Abstract
Pulmonary arterial hypertension (PAH) is a disease characterized by increased pulmonary vascular resistance and remodeling. Increase in the population of vascular smooth muscle cells is among the key events contributing to the remodeling. Endothelin-1 (ET-1), a potent vasoconstrictor, is linked to the etiology and progression of PAH. Here we analyze changes in protein expressions in response to ET-1 in pulmonary arterial smooth muscle cells (PASMC) from a healthy Control (non-PAH) and a PAH subject presenting a bone morphogenetic protein type II receptor (BMPR2) mutation with exon 1–8 deletion. Protein expressions were analyzed by proteomic mass spectrometry using label-free quantitation and the correlations were subjected to Ingenuity™ Pathway Analysis. The results point to eIF2/mTOR/p70S6K, RhoA/actin cytoskeleton/integrin and protein unbiquitination as canonical pathways whose protein expressions increase with the development of PAH. These pathways have an intimal function in the PAH-related physiology of smooth muscle proliferation, apoptosis, contraction and cellular stress. Exposure of the cells to ET-1 further increases protein expression within these pathways. Thus our results show changes in signaling pathways as a consequence of PAH and the effect of ET-1 interference on Control and PAH-affected cells.
Keywords: label-free quantitation, proteomic mass spectrometry, pulmonary arterial hypertension, signal transduction, endothelin-1, BMPR2
1. Introduction
Pulmonary arterial hypertension (PAH) is a fatal disorder of the pulmonary vasculature. PAH is progressive with very limited therapeutic success. Although of variable etiology including idiopathic and heritable forms of PAH, such as that characterized by deletions in the bone morphogenic protein type II receptor exon, the histological appearance of the lung tissue in all forms of PAH is generally similar, involving intimal fibrosis, increased medial thickness, increased proliferation and constriction of smooth muscle cells (SMC).1 The pathogenesis exhibits a combination of vasoconstriction and inward vascular wall remodeling.
The gene for the bone morphogenetic protein type II receptor (BMPR2) is part of a family of genes which regulate growth and differentiation. The role of BMP in vascular diseases has been reviewed recently.2 Although this family clearly regulates the growth and differentiation of numerous types of cells in various tissues, little is yet understood as to how mutations within the BMPR2 gene lead to pulmonary arterial hypertension. Approximately 10% of PAH cases have a familial origin. In these families, the disease segregates in an autosomal dominant pattern. Approximately 20–30% of individuals who carry BMPR2 mutations develop PAH. Although many patients who carry the BMPR2 deletion allele do not manifest clinical PAH, the relative risk for PAH in individual carrying a BMPR2 mutation is, nevertheless, in the order of 105 compared with a carrier of the wild-type allele. Clearly an additional factor is required for the disease to manifest itself in these cases. At this time, little is understood regarding the underlying requirements for this manifestation.
Endothelin-1 receptor ETA/ETB blockers have been utilized clinically to combat PAH.3 As is usual for receptor antagonists, they block all receptor signaling, regardless of positive or negative physiologic contribution. Little is yet understood as to the specific signaling cascades directly engaged in the response to treatment of PAH by the antagonists. An additional problem is that the ET-1 antagonists contribute to congestive heart failure. Thus, determination of factors and their pathways participating in the etiology of PAH is critical.
Herein we explore and compare protein expression in PASMCs derived from a Control (non PAH) lung donor and a PAH-afflicted individual with a BMPR2 mutation. We investigate the intercalation of this condition with the effects of ET-1, in terms of protein expression. We utilize a combination of gel-free enzyme digestion and label-free quantitative mass spectrometry analysis. Until very recently such studies employed proteomic approaches which included a combination of 2D-gel electrophoresis, image analysis, labeling strategies and mass spectrometry.4–9 These techniques require expensive isotope labels, specific software, and a high level of expertise to analyze the data. In addition, the number of samples that can be analyzed by this procedure in a single experiment is limited and labeling strategies are difficult to apply. For the study whose results are reported in this communication, we used a newly developed system that incorporates an unbiased screening procedure based on label-free quantitative liquid chromatography linked with mass spectrometry. This approach provides us with a much broader map of protein expression. The procedure is applicable to tissues, cultured cells, and fluids.10–12 Using this technique, we find a number of signaling changes taking place which are related to pulmonary arterial smooth muscle cell migration, apoptosis, proliferation and contraction (all PAH-associated functions).
2. Materials and Methods
2.1. Isolation and culture of human pulmonary artery smooth muscle cells (PASMC)
Control and PAH PASMCs were derived at the Cleveland Clinic Foundation. The Control PASMCs were from a 36-year-old male donor with no history of pulmonary or cardiac disease symptoms; the PAH PASMCs were from a 41-year-old male patient with a defined BMPR2 mutation (Exon 1–8 Deletion).13 The cells were isolated from elastic pulmonary arteries (>500-µm diameter) dissected from lungs obtained at explantation during lung transplant. Briefly, after removal of endothelial cells, PASMCs were dissociated by digestion with collagenase type II/DNase I solution overnight at 37 °C.14 Cells were cultured in 15 mM HEPES buffered DMEM/F12 (50:50) media (Mediatech, Manassas, VA) containing 10% fetal bovine serum (FBS) (Lonza), and 2.5% Antibiotic-Antimycotic from GIBCO (Invitrogen, Grand Island, NY, cat. no. 15240). Cells were passaged at 60–90% confluence by dissociation from plates with 0.05% trypsin and 0.53 mM EDTA. The smooth muscle phenotype of cultured cells was confirmed (>97% purity) by immunohistochemistry and flow cytometric analysis with antibodies against smooth muscle α-actin and calponin.15 Primary cultures of passages 6–10 were used in experiments. Supplementary Figure 1A demonstrates the alpha smooth muscle actin staining in the cultured cells.
The designations Control (+/−) ET-1 and PAH (+/−) ET-1 have been used throughout the text, as sample names for four groups of PASMCs derived from the healthy control and BMPR2 mutation PAH patient, treated with/without ET-1 stimulation. Approval to use these human cells was granted by the Boston University Institutional Review Board.
2.2 Sample Preparation
PASMCs derived from the Control and PAH subjects were treated with and without 10 nM endothelin-1 overnight (16 h). The cells were washed twice with ice-cold PBS. Cell lysates were prepared by addition of Urea Lysis Buffer (8 M urea, 50 mM HEPES buffer pH 7.5, 75 mM NaCl supplemented with protease and phosphatase inhibitors) and centrifuged at 25,000 rpm in a microcentrifuge at 4 oC for 20 min. The proteins were quantified with the BCA assay from Pierce (Rockford, IL). Cell lysates were then subjected to trypsin digestion. The lysate aliquots containing 50 µg of protein were reduced with 5 µL of 200 mM dithiothreitol (DTT) in 50 mM ammonium bicarbonate at room temperature for 1 h with gentle vortexing. For alkylation, 4 µL of 1 M iodoacetamide in 50 mM ammonium acetate was added and the mixture was incubated in the dark at room temperature for 1 h, and then excess iodoacetamide was reacted with DTT for 1 h at room temperature. After the urea concentration was reduced to 0.8 M, 1 µg of trypsin (Promega, Madison, WI) was introduced into the PASMC solution in the presence of 1 mM CaCl2, and the mixture was incubated overnight at 37 oC, followed by addition of 0.5% trifluoro acetic acid (TFA) to terminate the reaction. The peptide mixture was dried and then was desalted using an OMIX C18 tip (100 µL, Varian Medical System, Palo Alto, CA) with 100 µL elution buffer of 70% acetonitrile and 0.1 % TFA. The treated samples were dried, then stored at −20 oC for future analysis. All samples were simultaneously processed through all the steps including cell lysis, protein concentration assay, protein digestion and quantitative analysis, to minimize the variation between samples.
2.3 Mass spectrometry analysis
Lyophilized peptides from digested PASMC cell lysates were reconstituted in 40 µL of 1% acetonitrile and 0.1% formic acid solution, and 1 µL of this solution was analyzed with an LC-MS/MS system consisting of a nanoAcquity ultra-performance liquid chromatograph (Waters Inc., Milford, MA), coupled to an LTQ-Orbitrap hybrid mass spectrometer (ThermoFisher Scientific, San Jose, CA) equipped with a TriVersa NanoMate ion source (Advion, Ithaca, NY). Sample concentration and desalting were performed online using an in-house-packed polymicro trapping column (2 cm, packed with 5-µm particle-size, 200-Å pore size, reversed phase MagicC18AT material) at a flow rate of 15 µL/min for 1 min. Separation was carried out in an in-house-packed UPLC® capillary column (10 cm, packed with 3-µm particle size, 100-Å pore size, reversed phase MagicC18AT material), equipped with frits made with Kasil:formamide at ratio of 5:1. A linear gradient of A and B buffers (A: 99.9% acetonitrile−0.1% formic acid; B: 99.9% H2O−0.1% formic acid) from 3% to 40% buffer B over 87 min was used at a flow rate of 0.5 µl/min to elute peptides into the mass spectrometer. The columns were washed and re-equilibrated between runs. Electrospray ionization was carried out at 1.7 kV using the NanoMate, with the LTQ heated capillary set to 150 °C. Mass spectra were acquired in the Orbitrap in the positive-ion mode over the range m/z 300–2000. Mass accuracy after internal calibration was within 4 ppm. Simultaneously, tandem MS spectra were acquired using the LTQ for the three most abundant multiply-charged ions in each mass spectrum with signal intensity threshold > 50,000 x noise level. MS/MS collision energies were set at 35%, and helium was used as the collision gas. The MS/MS spectra were scanned over an m/z range dependent on the precursor ion. Dynamic exclusion was set such that the MS/MS data for each species were acquired a maximum of two times. Each Control (+/−) ET-1 and PAH (+/−) ET-1 sample was run with three analytical replicates for the purpose of label-free quantitation. LC-MS/MS spectra were recorded in the .raw profile mode for further processing and analysis. MS/MS spectra were then searched against the SwissProt human database (version 57.15) by Mascot 2.3.2. (Matrix Science). The search was performed using the following parameters: (1) enzyme: trypsin; (2) maximum number of missed cleavages: 2; (3) peptide ion mass tolerance: 5 ppm; fragment ion mass tolerance 0.5 Da. (4) variable modification: carbamidomethylation of cysteine (C); deamidation of asparagine and glutamine (NQ); oxidation of methionine (M). The search results for protein identification were validated using the post-analysis software package Scaffold_3 (Proteome Software Inc. Portland, OR, USA). Peptide identifications were accepted for ions with Mascot scores ≥ 20. Protein identifications were accepted when at least two unique peptides were identified and the significance threshold was p < 0.05.
2.4 Label-free quantitation of proteins by Progenesis LC-MS
All 12 LC-MS/MS data sets from Control (+/−) ET-1 and PAH (+/−) ET-1, each with three analytical replicates, were loaded and analyzed for label-free profiling and quantification in Progenesis LC-MS/MS (Nonlinear, version 4.0). The MS spectra from different experimental conditions were aligned over time to maximize overlap of the 2D feature maps, and then were mined for features in the data (m/z and retention time) that differed across the experimental conditions. Peptides observed within the range m/z 300 – 1700, elution time between 14 – 92 min with charge states of 2+ – 7+, were selected for quantitation and identification. The technical replicates of each sample were then grouped: Control (−) ET-1, Control (+) ET-1, PASM (−) ET-1 and PASM (+) ET-1. The statistical analyses were performed using normalized abundances for one-way analysis of variance (ANOVA) calculations of all the detected features. A combined peak list of peptides was generated containing MS/MS spectra from all 12 LC-MS/MS runs, and submitted for peptide identification search in Mascot, when their abundances showed statistically significant differences. Tandem mass spectra from these peptides were then searched using Mascot as described above. Search results were then analyzed in Progenesis LC-MS. Identified peptides (with Mascot ion score ≥ 20, and mass error < 5 ppm) of an identified protein were included and the total cumulative abundances were calculated by summing the abundances of all peptides assigned to the respective protein. All features identified as part of a given protein were used for each respective protein quantification. Calculations of the protein p-value (one-way ANOVA) were then performed on the sum of the normalized abundances across all runs. Fold changes were converted from protein abundances in pair-comparison mode, and the up/down regulations of protein expression were indicated by positive/negative values of fold changes. Proteins with ANOVA p-value < 0.05, and net changes of > 1.33-fold across two samples were regarded as significantly changed for further analysis.
