Abstract
Liver and kidney cancers are notorious for drug resistance. Due to the complexity, redundancy and interpatient heterogeneity of resistance mechanisms, most efforts targeting a single pathway were unsuccessful. Novel personalized therapies targeting multiple essential drug resistance pathways in parallel hold a promise for future cancer treatment. Exploiting the multitarget characteristic of microRNAs (miRNAs), we developed a new therapeutic strategy by the combinational use of miRNA and anticancer drugs to increase drug response. By a systems approach, we identified that miR-27b, a miRNA deleted in liver and kidney cancers, sensitizes cancer cells to a broad spectrum of anticancer drugs in vitro and in vivo. Functionally, miR-27b enhances drug response by activating p53-dependent apoptosis and reducing CYP1B1-mediated drug detoxification. Notably, miR-27b promotes drug response specifically in patients carrying p53-wild-type or CYP1B1-high signature. Together, we propose that miR-27b synergizes with anticancer drugs in a defined subgroup of liver and kidney cancer patients.
Keywords: liver cancer, kidney cancer, miR-27b, drug resistance, p53, CYP1B1
Introduction
Anticancer drugs, including cytotoxic agents and molecularly targeted compounds are the standard treatment regimen for most cancers, especially for late-stage cancers in which surgery is not an option1. According to clinical observations, liver and kidney cancers are especially notorious for drug resistance, which results in eventual treatment failure2. In these cancers, the use of sorafenib, a compound targeting multiple kinases, resulted in a 3-month improvement in median overall survival3,4; however, refractory cancers eventually developed in most patients. In addition, the high cost of sorafenib precludes it from widespread use in developing counties5. Chemotherapies are affordable and still used in clinical practice, especially in Asia6. Notably, chemotherapies showed effectiveness in treating liver and kidney cancers in many clinical studies6,7 and are recommended in the guidelines for the diagnosis and treatment of primary liver cancer compiled by the Ministry of Health of China8. However, for patients who have impaired liver and kidney functions, chemotherapy is less effective likely because the drug doses must be reduced in these cases to avoid intolerable side effects6,9. The same limitation also applies to molecularly targeted drugs. Therefore, it is desirable that a novel therapeutic strategy could be developed to enhance the effectiveness of low doses of chemotherapeutic drugs as well as molecularly targeted drugs.
It is noteworthy that tumors resistant to one drug also exhibit broad resistance to other chemically and functionally unrelated drugs10. These clinical observations suggest that tumor cells likely adopt common resistance mechanisms, such as ATP-binding cassette (ABC) transporter-dependent drug efflux, increased detoxification capacity, and evasion of drug-induced cell death, to escape the therapeutic impacts of different anticancer drugs10. For example, a recent study identified that the loss of MED12 confers resistance to multiple chemotherapeutic compounds and molecularly targeted drugs on a variety of cancer types by the same mechanism, i.e., activating the TGF-β signaling to reduce drug-induced cell death11. Theoretically, targeting any one of these essential pathways could increase the response to a wide range of anticancer drugs. However, due to the complexity and redundancy of drug resistance mechanisms, strategies targeting a single pathway failed to improve the response to anticancer drugs in clinical translations12. It becomes apparent that antagonizing multiple resistance pathways would be a new solution to improve the efficacy of anticancer drugs. Moreover, a comprehensive understanding of the resistance mechanisms targeted by such a strategy is critical for designing personalized therapies1.
MicroRNAs (miRNAs) are important posttranscriptional regulators involved in various biological processes, including tumorigenesis13. Notably, the first cancer-targeted miRNA drug MRX34, a liposome-based miR-34 mimic, has entered clinical trials in patients with advanced liver cancers14. Encouraged by this progress, miRNA-based therapies are attracting special attention nowadays. The most important advantage of miRNA-based therapies is that miRNAs are capable of concurrently targeting multiple effectors to fulfill biological functions15. Apparently, the combinational therapy of miRNA and anticancer drugs would be a good solution to achieve potent drug responses if multiple essential drug resistance pathways are targeted by miRNAs16.
Accumulating evidence has shown that miRNAs play important roles in drug resistance of human cancers17. For example, in liver cancers, miR-221/222 induce TNF-related apoptosis-inducing ligand (TRAIL) resistance by targeting tumor suppressors PTEN and TIMP318. miR-199a-3p enhanced cell sensitivity to doxorubicin by antagonizing mTOR and c-Met19. These pioneered studies have laid the ground for understanding the roles of miRNAs in drug resistance. The current challenge is to identify master miRNAs, which by targeting multiple essential drug resistance pathways are capable of improving the sensitivity to a broad spectrum of anticancer drugs, including chemotherapeutic drugs and molecularly targeted drugs. Ideally, such miRNAs should have genetic or epigenetic alterations in cancer cells so that the combinational therapy would spare normal cells. In addition, thorough characterization of the drug resistance pathways targeted by such master miRNAs would facilitate the application of the combinational therapy of miRNAs and anticancer drugs in a personalized manner.
To discover such master miRNAs, we took a systems approach by screening human miRNA libraries followed by comprehensive bioinformatic analysis of miRNA-pathway networks and clinical cancer data. By focusing on the most resistant liver and kidney cancers, we found that miR-27b, genetically deleted in human cancers, is a master miRNA capable of enhancing the response to multiple drugs in vitro and in vivo. miR-27b reduces drug resistance via two essential mechanisms, i.e., enhancing p53-dependent cell death and suppressing CYP1B1-mediated drug detoxification. Notably, clinical data indicate that high miR-27b levels together with drug treatment would specifically benefit cancer patients with a p53-wild-type or a CYP1B1-high signature, suggesting a personalized application of miR-27b and anticancer drugs to improve therapeutic effects.
Results
Identification of miRNAs that enhance drug sensitivity by functional screen and systems analysis
To identify master miRNAs that can improve the response of human cancer cells to anticancer drugs in a comprehensive manner, we designed and optimized a high-throughput miRNA screen including 828 human miRNA mimics20 (Figure 1A and Supplementary information, Figure S1A-S1E). The hepatoblastoma cell line HepG2 was employed in the screen, because it retains better hepatic characteristics, has been widely used for cytotoxicity studies21, and shares some important genetic alterations with hepatocellular carcinoma (HCC)22. In addition, doxorubicin, a widely used front-line chemotherapeutic drug, was used in the screen. High transfection efficiencies of small RNAs were confirmed in pilot experiments (Supplementary information, Figure S1A-S1E). Also, siRNA-mediated silence of multidrug resistance-associated protein-1 (MRP1), a major ABC transporter family member, was used as a positive control throughout the screen. Comparison of the data derived from duplicated experiments using 146 randomly selected miRNAs yielded a Pearson correlation coefficient of 0.67, indicating that the screening system was robust (Supplementary information, Figure S1F).
Figure 1.
