale brain systems, as well as studies of their functional dynamics and interactions. A major motivation for human connectomics derives from the theoretical idea that network maps are fundamental for understanding the brain's structural and dynamic organization. The objectives of connectomics are complementary to those of cerebral cartography—while connectomics charts the topology of brain networks, leveraging the extensive tools and methods of network science and graph theory, cartography is mainly concerned with building spatial maps that record brain topography. Together, cerebral cartography and connectomics are natural adjuncts in the on-going quest to make maps of the brain that reveal the principles of its architecture and organization, its evolutionary origins and its capacity to create complex dynamics that underpin behaviour and cognition [21].
Figure 1.
Maps of structural brain connectivity from nineteenth century cerebral cartography and present day connectomics. (a) A depiction of major white matter association pathways by the Belgian anatomist Arthur Van Gehuchten [18]. (b) A similar lateral view by the Austrian neurologist Heinrich Obersteiner [19]. (c) An early map of the brain's white matter connections represented as a network. Spheres mark cortical parcels (network nodes) and lines represent their connections (network edges) as mapped with diffusion imaging and tractography [20]. Only the strongest connections are included in the plot. The network is shown with nodes placed into their anatomical coordinates, hence preserving the spatial embedding of the network. (d) The same network as in (c), but with nodes placed by a widely used visualization algorithm that projects the network into two dimensions and places nodes such that densely connected regions are located nearby. Most highly connected parts of the brain, corresponding to portions of the cingulate and posterior parietal cortex, are placed in the centre of the plot. See Hagmann et al. [20] for details.
The goal of this article is to identify critical challenges and future developments at the intersection of cerebral cartography and connectomics. The article begins with a brief definition of core terms and concepts in brain connectivity and brain networks, drawing an important distinction between networks that record structural versus functional relations. The next section focuses on the key role of connectivity for defining anatomical and functional regions and sub-systems in the brain. Core themes here are the use of anatomical and/or functional connections to delineate the borders of coherent brain regions, and the application of various clustering and module detection methods to parse the brain into larger anatomical and/or functional sub-systems. The following three sections concentrate on three different aspects of how structural connectivity relates to brain function: the important role of model organisms for understanding this relationship at the level of individual neurons and circuits, the role of structural information in predicting patterns of neural dynamics and the prospect of building increasingly realistic network-based computational models of the brain.
2. Mapping dynamic brain networks
One of the most fundamental challenges for the study of brain networks is the appropriate definition of network elements (nodes and edges) and an explicit consideration of the neurobiological underpinnings of neural measures and signals that are employed in network construction. Especially important is the distinction between two main modes of brain connectivity—structural connectivity and functional connectivity [21,22]. Structural connectivity refers to maps of anatomical connections between pairs of nodes, corresponding to the popular notion of the ‘wiring diagram’. In aggregate, such connections define structural networks, and these networks are generally sparse (i.e. only a small number out of all possible anatomical connections exist). The strengths of anatomical connections are often expressed as a connection density or weight. Structural networks can be annotated with physiological data on synaptic efficacy or biophysical data on neurotransmitter systems. The complete set of all structural connections is the ‘connectome’, a network map of the synaptic connections and projections comprising a nervous system [14,15,21].
Functional connectivity is defined quite differently. It refers to estimates of pairwise statistical dependencies between time courses of neuronal activity [23,24], in the simplest case expressed as the system's covariance or correlation matrix. Functional networks can be dense (as for cross-correlations) or considerably sparser (as for partial correlations, or for measures that extract directed or causal influences). A major difference between structural and functional networks is that the former are relatively stable (at least on time scales of seconds to minutes) while the latter are inherently dynamic and time-dependent. Indeed, electrophysiological recordings indicate that changes in dynamic couplings among neurons or neuronal populations can occur as fast as tens or hundreds of milliseconds. Functional connectivity can be measured with a wide variety of recording or imaging methodologies—hence, estimates of functional connectivity are strongly dependent on acquisition techniques and parameters. In humans the majority of studies examining functional connectivity over the past several years have been based on fluctuations in blood oxygenation level dependent (BOLD) signals observed during a task-free or ‘resting’ state [25–27]. Despite its unconstrained nature, numerous studies have shown that spatial and temporal patterns of resting-brain activity are robust and reproducible, and that they provide rich information about the brain's functional organization [28,29]. Resting-state functional connectivity is generally expressed as the cross-correlation of time series of BOLD signals recorded with functional magnetic resonance imaging (fMRI) across the whole brain.
Once translated into network form, both structural and functional connectivity can be analysed and modelled using a large set of quantitative tools from graph theory and network science [22,30,31]. Method development is an active area of human connectomics, as is the development of more refined and sensitive approaches to mapping of structural and functional connectivity [32], currently subject to numerous methodological biases and shortcomings. Among the most prominent are the inability to extract directionality of projections from non-invasive diffusion imaging data [33], the uncertain biological basis and validity of measures expressing the density or magnitude of anatomical projections [34], and the failure of simple functional connectivity measures such as cross-correlation to distinguish between direct and indirect influences [35]. The latter shortcoming is addressed by measures of ‘effective connectivity’ that estimate networks of causal dependencies [36] and can be powerful tools for network discovery [37]. Other methodological issues surrounding the construction of brain networks involve the compatibility of different acquisition parameters and pre-processing methods [38,39], test-retest reliability for estimating connection maps in both structural [40] and functional modalities [41–43] and comparisons of patterns of connectivity estimated with different time-series analysis tools [44].
A major challenge arises from the observation that functional connectivity is both time- and task-dependent. For example, a comparison of resting-state functional connectivity with task-evoked functional connectivity, estimated across a broad range of task domains, has shown characteristic changes in subsets of functional connections as the brain switches between task-related states [45]. Even the so-called ‘resting-state’ appears to exhibit dynamic patterns involving changes in the strength and topology of functional networks, and characterizing these dynamic time-varying patterns represents a challenging new frontier for studies of functional connectivity [46,47]. For example, a sliding window analysis of resting-brain functional connectivity has shown that variability exhibits regional differences, with a ‘zone of instability’ that includes several highly connected and highly central regions [48]. Other studies have presented evidence for non-stationary fluctuations in the strengths of functional connections between specific pairs of brain regions [49,50]. While the precise nature and extent of these fluctuations in resting-brain fMRI functional connectivity remains under investigation, much evidence suggests that, at faster time scales accessible with electroencephalography/magnetoencephalography (EEG/MEG) or electrocorticography, dynamic interactions among neuronal populations are highly variable [51], engage in sets of recurrent network patterns or motifs [52], respond sensitively to momentary demands imposed by the external environment [53] and are related to behavioural performance [54]. Theoretical studies point to potential functional roles for dynamic variability of functional connectivity in the resting brain. For example, this variability resembles a so-called ‘critical state’, a dynamic regime poised between highly random and highly regular patterns characterized by series of noisy fluctuations [55]. These noisy dynamics define a repertoire of network states that is continually rehearsed and revisited in the resting brain.
Taken together, these studies paint a complex picture of time-based (functional connectivity) brain maps, the brain's ‘chronoarchitecture’ [56]. On the one side, the extended web of statistical dependencies among the neural time courses of remote brain regions is highly consistent when measured and averaged over many minutes of brain activity. Over such long sampling periods, functional connectivity may be interpreted as a reflection of anatomical constraints, rather than of specific brain responses or computations. However, over shorter time periods, functional connectivity appears highly variable as it reflects neural activations and interactions that occur in response to acute changes in internal state, tasks and stimuli. Characterizing the dynamic nature of functional connectivity represents a key challenge for cerebral cartography—creating dynamic brain maps will require developing new ways to capture temporal variability and context dependence of functional networks.
3. Parcellation and community detection
Since the beginnings of cerebral cartography, anatomists have expended extraordinary efforts to discern and map structural features of cortical micro-anatomy in order to define architectonic boundaries between brain areas. Increasingly, such areal maps are enriched by addition of multi-modal information such as data on gene expression patterns [57], energy metabolism [58] and meta-analyses of databases on regional brain activation patterns [59,60]. This multi-modality may ultimately result in maps that, like ‘Google Earth’, will allow the user to ‘zoom in’ on anatomical locations and view different layers of data types, with each layer representing distinct aspects of brain anatomy, physiology or function. Such maps would not only represent the brain in new ways but also aid in integrating multiple data domains, revealing multi-level interactions from molecules to structural anatomy and activity patterns of cells and circuits.