2.5. Western Blot Analysis
PASMCs derived from the Control and PAH subjects were treated with and without 10 nM endothelin-1 overnight (16 h). To demonstrate the variation among biological repeats, all samples were processed and analyzed in triplicate. After treatment, the cells were washed twice with ice-cold PBS. Cell lysates were prepared by addition of ice-cold RIPA buffer, 150 mM NaCl, 1.0% Igepal CA-630, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris, pH 8.0 and 1x complete protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN) and centrifuged at 12,000 rpm in a microcentrifuge at 4 oC for 20 min. The proteins were quantified with the BCA assay from Pierce (Rockford, IL). The proteins (15 µg) were fractionated on 10% SDS-PAGE gels and Western blots were carried out using antibodies against Ezrin (Cell Signaling Technologies, Beverly, MA), or Rho (Millipore, Billerica, MA). Proteins were detected by chemiluminescence and the film scanned with an Epson Perfection 3170 scanner using Epson Scan (version 1.22A) software. The image was then analyzed using the NIH ImageJ image analysis software to determine the intensity of each band. The blot was stained with Ponceau S (Sigma, St Louis, MO) to demonstrate even loading (Supplementary Figure 1B).
2.6. Signaling pathway analysis
Functional and canonical Pathway Analysis was performed using Ingenuity Pathway Analysis software (IPA) (Ingenuity® Systems, www.ingenuity.com). From label-free quantitation LC-MS/MS analysis of Control and PAH PASMC with/without ET-1 stimulation, a fold change cut-off of ±1.33 in expression level was selected to define up/down regulated proteins. Proteins from the dataset were filtered with this standard and were considered for analysis. The dynamic Canonical Pathways generated by IPA have been curated and hand-drawn from specific journal articles, review articles, text books, and KEGG Ligand. The significance of the association between the dataset and the canonical pathway was measured in two ways: 1) A ratio of the number of proteins from the dataset that meet the expression value cutoff and map to the pathway, divided by the total number of molecules that exist in the canonical pathway, is displayed. The ratio indicates the percentage of genes in a pathway that are found in the uploaded mass spectrometry data set. The ratio is therefore useful for determining which pathways show the greatest overlap with the proteins in the dataset. 2) Fisher‘s exact test is used to calculate a p-value determining the probability that the association between the proteins in the dataset and the canonical pathway is explained by chance alone. This p-value is a measure of the likelihood that the association between a set of Pathway Analysis proteins in the mass spectrometry experiment and a given pathway is due to random chance. The smaller the p-value, the less likely it is that the association is random and the more significant the association. The canonical pathways were combined into one network diagram to simplify the representation, as described in the results section.
3. Results
This is a study of protein expression by human Control and PAH pulmonary artery smooth muscle cells (PASMCs) under basal and endothelin-1 stimulated conditions. Under the ET-1 unstimulated conditions, this approach provided the opportunity to compare the protein expressions in Control and patient PASM cells to detect alterations that may be attributed to PAH. Stimulation with ET-1 then provided the further opportunity to determine the effect of endothelin-1 on this process. ET-1 is an important component in the treatment of PAH. Thus, comparison of the global effects of ET-1 on protein expression by Control and BMR2 PASMC provides important insights which could prove clinically useful. Label-free differential quantitation identified 561 proteins from tryptic digest lysates of PASMC with LC-MS/MS proteomic analysis. Differences in the relative fold change (>1.33 or <−1.33) of these proteins were used in pair-comparisons of Control and PAH cells (+/−) ET-1. IPA pathway analysis was chosen to facilitate interpretation of the protein expression changes detected by the LC-MS/MS analyses.
3.1. Proteomic profiling in human pulmonary arterial smooth muscle cells
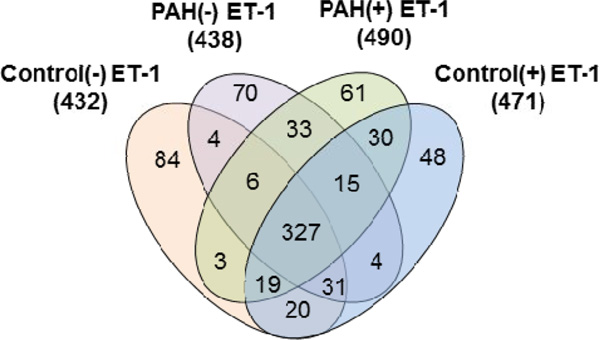
With triplicate LC-MS/MS runs for each Control (+/−) ET-1 and PAH (+/−) ET-1 sample, an average of 7500 MS/MS spectra was acquired from each sample, and these were searched for human proteins in Mascot. From the tryptic PASMC digests, 430–490 proteins (without grouping) were observed with 0.1% protein false discovery rate (FDR). As shown in Figure 1, the majority of these proteins were found in Control and PAH, regardless of ET-1 treatment. Three hundred twenty-seven proteins are represented in the 4-way Venn diagram (Figure 1), whereas 10–20% of the identified proteins were unique to each sample. Exposure of both Control and PAH cells to ET-1 resulted in the detection of about 10% more proteins specific to this stimulation. Apparently, ET-1 also caused 19 more proteins to be expressed in the cells from the PAH patient, as compared to Control cells. Each protein detected was linked to a biological process, cellular location and molecular function based on information obtained from the SwissProt, Entrez Gene, Genome Ontology (GO) databases and in-house-built annotation software, STRAP 16.
Figure 1.
Comparison of MS-identified proteins in Control/PAH PASMCs with the effect of ET-1 stimulation.
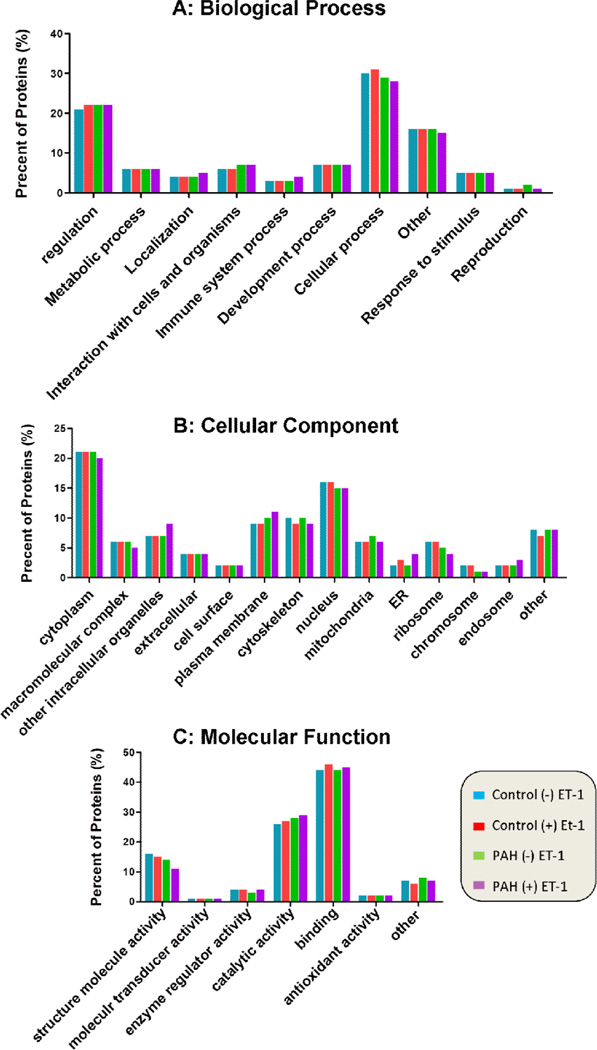
Figures 2A to 2C show similar patterns of protein distributions in PASMC from Control (+/−) ET-1 and PAH (+/−) ET-1. The majority of proteins are involved in cellular process (30%) and regulation (20%). A wide range of cellular locations was assigned to those proteins identified from the trypsin-digested PASMC lysate under study, including cytoplasm (20%), nucleus (17%), plasma membrane (10%) and cytoskeleton (10%). It was found that dominant proteins identified might be involved in binding and catalytic activity, and about 15% of these proteins may play structural roles.
Figure 2.
Distribution of the proteins identified in PASMCs according to (A) their biological processes, (B) their cellular components, and (C) their molecular
3.2. Label-free differential quantitation
The digested PASMC proteins, from Control and PAH with (+/−) ET-1, were analyzed by LC-MS/MS in the top-3 ion mode MS/MS profile to get sufficient data points to evaluate ion abundances in the MS1 scan. The peptide ion intensity measurements integrate the peak area, which is proportional to the relative peptide concentration in the sample. The signal intensities of ions assigned to the defined peptides have been shown to correlate with the abundance of a given protein in that sample. Determining the area for each mass-extracted peptide ion chromatogram/ retention time pair and comparing the areas between multiple LC-MS runs of different samples provides comprehensive differential quantification of thousands of peptides within a sample. Label-free quantitation by the Progenesis LC-MS program provided analysis and identification of differential protein changes correlated to ET-1 stimulation. Fold changes were calculated in four pair comparisons, to evaluate: 1) ET-1 stimulation in Control PASMC (Control (+) ET-1 vs. Control (−) ET-1): this comparison profiles the effect of ET-1 in the Control PASMC; 2) ET-1 effect on PAH PASMC (PAH (+) ET-1 vs. PAH (−) ET-1): this measures the effect of ET-1 in the BMPR2 mutated PAH cells; 3) protein expression difference in PAH and Control (PAH (−) ET-1 vs. Control (−) ET-1): this measures the differences of protein expression in PAH and Control PASMC under basal conditions; and 4) difference in ET-1 effect in PAH against Control (PAH (+) ET-1 vs. Control (+) ET-1): this comparison illustrates the combining effects of different cell types and ET-1 stimulation.
After alignment and normalization in Progenesis LC-MS, 38,113 features with fold change ≥ 2 and ANOVA p-value ≤ 0.05, were selected for LC-MS quantitation. The relevant data (51,232 MS/MS spectra) were exported for searching by Mascot, and 4,778 peptide sequences were identified as good matches according to the search criteria (mass errors: MS1 ≤ 5 ppm, MS2 ≤ 0.5 Da, ion score ≥ 20). After peptide conflicts in protein origins were resolved by manual inspection, 580 out of 1,137 proteins were identified on the basis of detection of at least 2 unique peptides. The assigned proteins were further analyzed for their interactions and correlation with ET-1 using Ingenuity pathway analysis (IPA). Protein expression differences were calculated as fold changes in pairwise comparisons, based on the sum of their assigned peptide ion abundances. In IPA analyses, 561 out of the 580 differentially expressed proteins were determined to show a minimum fold change of ± 1.33 or greater. As shown in Table 1, ET-1 stimulation in the Control cells resulted in the up-regulation of 472 proteins. Sixty of these were up-regulated by more than 2-fold. The most significantly changed protein in the Control cells was chromosome 14 open reading frame 166 (C14orf166), with the fold change of 4.52 following ET-1 stimulation. In comparison, ET-1 treatment induced up-regulation of 552 proteins in the PAH cells. Three hundred ninety of these showed a fold change > 2. The WD repeat domain 33 protein (WDR33) showed the greatest change at 7.18-fold. In the pairwise comparison between PAH (−) ET-1 vs. Control (−) ET-1, about half of the identified proteins showed no change, while 210 were up-regulated and 61 were down-regulated in the PAH cells.
Table 1.