Identification of miRNAs that enhance drug sensitivity by functional screen and systems analysis. (A) Schematic outline of the high-throughput miRNA screen. HepG2 cells were seeded in 96-well plates and transfected with miRNAs 12 h later. Forty-eight hours after miRNA transfection, the cells were treated with 0.1 μM doxorubicin for 3 days, which killed ∼25% of HepG2 cells transfected with scrambled miRNA control (Scr-miR). siRNA oligos antagonizing MRP1 were served as a positive control. The relative cell number was calculated to assess the effect of each miRNA on doxorubicin response. (B) Scatter plot of Z-scores. Each dot represents the Z-score of an individual miRNA. Thresholds with Z ≤ −1.65 or Z ≥ 1.65 were set for selecting miRNAs that enhanced or reduced drug responses, respectively (P < 0.1). miR-27a (blue dot) and miR-27b (red dot) are marked. (C) Validation of miRNAs sensitizing drug response from the primary screen. Out of these 33 miRNAs, 22 (closed bars) were confirmed to significantly enhance the sensitivity to doxorubicin when compared with MRP1 inactivation (*P< 0.05). (D) Predicted target genes of each miRNA were used to generate enriched pathways in Reactome. (E) miRNAs and their predicted target pathways were plotted into an interaction network using the Cytoscape program.
The Z-score was used to normalize the raw data in a way that provides explicit information regarding the strength of each miRNA relative to the rest of the sample distribution23. The distribution of Z-score values approximately recapitulated the normal distribution (Supplementary information, Figure S1G). 33 miRNAs with Z-scores below −1.65 enhanced the sensitivity of HepG2 cells to doxorubicin (P< 0.1), whereas 55 miRNAs increased the resistance of HepG2 cells to doxorubicin (Z-score ≥ 1.65, P< 0.1; Figure 1B and Supplementary information, Table S1). Although both the sets of miRNAs deserve in-depth investigation, we focused on the miRNAs that enhanced drug sensitivity in this study.
Twenty-two out of the thirty-three miRNAs were individually validated and shown to increase the sensitivity to doxorubicin in HepG2 cells (Figure 1C). Next, we adopted a network-based strategy, which provided a systems view of the functions of the 22 miRNAs. The predicted target genes of each miRNA were mapped in Reactome to reveal functional pathways (Figure 1D and Supplementary information, Table S3). The interactions among the miRNAs and target pathways were then projected into an integrated network (Figure 1E). These miRNAs were found to target essential biological functions, most of which could be categorized into cell proliferation, cell death, transportation, and detoxification of compounds (Figure 1E). Previous studies have demonstrated that the deregulation of these essential biological functions results in multidrug resistance1, which explains why these miRNAs could enhance the drug response.
Out of the 22 miRNAs identified in our analysis, 5 miRNAs (miR-20a, miR-27b, miR-20b, miR-183, and miR-367) were located in the center of the miRNA-pathway network and predicted to regulate 84% of the functional pathways. It is conceivable that these five miRNAs would have key roles in facilitating the response to multiple anticancer drugs.
miR-27b is a master miRNA capable of enhancing drug sensitivity in liver cancers
In addition to HepG2, the drug sensitizing effect of miR-27b could be extended to three additional HCC cell lines, SNU-182, SNU-739, and Tong (Figure 2A). miR-20a, miR-20b, and miR-367 only enhanced drug response in SNU-739 cells (Supplementary information, Figure S2A-S2L), and miR-183 exhibited no sensitizing effect in these cells lines (Supplementary information, Figure S2A-S2L). To further characterize the function of these miRNAs, we analyzed the drug sensitizing effect on primary cultured human HCC cells, which retained the properties of original cancers24. Remarkably, miR-27b significantly enhanced the response of liver cancer cells to doxorubicin in three out of five patient-derived primary cultures (Figure 2B). It is also worth mentioning that miR-27b enhanced the sensitivity to doxorubicin to the extent comparable to MRP1 silencing (Figure 1C and Supplementary information, Figure S1C). Together, these results suggest that miR-27b possesses the strongest capability to improve drug sensitivity among the five miRNA hits identified from the screen.
Figure 2.
miR-27b is a master miRNA capable of enhancing drug sensitivity in liver cancers. (A) Enhanced response to doxorubicin by miR-27b overexpression was shown in multiple liver cancer cell lines, including HepG2, SNU-182, SNU-739 and Tong. (B) The effects of miRNA overexpression on cell response to doxorubicin were determined in primary cultured liver cancer cells. Cancer cells from patients 1, 4, and 5 were used at passage 1, and cancer cells from patients 2 and 3 were used at passage 4. These primary cultured liver cancer cells were treated with 0.5 μM doxorubicin for 72 h after miRNA transfection. The number of viable cells transfected with the indicated miRNAs followed by doxorubicin treatment was normalized to that of the Scr-miR-treated control group. (C) miR-27b enhances the sensitivity of HepG2 cells to multiple anticancer drugs. IC50 for each treatment was presented. (D) miR-27b expression levels were reduced in human liver cancer samples (n = 49, P < 0.001) compared with paired non-cancerous tissues. (E) Loss of mir-27b alleles in human liver cancers (12 out of 49) as determined by the copy number variation analysis of the mir-27b gene locus. The relative log2 copy number ratio < −0.25 represents copy number loss. (F) The promoter methylation level of the mir-27b gene was unchanged in human liver cancers (n = 41). β value was applied to indicate the levels of DNA methylation at the promoter of the mir-27b gene.
Next, we asked whether miR-27b can enhance the response to other anticancer drugs besides doxorubicin. Notably, we found that miR-27b increased the response to Topoisomerase II poisons, including epirubicin, and etoposide (Figure 2C), as well as platinum compound cisplatin in HepG2 cells (Figure 2C). Also, we analyzed the combinational effect of miR-27b with sorafenib, the only targeted drug for liver cancers3, and gefitinib, an EGFR inhibitor in clinical trials for liver cancer treatment25. Strikingly, miR-27b enhanced the sensitivity to both molecularly targeted drugs (Figure 2C), echoing the clinical observation that tumor cells often adopt common resistance mechanisms to compromise the effects of different anticancer drugs10. These results further prove that miR-27b is a master miRNA increasing the sensitivity to multiple anticancer drugs.