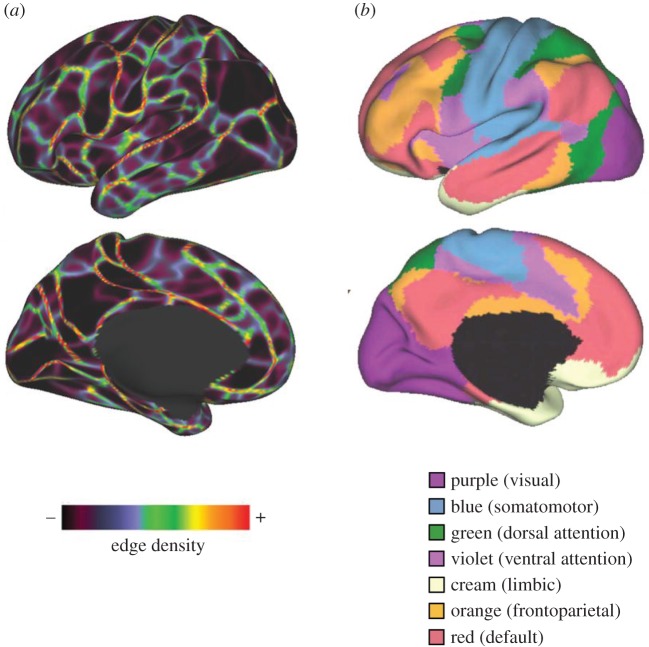
Creating accurate parcellations of the cerebral cortex remains challenging (figure 2). In this on-going endeavour, data on brain connectivity have turned out to be highly useful for defining the borders of cortical areas. Numerous strategies have been pursued in the past, including clustering of fibre bundles and tractography-derived streamlines [63], myelination patterns [64], boundary detection [65] and region-growing methods applied to resting-state functional connectivity [66], or combinations of functional connectivity and task-evoked regional activation profiles [67]. All of these strategies build on the notion that connectivity defines function. According to this idea, what and how a patch of cortex ‘computes’ (how it transforms inputs into outputs) depends crucially on its connectional relations with other patches of cortex and subcortex [68]. In fact, it has been consistently found that each anatomically distinct brain region maintains a specific and unique pattern of inputs and outputs. The similarities between these connection ‘fingerprints’ can be used to cluster regions into functionally related sub-groups [69,70]. Connections that are made over longer distances are generally sparser than short-distance connections [71], suggesting a strong role for long-distance projections in specialized cortical processing.
Figure 2.
Parcellation and network community detection. (a) A boundary map derived from group-averaged (n = 160) resting-state functional connectivity. ‘Hot colours’ (red/yellow) in edge density mark locations on the cortical surface where patterns of functional connections exhibit abrupt changes, corresponding to boundaries between relatively homogeneous (black/blue/purple) regions. (Adapted with permission from Gordon et al. [61].) (b) A network map of the cerebral cortex derived by clustering resting-state functional connectivity measured in a large number of participants (n = 1000) into seven network communities. These communities correspond to several well-studied resting-state or intrinsic connectivity networks. (Adapted with permission from Yeo et al. [62].)
Connectivity-based parcellation has begun to make important contributions in more refined mapping of some parts of the cerebral cortex. For example, recent studies using structural connectivity patterns have revealed new subdivisions of Broca's area [72], cingulate cortex [73], parietal cortex [74,75] and the frontal pole [76], among others. Some studies have suggested that regional borders defined through structural and functional parcellation significantly overlap [77], and others have shown that structural and functional organization closely parallel each other [78]. Yet, the generality and extent to which functionally established boundaries between regions agree with cytoarchitectonic or anatomically defined boundaries across the cortical surface is currently unknown.
While the aim of parcellation is to define relatively small-scale areas or regions, the related methodology of community detection is generally applied at the whole-brain level to define communities (clusters or ‘modules’) of areas that form extended networks [79,80]. It should be noted that the term ‘module’ is used in this context to refer to sets of densely interconnected cortical regions, and that its usage does not imply strict cognitive specialization or discrete ‘mental faculties’ [81]. Applications of community detection have yielded significant new insights about the organization and spatial arrangement of extended brain networks, especially coming from the use of resting-state functional connectivity [29]. Several systematic surveys have converged onto a taxonomy of resting-state (or intrinsic) networks that are reliably observed across large numbers of participants [62,82,83], can be robustly measured across different sites and acquisition protocols [28], resemble sets of areas that are co-active during a range of behavioural tasks [84], and exhibit topological features (e.g. modularity) that are directly relevant to cognitive function [85].
An interesting issue for the future concerns the relationship between areal parcellations and partitioning of the cortex into coherently active and spatially extended network communities. Are these network communities composed of discrete and non-overlapping sets of areas, or can areas straddle network boundaries and participate in multiple large-scale networks at different times? Recent work suggests that some parts of the cortex are more rigidly associated with a single network, while others can participate in multiple networks [86]. A related set of questions concerns the uniformity and temporal stability of both areal and network representations. For example, do task-evoked activations always respect areal boundaries, as established for example by connectivity-based gradient methods? Comparison of areal boundaries derived from resting-state functional connectivity with task-evoked areal activations and cytoarchitectonic boundaries suggests a high degree of spatial correspondence [61,87]. Other data suggest that, in addition to areal boundaries, supra-areal assemblies or ‘map clusters’ may also play an important functional role [88]. A related question concerns the extent to which intrinsic or resting-state networks represent coherent functional units. Recent studies suggest that resting-state networks can be further decomposed and hence may not always respond uniformly in diverse task contexts. For example, a convergent set of findings suggests variable task-dependent patterns of functional interactions among sub-components of specific resting-state networks, e.g. the default mode network [89–91], executive control networks [92] and saliency networks [93].
The emerging picture is considerably more complex than perhaps was anticipated in the early days of cerebral cartography, which were dominated by the idea that the cortical surface could be unambiguously subdivided into a mosaic of discrete and non-overlapping areas, each internally coherent and functionally specialized. Instead, the anatomical and functional units of the cortex may be more adequately represented as a nested hierarchy ranging from large-scale components or networks, to differentially engaged sub-components that can in turn be subdivided into finer and finer parcels. Classic brain areas may reside at a level that is somewhat distinct in terms of cortical microstructure and/or connectivity, but above and below this level areas blend into both finer and coarser partitions that manifest at different spatial scales. In line with this view, network analyses have provided evidence for hierarchical modularity in human structural [94] and functional brain networks [95]. Network analysis suggests that the theoretical framework of functional specialization and integration [96–98] requires extension to include multi-scale organization and dynamics [99,100]. This represents another future challenge for cerebral cartography—capturing the hierarchical arrangement of the brain's anatomical and functional units across multiple scales, from extended networks to local neuronal populations.
4. Mapping structure and function in model organisms
The remainder of the article will centre on how cerebral cartography and connectomics can make progress towards integrating structural and functional brain (network) maps. Structure/function relations are of fundamental importance in many biological systems—including the brain. One of the long-standing objectives of cerebral cartography is to link the physical arrangement of the brain's neural elements to their functional roles. Connectomics shares this integrative perspective on structure/function relations. Since the beginning, connectomics was explicitly motivated by the desire to provide mechanistic (structural) accounts of functional brain activity. This goal was a central objective in the original proposal for mapping the human connectome [14], and it was also inherent in the compilation of the Caenorhabditis elegans connectome, carried out at the level of individual neurons and synapses more than a quarter century ago [101]. In fact, model organisms such as C. elegans offer unique opportunities for understanding how network structure and function relate at the scale of cells and circuits.
At the microscale, modern advances in circuit mapping and manipulation [102] have begun to significantly expand our understanding of how circuit topology relates to neural computation and behaviour. Many important insights have come from studies in model organisms. For example, analysis of the ‘wiring diagram’ of the C. elegans adult male [103] has revealed network features such as multiple parallel pathways that link sensory neurons to effector neurons, some degree of recurrence within sensory systems, and structural modules—all features that can be related to specific aspects of sensorimotor processing and behaviour. In another study [104], differences in the synaptic connectivity of the pharyngeal nervous system of two nematode species could explain differences in their feeding behaviour. In Drosophila, the reconstruction of microscale wiring patterns in the optic medulla, a brain structure involved in motion detection, revealed specific connectional patterns of inter-neurons that were consistent with their specific roles in generating direction selectivity [105]. The Drosophila nervous system is also being mapped at the scale of the whole brain, and these mapping efforts are useful for demonstrating the structural basis of several well-studied behaviours. One approach involved the aggregation of thousands of single-neuron images into a map of functional subdivisions, so-called ‘local processing units’. The resulting mesoscale connectivity map comprised 41 nodes and their weighted interconnections [106]. Community detection methods revealed distinct network modules whose members were functionally specialized to carry out visual, olfactory, auditory and motor processing.