Summary of expression changes of 561 proteins identified in Control and PAH-BRMP2 PASMCs, in response to elevation in ET-1, as detected by label-free LC/MS quantitation.
| Fold change | Protein Numbers | |||
|---|---|---|---|---|
| Control(+) ET-1 vs. Control(–) ET-1 |
PAH (+) ET-1 vs. PAH (–) ET-1 |
PAH (–) ET-1 vs. Control(–) ET-1 |
PAH (+) ET-1 vs. Control(+) ET-1 |
|
| Up-regulated(>2) | 60 | 390 | 30 | 137 |
| Up-regulated (2 to1.33) | 412 | 162 | 180 | 312 |
| Down-regulated (−2 to −1.33) | 0 | 0 | 45 | 10 |
| Down-regulated (<−2) | 0 | 0 | 16 | 10 |
| No change (within ±1.33) | 89 | 9 | 290 | 92 |
Supplementary Table 1 shows fold differential expression of proteins calculated as pairwise comparisons. Clearly ET-1 treatment stimulates the expression of a wide range of proteins in PASMC. Additionally, their responses in the BMPR2 mutated PAH cells are quite different from those in the Control cells. For example, alkaline phosphatase (ALPL) in PAH was up-regulated 6.33 fold compared to that in the Control. ET-1 stimulation resulted in the up-regulation of this protein to a fold change of 9.75. In contrast, ET-1 had nearly no influence in the Control cells. The responses of glutaminase (GLS) to ET-1 stimulation were in the same level in either Control or PAH subject. The collagen type 1 protein (COL1A1) was down-regulated in PAH compared to Control alone, but ET-1 increased its expression to about 2-fold in the PAH cells, but only 1.24-fold in the Control. Most of the significantly changed proteins listed in Supplementary Table 1 are located in the cytoplasm.
3.3. Western blot to validate proteomics findings
To confirm protein changes found by Mass Spectrometry, Western blots were performed. We chose two important proteins, ezrin and RhoA, which are significant to the biology of this system (as discussed in the Discussion section). The Western blot results are shown in Figure 3. To illustrate reproducibility, each sample was run in triplicate as a separate cell incubation. We used ImageJ to quantify the band density. To ensure even loading on the gel, the BCA method was used to measure protein concentration, and 15 µg protein was loaded into each well. In addition, Ponceau S staining was used to normalize the protein level. Table 2 shows the amount of each protein determined by both Western blot and Mass Spectrometry. The amount of protein in Control cells under basal condition (no ET-1 stimulation) was set as 1. Figure 3 illustrates good reproducibility among the three biological repeats. Table 2 confirms that the same trend is detected by Western blot and Mass Spectrometry analyses.
Figure 3.
Erzin and RhoA protein expression in PASMC from normal Controls and PAH-BRMP2 subjects (HPAH) treated with or without 10 nM ET-1. Ponceau S staining was used to normalize the protein level (Supplementary Figure 1B).
Table 2.
Comparison of the results from Western blot and proteomic Mass Spectrometry. The density of the protein bands was quantitated using ImageJ software. The table shows the mean of the density determined from three biological repeated samples.
| Control Basal |
Control ET-1 |
PAH-BRMP2 Basal |
PAH-BRMP2 ET-1 |
||
|---|---|---|---|---|---|
| Ezrin | Western | 1 | 1.78 * | 2.80# | 3.93 * |
| Proteomics | 1 | 1.75 * | 1.66# | 3.39 * | |
| RhoA | Western | 1 | 1.94 * | 1.33 | 6.25 * |
| Proteomics | 1 | 1.38 * | 1.13 | 2.38 * | |
Significant difference between ET-1 and basal, (p<0.05)
Significant difference between PAH-BMPR2 and Control under basal condition (p<0.05)
3.4. Biological pathways
To gain insight into the complex pathophysiology of the BMPR2-related PAH, we analyzed the proteomic data using the Ingenuity Pathway Analysis software. Reciprocal fold changes of protein expression were computed for the entire expression data set and uploaded for IPA core analysis. Four comparisons with assessed biological importance were conducted for the canonical pathway analysis, as described in the previous section.
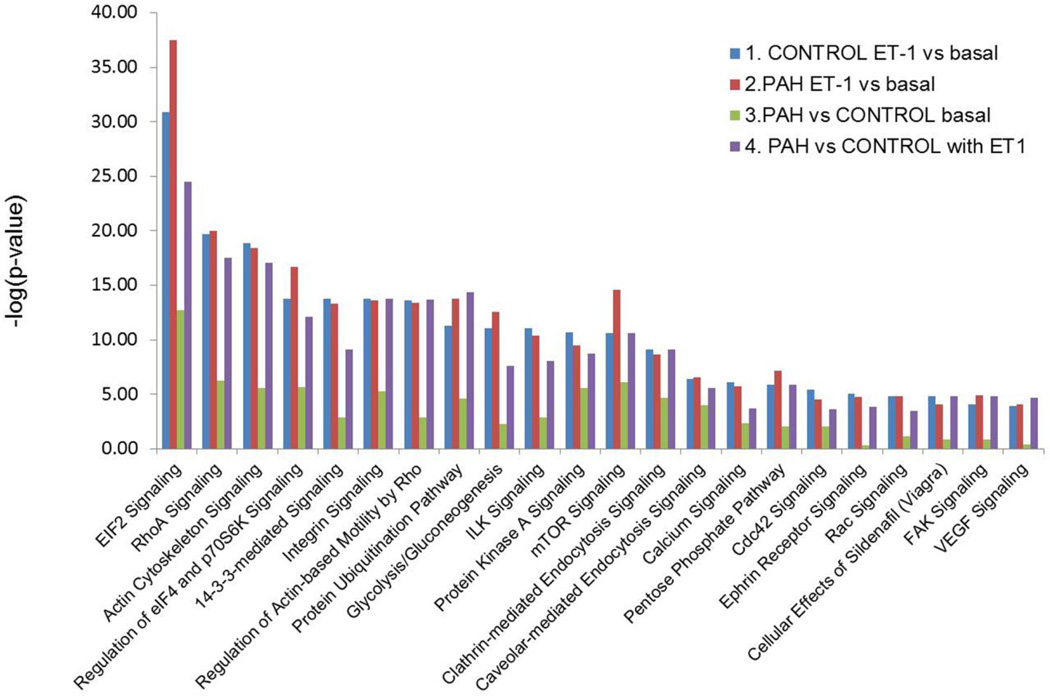
As illustrated in Figure 4, the most statistically significant canonical signaling pathways perturbed in PAH or responding to ET-1 stimulation, are EIF2, RhoA and actin cytoskeleton associated signaling. Other PAH-participating pathways are regulation of eIF4 and p70S6K signaling, 14-3-3-mediated signaling, integrin signaling, regulation of actin-based motility by Rho, protein ubiquitination, glycolysis/gluconeogenesis, ILK signaling, protein kinase A signaling, mTOR signaling, clathrin-mediated endocytosis signaling, caveolar-mediated endocytosis signaling, calcium signaling, pentose phosphate pathway, cdc42 signaling, ephrin receptor signaling, Rac signaling, cellular effects of sildenafil, FAK (focal adhesion kinase-1) signaling and VEGF (vascular endothelial growth factor) signaling. The number of proteins involved in each of these signaling pathways is summarized in Table 3. The total numbers of proteins participating in these signaling pathways based on the Ingenuity knowledge base are also listed in the table. Detailed expression changes of the proteins involved in these pathways are illustrated in Supplementary Table 2. The primary PAH and ET-1 related pathways are described in the discussion section. Based on this comparison analysis of the canonical pathways (Figure 3) when PAH vs. Control are compared in the absence of ET-1, their difference is not as pronounced as when ET-1 stimulation is compared (comparison 3 vs. comparison 1 or comparison 2). When the cells are stimulated with ET-1, the phenotype of PAH becomes marked, in comparison to Control (comparison 4 vs. comparison 3). This outcome indicates a compounding effect of ET-1 stimulation on PAH development.
Figure 4.
Summary of the canonical pathways predicted by Ingenuity pathway analysis in the four-way comparisons. 1: ET-1 treated vs. basal in Control PASMC; 2. ET-1 treated vs. basal in PAH PASMC; 3. PAH cells vs. Control cell under basal conditions; 4. PAH vs. Control cells both with ET-1 stimulation. Canonical pathways analysis identified the pathways, from the Ingenuity Pathways Analysis™ library of canonical pathways, that were most significant to the dataset. Proteins from the dataset that met the fold change cutoff of 1.33 and were associated with a canonical pathway in the Ingenuity knowledge base were considered for analysis. The significance of the association between the dataset and the canonical pathway was determined as a p-value calculated by Fisher's exact test, a measure of the probability that the association between the proteins in the dataset and the canonical pathway is taking place by chance alone.
Table 3.
Number of proteins involved in the canonical signaling pathways examined in the four comparisons: 1: ET-1 treated vs. basal in Control PASMC; 2. ET-1 treated vs. basal in PAH-BMPR2 PASMC; 3. PAH-BMPR2 cells vs. Control cells under basal conditions; 4. PAH-BMPR2 vs. Control cells, both with ET-1 stimulation.
| Ingenuity Canonical Pathways |
Number of proteins identified |
Total number of proteins in IPA pathway |
|||
|---|---|---|---|---|---|
| Control ET-1vs. basal |
PAH-BMPR2 ET-1 vs. basal |
PAH-BMPR2 vs. Control basal (no ET-1) |
PAH-BMPR2 vs. Control with ET-1 |
||
| EIF2 Signaling | 47 | 56 | 22 | 41 | 222 |
| RhoA Signaling | 28 | 30 | 11 | 26 | 114 |
| Actin Cytoskeleton Signaling |
36 | 38 | 14 | 34 | 238 |
| Regulation of eIF4 and p70S6K Signaling |
26 | 31 | 12 | 24 | 179 |
| 14-3-3-mediated Signaling | 23 | 24 | 7 | 18 | 120 |
| Integrin Signaling | 29 | 31 | 13 | 29 | 210 |
| Regulation of Actin-based Motility by Rho |
20 | 21 | 6 | 20 | 91 |
| Protein Ubiquitination Pathway |
30 | 36 | 14 | 34 | 274 |
| Glycolysis/Gluconeogenesis | 20 | 23 | 6 | 16 | 134 |
| ILK Signaling | 25 | 26 | 9 | 21 | 193 |
| Protein Kinase A Signaling | 32 | 33 | 17 | 29 | 328 |
| mTOR Signaling | 25 | 32 | 14 | 25 | 210 |
| Clathrin-mediated Endocytosis Signaling |
21 | 22 | 11 | 21 | 172 |
| Caveolar-mediated Endocytosis Signaling |
12 | 13 | 7 | 11 | 85 |
| Calcium Signaling | 18 | 19 | 8 | 14 | 207 |
| Pentose Phosphate Pathway | 10 | 12 | 4 | 10 | 82 |
| Cdc42 Signaling | 16 | 16 | 7 | 13 | 180 |
| Ephrin Receptor Signaling | 16 | 17 | 3 | 14 | 200 |
| NRF2-mediated Oxidative Stress Response |
16 | 20 | 5 | 16 | 192 |
| Rac Signaling | 12 | 13 | 4 | 10 | 123 |
| Cellular Effects of Sildenafil (Viagra) |
14 | 14 | 4 | 14 | 151 |
| FAK Signaling | 10 | 12 | 3 | 11 | 102 |
| VEGF Signaling | 10 | 11 | 2 | 11 | 99 |
| Thrombin Signaling | 15 | 16 | 6 | 16 | 207 |
| PI3K/AKT Signaling | 11 | 12 | 3 | 9 | 140 |
| Hypoxia Signaling in the Cardiovascular System |
7 | 7 | 3 | 7 | 68 |
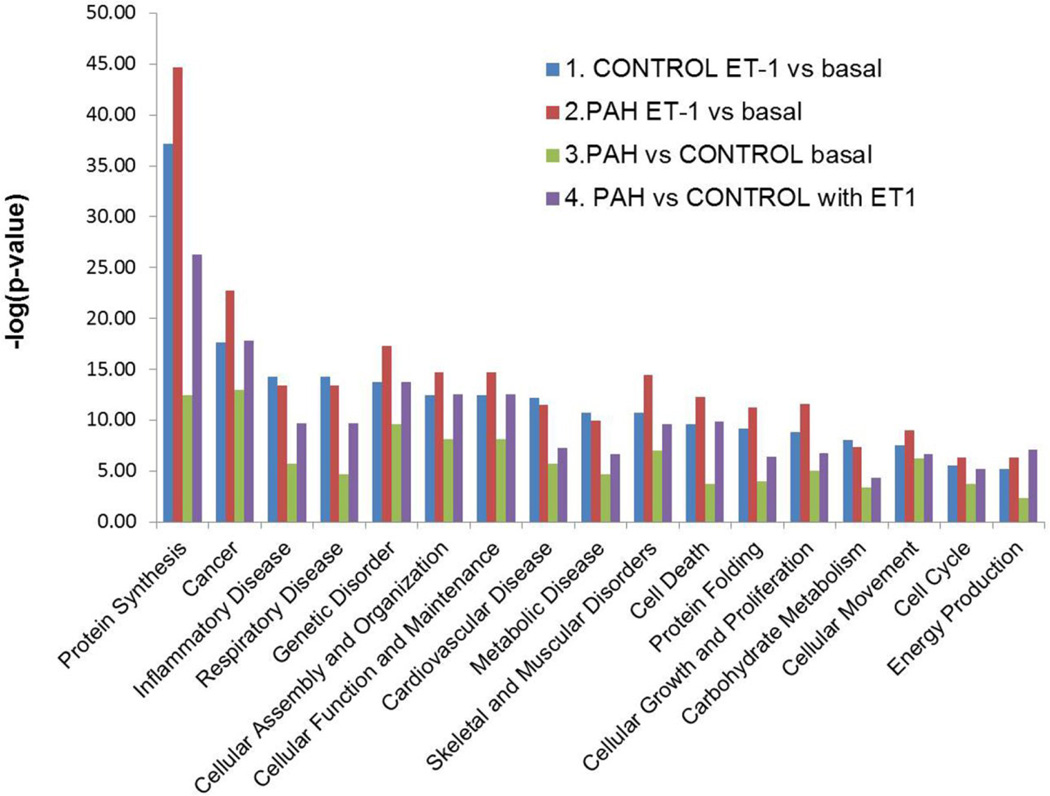
The IPA analysis of biological function also clearly shows that the protein expressions which change in the PAH cells affect cellular protein synthesis, death, proliferation, movement, and the production of free radicals and their scavengers (Figure 5). These events are all critical in the etiology of PAH and are related to respiratory disease and cardiovascular disease.