Genetic deletions of miR-27b in human liver cancers
Ideally, patients eligible for the combinational therapy of miRNA and anticancer drugs would be those with miRNA levels that are lower in tumors than in normal tissues, so that the combinational therapy would attack cancer cells with high specificity and spare normal cells in which the corresponding miRNA is already highly expressed17. To evaluate the miRNA expression levels in human liver cancers, we used data derived from the public domain, The Cancer Genome Atlas (TCGA), which has set up vigorous criteria for human cancer sample collection and information processing26. Out of the five miRNAs analyzed (Figure 1E), only miR-27b was abundantly expressed in non-tumor liver tissues but significantly reduced in 49 paired liver cancer samples (Figure 2D and Supplementary information, Figure S3A-S3H and Table S4). We additionally analyzed data of 10 pairs of human liver cancer samples from the public database ArrayExpress, and determined miR-27b expression in 15 pairs of collected liver cancer samples by quantitative real-time PCR (q-PCR). Both sets of data confirmed that miR-27b was significantly downregulated in liver cancers (Supplementary information, Figure S3E and S3F), suggesting that high miR-27b levels might confer a disadvantage on cancer cells.
Next, we assessed genetic and epigenetic alterations at the miR-27b gene locus using the same set of samples in TCGA. Strikingly, 12 out of 49 liver cancers displayed simultaneous loss of the miR-23b-27b-24-1 gene cluster and its tightly linked genes C9orf3 and FANCC as identified by copy number variation analysis (Figure 2E and Supplementary information, Figure S3I, relative log2 copy number ratio < −0.25). Also, the expression levels of these genes were decreased in liver cancers compared with paired non-cancerous tissues (Figure 2D and Supplementary information, Figure S3J). According to the DNA methylation data in TCGA, the promoter methylation status of the mir-27b gene was unchanged between liver cancers and paired non-tumor liver tissues (Figure 2F). These results suggest that genetic deletion likely accounts for reduced miR-27b levels in liver cancers.
The combinational therapy of miR-27b and doxorubicin shows synergistic therapeutic effect on liver cancers in vivo
Due to the strong side effect of doxorubicin, it is desirable in clinical practice to achieve therapeutic effect with low drug doses, especially for liver cancer patients who have impaired liver functions9. Therefore, we asked whether the combinational use of miR-27b and doxorubicin at low doses shows synergistic therapeutic effect in vivo (Figure 3A). HepG2 cells were orthotopically transplanted into the livers of immunodeficient mice to form liver tumors. The tumor-bearing mice were then treated with miR-27b and doxorubicin, either alone or in combination. Based on pilot experiments (data not shown), 3 mg/kg doxorubicin was applied, which was around 3-fold lower than the doses used in the clinic27 and showed minimal toxicity. miR-27b was efficiently delivered into the transplanted liver tumors as determined by q-PCR (Figure 3B). Significant reduction in liver cancer growth was observed with combinational miR-27b and doxorubicin treatment (Figure 3C and 3D), while miR-27b or doxorubicin treatment alone showed minimal effect on liver tumor size. The combinational treatment significantly reduced cancer cell proliferation and enhanced cancer cell death as shown by the decreased number of Ki67-positive cells (Figure 3E and Supplementary information, Figure S4A) and increased cleaved-caspase-3 levels (Figure 3F), respectively. It is noteworthy that liver damage was undetectable after the combinational therapy, as demonstrated by normal liver histology (Figure 3G), unchanged hepatocyte proliferation (Figure 3G), and unchanged serum levels of liver injury markers ALT, AST, and total bilirubin (Figure 3H-3J), suggesting that the combinational treatment specifically attacks liver cancer cells.
Figure 3.
The combinational use of miR-27b and doxorubicin shows synergistic therapeutic effect on liver cancers in vivo. (A) Schematic outline of the combinational therapy in a liver cancer mouse model. HepG2 cells mixed with matrigel were orthotopically transplanted into the livers of immunodeficient mice. Cancers were allowed to develop for 3 weeks before three rounds of combined treatment of miR-27b and doxorubicin. (B) Levels of miR-27b in transplanted tumors were determined by q-PCR. (C, D) After three rounds of sequential treatment, cancer volumes were quantified (n = 5 for each group). NS: not significant. (E) Proliferation of tumor cells was characterized by Ki67 staining (n = 5 for each group). (F) Full-length caspase-3 and cleaved caspase-3 levels of tumor cells were detected by western blot analysis. Three independent replicates were shown. (G) Tissue damages in mouse normal livers were undetectable after the combinational treatment as characterized by Hematoxylin and eosin (H&E) staining. The proliferation of hepatocytes in non-tumor tissues were characterized by Ki67 staining. Scale bar, 50 μm. (H-J) Serum levels of liver injury markers ALT (H), AST (I) and total bilirubin (J) were determined. The normal serum levels of these markers for mice are as follows: ALT (24-140 U/L), AST (72-288 U/L), total bilirubin (5-17 μM).
miR-27b synergizes with anticancer drugs in kidney cancers
We next asked whether miR-27b could enhance responses of other types of drug-resistant cancers, such as kidney cancers2. Markedly, we found that forced miR-27b expression sensitized kidney cancer cell line ACHN to different anticancer drugs (Figure 4A). Moreover, the drug sensitizing effect of miR-27b could be extended to three additional kidney cancer cell lines, 786-O, 769-P, and Caki-1 (Figure 4B).
Figure 4.
miR-27b synergizes with anticancer drugs in kidney cancers. (A) miR-27b enhances the sensitivity of kidney cancer cell line ACHN to multiple anticancer drugs. IC50 for each treatment was presented. (B) Enhanced response to doxorubicin by miR-27b overexpression was shown in multiple kidney cancer cell lines, including 786-O, 769-P and Caki-1. IC50 for each treatment was presented. (C) Schematic outline of the combinational therapy in a kidney cancer mouse model. ACHN cells were subcutaneously transplanted into immunodeficient mice. Cancers were allowed to develop for 4 weeks before four rounds of combined treatment of miR-27b and doxorubicin. (D) Levels of miR-27b in transplanted tumors were determined by q-PCR. (E) Tumor volumes were quantified (n = 5 for each group; *P < 0.05). (F) Proliferation of tumor cells was characterized by Ki67 staining (n = 5 for each group). (G) Full-length caspase-3 and cleaved caspase-3 levels of tumor cells were detected by western blot analysis. Three independent replicates were shown. (H) miR-27b expression levels were reduced in human kidney cancer samples (n = 66, P< 0.001) compared with paired non-cancerous tissues. (I) Loss of mir-27b alleles in human kidney cancers (21 out of 66) as determined by copy number variation analysis of the mir-27b gene locus. The relative log2 copy number ratio < −0.25 represents copy number loss. (J) Hypermethylation of the mir-27b promoter in human kidney cancers (n = 24). β value was applied to indicate the levels of DNA methylation at the promoter of the mir-27b gene.
To confirm the synergistic effect in vivo, miR-27b was delivered into subcutaneously transplanted ACHN kidney tumors by intratumoral injection 2 days before doxorubicin treatment (Figure 4C and 4D). Combinational treatment of miR-27b and doxorubicin significantly suppressed tumor growth (Figure 4E and Supplementary information, Table S5). Remarkably reduced cell proliferation and increased cell death were observed in kidney tumors after combinational treatment (Figure 4F, 4G and Supplementary information, Figure S4B and S4C).