Microscale structure/function relations are also increasingly explored in vertebrate animals, particularly in the mouse brain. Using serial block-face electron microscopy, a recent study carried out in the mouse retina [107] showed specific patterns of structural connectivity between amacrine and ganglion cells that coincided with physiologically measured direction selectivity of individual neurons. These ‘dense reconstructions’ at the sub-micrometer scale are continually growing in size and scale. Most recently, the use of a combination of manual annotation and machine learning has resulted in the construction of a synaptic ‘contact matrix’ among approximately a thousand neurons in the inner plexiform layer of the mouse retina [108]. Microscale connection motifs in this matrix revealed circuit mechanisms underlying motion detection and other aspects of visual function. A related study has shown that specific wiring patterns among individual amacrine and bipolar cells can account for the timing and receptive field properties of these neurons in motion detection [109]. These findings strongly underscore the importance of circuit structure for circuit function [110].
Outside of the retina, similar dense reconstruction approaches combined with physiological recordings are beginning to reveal structure/function relations in specific regions of mouse cerebral cortex. An example is an analysis of anatomy and physiology of a subset of neurons in primary visual cortex of the mouse [111]. First, functional properties of neurons such as their preferred stimulus orientation were established by using optical imaging. This was followed by serial sectioning electron microscopy of the same tissue volume with the aim of mapping and reconstructing synaptic interconnections among these neurons. Detailed analysis of the structural connection pattern revealed some specific connectional features such as convergence of inputs from multiple pyramidal cells with diverse orientation preference onto inhibitory neurons.
Structure/function relations are also seen at the larger scale of whole-brain mouse connectome maps. For example, a community detection analysis of the anatomical connections of 10 areas of mouse visual cortex demonstrated a division of mouse cortex into two processing streams somewhat analogous to the dorsal/ventral streams found in primate visual cortex [112]. In addition, several systematic efforts to compile whole-brain mouse connectomes have been carried out. Aggregation of data from hundreds of tracer injections into a single network representation resulted in a directed connectivity network that was shown to contain several modules corresponding to sets of areas jointly involved in various sensory, motor and integrative functions [113]. A parallel effort [114], involving high-resolution optical imaging and tracing of projections across the entire mouse brain, has resulted in an even more comprehensive mouse connectome map which charts the directed and weighted anatomical links among 295 grey-matter regions. Initial network analysis of this map indicates the presence of high clustering as well as a number of highly connected network hubs, network features that are consistent with those found in a number of other mammalian species [115].
These brain mapping studies in model organisms will provide new methodological tools and approaches that eventually may become important for use in humans. While most human cerebral cartography has so far been carried out at the macroscale of regions and interregional projections, the next few decades will likely see strong efforts to link macro-scale human brain mapping to microscale circuits and maps of connectivity at the level of single cells. For example, future studies may capitalize on surgical interventions that offer the opportunity to carry out single-cell physiology and micro-anatomy in resected human tissue. This would allow more detailed representation of regional ‘nodes’ as composed of heterogeneous cell types and differentiated circuits. ‘Edges’ of connectivity maps at macro- and microscales will also become more multi-dimensional, going beyond merely reporting adjacency (‘what connects to what’) by including data on additional anatomical or physiological parameters of connections, for example, axonal microstructure, myelination, conduction velocity, neurotransmitter receptors, plasticity and neuromodulators. Jointly, these more detailed node/edge features reflect structural features of connectivity that contribute to neuronal signalling and dynamic interactions reflected in functional connectivity.
5. Predicting brain dynamics
The observation of large populations of neurons, potentially including the whole brain, results in very rich datasets of neuronal time series, sometimes referred to as the brain's ‘functional connectome’ [116]. Studies in model organisms are beginning to provide unprecedented insights into functional connectomes imaged at the microscale of single-cell resolution. Large-scale recording methods applied to organisms such as the zebrafish larva yield whole-brain recordings of highly resolved neural population activity [117], and these data can be decomposed into functionally coherent circuits forming clusters or modules [118]. The use of optogenetics may soon open the possibility to not only monitor but also manipulate circuit activity in behaving model organisms (e.g. [119]). The complexity of cellular-scale whole-brain recordings presents many challenges, which can be addressed for example by reducing the dimensionality of the datasets through various clustering or component mapping techniques [120–122].
Studies in mouse and macaque combining structural connectome data and non-invasive recording of functional brain activity are adding to our understanding of how anatomy shapes dynamic functional connectivity at the meso- and macro-scales. A recent study of monkey somatosensory cortex [123] examined the relationship between structural and functional connectivity at high spatial resolution. The study focused on connectivity within two specialized areas of the squirrel monkey somatosensory cortex (areas 3b and 1), both containing representations of the digits of the monkey's hand. Resting-state functional connectivity was recorded with high-field strength fMRI and exhibited topographically precise coupling between corresponding digits across both areas, as well as within area 3b. This pattern corresponded closely to anatomical connectivity patterns observed after injections of anatomical tracers. Overall, these findings suggest that connectivity within the squirrel monkey somatosensory cortex appears to be organized anatomically and functionally in highly similar patterns.
Correspondence between patterns of structural and functional connectivity has also been observed at the whole-brain level. Studies carried out in the macaque cortex found that patterns of coherent spontaneous BOLD fluctuations are similar to patterns of anatomical connectivity derived from tract tracing studies [124–126]. The relationship between patterns of structural and functional connections extends to pairs of regions that are not directly structurally linked. A detailed analysis of macaque cortex functional connectivity patterns demonstrated that strong coupling among brain regions could be observed even if no direct anatomical connection was present [125]. These functional connections were predicted by indirect structural paths and other more complex network-wide coupling effects, strongly suggesting that functional connectivity is due to a mixture of direct and indirect effects emerging from the underlying structural network. In support of this notion, both direct and indirect couplings could be successfully captured in computational models that were based on simulating dynamics constrained by structural connectivity. Taken together, these studies suggest a mechanistic role of structural connections in generating organized patterns of neural dynamics.
In the human brain, direct comparisons of resting-state functional connectivity and structural connectivity (connectome) networks have revealed robust and reproducible statistical relationships, lending further support to the idea that structural connections shape functional connectivity. An early example came from a systematic analysis of structural and functional connectivity in a small cohort of human participants [20], which reported robust correlations between the strengths of structural and functional connectivity across the cortex. A more detailed analysis of the same dataset [127] demonstrated that this correlation persisted even after potential confounds such as spatial proximity between regions were taken into account. The analysis also showed that indirect structural connections could account for a significant proportion of the functional connectivity observed between node pairs lacking direct linkage. This finding strongly suggested that functional connectivity may be partly due to the passing on of indirect influence along multi-step paths in the connectome [125].
More recent studies combining analyses of structural and functional connectivity have confirmed the existence of robust and significant statistical relationships between structural and (resting state) functional connectivity in the human brain (e.g. [128,129]). Computational analyses have addressed how anatomical connections constrain not only resting-brain functional connectivity, but also task-dependent correlations in neural activity [130], suggesting that the separation between distinct cognitive states is reflected in the topology of the human connectome. This interpretation is compatible with findings that suggest the existence of a ‘core functional architecture’ that is common to numerous task-dependent and reconfigurable modes of functional connectivity [131]. Anatomical connectivity may constrain this shared core, thus effectively reducing the dimensionality of the patterns of functional connectivity that emerge in response to momentary task demands. The notion that structural connections shape and/or constrain functional connections is further reinforced by interventional studies that have reported changes in functional connectivity resulting from targeted manipulations of the anatomical substrate, e.g. callosotomy carried out in both humans and non-human primates [132,133].
An important extension of structure/function relations is in clinical and translational research that examines brain and mental disorders. A large number of studies have attempted to link dysregulation of functional connectivity patterns to underlying disturbances of structural connectivity, e.g. in disruptions of highly central nodes or edges [134,135]. Disturbed structure/function relationships have been detected in schizophrenia [136], and disruptions of both structural and functional connectivity appear to be associated with different neurodegenerative conditions [137]. These clinical findings reinforce the need for cerebral cartography to capture not only normative patterns or population averages but also individual differences in brain connectivity. Mapping brain networks in individuals may provide new diagnostic tools or measures, and may even become important for developing new network-based interventions and therapies that capitalize on charting the network architecture of an individual's brain. New approaches may involve targeted manipulations of nodes or edges, e.g. with tools of brain stimulation [138], in order to restore network function. Of particular importance in this endeavour are new developments in computational neuroscience that combine connectome data with sophisticated computer models that can reproduce and predict the dynamic activity of human brain networks.