Figure 5.
Summary of the biological functions generated by Ingenuity Pathway Analysis in the 4-way comparisons. The Biological Functional Analysis™ identified the biological functions and/or diseases that are most significant to the data set. Fischer's exact test was used to calculate a p-value determining the probability that each biological function and/or disease assigned to that data set is
4. Discussion
We have examined protein expression differences and their relevant signaling pathways in PASMCs derived from a healthy control (Control) and a BMPR2 mutation PAH-afflicted patient (PAH), using label-free quantitative proteomic mass spectrometry and IPA analysis.
4.1. Detection of PASMC proteins
With our gel-free approach, we could assign 430–490 proteins in the trypsin-digested human PASMC lysate from Control and PAH human cells to evaluate the effect of ET-1 stimulation. For their ET-1 relevant study that used 2D-gels, in-gel digestion of spots that showed ≥ 5-fold intensity changes, and MALDI-TOF MS for peptide mapping, Yang et al. had reported that 27 proteins were up- or down-regulated by ET-1 and then differentially affected by the selective KATP channel opener iptakalim in normal human pulmonary arterial smooth muscle cells, but these authors did not provide a list of the proteins that differed between untreated and ET-1 stimulated cells.6 As shown in Figure 2, Dupont et al. characterized 50 proteins as intracellular proteins expressed in human arterial smooth muscle cells through gel-based protein analysis.8 Ten percent of the proteins identified in the latter study fall into the cytoskeletal class. For example, gelsolin (GSN) controls the polymerization/depolymerization of architectural filaments, and is an effector of apoptosis in smooth muscle cells.17 Caldesmon (CALD1) is an actin- and myosin-binding protein which regulates vascular smooth muscle tone.18 Calponin H2 (CNN2) is a thin filament-associated protein identified in the regulation and modulation of smooth muscle contraction. Heat shock protein β-1 (HSPB1) is expressed in human PASMCs and is involved in stress resistance and actin organization.19,20 Additionally, the cytoskeletal structure proteins moesin (MSN), a marker of SMC migration,21 and the contractile and adhesion protein transgelin (TAGLN) were detected in human PASMCs. These proteins are potential markers for the characterization of PASMC during PAH development.
In their study, Godovac-Aimmermann et al. used pulsed [35S] methionine labeling and 2D electrophoresis proteomic analysis to look at the response of newly synthesized proteins in human lung fibroblasts upon stimulation by endothelin-1.7 They found about 70 proteins responsive to ET-1 stimulation. Among the 35 proteins they reported to exhibit the most pronounced change in expression, we found that 21 proteins were upregulated in the Control PASMC responding to ET-1, such as tubulin (tubulin β-1 chain), intermediate filaments (vimentin), and microfilaments (vinculin, filamin, α-actin), all proteins involved in cytoskeletal actions. It is known that cytoskeletal actions are important in a wide variety of cellular process including signaling systems, cell-cell interactions, and mitosis. The structural protein, vinculin, has been used as a standard in proteomic analysis of entire lung tissue from various patients with PAH.22 Our data, reported in this communication, indicated that the abundance of vinculin was similar in Control and PAH PASMC cells (fold change: 1.17). ET-1 stimulation overnight caused vinculin upregulation at 1.67-fold in Control and 2.05-fold in PAH cells. Two proteins, PDZ and LIM domain 1 (PDLIM1, fold change: −17.37) and alcohol dehydrogenase 1B (ADH1B, fold change: −41.79) showed extremely low abundances in PAH as compared to Control cells. These proteins also showed little response to ET-1 stimulation in both cell populations. Detailed data can be seen in Supplementary Table 1.
A number of the identified proteins are chaperones known to mediate folding of other proteins. For example, disulfide-isomerase A6 (PDIA6) inhibits aggregation of misfolded proteins in the platelet activation pathway.23 The 60-kDa heat shock protein, HSPD1, has been identified in vascular diseases. Puccetti and coworkers24 reported that inflammatory immunological reactions against HSPD1 activate human arterial smooth muscle cell functions relevant to the development of atherosclerosis. Human PASMCs also express serpin H1 (SERPINH1) which functions as a chaperone in the intracellular processing and secretion of procollagens. Human PASMCs express enzymes involved in metabolic processes, such as glycolytic enzymes including phosphoglycerate mutase 1 (PGAM1) and UDP-glucose-6-dehydrogenase (UGDH); and 26S protease regulatory subunits which are involved in the ATP-dependent degradation of ubiquitinated proteins. Oxidative stress is an important factor in the pathogenesis of vascular diseases.25 Proteins involved in cellular antioxidant activity were detected in the human PASMCs, including enzymes which regulate cellular redox state, such as peroxiredoxin 1 (PRDX1), thioredoxin reductase 1 (TXNRD1) and glutathione-S-transferase omega-1 (GSTO1).
4.2. Signaling pathways and individual proteins likely involved in the etiology of PAH
The results from Ingenuity analysis of the two most prominent signaling pathways involved in PAH, the eIF2, p70S6K/eIF4, mTOR cascades and RhoA signaling, Cytoskeleton organization and muscle contraction were selected for in-depth presentation here. Several other PAH-altered pathways, Protein Ubiqitination, Glycolysis/Gluconeogenesis and the Pentose Phosphate Pathway were included in the analysis. We also examined expression of certain individual proteins such as the HSPs (heat shock proteins) and Heme oxygenase 1 (HMOX1).
4.2.1 eIF2, p70S6K/eIF4 and mTOR cascades
This group includes eIF2 signaling, p70S6K/eIF4 signaling and mTOR signaling. The three canonical pathways are closely related and share many common proteins. The initiation of protein synthesis requires eIFs (eukaryotic translation initiation factors). eIF2 is a GTP-binding protein that escorts the initiation-specific form of met-tRNA onto the ribosome and identifies the translational start site. The serine/threonine kinase, p70S6K, participates in the control of protein synthesis. It phosphorylates the 40S ribosomal protein S6 which is involved in the translation of certain mRNAs, the so-called 5'-TOP mRNAs encoding ribosomal proteins and elongation factors. A principal pathway that signals through mTOR is the PI3K/Akt signalling pathway which is involved in cell survival and proliferation. The activation of mTOR results in phosphorylation of several downstream targets. For example, activated mTOR mediates the phosphorylation of eIF4EBP1 and the ribosomal protein p70S6K or S6K1. These pathways are involved in the regulation of protein synthesis. They are also closely related to the proliferation and hypertrophy of pulmonary arterial smooth muscle cells which play important roles in the pathophysiology of PAH. Two decades ago, endothelin-1 was shown to stimulate protein synthesis in smooth muscle cells26 and promote the proliferation of the pulmonary arterial smooth muscle cells.27 It has recently been shown that p70S6K and mTOR are involved in the hypertrophy of pulmonary artery smooth muscle cells.28
As shown in Figure 6, a cascade of molecules involving eIF2, p70S6K and mTOR illustrates a general increase in cellular protein content following exposure of Control cells to ET-1. We observed a very similar pattern of increase in ET-1 treated PAH cells (Supplementary Figure 2A). When examining basal changes in PAH in comparison to Control (Supplementary Figure 2B), we found a general decline in expression of proteins which support protein synthesis. However, this is not the case when ET-1 stimulated Control and PAH cells are compared (Supplementary Figure 2C). In this case, ET-1 becomes considerably more active in increasing the general concentrations of proteins that lead to protein synthesis.
Figure 6.
Diagram of the differentially expressed proteins related to protein synthesis in ET-1 treated vs. basal in Control PASMC. The nodes represent different proteins. Green indicates down-regulation, red indicates up-regulation, and color intensity is proportional to fold change. The full name and fold changes of these proteins can be found in Supplementary Table 3. The annotations are supported by at least one reference from the literature, a textbook, or canonical information stored in the Ingenuity knowledge base.