Furthermore, we found that miR-27b levels were reduced in 66 kidney cancers compared with paired non-cancerous tissues in TCGA (Figure 4H and Supplementary information, Table S4). Twenty-one out of the sixty-six kidney cancers displayed simultaneous loss of C9orf3 and FANCC as well as the miR-23b-27b-24-1 gene cluster by analyzing the copy number variation (relative log2 copy number ratio < −0.25; Figure 4I and Supplementary information, Figure S4D and S4E). In addition, the level of DNA methylation at the promoter of the mir-27b gene was increased in human kidney cancers (Figure 4J). Because genetic deletion and DNA hypermethylation account for decreased expression of tumor suppressor genes28, attenuated miR-27b expression in kidney cancers may be caused by both genetic and epigenetic aberrations. Together, our data indicate that miR-27b is a master miRNA that synergizes with anticancer drugs in the treatment of liver cancers and kidney cancers, both of which are notorious for multidrug resistance.
miR-27b enhances the drug response by activating p53
We next analyzed the mechanisms by which miR-27b synergizes with multiple anticancer drugs. By comparing the whole-genome expression profiles of miR-27b-overexpressing cells with those of scrambled miRNA-overexpressing cells after doxorubicin treatment, we found that levels of the ABC transporter family genes and doxorubicin target genes, e.g., Topoisomerase IIα, were unchanged (Supplementary information, Table S6). Intracellular concentrations of doxorubicin as revealed by its autofluorescence detected by FACS were not altered in cells with forced miR-27b expression (Supplementary information, Figure S5A). Because both doxorubicin and its major metabolites emit red fluorescence and, importantly, both of them are substrates of ABC transporters29, therefore, it is unlikely that miR-27b promotes drug sensitivity by reducing drug efflux.
Expression profile analysis revealed that the expression levels of genes associated with cell cycle transition and cell death were dramatically changed upon miR-27b overexpression (Supplementary information, Table S6), which were in line with the bioinformatic prediction (Figure 1E). Indeed, miR-27b enhanced doxorubicin-induced S phase arrest (Figure 5A) and cell death (Figure 5B and Supplementary information, Figure S5B and S5C), while miR-27b alone showed no significant effect on cell proliferation or cell death (Figure 5A, 5B and Supplementary information, Figure S1H, S5C, S5D and Table S2). By performing Gene Set Enrichment Analysis (GSEA)30, we found that the p53 pathway was listed as the most enriched functional pathway (Figure 5C and Supplementary information, Figure S5E). miR-27b-mediated increase in phosphorylated p53 and total p53 levels was confirmed in three p53-wild-type cell lines after doxorubicin treatment (Figure 5D). Knockdown of p53 reversed the miR-27b-mediated sensitizing effect in these cells (Figure 5E and Supplementary information, Figure S5F). Together, these results demonstrate that miR-27b enhances anticancer drug-induced cell death by promoting p53 activity.
Figure 5.
miR-27b enhances drug responses by activating p53. (A) miR-27b enhances doxorubicin-induced S phase arrest as assessed by propidium iodide (PI) staining. (B) miR-27b increases doxorubicin-induced apoptosis as determined by Annexin V staining. (C) Eight top-listed pathways regulated by miR-27b. Expression profiles were analyzed between cells transfected with miR-27b and scrambled miRNA control after doxorubicin treatment. Signaling pathways enriched in cells with forced miR-27b expression were identified by GSEA. (D) Phosphorylated p53 (pp53) and total p53 levels were determined by western blot analysis in HepG2, ACHN and 769-P cells. (E) miR-27b-induced sensitizing effect was reverted by shRNA-mediated p53 knockdown in HepG2, ACHN and 769-P cells.
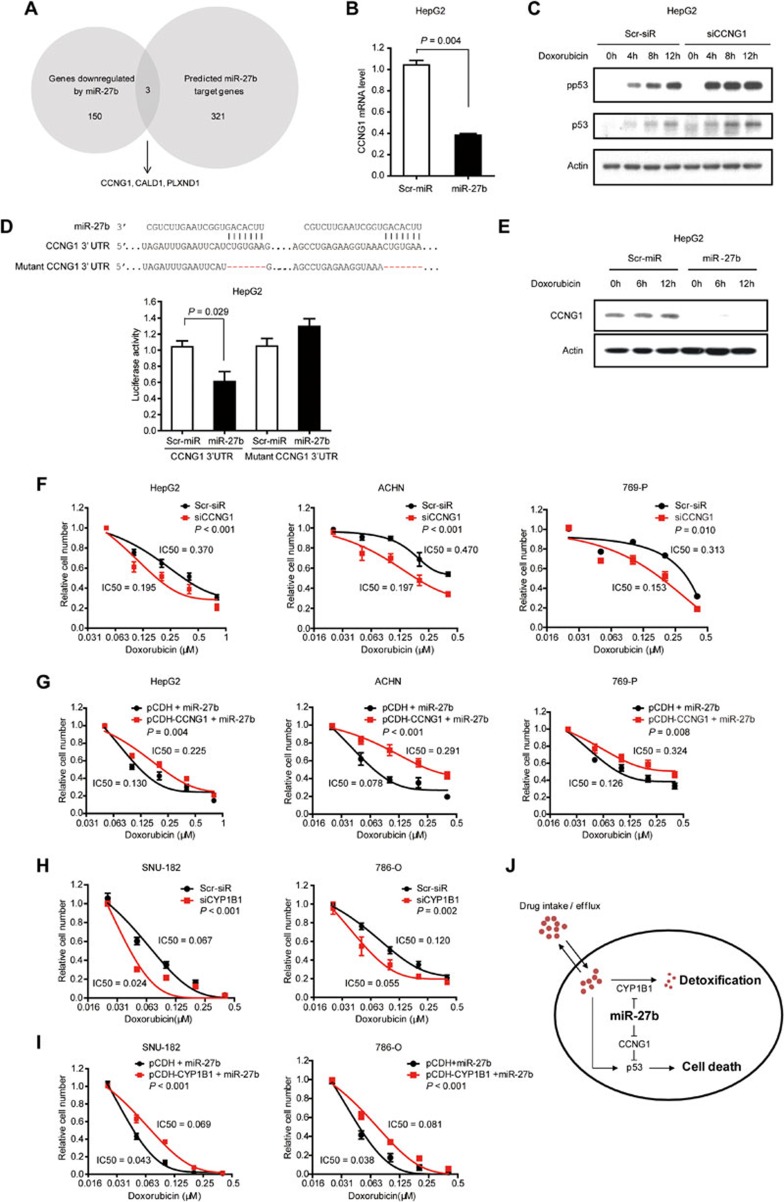
miR-27b targets CCNG1 to regulate p53 activity
Because miRNAs suppress the expression of their target genes, it is unlikely that miR-27b increases p53 activity directly. We predicted miR-27b target genes using Targetscan31, Pictar32, and miRanda33. By comparing 321 predicted miR-27b target genes with miR-27b-downregulated genes from microarray assays (Supplementary information, Table S7), we identified three potential miR-27b targets, CCNG1, CALD1, and PLXND1 (Figure 6A). The mRNA levels of these three genes were reduced upon miR-27b overexpression as confirmed via q-PCR (Figure 6B and Supplementary information, Figure S6A and S6B). Notably, only CCNG1 knockdown enhanced the levels of phosphorylated p53 and total p53 (Figure 6C and Supplementary information, Figure S6C-S6E), therefore CALD1 and PLXND1 were not followed in the current study. Consistent with this result, previous studies have shown that CCNG1 is a negative regulator of p53 in fibroblasts34 and liver cells35.