6. Network-based computational models of the brain
Linking brain structure to brain function is not only a major challenge for cerebral cartography—it also offers unprecedented opportunities for building computational models of the brain. Propelled by rapid progress in information technology, high-performance computing and chip design, sophisticated computational models aim at constructing large network models of cortical micro- and macro-circuits capable of replicating realistic neuronal firing patterns. The sheer complexity of the task and limitations on currently available anatomical and physiological data may prevent the construction of cell-based models of the human brain for some time to come. But these difficulties notwithstanding, ‘building a model of the human brain in a computer’ will almost certainly remain an ambitious and alluring goal for generations.
Modest and yet promising beginnings have been made, especially in attempts to computationally reproduce empirically observed features and patterns in resting-brain dynamics. A series of models have addressed the structural basis of spontaneous or resting-brain functional connectivity as recorded with fMRI (reviewed in [139]). The design of these models generally combines sets of biophysical equations that specify the ‘nodal dynamics’ of neurons or neuronal populations with sets of coupling terms that define their ‘inter-nodal edges’. These coupling terms are specified by a connectome map of structural connectivity. The model generates time series that can then be analysed using the same time series measures (e.g. cross-correlations or measures of directed neuronal interactions) that are employed in empirical studies. The approach has been elaborated within the framework of the ‘Virtual Brain’, a set of modelling, analysis and visualization tools that allow users to upload anatomical and physiological data, perform simulations, and compare model results with empirical observations [140].
Models of resting-brain dynamics contribute to our understanding of the structural basis of functional connectivity. For example, modelling work has established robust relations between empirical and simulated functional networks [125,127,141], as well as an important role for conduction delays and noise in generating realistic resting-brain dynamics [142]. Computational models have also proved useful for understanding the network dynamics underlying non-stationary fluctuations in functional connectivity [143]. While most studies so far have focused on ‘normal’ brain dynamics, the framework can be extended to include anatomically detailed models of dynamic effects induced by focal brain lesions [144], degeneration [145] or psychiatric disorders [146]. Another intriguing application of network-based computational modelling is the use of such models to capture spreading dynamics of epilepsy, which may eventually become a tool for data-driven surgical mapping and selection of stimulation sites [147].
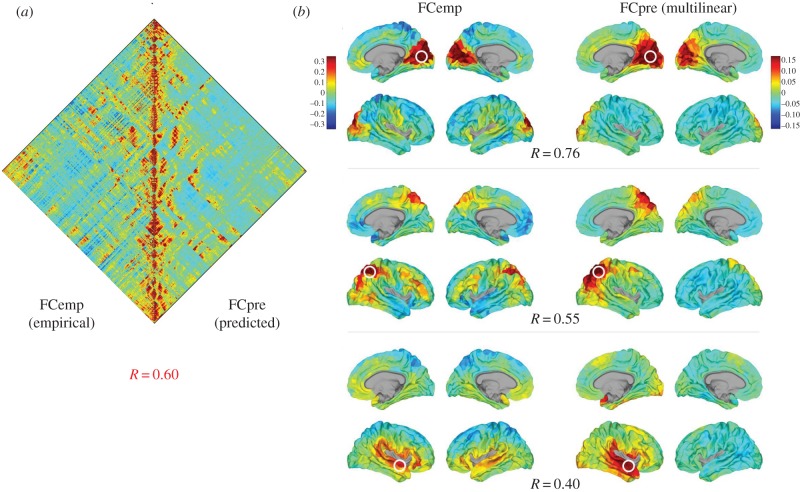
Computational models are also increasingly useful for understanding the mechanistic underpinnings of functional connectivity. This is important since functional connectivity has become a prominent mode for measuring brain dynamics, despite the fact that its neurobiological origin and meaning are far from clear. It is generally assumed that functional connectivity reflects ‘dynamic interactions’ among nodes in the brain, i.e. is driven by the temporal dynamics of neural signalling and communication. However, it is worth noting that neuronal communication is difficult, if not impossible, to observe and measure directly—usually, only the consequences of numerous communication events are accessible, for example, in the temporal alignment of neural time series expressed in their covariance. Our understanding of how functional connectivity emerges from distributed communication processes unfolding in structural brain networks is hence severely limited. Modelling can help in this regard, for example through the application of deliberately simple physics models of communication processes. Such models are complementary to more detailed physiological models that can generate simulations of rich temporal patterns of brain dynamics, but that are also computationally costly and difficult to implement. Physics-based models of communication processes combine computational simplicity with analytic transparence. Despite their simplicity, models that are based on structural graph measures [148] and/or models of diffusive processes [149] and routing [150] have proved capable of reproducing observational data on brain dynamics, for example, the topography of resting-brain functional connectivity (figure 3).
Figure 3.
A network-based computational model of functional connectivity [148]. (a) The left triangular half of the plot shows a functional connectivity (cross-correlation) map of 500 parcels comprising the right cortical hemisphere, computed from empirical resting-state fMRI (n = 5 participants, [20]). The right triangular half shows a prediction derived from a computational model, based entirely on network measures of communication applied to the underlying structural connectivity matrix. The model uses direct connections as well as (mostly indirect) shortest paths within the structural connectivity to predict how strongly each node pair should be correlated. The two halves of the plot are significantly correlated (R = 0.60). (b) Seed-based cross-correlation maps projected onto the surface of the cortex, with each plot showing lateral and medial surfaces. Plots on the left (‘FCemp’) depict correlation maps from empirical data [20]. Plots on the right (‘FCpre’) depict maps from model predictions. Three example seeds are shown, placed in the visual cortex (top), superior parietal cortex (middle) and the superior temporal cortex (bottom). Note loss of cross-hemispheric functional connectivity in model prediction for temporal cortex, likely due to inter-hemispheric pathways that were missed in diffusion imaging and tractography.
In summary, computational models of the human brain that can reproduce and predict patterns of brain activity at rest and in response to stimulus- and task-evoked perturbations have come within reach, not least because of significant progress in human connectomics and cerebral cartography. It seems certain that network maps will remain important ingredients for building ever more detailed computational brain models in the future.
7. Conclusion
Cerebral cartography and connectomics pursue complementary goals. Cerebral cartography is directed at map-making of the anatomical and functional topography of the cerebral cortex, using an ever more sensitive and sophisticated array of molecular, histological, physiological and imaging tools. Connectomics aims to chart network maps of the brain's structural and functional connections, across scales, from cells to systems, and these maps have begun to reveal principles of organization (e.g. modules and hubs) that are shared across nervous systems of different species. Challenges arise in the intersection of these two complementary endeavours, and this review attempted to sketch how some of these challenges might be addressed in the near future. Principal challenges concern the temporal dynamics of functional brain connectivity, the definition of areal parcellations and their hierarchical organization into large-scale networks, the extension of whole-brain connectivity to cellular-scale networks, and the mapping of structure/function relations in empirical recordings and computational models.
The long-term future of cerebral cartography and connectomics is difficult to predict. As in the past, new technologies are likely to open entirely new vistas on brain structure and function. Almost certainly, the future of both fields will bring a flood of data (‘big data’ as it is fashionably called) that will require a whole new set of analytic and visualization tools. But one hopes that, in addition to more data, there will also be progress in creating a theoretical framework for understanding the brain [151]—a framework that serves to organize our rapidly increasing knowledge and reveals fundamental principles of operation. If current trends prevail, network science will be a cornerstone of this framework, paving the way towards a more complete understanding of the brain as a complex networked system.
Funding statement
O.S. was supported by the J.S. McDonnell Foundation.