4.2.2 Cytoskeleton organization and muscle contraction
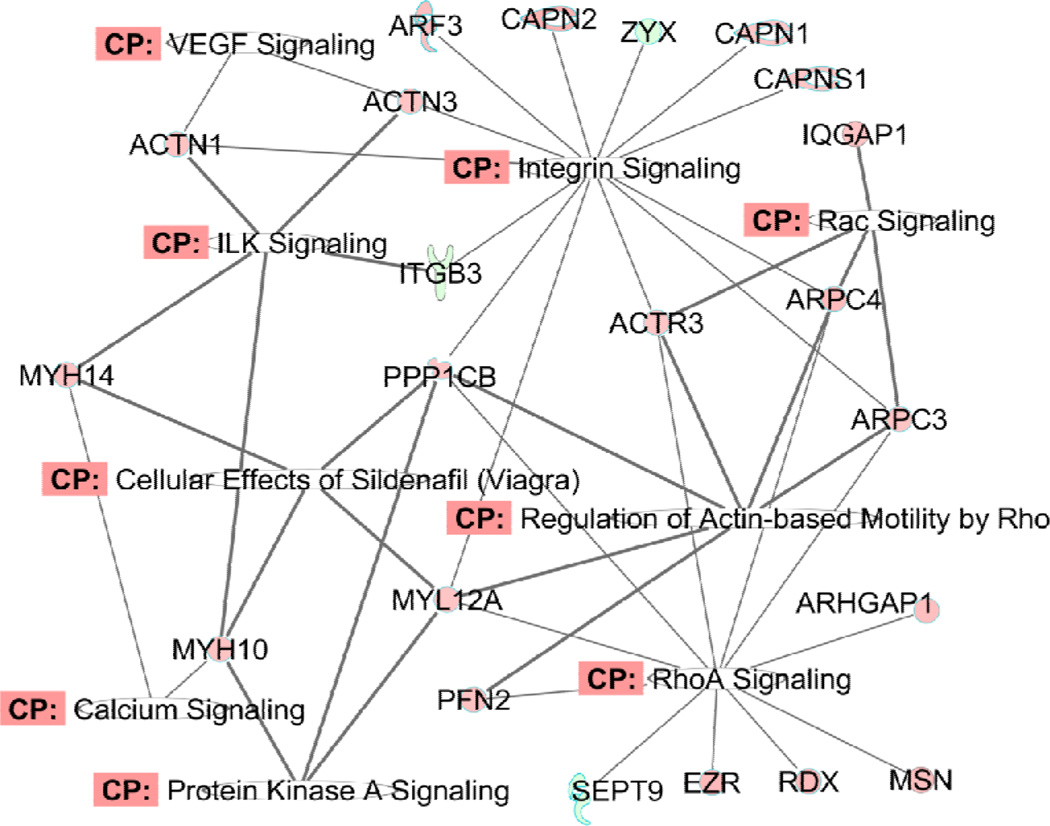
The second group of pathways is the combination of RhoA signaling, actin cytoskeleton signaling, integrin signaling and the regulation of actin-based motility by RhoA. These pathways share many common components that are involved in actin cytoskeleton organization, cell cycling, cellular movement, cellular cytoskeleton assembly and organization, and muscle contraction. One annotated protein network which demonstrates the involvement of proteins in those pathways in PAH cells (compared with Control cells) under basal conditions is shown in Figure 7 and the diagrams for the other three comparisons are shown in Supplementary Figure 3A, B and C. The metadata are shown in Supplementary Table 4. The patterns among the four comparisons are consistent with the changes in the protein synthesis pathway described above. ET-1 strongly promotes RhoA signaling-related pathways in both Control and PAH cells, whereas the basal level expressions of proteins in this pathway do not change dramatically in PAH compared to Control PASMC. In the presence of ET-1, the difference becomes much more obvious, confirming that ET-1 is an important constituent in PAH pathogenesis. ET-1 signaling seems to exaggerate the RhoA pathway and cytoskeleton organization in the defective PASMC with BMPR2 mutation. In fact, it was shown recently that the cytoskeleton is defective in BMPR2-associated pulmonary arterial hypertension.29 RhoA and Rho kinase have long been associated with the development of PAH and this pathway has been proposed as a potential target for PAH therapy.30
Figure 7.
Diagram of the differentially expressed proteins related with RhoA signaling, cytoskeleton organization and muscle contraction in PAH cells compared with Control cells under basal condition. The full name and fold changes of these proteins can be found in Supplementary Table
Different expression patterns for these proteins were observed in Control and PAH PASMCs, with or without ET-1. For example, the ERM protein family in the RhoA pathway exhibited marked changes. The ERM family consists of three closely related proteins, ezrin (EZR), radixin (RDX) and moesin (MSN). The levels of all three proteins showed increase upon ET-1 stimulation. ERM proteins crosslink actin filaments with plasma membranes. They are a substrate for Rho-kinase. This protein family is involved in smooth muscle cell movement and differentiation.31 These proteins also act as inflammatory stimuli which are upregulated by Rho-kinase in human coronary vascular smooth muscle cells.32 ET-1 induces expression of the proteins ezrin and moesin which contribute to a contractile phenotype in lung fibroblasts.33 For Wb data on ezrin, see Figure 3 and Supplementary Table 5.
Also of interest is calpain (CAPN2). CAPN2 belongs to the family of calcium-dependent, non-lysosomal cysteine proteases. It is apparently involved in vascular smooth muscle cell proliferation34 and has been shown to counteract apoptosis of vascular smooth muscle cells in vitro and in vivo.35 Inhibition of calpain was shown to attenuate the pathological effect of PAH in rat models.36 As illustrated in Supplementary Table 4, ET-1 increased the expression of calpain in PAH cells but not in the Control cells.
Myosin heavy chain, myosin light chain and vinculin were all increased in the PAH PASMC compared to the Control cells. ET-1 stimulates the expression of those proteins in both cell types (Supplementary Table 5). Myosin heavy chain (MYH) and light chain (MYL) are known for their role in smooth muscle contraction, and the phosphorylation of myosin light chain is the fulcrum for the regulation of this process. Vinculin (VCL) is a cytoskeletal protein associated with cell-cell and cell-matrix junctions, where it is thought to function as one of several interacting proteins involved in anchoring F-actin to the membrane.
ET-1 increased RhoA expression in both Control (1.38-fold) and PAH (2.10-fold) cells (Figure 3 and Supplementary Table 5). There was no significant difference in RhoA expression between Control and PAH cells under basal conditions; however, in the presence of ET-1, RhoA expression increased to 1.73-fold in PAH cells compared to Control cells. This suggests that regulation RhoA expression by ET-1 plays an important role in the PAH pathology. Consideration of interference with RhoA expression and signaling is clearly warranted as a strategy toward the treatment of PAH.
4.2.3 Protein ubiquitination
The protein ubiquitination pathway plays a major role in the degradation of short-lived or regulatory proteins involved in a variety of cellular processes, including cell cycle, cell proliferation, apoptosis, and cell surface receptors and regulation of ion channels. Degradation of proteins via the protein ubiquitination pathway involves two successive steps: (1). Conjugation of multiple ubiquitin moieties (Ub) to the target protein. (2). Degradation of the polyubiquitinated protein by the 26S proteasome complex. Biologically, this process is related to the apoptosis of the SMCs. The proteins and the expression changes of those proteins observed in our dataset are presented in Table 3A. Our data confirmed the results reported for a DNA microarray study.37 Indeed, recent experimental evidence supports a potential involvement of the ubiquitin–proteasome system in the initiation, progression, and complication stage of atherogenesis.61 Again, as with RhoA signaling, protein changes in PAH cells relative to Control were evident under basal conditions. Upon exposure to ET-1, the levels of the Ubiquitin cascade proteins increased markedly in the PAH cells, as compared to the Control cells.
4.2.4. Glycolysis/Gluconeogenesis and the Pentose Phosphate Pathway
PAH is associated with a glycolytic shift in metabolism.38 The PASMCs in PAH are resistant to apoptosis due to the increase in their rate of glycolysis and the impairment of glucose oxidation in those cells.39 Also, pentose phosphate pathway-derived NADPH may play important role in hypoxic pulmonary vasoconstriction.40 Our proteomic data indicated ET-1 promotes the gylcolysis/gluconeogenesis pathways in both Control and BMPR2 mutant PAH PASMC. The key proteins are listed in Tables 4B and 4C.
Table 4.
Additional assignments for differentially expressed proteins. Fold changes were generated for four comparisons: A1: ET-1 treated vs. basal in Control PASMC; A2. ET-1 treated vs. basal in PAH-BRMP2 PASMC; A3. PAH-BRMP2 cells vs. Control cells under basal condition; A4. PAH-BRMP2 vs. Control cells, both with ET-1 stimulation.
| A. Differentially expressed proteins involved in protein ubiquination. | |||||
|---|---|---|---|---|---|
| Symbol | Entrez Gene Name | Fold Change (A1) |
Fold Change (A2) |
Fold Change (A3) |
Fold Change (A4) |
| DNAJB4 | DnaJ (Hsp40) homolog, subfamily B, member 4 |
1.70 | 2.65 | 4.02 | 6.29 |
| HLA-B | major histocompatibility complex, class I, B |
1.64 | 2.53 | 1.69 | 2.60 |
| HSP90AA1 | heat shock protein 90 kDa alpha (cytosolic), class A member 1 |
1.91 | 2.48 | 1.33 | 1.73 |
| HSP90AB1 | heat shock protein 90 kDa alpha (cytosolic), class B member 1 |
1.91 | 2.40 | 1.31 | 1.65 |
| HSP90B1 | heat shock protein 90 kDa beta (Grp94), member 1 |
1.78 | 2.48 | 1.14 | 1.59 |
| HSPA2 | heat shock 70 kDa protein 2 | 1.83 | 2.49 | 1.12 | 1.52 |
| HSPA5 | heat shock 70 kDa protein 5 (glucose-regulated protein, 78 kDa) | 1.80 | 2.37 | −1.02 | 1.29 |
| HSPA6 | heat shock 70 kDa protein 6 (HSP70B') |
1.85 | 2.16 | 1.53 | 1.79 |
| HSPA8 | heat shock 70 kDa protein 8 | 1.65 | 2.34 | −1.06 | 1.33 |
| HSPA9 | heat shock 70 kDa protein 9 (mortalin) |
1.35 | 1.59 | 1.44 | 1.70 |
| HSPA1L | heat shock 70 kDa protein 1-like | 1.66 | 1.88 | 1.62 | 1.83 |
| HSPD1 | heat shock 60 kDa protein 1 (chaperonin) |
1.89 | 2.36 | 1.31 | 1.63 |
| HSPE1 | heat shock 10 kDa protein 1 (chaperonin 10) |
1.73 | 2.51 | 1.24 | 1.80 |
| PSMA1 | proteasome (prosome, macropain) subunit, alpha type, 1 |
2.10 | 2.89 | 1.30 | 1.79 |
| PSMA2 | proteasome (prosome, macropain) subunit, alpha type, 2 |
1.72 | 1.97 | 1.32 | 1.51 |
| PSMA3 | proteasome (prosome, macropain) subunit, alpha type, 3 |
1.52 | 2.28 | 1.01 | 1.52 |
| PSMA5 | proteasome (prosome, macropain) subunit, alpha type, 5 |
1.99 | 2.45 | 1.36 | 1.68 |
| PSMA6 | proteasome (prosome, macropain) subunit, alpha type, 6 |
1.39 | 2.56 | −1.13 | 1.63 |
| PSMA7 | proteasome (prosome, macropain) subunit, alpha type, 7 |
1.80 | 2.14 | 1.21 | 1.44 |
| PSMB1 | proteasome (prosome, macropain) subunit, beta type, 1 |
1.60 | 1.92 | 1.21 | 1.45 |
| PSMB2 | proteasome (prosome, macropain) subunit, beta type, 2 |
1.42 | 2.57 | −1.01 | 1.80 |
| PSMB3 | proteasome (prosome, macropain) subunit, beta type, 3 |
2.02 | 2.38 | 1.40 | 1.65 |
| PSMC2 | proteasome (prosome, macropain) 26S subunit, ATPase, 2 |
1.50 | 1.97 | 1.07 | 1.40 |
| PSMD8 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 8 |
1.91 | 1.68 | 1.88 | 1.65 |
| PSME1 | proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
2.56 | 3.69 | 1.56 | 2.24 |
| PSME2 | proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
1.72 | 2.17 | 1.30 | 1.64 |
| UBA1 | ubiquitin-like modifier activating enzyme 1 |
1.67 | 2.05 | 1.01 | 1.24 |
| UBE2N | ubiquitin-conjugating enzyme E2N | 1.78 | 2.29 | 1.28 | 1.65 |
| USO1 | USO1 vesicle docking protein homolog (yeast) |
1.97 | 3.58 | −1.10 | 1.65 |
| USP5 | ubiquitin specific peptidase 5 (isopeptidase T) |
1.52 | 1.72 | 1.58 | 1.78 |
| B. Differentially expressed proteins involved in the Glycolysis/Gluconeogenesis pathway. | |||||
|---|---|---|---|---|---|
| Symbol | Entrez Gene Name | Fold Change (A1) |
Fold Change (A2) |
Fold Change (A3) |
Fold Change (A4) |
| ADH5 | alcohol dehydrogenase 5 (class III), chi polypeptide | 1.63 | 2.14 | 1.24 | 1.64 |
| ALDH9A1 | aldehyde dehydrogenase 9 family, member A1 |