Figure 6.
miR-27b enhances drug sensitivity by targeting CCNG1 and CYP1B1. (A) Strategies to identify miR-27b target genes. Microarray analysis showed that 150 genes were downregulated (> 2.5-fold) upon forced miR-27b expression. Three hundred and twenty-one miR-27b target genes were predicted by Targetscan, Pictar, and miRanda in common. By comparing genes downregulated by miR-27b with predicted targets, CCNG1, CALD1 and PLXND1 were identified as potential candidate targets. (B) miR-27b overexpression reduced CCNG1 mRNA level as measured by q-PCR in doxorubicin-treated HepG2 cells. (C) siRNA-mediated CCNG1 knockdown increased phosphorylated and total p53 levels as shown by western blot analysis. (D) miR-27b reduced the activity of the luciferase reporter with CCNG1 wild-type 3′-UTR but not that with the mutant 3′-UTR. Sequence alignment of miR-27b and CCNG1 3′-UTR was shown. (E) Forty-eight hours after miRNA transfection, HepG2 cells were treated with doxorubicin for the indicated time points. Protein levels of CCNG1 were determined by western blot analysis. miR-27b efficiently reduced CCNG1 protein levels. (F) siRNA-mediated CCNG1 knockdown conferred drug sensitivity on HepG2, ACHN and 769-P cells. (G) CCNG1 overexpression attenuated the sensitizing effect induced by miR-27b in HepG2, ACHN, and 769-P cells. CCNG1-overexpressing cells were transfected with miR-27b mimics and then treated with doxorubicin. (H) siRNA-mediated CYP1B1 knockdown conferred sensitivity on p53-deficient cancer cell lines, SNU-182 and 786-O. (I) Ectopic expression of CYP1B1 restored resistance to doxorubicin in miR-27b-overexpressing SNU-182 and 786-O cells. (J) Schematic model of miR-27b in regulating drug response. miR-27b increases drug response in cancer cells through increasing p53-mediated cell death in a CCNG1-dependent manner. In addition, miR-27b improves drug sensitivity by reducing CYP1B1 expression levels, which is a key enzyme controlling detoxification of anticancer drugs. In cancer cells with wild-type p53 or high CYP1B1 activities, forced expression of miR-27b would enhance the response to anticancer drugs.
We determined whether miR-27b reduces CCNG1 levels via direct binding to its 3′-UTR. Wild-type or mutant 3′-UTR of CCNG1 was cloned into a luciferase reporter construct. miR-27b significantly reduced the luciferase activity of the reporter with wild-type 3′-UTR but not that harboring the mutant 3′-UTR (Figure 6D). In addition, miR-27b markedly reduced CCNG1 protein levels, suggesting that CCNG1 is a bona fide target of miR-27b (Figure 6E).
Next, we analyzed whether reduced CCNG1 levels are responsible for miR-27b-enhanced drug sensitivity. Similar to forced miR-27b expression, siRNA-mediated CCNG1 knockdown promoted the response to doxorubicin in HepG2, ACHN, and 769-P cells (Figure 6F). By contrast, CCNG1 overexpression attenuated the sensitizing effect to doxorubicin caused by miR-27b (Figure 6G). It is noteworthy that the miR-27b-CCNG1-p53 mechanism also accounted for the regulation of sensitivity to sorafenib (Supplementary information, Figure S7A-S7C). Taken together, these results showed that miR-27b augments p53-dependent drug sensitivity via targeting CCNG1.
miR-27b increases drug sensitivity by additionally reducing CYP1B1 levels
Intriguingly, of the cell lines characterized, HepG2, ACHN, Caki-1, and 769-P cells carry wild-type p53 alleles, whereas SNU-182, SNU-739, Tong, and 786-O cells harbor missense p53 mutations (Supplementary information, Table S8). Although miR-27b attenuated CCNG1 expression in p53-mutant cell lines (Supplementary information, Figure S8A), we found that knockdown of CCNG1 alone did not confer drug sensitivity on these cancer cells (Supplementary information, Figure S8B). These results indicate that miR-27b promotes drug sensitivity via additional mechanisms in p53-mutant cancer cells.
We focused on one of the top potential targets of miR-27b predicted by Targetscan, Pictar and miRanda, CYP1B136, because CYP1B1 is a major P450 enzyme that inactivates and detoxifies a large panel of anticancer drugs, including doxorubicin37. miR-27b markedly decreased doxorubicin-induced CYP1B1 protein levels both in cultured cancer cells and in orthotopically transplanted liver cancers, but the mRNA levels of CYP1B1 were unchanged (Supplementary information, Figure S9A-S9C). Accordingly, miR-27b reduced the activity of the luciferase reporter with wild-type 3′-UTR of CYP1B1, but not the reporter with mutant 3′-UTR (Supplementary information, Figure S9D).
In addition, we further analyzed the function of CYP1B1 in miR-27b-enhanced drug sensitivity. CYP1B1 knockdown (Supplementary information, Figure S10A) increased the sensitivity to doxorubicin in p53-mutant cells SNU-182 and 786-O (Figure 6H) as well as in p53-wild-type cells HepG2 and ACHN (Supplementary information, Figure S10B). Furthermore, overexpression of CYP1B1 attenuated the sensitizing effect to doxorubicin caused by miR-27b in both p53-wild-type and -mutant cells (Figure 6I and Supplementary information, Figure S10C). Interestingly, forced CYP1B1 expression also reduced the miR-27b-mediated sensitivity to sorafenib (Supplementary information, Figure S10D). Together, these data prove that miR-27b synergizes with anticancer drugs via at least two essential mechanisms, i.e., promoting wild-type p53-dependent cell death by targeting CCNG1 and reducing drug detoxification by targeting CYP1B1 (Figure 6J).