References
- 1.Zeki S. 2005. Introduction: cerebral cartography 1905–2005. Phil. Trans. R. Soc. B 360, 651–652. ( 10.1098/rstb.2005.1632) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lima M. 2011. Visual complexity: mapping patterns of information. New York, NY: Princeton Architectural Press. [Google Scholar]
- 3.Börner K. 2010. Atlas of science. Cambridge, MA: MIT Press. [Google Scholar]
- 4.Newman M. 2010. Networks: an introduction. Oxford, UK: Oxford University Press. [Google Scholar]
- 5.Rual JF, et al. 2005. Towards a proteome-scale map of the human protein–protein interaction network. Nature 437, 1173–1178. ( 10.1038/nature04209) [DOI] [PubMed] [Google Scholar]
- 6.Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabási AL. 2007. The human disease network. Proc. Natl Acad. Sci. USA 104, 8685–8690. ( 10.1073/pnas.0701361104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vespignani A. 2012. Modelling dynamical processes in complex socio-technical systems. Nat. Phys. 8, 32–39. ( 10.1038/nphys2160) [DOI] [Google Scholar]
- 8.Montoya JM, Pimm SL, Solé RV. 2006. Ecological networks and their fragility. Nature 442, 259–264. ( 10.1038/nature04927) [DOI] [PubMed] [Google Scholar]
- 9.Börner K, Sanyal S, Vespignani A. 2007. Network science. Annu. Rev. Inf. Sci. Technol. 41, 537–607. ( 10.1002/aris.2007.1440410119) [DOI] [Google Scholar]
- 10.Brandes U, Robins G, McCranie A, Wasserman S. 2013. What is network science? Netw. Sci. 1, 1–15. ( 10.1017/nws.2013.2) [DOI] [Google Scholar]
- 11.Zeki S, Shipp S. 1988. The functional logic of cortical connections. Nature 335, 311–317. ( 10.1038/335311a0) [DOI] [PubMed] [Google Scholar]
- 12.Felleman DJ, van Essen DC. 1991. Distributed hierarchical processing in the primate cerebral cortex. Cereb. Cortex 1, 1–47. ( 10.1093/cercor/1.1.1) [DOI] [PubMed] [Google Scholar]
- 13.Scannell JW, Burns GAPC, Hilgetag CC, O'Neil MA, Young MP. 1999. The connectional organization of the cortico-thalamic system of the cat. Cereb. Cortex 9, 277–299. ( 10.1093/cercor/9.3.277) [DOI] [PubMed] [Google Scholar]
- 14.Sporns O, Tononi G, Kötter R. 2005. The human connectome: a structural description of the human brain. PLoS Comput. Biol. 1, 245–251. ( 10.1371/journal.pcbi.0010042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sporns O. 2013. The human connectome: origins and challenges. Neuroimage 80, 53–61. ( 10.1016/j.neuroimage.2013.03.023) [DOI] [PubMed] [Google Scholar]
- 16.Behrens TEJ, Sporns O. 2012. Human connectomics. Curr. Opin. Neurobiol. 22, 144–153. ( 10.1016/j.conb.2011.08.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sporns O. 2014. Contributions and challenges for network models in cognitive neuroscience. Nat. Neurosci. 17, 652–660. ( 10.1038/nn.3690) [DOI] [PubMed] [Google Scholar]
- 18.Van Gehuchten A. 1894. Le Système Nerveux de L'Homme. Louvain, Belgium: Uystpuyst-Dieudonné. [Google Scholar]
- 19.Obersteiner H. 1890. The anatomy of the central nervous organs in health and disease. Philadelphia, PA: Blakiston. [Google Scholar]
- 20.Hagmann P, Cammoun L, Gigandet X, Meuli R, Honey CJ, Wedeen V, Sporns O. 2008. Mapping the structural core of human cerebral cortex. PLoS Biol. 6, e159 ( 10.1371/journal.pbio.0060159) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sporns O. 2011. Networks of the brain. Cambridge, MA: MIT Press. [Google Scholar]
- 22.Bullmore E, Sporns O. 2009. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat. Rev. Neurosci. 10, 186–198. ( 10.1038/nrn2575) [DOI] [PubMed] [Google Scholar]
- 23.Friston KJ. 1994. Functional and effective connectivity in neuroimaging: a synthesis. Hum. Brain Mapp. 2, 56–78. ( 10.1002/hbm.460020107) [DOI] [Google Scholar]
- 24.Friston KJ. 2011. Functional and effective connectivity: a review. Brain Connect. 1, 13–36. ( 10.1089/brain.2011.0008) [DOI] [PubMed] [Google Scholar]
- 25.Fox MD, Raichle ME. 2007. Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat. Rev. Neurosci. 8, 700–711. ( 10.1038/nrn2201) [DOI] [PubMed] [Google Scholar]
- 26.Raichle ME. 2011. The restless brain. Brain Connect. 1, 3–12. ( 10.1089/brain.2011.0019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Den Heuvel MP, Hulshoff Pol HE. 2010. Exploring the brain network: a review on resting-state fMRI functional connectivity. Eur. Neuropsychopharmacol. 20, 519–534. ( 10.1016/j.euroneuro.2010.03.008) [DOI] [PubMed] [Google Scholar]
- 28.Biswal BB, et al. 2010. Toward discovery science of human brain function. Proc. Natl Acad. Sci. USA 107, 4734–4739. ( 10.1073/pnas.0911855107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buckner RL, Krienen FM, Yeo BT. 2013. Opportunities and limitations of intrinsic functional connectivity MRI. Nat. Neurosci. 16, 832–837. ( 10.1038/nn.3423) [DOI] [PubMed] [Google Scholar]
- 30.Rubinov M, Sporns O. 2010. Complex network measures of brain connectivity: uses and interpretations. Neuroimage 52, 1059–1069. ( 10.1016/j.neuroimage.2009.10.003) [DOI] [PubMed] [Google Scholar]
- 31.Lohmann G, Stelzer J, Neumann J, Ay N, Turner R. 2013. ‘More is different’ in functional magnetic resonance imaging: a review of recent data analysis techniques. Brain Connect. 3, 223–239. ( 10.1089/brain.2012.0133) [DOI] [PubMed] [Google Scholar]
- 32.Craddock RC, et al. 2013. Imaging human connectomes at the macroscale. Nat. Methods 10, 524–539. ( 10.1038/nmeth.2482) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jbabdi S, Johansen-Berg H. 2011. Tractography: where do we go from here? Brain Connect. 1, 169–183. ( 10.1089/brain.2011.0033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones DK, Knösche TR, Turner R. 2013. White matter integrity, fiber count, and other fallacies: the do's and don'ts of diffusion MRI. Neuroimage 73, 239–254. ( 10.1016/j.neuroimage.2012.06.081) [DOI] [PubMed] [Google Scholar]
- 35.Zalesky A, Fornito A, Bullmore E. 2012. On the use of correlation as a measure of network connectivity. Neuroimage 60, 2096–2106. ( 10.1016/j.neuroimage.2012.02.001) [DOI] [PubMed] [Google Scholar]
- 36.Valdes-Sosa PA, Roebroeck A, Daunizeau J, Friston K. 2011. Effective connectivity: influence, causality and biophysical modeling. Neuroimage 58, 339–361. ( 10.1016/j.neuroimage.2011.03.058) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Friston KJ, Li B, Daunizeau J, Stephan KE. 2011. Network discovery with DCM. Neuroimage 56, 1202–1221. ( 10.1016/j.neuroimage.2010.12.039) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bassett DS, Brown JA, Deshpande V, Carlson JM, Grafton ST. 2011. Conserved and variable architecture of human white matter connectivity. Neuroimage 54, 1262–1279. ( 10.1016/j.neuroimage.2010.09.006) [DOI] [PubMed] [Google Scholar]
- 39.Bastiani M, Shah NJ, Goebel R, Roebroeck A. 2012. Human cortical connectome reconstruction from diffusion weighted MRI: the effect of tractography algorithm. Neuroimage 62, 1732–1749. ( 10.1016/j.neuroimage.2012.06.002) [DOI] [PubMed] [Google Scholar]
- 40.Besson P, Lopes R, Leclerc X, Derambure P, Tyvaert L. 2014. Intra-subject reliability of the high-resolution whole-brain structural connectome. NeuroImage 102, 283–293. ( 10.1016/j.neuroimage.2014.07.064) [DOI] [PubMed] [Google Scholar]
- 41.Van Dijk KR, Hedden T, Venkataraman A, Evans KC, Lazar SW, Buckner RL. 2010. Intrinsic functional connectivity as a tool for human connectomics: theory, properties, and optimization. J. Neurophysiol. 103, 297–321. ( 10.1152/jn.00783.2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zuo XN, Xing XX. 2014. Test-retest reliabilities of resting-state FMRI measurements in human brain functional connectomics: a systems neuroscience perspective. Neurosci. Biobehav. Rev. 45, 100–118. ( 10.1016/j.neubiorev.2014.05.009) [DOI] [PubMed] [Google Scholar]
- 43.Cao H, et al. 2014. Test–retest reliability of fMRI-based graph theoretical properties during working memory, emotion processing, and resting state. Neuroimage 84, 888–900. ( 10.1016/j.neuroimage.2013.09.013) [DOI] [PubMed] [Google Scholar]
- 44.Smith SM, Miller KL, Salimi-Khorshidi G, Webster M, Beckmann CF, Nichols TE, Ramsey JD, Woolrich MW. 2011. Network modelling methods for FMRI. Neuroimage 54, 875–891. ( 10.1016/j.neuroimage.2010.08.063) [DOI] [PubMed] [Google Scholar]