1.55 | 2.27 | 2.62 | |
| ALDOA | aldolase A, fructose-bisphosphate | 1.45 | 1.99 | −1.16 | 1.19 |
| ENO1 | enolase 1, (alpha) | 1.46 | 1.87 | 1.03 | 1.32 |
| ENO2 | enolase 2 (gamma, neuronal) | 1.59 | 1.72 | 1.28 | 1.38 |
| ENO3 | enolase 3 (beta, muscle) | 1.60 | 1.71 | 1.27 | 1.37 |
| GPI | glucose-6-phosphate isomerase | 1.57 | 1.98 | 1.31 | 1.66 |
| HK1 | hexokinase 1 | 1.43 | 1.62 | 1.69 | 1.92 |
| LDHA | lactate dehydrogenase A | 1.70 | 2.88 | 1.33 | 2.26 |
| LDHAL6A | lactate dehydrogenase A-like 6A | 1.41 | 2.70 | 1.11 | 2.13 |
| LDHAL6B | lactate dehydrogenase A-like 6B | 2.33 | 3.30 | 1.86 | 2.64 |
| LDHB | lactate dehydrogenase B | 1.49 | 2.59 | 1.18 | 2.05 |
| PFKL | phosphofructokinase, liver | 1.74 | 2.37 | 1.17 | 1.60 |
| PGAM1 | phosphoglycerate mutase 1 (brain) |
1.33 | 2.12 | −1.28 | 1.24 |
| PGK1 | phosphoglycerate kinase 1 | 1.64 | 2.40 | −1.21 | 1.21 |
| PGK2 | phosphoglycerate kinase 2 | 1.75 | 2.42 | −1.15 | 1.20 |
| PGM1 | phosphoglucomutase 1 | 2.09 | 1.92 | 1.97 | 1.81 |
| PKM2 | pyruvate kinase, muscle | 1.69 | 1.74 | 1.10 | 1.13 |
| TPI1 | triosephosphate isomerase 1 | 1.43 | 2.36 | −1.23 | 1.34 |
| C. Differentially expressed proteins involved in the Pentose Phosphate Pathway. | |||||
|---|---|---|---|---|---|
| Symbol | Entrez Gene Name | Fold Change (A1) |
Fold Change (A2) |
Fold Change (A3) |
Fold Change (A4) |
| ALDOA | aldolase A, fructose-bisphosphate | 1.45 | 1.99 | −1.16 | 1.19 |
| G6PD | glucose-6-phosphate dehydrogenase |
1.59 | 2.56 | 1.00 | 1.61 |
| GPI | glucose-6-phosphate isomerase | 1.57 | 1.98 | 1.31 | 1.66 |
| PFKL | phosphofructokinase, liver | 1.74 | 2.37 | 1.17 | 1.60 |
| PGLS | 6-phosphogluconolactonase | 1.38 | 1.90 | 1.41 | 1.94 |
| PGM1 | phosphoglucomutase 1 | 2.09 | 1.92 | 1.97 | 1.81 |
| PRPS1 | phosphoribosyl pyrophosphate synthetase 1 |
1.80 | 2.44 | 1.17 | 1.58 |
| PRPS1L1 | phosphoribosyl pyrophosphate synthetase 1-like 1 |
1.63 | 2.13 | 1.17 | 1.53 |
| TALDO1 | transaldolase 1 | 1.55 | 2.21 | 1.39 | 1.98 |
| TKT | transketolase | 1.52 | 1.67 | 1.33 | 1.46 |
| D. Differentially expressed heat shock proteins. | |||||
|---|---|---|---|---|---|
| Symbol | Entrez Gene Name | Fold Change (A1) |
Fold Change (A2) |
Fold Change (A3) |
Fold Change (A4) |
| HSP90AA1 | heat shock protein 90 kDa alpha (cytosolic), class A member 1 |
1.91 | 2.48 | 1.33 | 1.73 |
| HSP90AB1 | heat shock protein 90 kDa alpha (cytosolic), class B member 1 |
1.91 | 2.39 | 1.31 | 1.65 |
| HSP90B1 | heat shock protein 90 kDa beta (Grp94), member 1 |
1.78 | 2.48 | 1.14 | 1.59 |
| HSPA1L | heat shock 70 kDa protein 1-like | 1.66 | 1.88 | 1.62 | 1.83 |
| HSPA4 | heat shock 70 kDa protein 4 | 1.28 | 1.84 | 1.24 | 1.77 |
| HSPA6 | heat shock 70 kDa protein 6 (HSP70B') |
1.85 | 2.16 | 1.53 | 1.79 |
| HSPA8 | heat shock 70 kDa protein 8 | 1.65 | 2.33 | −1.06 | 1.33 |
| HSPA9 | heat shock 70 kDa protein 9 (mortalin) |
1.35 | 1.59 | 1.44 | 1.70 |
| HSPB1 | heat shock 27 kDa protein 1 | 1.15 | 1.69 | 1.34 | 1.97 |
| HSPB6 | heat shock protein, alpha-crystallin-related, B6 | 1.33 | 1.73 | 2.25 | 2.93 |
| HSPD1 | heat shock 60 kDa protein 1 (chaperonin) |
1.89 | 2.36 | 1.31 | 1.63 |
| HSPE1 | heat shock 10 kDa protein 1 (chaperonin 10) |
1.73 | 2.51 | 1.24 | 1.80 |
| HYOU1 | hypoxia up-regulated 1 | 2.02 | 2.34 | 1.42 | 1.65 |
| SERPINH1 | serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1) |
1.64 | 1.80 | −1.49 | −1.36 |
| E. Expression change of the 14-3-3 family proteins. | |||||
|---|---|---|---|---|---|
| Symbol | Entrez Gene Name | Fold Change (A1) |
Fold Change (A2) |
Fold Change (A3) |
Fold Change (A4) |
| YWHAZ | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide |
1.85 | 2.76 | 1.23 | 1.84 |
| YWHAQ | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide |
1.63 | 2.14 | −1.07 | 1.22 |
| YWHAG | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide |
1.68 | 2.40 | −1.19 | 1.20 |
| YWHAE | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide |
1.88 | 3.85 | −1.04 | 1.97 |
| YWHAB | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide |
1.80 | 2.68 | 1.13 | 1.68 |
Glucose-6-phosphate dehydrogenase (G6PD) is the rate-limiting enzyme in the pentose phosphate pathway and represents a major source of reduced nicotinamide adenine dinucleotide phosphate (NADPH), which regulates numerous enzymes, including glutathione reductase and NADPH oxidase. G6PD has been shown to regulate the contraction of SMCs and the inhibition of G6PD ameliorates hypoxia-induced pulmonary hypertension.41 ET-1 increased the expression of this enzyme 1.6- and 2.6-fold in the Control and PAH PASMC, respectively. There was no significant change in the expression of this enzyme when Control and PAH cells were compared under basal conditions. However, upon treatment with ET-1, G6PD levels increased 1.6-fold in the PAH cells compared to Control cells.
4.2.5. Additional PAH-related Proteins
HSPs (heat shock proteins)
We observed a substantial heat shock protein response with ET-1 stimulation. The detailed assignments for the HSPs and their fold changes under different conditions are shown in Table 4D. Heat shock proteins are clearly involved in PAH.
Heat shock protein 90 (HSP90) functions as a molecular chaperone to ensure the correct conformation, activity, intracellular localization and proteolytic turnover of a range of proteins related to cell growth, differentiation, activity and survival.42 Hsp90 influences both prostanoid and eNOS signaling in the pulmonary circulation of newborn piglets and the impact of pharmacological inhibition of Hsp90 on these signaling pathways is altered during chronic hypoxia.43 Hsp90 has also been found to protect pulmonary artery smooth muscle cells from undergoing apoptosis.44 Our data show that some heat shock proteins are very responsive to ET-1 stimulation. After 16-h incubation with ET-1, Hsp90 was up-regulated by 2-fold in Control cells, and markedly more in PAH cells. Hsp60 and Hsp10 also increased in both Control and PAH cells in response to ET-1 treatment. Hsp60 forms complexes with Bax, Bak, and Bcl-XL; when Hsp-60 decreases, both Bax and Bak increase, while Bcl-2 decreases. Whereas overexpression of Hsp60 leads to the increase of Bcl-XL and Bcl-2 and decreased expression of Bax, the down-regulation of Hsp60 or interference with the binding of Hsp60 to Bax, Bak, or Bcl-XL induces apoptosis.45–48 CRYAB (crystallin, alpha B) also belongs to the heat shock protein family and is involved in the protein ubiquination pathway. CRYAB binds to BMPR2.49 Our data show that the CRYAB level increased more than 3-fold in the PAH PASMC compared to Control. Overall, ET-1 increases the HSPs to higher levels in PAH than in Control cells. The data suggest that increasing ET-1 elevates the expression of HSPs in PAH PASMC.
Heme oxygenase 1 (HMOX1) is a stress-responsive protein. HMOX1 expression was increased by almost 5-fold in PAH cells compared to Control under basal conditions. In the presence of ET-1, its expression increased 7.2-fold in PAH cells compared to Control. Thus HMOX1 may be a key molecule in the development of PAH in the BMPR2 mutation. The IPA knowledgebase points out that HMOX1 is associated with mTOR, phospholipase C and endothelin-1 signaling and phospholipid degradation. The lungs of MCT-treated experimental animals that develop PAH have shown increases in their HMOX1 levels.50
NAD (PH) dehydrogenase 1 (NQO1) level was elevated 2.24-fold in PAH cells compared to Control cells and ET-1 increased its expression in PAH cells another 2.35-fold. NQO1 is involved in hypoxia signaling in the cardiovascular system and is related to the NRF2-mediated oxidative stress response.51, 52
ITGB1(integrin β1)
ET-1 increased the expression of ITGB1 2.41-fold in the PAH cells. This suggests that part of the effect of ET-1 occurs through the regulation of integrin. Integrin β1 is involved in the attachment and spreading of smooth muscle cells. Antibodies against β1 integrin receptors prevent attachment and migration of SMCs by bovine collagen Type VIII complexes.53 In cell culture, interference by human ITGB1 protein by activating antibody increases spreading of primary smooth muscle cells from the human coronary artery. Integrin β1 is also involved in protein kinase cascades54 and pulmonary fibroblast proliferation.55 Therefore, ET-1 may promote SMC spreading in PAH partly through increased expression of integrin β1.
Alp (alkaline phenyl phosphatase) expression was increased markedly in the PAH cells. Interestingly, alkaline phenyl phosphatase has been shown to be involved in BMPR2 signaling. For example, BMP-2-induced recruitment of BMPR2s results in the induction of alkaline phosphatase activity via p38 MAPK.56 In PASMC, BMPR2 knockout decreases the expression of mouse Alp mRNA(s) that is increased by BMP4 protein.57
14-3-3 family
Expression of the proteins in the 14-3-3 family (14-3-3 α, β, γ, δ and ε) was increased by ET-1 in both Control and PAH PASMCs (Table 3E). This protein family mediates signal transduction by binding to phosphoserine-containing proteins. 14-3-3 β has been shown to bind to myosin phosphatase target subunit 1 (MYPT1). This interaction diminishes the direct binding of MYPT1 to myosin II, and 14-3-3 β overexpression abolishes MYPT1 localization within the stress fiber. This indicates that 14-3-3 β is involved in the regulation of the RhoA/Rho-kinase-dependent regulatory mechanism of myosin II phosphorylation by dissociating MLCP from myosin II and attenuates MLCP activity.58 Autieri et al. have shown that 14-3-3 γ expression is induced in arterial trauma by cytokines and suggests that this protein may play an important role in progression of vascular proliferative diseases.59,60 Our data show that ET-1 increased 14-3-3 γ expression in both Control and PAH cells, although its expression in PAH cells was similar to Control cells.