miR-27b promotes drug responses in patients with wild-type p53 or high CYP1B1 levels
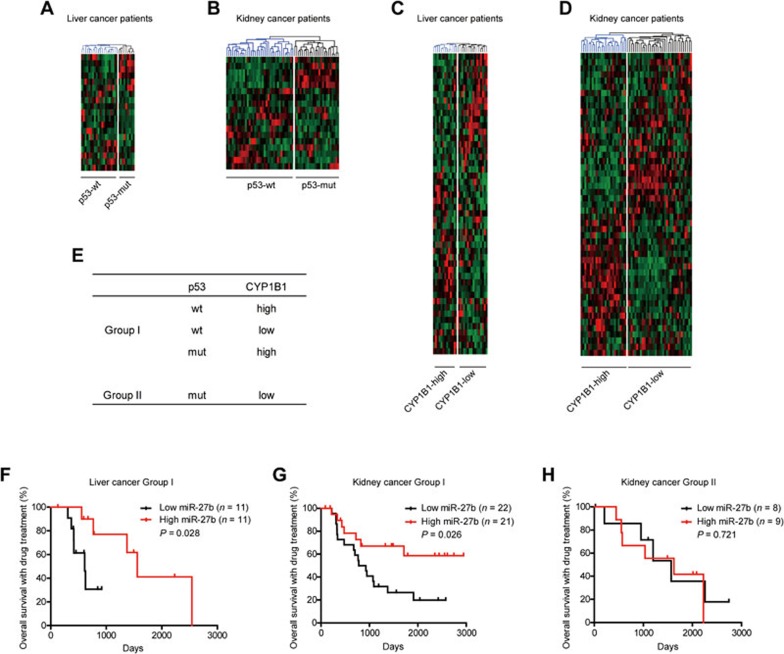
The lack of clear molecular classification impedes the development of personalized therapies, especially for liver cancers38. Our findings (Figure 6J) suggest that the p53 mutation status and CYP1B1 expression levels are key factors determining miR-27b-mediated drug sensitivity. These results also infer that upon anticancer drug treatment, high miR-27b levels would be beneficial for patients with either wild-type p53 or high CYP1B1 levels but not those with both mutated p53 and low CYP1B1 levels. To verify this hypothesis, we analyzed the data of cancer patients in TCGA with annotated information of drug treatment. However, the p53 mutation status and CYP1B1 protein levels were unavailable in TCGA. Instead, we stratified patients using gene expression signature, which has been shown to provide an accurate measurement of the function of a particular gene in cancer cells39.
Previous studies have proven that a 32-gene signature robustly distinguishes p53-wild-type from p53-mutant cancers40. To establish a gene signature representing CYP1B1 protein levels (Supplementary information, Figure S11A), we analyzed 14 cancer cell lines with known CYP1B1 protein levels from the Human Protein Atlas41,42 and known whole-genome expression profiles from the Cancer Cell Line Encyclopedia (CCLE)43. These cell lines were first divided into two groups according to their CYP1B1 protein levels, and genes differentially expressed between CYP1B1-high and -low cell lines were identified using the GenePattern analysis44. A classifier constituting 57 genes (Supplementary information, Table S9), which perfectly distinguished the 14 cell lines according to CYP1B1 protein levels, was referred to as the CYP1B1 signature (Supplementary information, Figure S11B). To evaluate the performance of the CYP1B1 signature, 64 liver and kidney cancer cell lines with expression profile data available in CCLE were clustered into two groups, CYP1B1-high and CYP1B1-low, according to their CYP1B1 signatures (Supplementary information, Figure S11C). Notably, the predicted CYP1B1 protein levels in eight cell lines available in our laboratory were successfully validated via western blot analysis, supporting a robust discriminative power of the CYP1B1 signature genes (Supplementary information, Figure S11D).
We retrieved data of all liver (29 patients) and kidney cancer patients (60 patients) from TCGA, who had annotated information of treatment with cytotoxic drugs or multikinase inhibitors (Supplementary information, Table S10). According to the p53 and CYP1B1 signatures, these patients were stratified into the two groups via hierarchical clustering (Figure 7A-7E and Supplementary information, Table S10). Group I patients have either a p53-wild-type signature or a CYP1B1-high signature and Group II patients have both p53-mutant and CYP1B1-low signatures. Twenty-two out of the twenty-nine liver cancer patients were classified as Group I. Patients with high levels of miR-27b in this group (11 out of 22) showed significantly extended overall survival time after anticancer drug treatment (Figure 7F). Forty-three out of sixty kidney cancer patients were classified as Group I. Remarkably, patients with high miR-27b levels also showed good response to anticancer drug treatment in Group I (Figure 7G), but not in Group II kidney cancer patients (Figure 7H). These data suggest that miR-27b improves the responses to anticancer drugs specifically in patients with wild-type p53 or high CYP1B1 levels.
Figure 7.
miR-27b promotes drug responses in patients with either wild-type p53 or high CYP1B1 levels. (A-D) Expression profiles of 29 liver cancer patients and 60 kidney cancer patients were retrieved from TCGA and clustered according to the p53 signature (A, B) or the CYP1B1 signature (C, D). (E) The patients were stratified according to the p53 and CYP1B1 signatures. Patients with either a p53-wild-type (p53-wt) or a CYP1B1-high signature were classified as Group I. Patients with both p53-mutant (p53-mut) and CYP1B1-low signatures were classified as Group II. (F) Liver cancer patients who were treated with cytotoxic drugs and/or multikinase inhibitors were classified into Group I (n = 22) and Group II (n = 7). Kaplan-Meier analysis showed that patients with high miR-27b levels showed prolonged the overall survival in Group I liver cancer patients (log-rank test). The sample size of Group II is too small and thus this group cannot be analyzed using Kaplan-Meier method. (G, H) Kidney cancer patients who were treated with cytotoxic drugs and/or multikinase inhibitors were classified into Group I (G, n = 43) and Group II (H, n = 17). Kaplan-Meier analysis showed that patients with high miR-27b levels showed prolonged overall survival in Group I kidney cancer patients (G, log-rank test), but not in Group II patients (H, log-rank test). Original raw data were all derived from TCGA database.
We also extended the analysis to other types of human cancers. Remarkably, in lung cancers and head and neck cancers, Group I patients with high miR-27b levels showed prolonged overall survival after anticancer drug treatment (Supplementary information, Figure S12A and S12C), whereas high miR-27b levels had no effect on drug response in Group II patients (Supplementary information, Figure S12B and S12D). Data from these four types of cancers, comprising 215 patients in total, support that miR-27b synergizes with anticancer drugs largely through p53 and CYP1B1. More importantly, these results suggest a selection strategy of potential responsive patients for personalized combinational therapy of miR-27b and anticancer drugs.