- 45.Cole MW, Bassett DS, Power JD, Braver TS, Petersen SE, Cole MW. 2014. Intrinsic and task-evoked network architectures of the human brain. Neuron 83, 238–251. ( 10.1016/j.neuron.2014.05.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hutchison RM, et al. 2013. Dynamic functional connectivity: promise, issues, and interpretations. Neuroimage 80, 360–378. ( 10.1016/j.neuroimage.2013.05.079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Calhoun VD, Miller R, Pearlson G, Adali T. 2014. The chronnectome: time-varying connectivity networks as the next frontier in fMRI data discovery. Neuron 84, 262–274. ( 10.1016/j.neuron.2014.10.015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Allen EA, Damaraju E, Plis SM, Erhardt EB, Eichele T, Calhoun VD. 2014. Tracking whole-brain connectivity dynamics in the resting state. Cereb. Cortex 24, 663–676. ( 10.1093/cercor/bhs352) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zalesky A, Fornito A, Cocchi L, Gollo LL, Breakspear M. 2014. Time-resolved resting-state brain networks. Proc. Natl Acad. Sci. USA 111, 10 341–10 346. ( 10.1073/pnas.1400181111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gonzalez-Castillo J, Daniel HA, Robinson ME, Hoy CW, Buchanan LC, Saad ZS, Bandettini PA. 2014. The spatial structure of resting state connectivity stability on the scale of minutes. Front. Neurosci. 8, 138 ( 10.3389/fnins.2014.00138) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Palva S, Palva JM. 2012. Discovering oscillatory interaction networks with M/EEG: challenges and breakthroughs. Trends Cogn. Sci. 16, 219–230. ( 10.1016/j.tics.2012.02.004) [DOI] [PubMed] [Google Scholar]
- 52.Chu CJ, Kramer MA, Pathmanathan J, Bianchi MT, Westover MB, Wizon L, Cash SS. 2012. Emergence of stable functional networks in long-term human electroencephalography. J. Neurosci. 32, 2703–2713. ( 10.1523/JNEUROSCI.5669-11.2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kitzbichler MG, Henson RN, Smith ML, Nathan PJ, Bullmore ET. 2011. Cognitive effort drives workspace configuration of human brain functional networks. J. Neurosci. 31, 8259–8270. ( 10.1523/JNEUROSCI.0440-11.2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bassett DS, Bullmore ET, Meyer-Lindenberg A, Apud JA, Weinberger DR, Coppola R. 2009. Cognitive fitness of cost-efficient brain functional networks. Proc. Natl Acad. Sci. USA 106, 11 747–11 752. ( 10.1073/pnas.0903641106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deco G, Jirsa VK, McIntosh AR. 2013. Resting brains never rest: computational insights into potential cognitive architectures. Trends Neurosci. 36, 268–274. ( 10.1016/j.tins.2013.03.001) [DOI] [PubMed] [Google Scholar]
- 56.Bartels A, Zeki S. 2005. The chronoarchitecture of the cerebral cortex. Phil. Trans. R. Soc. B 360, 733–750. ( 10.1098/rstb.2005.1627) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hawrylycz MJ, et al. 2012. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 489, 391–399. ( 10.1038/nature11405) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vaishnavi SN, Vlassenko AG, Rundle MM, Snyder AZ, Mintun MA, Raichle ME. 2010. Regional aerobic glycolysis in the human brain. Proc. Natl Acad. Sci. USA 107, 17 757–17 762. ( 10.1073/pnas.1010459107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Laird AR, Lancaster JJ, Fox PT. 2005. Brainmap. Neuroinformatics 3, 65–77. ( 10.1385/NI:3:1:065) [DOI] [PubMed] [Google Scholar]
- 60.Yarkoni T, Poldrack RA, Nichols TE, Van Essen DC, Wager TD. 2011. Large-scale automated synthesis of human functional neuroimaging data. Nat. Methods 8, 665–670. ( 10.1038/nmeth.1635) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gordon EM, Laumann TO, Adeyemo B, Huckins JF, Kelley WM, Petersen SE. In press. Generation and evaluation of a cortical area parcellation from resting-state correlations. Cereb. Cortex. ( 10.1093/cercor/bhu239) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yeo BTT, et al. 2011. The organization of the human cerebral cortex estimated by functional connectivity. J. Neurophysiol. 106, 1125–1165. ( 10.1152/jn.00338.2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rushworth MFS, Behrens TEJ, Johansen-Berg H. 2006. Connection patterns distinguish 3 regions of human parietal cortex. Cereb. Cortex 16, 1418–1430. ( 10.1093/cercor/bhj079) [DOI] [PubMed] [Google Scholar]
- 64.Glasser MF, Van Essen DC. 2011. Mapping human cortical areas in vivo based on myelin content as revealed by T1- and T2-weighted MRI. J. Neurosci. 31, 11 597–11 616. ( 10.1523/JNEUROSCI.2180-11.2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cohen AL, Fair DA, Dosenbach NUF, Miezin FM, Dierker D, Van Essen DC, Schlaggar BL, Petersen SE. 2008. Defining functional areas in individual human brains using resting functional connectivity MRI. Neuroimage 41, 45–57. ( 10.1016/j.neuroimage.2008.01.066) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wig GS, et al. 2013. Parcellating an individual subject's cortical and subcortical brain structures using snowball sampling of resting-state correlations. Cereb. Cortex 24, 2036–2054. ( 10.1093/cercor/bht056) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nelson SM, et al. 2010. A parcellation scheme for human left lateral parietal cortex. Neuron 67, 156–170. ( 10.1016/j.neuron.2010.05.025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Passingham RE, Stephan KE, Kötter R. 2002. The anatomical basis of functional localization in the cortex. Nat. Rev. Neurosci. 3, 606–616. ( 10.1038/nrn893) [DOI] [PubMed] [Google Scholar]
- 69.Stephan KE, Kamper L, Bozkurt A, Burns GA, Young MP, Kötter R. 2001. Advanced database methodology for the collation of connectivity data on the macaque brain (CoCoMac). Phil. Trans. R. Soc. Lond. B 356, 1159–1186. ( 10.1098/rstb.2001.0908) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hilgetag CC, Kaiser M. 2004. Clustered organization of cortical connectivity. Neuroinformatics 2, 353–360. ( 10.1385/NI:2:3:353) [DOI] [PubMed] [Google Scholar]
- 71.Markov NT, et al. 2013. The role of long-range connections on the specificity of the macaque interareal cortical network. Proc. Natl Acad. Sci. USA 110, 5187–5192. ( 10.1073/pnas.1218972110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Anwander A, Tittgemeyer M, von Cramon DY, Friederici AD, Knösche TR. 2007. Connectivity-based parcellation of Broca's area. Cereb. Cortex 17, 816–825. ( 10.1093/cercor/bhk034) [DOI] [PubMed] [Google Scholar]
- 73.Beckmann M, Johansen-Berg H, Rushworth MF. 2009. Connectivity-based parcellation of human cingulate cortex and its relation to functional specialization. J. Neurosci. 29, 1175–1190. ( 10.1523/JNEUROSCI.3328-08.2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mars RB, et al. 2011. Diffusion-weighted imaging tractography-based parcellation of the human parietal cortex and comparison with human and macaque resting-state functional connectivity . J. Neurosci. 31, 4087–4100. ( 10.1523/JNEUROSCI.5102-10.2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ruschel M, Knösche TR, Friederici AD, Turner R, Geyer S, Anwander A. 2013. Connectivity architecture and subdivision of the human inferior parietal cortex revealed by diffusion MRI. Cereb. Cortex 24, 2436–2448. ( 10.1093/cercor/bht098) [DOI] [PubMed] [Google Scholar]
- 76.Liu H, Qin W, Li W, Fan L, Wang J, Jiang T, Yu C. 2013. Connectivity-based parcellation of the human frontal pole with diffusion tensor imaging. J. Neurosci. 33, 6782–6790. ( 10.1523/JNEUROSCI.4882-12.2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Johansen-Berg H, Behrens TEJ, Robson MD, Drobnjak I, Rushworth MFS, Brady JM, Smith SM, Higham DJ, Matthews PM. 2004. Changes in connectivity profiles define functionally distinct regions in human medial frontal cortex. Proc. Natl Acad. Sci. USA 101, 13 335–13 340. ( 10.1073/pnas.0403743101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jeon HA, Anwander A, Friederici AD. 2014. Functional network mirrored in the prefrontal cortex, caudate nucleus, and thalamus: high-resolution functional imaging and structural connectivity. J. Neurosci. 34, 9202–9212. ( 10.1523/JNEUROSCI.0228-14.2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Newman MEJ, Girvan M. 2004. Finding and evaluating community structure in networks. Phys. Rev. E 69, 026113 ( 10.1103/PhysRevE.69.026113) [DOI] [PubMed] [Google Scholar]