5. Conclusions
Through label free quantitative proteomic mass spectrometry and IPA pathway analysis, methods chosen for this study had sufficient sensitivity and specificity to provide a detailed analysis of the global protein expression in PASMCs from Control and PAH (with BMPR2 mutation) lung explants, at basal and ET-1 stimulated levels. The proteins we found to be differentially expressed in PAH cells are participants in biological pathways such as the regulation of protein synthesis, RhoA signaling, cytoskeleton organization and protein degradation. Changes in the regulation of pathways bear witness to alterations taking place in the physiology of PAH and are likely involved in the etiology and progress of the disease. Furthermore, our data suggest that endothelin-1 exacerbates these physiologic malfunctions. Thus, even these preliminary results can help to illuminate routes for delineation of the biology of PAH and the role of ET-1. Further studies with samples from additional PAH patients, with and without BMPR2R mutations, having various manifestations of PAH, will enrich the dataset and provide deeper insight into PAH pathogenesis. The properties related specifically to PAH or the gene mutation should be able to be decoupled and the significance of their interplay determined.
Supplementary Material
Highlights.
Pulmonary arterial hypertension (PAH) increases vascular resistance and smooth muscle cells.
Endothelin-1 (ET-1), a potent vasoconstrictor, is linked to etiology and progression of PAH.
Proteomic mass spectrometry reveals PAH pathway changes and ET-1 effects.
PAH increases multiple signalling and protein degradation pathways; enhances response to ET-1.
Bone morphogenetic protein type II receptor (BMPR2) mutation likely affects PAH pathology.
ACKNOWLEDGMENTS
We thank Dr. Serpil Erzurum from Cleveland Clinic for pulmonary smooth muscle cell samples. We thank Dr. Nancy Leymarie and Dr. Deborah Francoleon for their technical support in the use of LTQ-Orbitrap mass spectrometer. Thanks to Xiaobin Xu and Han Hu for their help with the data analysis. This work was supported by NIH grant R01 HL025776 (to P.P.), grants P41 GM104603, S10 RR020946, and NHLBI Contract HHSN2682101000031C (to C.E.C.).
Abbreviations
- Alp
alkaline phenyl phosphatase
- ALPL
alkaline phosphatase
- BMPR2
Bone morphogenic type II receptor
- CALD1
caldesmon
- CAPN2
calpain
- CNN2
calponin H2
- COL1A1
Collagen protein 1
- CRYAB
crystallin, alpha B
- eIFs
eukaryotic translation initiation factors
- DTT
dithiothreitol
- ET-1
endothelin-1
- EZR
ezrin
- FAK
focal adhesion kinase-1
- G6PD
glucose-6-phosphate dehydrogenase
- GLS
glutaminase
- GSN
Gelsolin
- GSTO1
S-transferase omega-1
- HMOX1
Heme oxygenase 1
- HSP90
Heat shock protein 90
- HSPB1
heart shock protein β-1
- HSPD1
60-kDa heat shock protein
- IPA
Ingenuity Pathway Analysis
- ITGB1
integrin β1
- MSN
moesin
- MYPT1
myosin phosphatase target subunit 1
- NADPH
of reduced nicotinamide adenine dinucleotide phosphate
- NQO1
NAD (PH) dehydrogenase 1
- PAH
pulmonary arterial hypertension
- PASMC
pulmonary arterial smooth muscle cells
- PDIA6
disulfide-isomerase A6
- PDLIM1
PDA and LIM domain 1
- PGAM1
phosphoglycerate mutase 1
- PRDX1
peroxiredoxin 1
- RDX
radixin
- SERPINH1
serpin H1
- SMC
smooth muscle cell
- TAGLN
tansgelin
- TXNRD1
thioredoxin reductase 1
- UGDH
UDP-glucose-6-dehydrogenase
- VCL
vinculin
- VEGF
vascular endothelial growth factor
- WDR33
WD repeat domain 33 protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Farber HW, Loscalzo J. Pulmonary arterial hypertension. N. Engl. J. Med. 2004;351(16):1655–1665. doi: 10.1056/NEJMra035488. [DOI] [PubMed] [Google Scholar]
- 2.Cai J, Pardali E, Sánchez-Duffhues G, ten Dijke P. BMP signaling in vascular diseases. FEBS Lett. 2012;586(14):1993–2002. doi: 10.1016/j.febslet.2012.04.030. [DOI] [PubMed] [Google Scholar]
- 3.Rich S, McLaughlin VV. Endothelin receptor blockers in cardiovascular disease. Circulation. 2003;108(18):2184–2190. doi: 10.1161/01.CIR.0000094397.19932.78. [DOI] [PubMed] [Google Scholar]
- 4.Abdul-Salam VB, Wharton J, Cupitt J, Berryman M, Edwards RJ, Wilkins MR. Proteomic analysis of lung tissues from patients with pulmonary arterial hypertension. Circulation. 2010;122(20):2058–2067. doi: 10.1161/CIRCULATIONAHA.110.972745. [DOI] [PubMed] [Google Scholar]
- 5.Casey TM, Arthur PG, Bogoyevitch MA. Proteomic analysis reveals different protein changes during endothelin-1- or leukemic inhibitory factor-induced hypertrophy of cardiomyocytes in vitro. Mol. Cell. Proteomics. 2005;4(5):651–661. doi: 10.1074/mcp.M400155-MCP200. [DOI] [PubMed] [Google Scholar]
- 6.Yang MX, Liu ZX, Zhang S, Jing Y, Zhang SJ, Xie WP, Ma L, Zhu CL, Wang H. Proteomic analysis of the effect of iptakalim on human pulmonary arterial smooth muscle cell proliferation. Acta Pharmacol. Sin. 2009;30(2):175–183. doi: 10.1038/aps.2008.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Predic J, Soskic V, Bradley D, Godovac-Zimmermann J. Monitoring of gene expression by functional proteomics: response of human lung fibroblast cells to stimulation by endothelin-1. Biochemistry. 2002;41(3):1070–1078. doi: 10.1021/bi0117854. [DOI] [PubMed] [Google Scholar]
- 8.Dupont A, Corseaux D, Dekeyzer O, Drobecq H, Guihot A-L, Susen S, Vincentelli A, Amouyel P, Jude B, Pinet F. The proteome and secretome of human arterial smooth muscle cells. Proteomics. 2005;5(2):585–596. doi: 10.1002/pmic.200400965. [DOI] [PubMed] [Google Scholar]
- 9.Laudi S, Steudel W, Jonscher K, Schoning W, Schniedewind B, Kaisers U, Christians U, Trump S. Comparison of lung proteome profiles in two rodent models of pulmonary arterial hypertension. Proteomics. 2007;7(14):2469–2478. doi: 10.1002/pmic.200600848. [DOI] [PubMed] [Google Scholar]
- 10.Patel VJ, Thalassinos K, Slade SE, Connolly JB, Crombie A, Murrell JC, Scrivens JH. A comparison of labeling and label-free mass spectrometry-based proteomics approaches. J. Proteome Res. 2009;8(7):3752–3759. doi: 10.1021/pr900080y. [DOI] [PubMed] [Google Scholar]
- 11.Katz E, Fon M, Eigenheer R, Phinney B, Fass J, Lin D, Sadka A, Blumwald E. A label-free differential quantitative mass spectrometry method for the characterization and identification of protein changes during citrus fruit development. Proteome Sci. 2010;8(1):68. doi: 10.1186/1477-5956-8-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neilson KA, Ali NA, Muralidharan S, Mirzaei M, Mariani M, Assadourian G, Lee A, van Sluyter SC, Haynes PA. Less label, more free: approaches in label-free quantitative mass spectrometry. Proteomics. 2011;11(4):535–553. doi: 10.1002/pmic.201000553. [DOI] [PubMed] [Google Scholar]
- 13.Aldred MA, Comhair SA, Varella-Garcia M, Asosingh K, Xu W, Noon GP, Thistlethwaite PA, Tuder RM, Erzurum SC, Geraci MW, Coldren CD. Somatic chromosome abnormalities in the lungs of patients with pulmonary arterial hypertension. Am. J. Respir. Crit. Care Med. 2010;182(9):1153–1160. doi: 10.1164/rccm.201003-0491OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu W, Kaneko FT, Zheng S, Comhair SA, Janocha AJ, Goggans T, Thunnissen FB, Farver C, Hazen SL, Jennings C, Dweik RA, Arroliga AC, Erzurum SC. Increased arginase II and decreased NO synthesis in endothelial cells of patients with pulmonary arterial hypertension. FASEB J. 2004;18(14):1746–1748. doi: 10.1096/fj.04-2317fje. [DOI] [PubMed] [Google Scholar]
- 15.Aytekin M, Comhair SAA, de la Motte C, Bandyopadhyay SK, Farver CF, Hascall VC, Erzurum SC, Dweik RA. High levels of hyaluronan in idiopathic pulmonary arterial hypertension. Am. J. Physiol. Lung Cell Mol. Physiol. 2008;295(5):L789–L799. doi: 10.1152/ajplung.90306.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhatia VN, Perlman DH, Costello CE, McComb ME. Software Tool for Researching Annotations of Proteins: Open-source protein annotation software with data visualization. Anal. Chem. 2009;81(23):9819–9823. doi: 10.1021/ac901335x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geng Y-J, Azuma T, Tang JX, Hartwig JH, Muszynski M, Wu Q, Libby P, Kwiatkowski DJ. Caspase-3-induced gelsolin fragmentation contributes to actin cytoskeletal collapse, nucleolysis, and apoptosis of vascular smooth muscle cells exposed to proinflammatory cytokines. Eur.J. Cell Biol. 1998;77(4):294–302. doi: 10.1016/S0171-9335(98)80088-5. [DOI] [PubMed] [Google Scholar]
- 18.Katsuyama H, Wang CL, Morgan KG. Regulation of vascular smooth muscle tone by caldesmon. J. Biol. Chem. 1992;267(21):14555–14558. [PubMed] [Google Scholar]
- 19.Hino M, Kurogi K, Okubo MA, Murata-Hori M, Hosoya H. Small heat shock protein 27 (HSP27) associates with tubulin/microtubules in HeLa cells. Biochem. Biophys. Res. Commun. 2000;271(1):164–169. doi: 10.1006/bbrc.2000.2553. [DOI] [PubMed] [Google Scholar]
- 20.Leong WF, Chow VTK. Transcriptomic and proteomic analyses of rhabdomyosarcoma cells reveal differential cellular gene expression in response to enterovirus 71 infection. Cell. Microbiol. 2006;8(4):565–580. doi: 10.1111/j.1462-5822.2005.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blindt R, Zeiffer U, Krott N, Filzmaier K, Voss M, Hanrath P, vom Dahl J, Bosserhoff AK. Upregulation of the cytoskeletal-associated protein Moesin in the neointima of coronary arteries after balloon angioplasty: a new marker of smooth muscle cell migration? Cardiovasc. Res. 2002;54(3):630–639. doi: 10.1016/s0008-6363(02)00252-3. [DOI] [PubMed] [Google Scholar]
- 22.Rajkumar R, Konishi K, Richards TJ, Ishizawar DC, Wiechert AC, Kaminski N, Ahmad F. Genomewide RNA expression profiling in lung identifies distinct signatures in idiopathic pulmonary arterial hypertension and secondary pulmonary hypertension. Am. J. Physiol. Heart Circ. Physiol. 2010;298(4):1235–1248. doi: 10.1152/ajpheart.00254.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jordan PA, Stevens JM, Hubbard GP, Barrett NE, Sage T, Authi KS, Gibbins JM. A role for the thiol isomerase protein ERP5 in platelet function. Blood. 2005;105(4):1500–1507. doi: 10.1182/blood-2004-02-0608. [DOI] [PubMed] [Google Scholar]
- 24.Bason C, Corrocher R, Lunardi C, Puccetti P, Olivieri O, Girelli D, Navone R, Beri R, Millo E, Margonato A, Martinelli N, Puccetti A. Interaction of antibodies against cytomegalovirus with heat-shock protein 60 in pathogenesis of atherosclerosis. Lancet. 2003;362(9400):1971–1977. doi: 10.1016/S0140-6736(03)15016-7. [DOI] [PubMed] [Google Scholar]