Discussion
Because of the complexity and redundancy of drug resistance mechanisms, ablation of a single pathway is unable to produce sustained drug response. Novel approaches that interfere with multiple essential drug resistance mechanisms are believed to promote the outcome of anticancer drug treatment12. In this study, by functional screening and bioinformatic analysis followed by exploring publically available human liver cancer and kidney cancer data, we identified miR-27b as a master miRNA antagonizing multiple drug resistance mechanisms, which remarkably increases drug responses to various anticancer drugs, including doxorubicin and sorafenib. Sorafenib is the only FDA-approved medicine for systemic liver cancer treatment. However, < 5% of the late-stage patients respond to sorafenib3,45. Intriguingly, based on the data retrieved from TCGA, patients with high miR-27b levels showed prolonged survival after treatment with sorafenib or other compounds in about 60%-75% cases (Figure 7F, 7G and Supplementary information, Figure S12A and S12C). In addition, we would like to emphasize that our findings indicate a promising approach for the better use of traditional chemotherapeutic compounds, such as doxorubicin and cisplatin, which, despite arguable therapeutic effect, are widely used in Asian clinical practices. Importantly, miR-27b improves the response to these compounds used at low concentrations. This is of specific interest because most advanced liver and kidney cancer patients are not tolerable to standard dosage of anticancer drugs.
Previous studies have identified master miRNAs in cancer initiation and promotion by whole-genome sequencing13 and integrated analysis of the miRNA-transcription factor interactions46,47. These studies lay the ground for using large-scale genomics or network biology to identify master miRNAs. In recent years, accumulating evidence has shown that miRNAs also play important roles in drug response of human cancers. While most studies on drug response focus on individual miRNAs, genome-wide evaluation of each miRNA in the background of the entire miRNome is largely uncharacterized48. By functional screening, we identified 22 miRNAs that promote drug sensitivity in liver cancer cells. Importantly, our results provide a network-based perspective to understand multiple drug resistance mechanisms and help identify master regulators of drug response. Interestingly, there are overlapping pathways predicted to be regulated by multiple miRNAs (Figure 1E). For example, both miR-20a and miR-27b induce cell apoptosis. On the other hand, miR-20b and miR-183 are predicted to target the AKT pathway, one of the most important cell survival pathways. Investigation of combination of multiple miRNAs may ensure promising therapeutic strategies in the future.
Notably, miR-27b is located in the center of the miRNA-pathway network. The function of miR-27b was characterized in detail in liver cancers and kidney cancers, both of which are refractory to anticancer drugs. We found that miR-27b sensitized cancer cells in vitro and in vivo to various types of drugs, including chemotherapeutic drugs and molecularly targeted drugs. In addition, miR-27b improves drug response not only in a large panel of cancer cell lines but also in patient-derived primary cancer cells. Besides the potent drug-sensitizing effect, the significance of miR-27b was further highlighted by the finding that the mir-27b gene locus is genetically deleted or hypermethylated in human liver and kidney cancers, suggesting that low miR-27b levels might confer an advantage on cancer cells and that restoring miR-27b might selectively kill cancer cells in combination with anticancer drugs. Taken together, we consider miR-27b as a master miRNA in enhancing drug sensitivity. Interestingly, miR-27a, which is a homolog of miR-27b and located on a different chromosome, was not identified in the screen (Figure 1B). Although the predicted target genes of these two miRNAs are largely overlapped, variations in mature sequences at the 3′ ends of miR-27a and miR-27b may lead to different binding capacities for target genes49. According to the data in miRTarBase, less than 15% of the experimentally validated targets are shared by miR-27a and miR-27b. Also, miR-27b, more abundantly expressed in the liver, is unlikely to be co-expressed with miR-27a in cancerous and non-cancerous liver tissues (Figure 2D and Supplementary information, Figure S3G and S3H). Taken together, miR-27a may not play a redundant effect in regulating drug sensitivity.
Pioneered studies have been successful in reintroducing miRNAs into cancer cells as single agents to inhibit tumor growth14,50. Interestingly, our study demonstrated the use of miRNA as an adjuvant regimen, because combination of miR-27b and doxorubicin, but not miR-27b alone, showed potent therapeutic effect on liver and kidney cancers. Combinational therapies of chemical compounds have already been shown as promising approaches to achieve a synergistic effect. For example, cetuximab with irinotecan and lapatinib with capecitabine have successfully prolonged progression-free survival in colon51 and breast cancer patients52, respectively. We think that miRNA-based combinational therapies have two apparent advantages: miRNAs are relatively safe for in vivo delivery, and miRNAs are powerful in overcoming drug resistance due to the multitarget characteristic15. In addition, miR-27b-based combinational therapy was effective in vivo even though the dose of doxorubicin was used much lower than that in clinical routine, representing an important advance for the rational use of traditional chemotherapeutic compounds, which are still the first choice in developing countries. In the future, miR-27b and anticancer drugs might be applied as an encapsulated “cocktail” to ensure delivery to the same tumor cell and achieve maximal synergistic effect53.
Remarkably, we showed that miR-27b improved the response to various types of anticancer drugs in multiple human cancer cell lines. We found that the forced expression of miR-27b strongly increased the levels of phosphorylated and total p53 proteins. We further revealed that miR-27b activates p53 by antagonizing CCNG1. Unlike the complete deletion of CCNG154, the partially attenuated CCNG1 expression by miR-27b overexpression has no effect on cell cycle progression in non-stimulated cells (Figure 5A). However, doxorubicin induces S phase arrest and miR-27b promotes the sensitivity of cancer cells to doxorubicin, leading to more severe replication defects (Figure 5A). CCNG1 can negatively regulate ATM-dependent p53 activation during DNA damage54. In addition, CCNG1 recruits the β-subunit of phosphatase 2A (PP2A) to dephosphorylate MDM2, thereby resulting in p53 degradation34. Intriguingly, p53 mutations are rarely identified in human kidney cancers55. pVHL, often mutated in kidney cancers, was found to stabilize p53 by suppressing Mdm2-mediated ubiquitination56. Therefore, attenuated p53 activity caused by VHL mutation not only explains why kidney cancers are resistant to anticancer drug therapy, but also supports our finding that enhancing p53 activity by miR-27b can improve drug response in kidney cancers.
miR-27b also promotes drug response in p53-mutated cancer cells. In these cancers, miR-27b targets CYP1B1 to enhance drug sensitivity. In addition to the posttranscriptional regulation of CYP1B1 by miR-27b36, we further uncovered the function of CYP1B1 in miR-27b-mediated drug sensitivity and constructed a CYP1B1 signature to stratify cancer patients, which might facilitate the personalized use of miR-27b-based combinational therapy. CYP1B1 belongs to the cytochrome P450 superfamily, which contains major enzymes controlling the detoxification of anticancer drugs57. CYP1B1 conjugates and inactivates anticancer agents with high affinity, which is facilitated by additional binding of cofactors37. Interestingly, although CYP1B1 is barely detected in non-cancerous tissues, the protein levels of CYP1B1 are elevated in various types of malignant cancers including liver cancers and kidney cancers58, which suggest that CYP1B1 may represent a tumor-specific target. Intriguingly, a recent study showed miR-27b-dependent regulation of dihydropyrimidine dehydrogenase, a rate-limiting enzyme in metabolism of 5-fluorouracil, further suggesting a key role of miR-27b in the detoxification of anticancer drugs59. Taken together, the combinational therapy of miR-27b and anticancer drugs greatly cuts off the “escape route” for cancer cells by both increasing p53-mediated cell death and suppressing CYP1B1-mediated drug detoxification (Figure 6J). Our approach fortifies the concept of “horizontal” targeting of parallel pathways, which has been widely applied in therapies using small compounds targeting essential survival pathways60.