- 80.Fortunato S. 2010. Community detection in graphs. Phys. Rep. 486, 75–174. ( 10.1016/j.physrep.2009.11.002) [DOI] [Google Scholar]
- 81.Fodor JA. 1983. The modularity of mind: an essay on faculty psychology. Cambridge, MA: MIT Press. [Google Scholar]
- 82.Power JD, et al. 2011. Functional network organization of the human brain. Neuron 72, 665–678. ( 10.1016/j.neuron.2011.09.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Doucet G, et al. 2011. Brain activity at rest: a multiscale hierarchical functional organization. J. Neurophysiol. 105, 2753–2763. ( 10.1152/jn.00895.2010) [DOI] [PubMed] [Google Scholar]
- 84.Smith SM, et al. 2009. Correspondence of the brain's functional architecture during activation and rest. Proc. Natl Acad. Sci. USA 106, 13 040–13 045. ( 10.1073/pnas.0905267106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Crossley NA, Mechelli A, Vertes PE, Winton-Brown TT, Patel AX, Ginestet CE, McGuire P, Bullmore ET. 2013. Cognitive relevance of the community structure of the human brain functional coactivation network. Proc. Natl Acad. Sci. USA 110, 11 583–11 588. ( 10.1073/pnas.1220826110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yeo BT, Krienen FM, Chee MW, Buckner RL. 2014. Estimates of segregation and overlap of functional connectivity networks in the human cerebral cortex. Neuroimage 88, 212–227. ( 10.1016/j.neuroimage.2013.10.046) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wig GS, Laumann TO, Petersen SE. 2014. An approach for parcellating human cortical areas using resting-state correlations. Neuroimage 93, 276–291. ( 10.1016/j.neuroimage.2013.07.035) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Buckner RL, Yeo BT. 2014. Borders, map clusters, and supra-areal organization in visual cortex. Neuroimage 93, 292–297. ( 10.1016/j.neuroimage.2013.12.036) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Andrews-Hanna JR, Reidler JS, Sepulcre J, Poulin R, Buckner RL. 2010. Functional-anatomic fractionation of the brain's default network. Neuron 65, 550–562. ( 10.1016/j.neuron.2010.02.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fornito A, Harrison BJ, Zalesky A, Simons JS. 2012. Competitive and cooperative dynamics of large-scale brain functional networks supporting recollection. Proc. Natl Acad. Sci. USA 109, 12 788–12 793. ( 10.1073/pnas.1204185109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kragel JE, Polyn SM. 2013. Functional interactions between large-scale networks during memory search. Cereb. Cortex 25, 667–679. ( 10.1093/cercor/bht258) [DOI] [PubMed] [Google Scholar]
- 92.Dosenbach NU, et al. 2007. Distinct brain networks for adaptive and stable task control in humans. Proc. Natl Acad. Sci. USA 104, 11 073–11 078. ( 10.1073/pnas.0704320104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Elton A, Gao W. 2014. Divergent task-dependent functional connectivity of executive control and salience networks. Cortex 51, 56–66. ( 10.1016/j.cortex.2013.10.012) [DOI] [PubMed] [Google Scholar]
- 94.Bassett DS, Greenfield DL, Meyer-Lindenberg A, Weinberger DR, Moore SW, Bullmore ET. 2010. Efficient physical embedding of topologically complex information processing networks in brains and computer circuits. PLoS Comput. Biol. 6, e1000748 ( 10.1371/journal.pcbi.1000748) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Meunier D, Lambiotte R, Fornito A, Ersche KD, Bullmore ET. 2009. Hierarchical modularity in human brain functional networks. Front. Neuroinf. 3, 37 ( 10.3389/neuro.11.037.2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zeki S. 1993. A vision of the brain. Oxford, UK: Oxford University Press. [Google Scholar]
- 97.Tononi G, Sporns O, Edelman GM. 1994. A measure for brain complexity: relating functional segregation and integration in the nervous system. Proc. Natl Acad. Sci. USA 91, 5033–5037. ( 10.1073/pnas.91.11.5033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tononi G, Edelman GM, Sporns O. 1998. Complexity and coherency: integrating information in the brain. Trends Cogn. Sci. 2, 474–484. ( 10.1016/S1364-6613(98)01259-5) [DOI] [PubMed] [Google Scholar]
- 99.Sporns O. 2013. Network attributes for segregation and integration in the human brain. Curr. Opin. Neurobiol. 23, 162–171. ( 10.1016/j.conb.2012.11.015) [DOI] [PubMed] [Google Scholar]
- 100.Park HJ, Friston K. 2013. Structural and functional brain networks: from connections to cognition. Science 342, 1238411 ( 10.1126/science.1238411) [DOI] [PubMed] [Google Scholar]
- 101.White JG, Southgate E, Thomson JN, Brenner S. 1986. The structure of the nervous system of the nematode Caenorhabditis elegans. Phil. Trans. R. Soc. Lond. B 314, 1–340. ( 10.1098/rstb.1986.0056) [DOI] [PubMed] [Google Scholar]
- 102.Deisseroth K. 2014. Circuit dynamics of adaptive and maladaptive behaviour. Nature 505, 309–317. ( 10.1038/nature12982) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jarrell TA, Wang Y, Bloniarz AE, Brittin CA, Xu M, Thomson JN, Albertson DG, Hall DH, Emmons SW. 2012. The connectome of a decision-making neural network. Science 337, 437–444. ( 10.1126/science.1221762) [DOI] [PubMed] [Google Scholar]
- 104.Bumbarger DJ, Riebesell M, Rödelsperger C, Sommer RJ. 2013. System-wide rewiring underlies behavioral differences in predatory and bacterial-feeding nematodes. Cell 152, 109–119. ( 10.1016/j.cell.2012.12.013) [DOI] [PubMed] [Google Scholar]
- 105.Takemura SY, et al. 2013. A visual motion detection circuit suggested by Drosophila connectomics. Nature 500, 175–181. ( 10.1038/nature12450) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chiang AS, et al. 2011. Three-dimensional reconstruction of brain-wide wiring networks in Drosophila at single-cell resolution. Curr. Biol. 21, 1–11. ( 10.1016/j.cub.2010.11.056) [DOI] [PubMed] [Google Scholar]
- 107.Briggman KL, Helmstaedter M, Denk W. 2011. Wiring specificity in the direction-selectivity circuit of the retina. Nature 471, 183–188. ( 10.1038/nature09818) [DOI] [PubMed] [Google Scholar]
- 108.Helmstaedter M, Briggman KL, Turaga SC, Jain V, Seung HS, Denk W. 2013. Connectomic reconstruction of the inner plexiform layer in the mouse retina. Nature 500, 168–174. ( 10.1038/nature12346) [DOI] [PubMed] [Google Scholar]
- 109.Kim JS, et al. 2014. Space-time wiring specificity supports direction selectivity in the retina. Nature 509, 331–336. ( 10.1038/nature13240) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Denk W, Briggman KL, Helmstaedter M. 2012. Structural neurobiology: missing link to a mechanistic understanding of neural computation. Nat. Rev. Neurosci. 13, 351–358. ( 10.1038/nrn3169) [DOI] [PubMed] [Google Scholar]
- 111.Bock DD, et al. 2011. Network anatomy and in vivo physiology of visual cortical neurons. Nature 471, 177–182. ( 10.1038/nature09802) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang Q, Sporns O, Burkhalter A. 2012. Network analysis of corticocortical connections reveals ventral and dorsal processing streams in mouse visual cortex. J. Neurosci. 32, 4386–4399. ( 10.1523/JNEUROSCI.6063-11.2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zingg B, et al. 2014. Neural networks of the mouse neocortex. Cell 156, 1096–1111. ( 10.1016/j.cell.2014.02.023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Oh SW, et al. 2014. A mesoscale connectome of the mouse brain. Nature 508, 207–214. ( 10.1038/nature13186) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sporns O, Bullmore ET. 2014. From connections to function: the mouse brain connectome atlas. Cell 157, 773–775. ( 10.1016/j.cell.2014.04.023) [DOI] [PubMed] [Google Scholar]
- 116.Alivisatos AP, Chun M, Church GM, Greenspan RJ, Roukes ML, Yuste R. 2012. The brain activity map project and the challenge of functional connectomics. Neuron 74, 970–974. ( 10.1016/j.neuron.2012.06.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ahrens MB, Orger MB, Robson DN, Li JM, Keller PJ. 2013. Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nat. Methods 10, 413–420. ( 10.1038/nmeth.2434) [DOI] [PubMed] [Google Scholar]