- 25.Ceconi C, Boraso A, Cargnoni A, Ferrari R. Oxidative stress in cardiovascular disease: myth or fact? Arch. Biochem. Biophys. 2003;420(2):217–221. doi: 10.1016/j.abb.2003.06.002. [DOI] [PubMed] [Google Scholar]
- 26.Chua BH, Krebs CJ, Chua CC, Diglio CA. Endothelin stimulates protein synthesis in smooth muscle cells. Am. J. Physiol. Endocrinol. Metab. 1992;262(4):E412–E416. doi: 10.1152/ajpendo.1992.262.4.E412. [DOI] [PubMed] [Google Scholar]
- 27.Zamora MA, Dempsey EC, Walchak SJ, Stelzner TJ. BQ123, An ETAreceptor antagonist, inhibits endothelin-1-mediated proliferation of human pulmonary artery smooth muscle cells. Am. J. Respir. Cell Mol. Biol. 1993;9(4):429–433. doi: 10.1165/ajrcmb/9.4.429. [DOI] [PubMed] [Google Scholar]
- 28.Deng H, Hershenson MB, Lei J, Anyanwu AC, Pinsky DJ, Bentley JK. Pulmonary artery smooth muscle hypertrophy: roles of glycogen synthase kinase-3β and p70 ribosomal S6 kinase. Am. J. Physiol. Lung Cell. Mol. Physiol. 2010;298(6):L793–L803. doi: 10.1152/ajplung.00108.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson JA, Hemnes AR, Perrien DS, Schuster M, Robinson LJ, Gladson S, Loibner H, Bai S, Blackwell TR, Tada Y, Harral JW, Talati M, Lane KB, Fagan KA, West J. Cytoskeletal defects in Bmpr2-associated pulmonary arterial hypertension. Am. J. Physiol. Lung Cell. Mol. Physiol. 2012;302(5):474–484. doi: 10.1152/ajplung.00202.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barman SA, Zhu S, White RE. RhoA/Rho-kinase signaling: a therapeutic target in pulmonary hypertension. Vasc. Health Risk Manag. 2009;5:663–671. doi: 10.2147/vhrm.s4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doevendans PA, van Eys G. Smooth muscle cells on the move: the battle for actin. Cardiovasc. Res. 2002;54(3):499–502. doi: 10.1016/s0008-6363(02)00395-4. [DOI] [PubMed] [Google Scholar]
- 32.Hiroki J, Shimokawa H, Higashi M, Morikawa K, Kandabashi T, Kawamura N, Kubota T, Ichiki T, Amano M, Kaibuchi K, Takeshita A. Inflammatory stimuli upregulate Rho-kinase in human coronary vascular smooth muscle cells. J. Mol. Cell. Cardiol. 2004;37(2):537–546. doi: 10.1016/j.yjmcc.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 33.Shi-Wen X, Chen Y, Denton CP, Eastwood M, Renzoni EA, Bou-Gharios G, Pearson JD, Dashwood M, du Bois RM, Black CM, Leask A, Abraham DJ. Endothelin-1 promotes myofibroblast induction through the ETA receptor via a rac/phosphoinositide 3-kinase/Akt-dependent pathway and iIs essential for the enhanced contractile phenotype of fibrotic fibroblasts. Mol. Biol. Cell. 2004;15(6):2707–2719. doi: 10.1091/mbc.E03-12-0902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ariyoshi H, Okahara K, Sakon M, Kambayashi J-i, Kawashima S-i, Kawasaki T, Monden M. Possible involvement of m-calpain in bascular smooth muscle cell proliferation. Arterioscler., Thromb., Vasc. Biol. 1998;18(3):493–498. doi: 10.1161/01.atv.18.3.493. [DOI] [PubMed] [Google Scholar]
- 35.Sedding DG, Homann M, Seay U, Tillmanns H, Preissner KT, Braun-Dullaeus RC. Calpain counteracts mechanosensitive apoptosis of vascular smooth muscle cells in vitro and in vivo. FASEB J. 2008;22(2):579–589. doi: 10.1096/fj.07-8853com. [DOI] [PubMed] [Google Scholar]
- 36.Ma W, Han W, Greer PA, Tuder RM, Toque HA, Wang KKW, Caldwell RW, Su Y. Calpain mediates pulmonary vascular remodeling in rodent models of pulmonary hypertension, and its inhibition attenuates pathologic features of disease. J. Clin. Invest. 2011;121(11):4548–4566. doi: 10.1172/JCI57734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rajkumar R, Konishi K, Richards TJ, Ishizawar DC, Wiechert AC, Kaminski N, Ahmad F. Genomewide RNA expression profiling in lung identifies distinct signatures in idiopathic pulmonary arterial hypertension and secondary pulmonary hypertension. A. J. Physiol. Heart Circ. Physiol. 2010;298(4):H1235–H1248. doi: 10.1152/ajpheart.00254.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rubin LJ. Metabolic dysfunction in the pathogenesis of pulmonary hypertension. Cell Metab. 2010;12(4):313–314. doi: 10.1016/j.cmet.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 39.Sutendra G, Bonnet S, Rochefort G, Haromy A, Folmes KD, Lopaschuk GD, Dyck JRB, Michelakis ED. Fatty acid oxidation and malonyl-CoA decarboxylase in the vascular remodeling of pulmonary hypertension. Sci. Trans. Med. 2010;2(44):44ra58. doi: 10.1126/scitranslmed.3001327. [DOI] [PubMed] [Google Scholar]
- 40.Gupte SA, Okada T, McMurtry IF, Oka M. Role of pentose phosphate pathway-derived NADPH in hypoxic pulmonary vasoconstriction. Pulm. Pharmacol. Ther. 2006;19(4):303–309. doi: 10.1016/j.pupt.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 41.Gupte RS, Ata H, Rawat D, Abe M, Taylor MS, Ochi R, Gupte SA. Glucose-6-phosphate dehydrogenase is a regulator of vascular smooth muscle contraction. Antioxid. Redox. Signal. 2011;14(4):543–558. doi: 10.1089/ars.2010.3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat. Rev. Cancer. 2005;5(10):761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 43.Fike CD, Pfister SL, Slaughter JC, Kaplowitz MR, Zhang Y, Zeng H, Frye NR, Aschner JL. Protein complex formation with heat shock protein 90 in chronic hypoxia-induced pulmonary hypertension in newborn piglets. Amer. J. Physiol. Heart Circ. Physiol. 2010;299(4):H1190–H1204. doi: 10.1152/ajpheart.01207.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang L, Ma J, Li Y, Guo L, Ran Y, Liu S, Jiang C, Zhu D. 15-Hydroxyeicosatetraenoic acid (15-HETE) protects pulmonary artery smooth muscle cells against apoptosis via HSP90. Life Sci. 2010;87(7–8):223–231. doi: 10.1016/j.lfs.2010.06.019. [DOI] [PubMed] [Google Scholar]
- 45.Ghosh JC, Dohi T, Kang BH, Altieri DC. Hsp60 regulation of tumor cell apoptosis. J. Biol. Chem. 2008;283(8):5188–5194. doi: 10.1074/jbc.M705904200. [DOI] [PubMed] [Google Scholar]
- 46.Shan Y-X, Liu T-J, Su H-F, Samsamshariat A, Mestril R, Wang PH. Hsp10 and Hsp60 modulate Bcl-2 family and mitochondria apoptosis signaling induced by doxorubicin in cardiac muscle cells. J. Mol. Cell. Cardiol. 2003;35(9):1135–1143. doi: 10.1016/s0022-2828(03)00229-3. [DOI] [PubMed] [Google Scholar]
- 47.Gupta S, Knowlton AA. HSP60, Bax, Apoptosis and the heart. J. Cell. Mol. Med. 2005;9(1):51–58. doi: 10.1111/j.1582-4934.2005.tb00336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gupta S, Knowlton AA. Cytosolic heat shock protein 60, hypoxia, and apoptosis. Circulation. 2002;106(21):2727–2733. doi: 10.1161/01.cir.0000038112.64503.6e. [DOI] [PubMed] [Google Scholar]
- 49.Hassel S, Eichner A, Yakymovych M, Hellman U, Knaus P, Souchelnytskyi S. Proteins associated with type II bone morphogenetic protein receptor (BMPR-II) and identified by two-dimensional gel electrophoresis and mass spectrometry. Proteomics. 2004;4(5):1346–1358. doi: 10.1002/pmic.200300770. [DOI] [PubMed] [Google Scholar]
- 50.Goto J, Ishikawa K, Kawamura K, Watanabe Y, Matumoto H, Sugawara D, Maruyama Y. Heme oxygenase-1 reduces murine monocrotaline-induced pulmonary inflammatory responses and resultant right ventricular overload. Antioxid. Redox. Signal. 2002;4(4):563–568. doi: 10.1089/15230860260220058. [DOI] [PubMed] [Google Scholar]
- 51.Asher G, Lotem J, Sachs L, Kahana C, Shaul Y. Mdm-2 and ubiquitin-independent p53 proteasomal degradation regulated by NQO1. Proc. Natl. Acad. Sci. USA. 2002;99(20):13125–13130. doi: 10.1073/pnas.202480499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kobayashi M, Yamamoto M. Molecular mechanisms activating the Nrf2-Keap1 pathway of antioxidant gene regulation. Antioxid. Redox. Signal. 2005;7(3–4):385–394. doi: 10.1089/ars.2005.7.385. [DOI] [PubMed] [Google Scholar]
- 53.Hou G, Mulholland D, Gronska MA, Bendeck MP. Type VIII collagen stimulates smooth muscle cell migration and matrix metalloproteinase synthesis after arterial injury. Am. J. Pathol. 2000;156(2):467–476. doi: 10.1016/S0002-9440(10)64751-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Campbell S, Otis M, Côté M, Gallo-Payet N, Payet MD. Connection between integrins and cell activation in rat adrenal glomerulosa cells: a role for Arg-Gly-Asp peptide in the activation of the p42/p44mapk pathway and iIntracellular calcium. Endocrinology. 2003;144(4):1486–1495. doi: 10.1210/en.2002-220903. [DOI] [PubMed] [Google Scholar]
- 55.Zhou Q, Pardo A, Königshoff M, Eickelberg O, Budinger GRS, Thavarajah K, Gottardi CJ, Jones J, Varga J, Selman M, Sznajder JI, Raj JU, Zhou G. Role of von Hippel-Lindau protein in fibroblast proliferation and fibrosis. FASEB J. 2011;25(9):3032–3044. doi: 10.1096/fj.10-177824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nohe A, Hassel S, Ehrlich M, Neubauer F, Sebald W, Henis YI, Knaus P. The mode of bone morphogenetic protein (BMP) receptor oligomerization determines different BMP-2 signaling pathways. J. Biol. Chem. 2002;277(7):5330–5338. doi: 10.1074/jbc.M102750200. [DOI] [PubMed] [Google Scholar]
- 57.Yu PB, Deng DY, Beppu H, Hong CC, Lai C, Hoyng SA, Kawai N, Bloch KD. Bone morphogenetic protein (BMP) type II receptor is required for BMP-mediated growth arrest and differentiation in pulmonary artery smooth muscle cells. J. Biol. Chem. 2008;283(7):3877–3888. doi: 10.1074/jbc.M706797200. [DOI] [PubMed] [Google Scholar]
- 58.Koga Y, Ikebe M. A novel regulatory mechanism of myosin light chain phosphorylation via binding of 14-3-3 to myosin phosphatase. Mol. Biol. Cell. 2008;19(3):1062–1071. doi: 10.1091/mbc.E07-07-0668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Autieri MV. Inducible expression of the signal transduction protein 14-3-3γ in injured arteries and stimulated human vascular smooth muscle cells. Exp. Mol. Pathol. 2004;76(2):99–107. doi: 10.1016/j.yexmp.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 60.Autieri M, Haines D, Romanic A, Ohlstein E. Expression of 14-3-3 gamma in injured arteries and growth factor- and cytokine-stimulated human vascular smooth muscle cells. Cell Growth Differ. 1996;7(11):1453–1460. [PubMed] [Google Scholar]
- 61.Herrmann J, Ciechanover A, et al. The ubiquitin–proteasome system in cardiovascular diseases—a hypothesis extended. Cardiovasc. Res. 2004;61(1):11–21. doi: 10.1016/j.cardiores.2003.09.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.