- 118.Portugues R, Feierstein CE, Engert F, Orger MB. 2014. Whole-brain activity maps reveal stereotyped, distributed networks for visuomotor behavior. Neuron 81, 1328–1343. ( 10.1016/j.neuron.2014.01.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Portugues R, Severi KE, Wyart C, Ahrens MB. 2013. Optogenetics in a transparent animal: circuit function in the larval zebrafish. Curr. Opin. Neurobiol. 23, 119–126. ( 10.1016/j.conb.2012.11.001) [DOI] [PubMed] [Google Scholar]
- 120.Sporns O. 2013. Making sense of brain network data. Nat. Methods 10, 491–493. ( 10.1038/nmeth.2485) [DOI] [PubMed] [Google Scholar]
- 121.Cunningham JP, Byron MY. 2014. Dimensionality reduction for large-scale neural recordings. Nat. Neurosci. 17, 1500–1509. ( 10.1038/nn.3776) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Freeman J, et al. 2014. Mapping brain activity at scale with cluster computing. Nat. Methods 11, 941–950. ( 10.1038/nmeth.3041) [DOI] [PubMed] [Google Scholar]
- 123.Wang Z, Chen LM, Négyessy L, Friedman RM, Mishra A, Gore JC, Roe AW. 2013. The relationship of anatomical and functional connectivity to resting-state connectivity in primate somatosensory cortex. Neuron 78, 1116–1126. ( 10.1016/j.neuron.2013.04.023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Vincent JL, et al. 2007. Intrinsic functional architecture in the anaesthetized monkey brain. Nature 447, 83–86. ( 10.1038/nature05758) [DOI] [PubMed] [Google Scholar]
- 125.Adachi Y, Osada T, Sporns O, Watanabe T, Matsui T, Miyamoto K, Miyashita Y. 2012. Functional connectivity between anatomically unconnected areas is shaped by collective network-level effects in the macaque cortex. Cereb. Cortex 22, 1586–1592. ( 10.1093/cercor/bhr234) [DOI] [PubMed] [Google Scholar]
- 126.Miranda-Dominguez O, Mills BD, Grayson D, Woodall A, Grant KA, Kroenke CD, Fair DA. 2014. Bridging the gap between the human and macaque connectome: a quantitative comparison of global interspecies structure-function relationships and network topology. J. Neurosci. 34, 5552–5563. ( 10.1523/JNEUROSCI.4229-13.2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Honey CJ, Sporns O, Cammoun L, Gigandet X, Thiran JP, Meuli R, Hagmann P. 2009. Predicting human resting-state functional connectivity from structural connectivity. Proc. Natl Acad. Sci. USA 106, 2035–2040. ( 10.1073/pnas.0811168106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Skudlarski P, Jagannathan K, Calhoun VD, Hampson M, Skudlarska BA, Pearlson G. 2008. Measuring brain connectivity: diffusion tensor imaging validates resting state temporal correlations. Neuroimage 43, 554–561. ( 10.1016/j.neuroimage.2008.07.063) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hermundstad AM, et al. 2013. Structural foundations of resting-state and task-based functional connectivity in the human brain. Proc. Natl Acad. Sci. USA 110, 6169–6174. ( 10.1073/pnas.1219562110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hermundstad AM, et al. 2014. Structurally-constrained relationships between cognitive states in the human brain. PLoS Comput. Biol. 10, e1003591 ( 10.1371/journal.pcbi.1003591) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Krienen FM, Yeo BT, Buckner RL. 2014. Reconfigurable task-dependent functional coupling modes cluster around a core functional architecture. Phil. Trans. R. Soc. B 369, 20130526 ( 10.1098/rstb.2013.0526) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Johnston JM, Vaishnavi SN, Smyth MD, Zhang D, He BJ, Zempel JM, Shimony JS, Snyder AZ, Raichle ME. 2008. Loss of resting interhemispheric functional connectivity after complete section of the corpus callosum . J. Neurosci. 28, 6453–6458. ( 10.1523/JNEUROSCI.0573-08.2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.O'Reilly JX, et al. 2013. Causal effect of disconnection lesions on interhemispheric functional connectivity in rhesus monkeys. Proc. Natl Acad. Sci. USA 110, 13 982–13 987. ( 10.1073/pnas.1305062110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.van den Heuvel MP, Sporns O. 2013. Network hubs in the human brain. Trends Cogn. Sci. 17, 683–696. ( 10.1016/j.tics.2013.09.012) [DOI] [PubMed] [Google Scholar]
- 135.Stam CJ. 2014. Modern network science of neurological disorders. Nat. Rev. Neurosci. 15, 683–695. ( 10.1038/nrn3801) [DOI] [PubMed] [Google Scholar]
- 136.Cocchi L, Harding IH, Lord A, Pantelis C, Yucel M, Zalesky A. 2014. Disruption of structure–function coupling in the schizophrenia connectome. Neuroimage Clin. 4, 779–787. ( 10.1016/j.nicl.2014.05.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhou J, Seeley WW. 2014. Network dysfunction in Alzheimer's disease and frontotemporal dementia: implications for psychiatry. Biol. Psychiatry 75, 565–573. ( 10.1016/j.biopsych.2014.01.020) [DOI] [PubMed] [Google Scholar]
- 138.Luft CDB, Pereda E, Banissy MJ, Bhattacharya J. 2014. Best of both worlds: promise of combining brain stimulation and brain connectome. Front. Syst. Neurosci. 8, 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Deco G, Jirsa VK, McIntosh AR. 2011. Emerging concepts for the dynamical organization of resting-state activity in the brain. Nat. Rev. Neurosci. 12, 43–56. ( 10.1038/nrn2961) [DOI] [PubMed] [Google Scholar]
- 140.Ritter P, Schirner M, McIntosh AR, Jirsa VK. 2013. The virtual brain integrates computational modeling and multimodal neuroimaging. Brain Connect. 3, 121–145. ( 10.1089/brain.2012.0120) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Honey CJ, Kötter R, Breakspear M, Sporns O. 2007. Network structure of cerebral cortex shapes functional connectivity on multiple time scales. Proc. Natl Acad. Sci. USA 104, 10 240–10 245. ( 10.1073/pnas.0701519104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Deco G, Jirsa V, McIntosh AR, Sporns O, Kötter R. 2009. Key role of coupling, delay, and noise in resting brain fluctuations. Proc. Natl Acad. Sci. USA 106, 10 302–10 307. ( 10.1073/pnas.0901831106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hansen ECA, Battaglia D, Spiegler A, Deco G, Jirsa VK. 2015. Functional connectivity dynamics: modeling the switching behavior of the resting state. Neuroimage 105, 525–535. ( 10.1016/j.neuroimage.2014.11.001) [DOI] [PubMed] [Google Scholar]
- 144.Alstott J, Breakspear M, Hagmann P, Cammoun L, Sporns O. 2009. Modeling the impact of lesions in the human brain. PLoS Comput. Biol. 5, e1000408 ( 10.1371/journal.pcbi.1000408) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.de Haan W, Mott K, van Straaten EC, Scheltens P, Stam CJ. 2012. Activity dependent degeneration explains hub vulnerability in Alzheimer's disease. PLoS Comput. Biol. 8, e1002582 ( 10.1371/journal.pcbi.1002582) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Cabral J, Fernandes HM, Van Hartevelt TJ, James AC, Kringelbach ML, Deco G. 2013. Structural connectivity in schizophrenia and its impact on the dynamics of spontaneous functional networks. Chaos 23, 046111 ( 10.1063/1.4851117) [DOI] [PubMed] [Google Scholar]
- 147.Taylor PN, Kaiser M, Dauwels J. 2014. Structural connectivity based whole brain modelling in epilepsy. J. Neurosci. Methods 236, 51–57. ( 10.1016/j.jneumeth.2014.08.010) [DOI] [PubMed] [Google Scholar]
- 148.Goñi J, et al. 2014. Resting-brain functional connectivity predicted by analytic measures of network communication. Proc. Natl Acad. Sci. USA 111, 833–838. ( 10.1073/pnas.1315529111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Abdelnour F, Voss HU, Raj A. 2014. Network diffusion accurately models the relationship between structural and functional brain connectivity networks. Neuroimage 90, 335–347. ( 10.1016/j.neuroimage.2013.12.039) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Mišić B, Sporns O, McIntosh AR. 2014. Communication efficiency and congestion of signal traffic in large-scale brain networks. PLoS Comput. Biol. 10, e1003427 ( 10.1371/journal.pcbi.1003427) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sejnowski TJ, Churchland PS, Movshon JA. 2014. Putting big data to good use in neuroscience. Nat. Neurosci. 17, 1440–1441. ( 10.1038/nn.3839) [DOI] [PMC free article] [PubMed] [Google Scholar]