Because human cancers develop with a high degree of molecular heterogeneity61, the responses to the same type of therapy are dramatically different among cancer patients. One challenge is to identify molecular subtypes of individuals that are susceptible to a particular treatment, thereby maximizing treatment efficacy in defined populations. Our molecular findings suggested that miR-27b could be used in a personalized manner by evaluating its major functional effectors. Indeed, by stratifying patients according to the p53- and CYP1B1-gene signatures, we found that high miR-27b levels confer good drug response specifically on patients with wild-type p53 or high CYP1B1 levels. It is notable that while our study took liver and kidney cancers as a starting point, the finding could be extended to human lung cancers and head and neck cancers. In conclusion, we provide a combinational use of miR-27b and anticancer drugs as a new therapeutic strategy that would be beneficial for p53-wild-type or CYP1B1-high patients.
Materials and Methods
High-throughput miRNA screen
The screen library includes 828 synthesized human miRNA mimics20. HepG2 cells were seeded in 96-well plates (1 × 104 cells per well). Twelve hours later, cells were transfected with miRNA mimics (final concentration 50 nM) using Lipofectamine RNAiMAX (Invitrogen). Each miRNA was transfected with two replicas in the same plate. Forty-eight hours after miRNA transfection, one was treated with 0.1 μM doxorubicin for 3 days and the other was still cultured in doxorubicin-free medium. To assess the effect of each miRNA on doxorubicin response, the number of viable cells was measured using CellTiter-Glo Luminescent Cell Viability Assay (Promega). “Relative cell number” for an individual miRNA = the number of viable cells in doxorubicin-containing medium / the number of viable cells in doxorubicin-free medium. “Normalized to Scr-miR” is defined as the ratio of “Relative cell number” for each miRNA to the scrambled miRNA control.
The Z-score was used to normalize these raw data in a way that provides explicit information regarding the strength of each miRNA relative to the rest of the sample distribution. In brief, Z-score = (sample value – sample mean) / sample SD, where “sample” represents the relative cell number of each miRNA that has been normalized to the scrambled miRNA control.
Liver cancer model for the combinational therapy of miR-27b and doxorubicin
HepG2 cells (3 × 106) mixed with matrigel were injected into the largest lobe of the liver in immunodeficient mice. Liver cancers were allowed to form for 3 weeks. Scrambled miRNA control and miR-27b (3.5 mg/kg) were encapsulated in liposomes using Inviofectamine 2.0 (Invitrogen) in a volume of 200 μl and were incubated for 30 min at 50 °C before tail vein injection. Doxorubicin was administered through tail vein injection at 3 mg/kg, which was around 3-fold lower than the doses used in the clinic27. After three rounds of sequential treatment, cancer volumes were calculated according to the formula: volume = a × b2/ 2, where “a” represents the largest diameter and “b” represents the perpendicular diameter. All animal experiments were performed according to institutional animal regulations.
Kidney cancer model for the combinational therapy of miR-27b and doxorubicin
ACHN cells (2 × 106) were injected subcutaneously into immunodeficient mice. Kidney cancers were allowed to form for 4 weeks. Scrambled miRNA control and miR-27b (15 μg/tumor) were encapsulated in liposomes using Inviofectamine 2.0 (Invitrogen) in a volume of 20 μl and were incubated for 30 min at 50 °C before intratumoral injection. Doxorubicin was administered at 6 mg/kg through tail vein injection. After four rounds of sequential treatment, cancer volumes were calculated according to the formula: volume = a × b2/ 2, where “a” represents the largest diameter and “b” represents the perpendicular diameter. All animal experiments were performed according to institutional animal regulations.
Cell culture
Human cancer cell lines, HepG2, HeLa, Tong, and HEK-293FT were cultured in DMEM medium. SNU-182, SNU-739, 769-P, and 786-O cells were cultured in RPMI1640 medium. ACHN cells were cultured in MEM medium. Caki-1 cells were cultured in McCoy's 5A medium. All the media were supplemented with 10% fetal bovine serum.
For cancer cell primary culture, all patients were from the Eastern Hepatobilliary Surgery Hospital in Shanghai, China. The liver cancer specimens were collected with patient consent. Samples were transported on ice to the laboratory within 1 h of collection. Solid tumor specimens were rinsed twice with PBS supplemented with penicillin and streptomycin and finely minced with scissors. Both necrotic tissues and apparently normal tissues were discarded. Tumor fragments were digested by 0.1% collagenase intravenously for 30 min at 37 °C and then filtered through a 70-μm nylon cell strainer. Cell suspension was centrifuged and the supernatant was then transferred to collagen-coated dishes. Cells were cultured in RPMI1640 medium supplemented with 10% fetal bovine serum, 10 mg/L insulin, 5.5 mg/L transferrin, 6.7 μg/L sodium selenite and 40 μg/L of epidermal growth factor.
All the above cell lines and primary cultured cancer cells were maintained at 37 °C in a humidified incubator at 5% CO2.
Drug sensitivity test and IC50 estimation
Cells were seeded in 96-well plates. Twelve hours later, cells were transfected with miR-27b or Scr-miR (final concentration 50 nM) using Lipofectamine RNAiMAX (Invitrogen). Forty-eight hours after miRNA transfection, the cells were treated with the indicated concentration of anticancer drugs as shown in the corresponding figures for 3 days. The number of viable cells was measured using CellTiter-Glo Luminescent Cell Viability Assay (Promega). “Relative cell number” = the number of viable cells in drug-containing medium / the number of viable cells in drug-free medium. “Relative cell number” was further fitted to a dose-response curve to estimate the IC50 by the SPSS software.
Cell cycle analysis
For cell cycle analysis, HepG2 cells (2 × 105 per well) were plated in 6-well plates. Twelve hours later, the cells were transfected with miR-27b or scrambled miRNA control. Two days after transfection, the cells were treated with 0.5 μM doxorubicin for 48 h and then cultured in the absence of doxorubicin for another 24 h. Next, the cells were trypsinized, washed in PBS, and fixed with ice-cold 70% ethanol for 30 min at 4 °C. After washing in cold PBS for three times, cells were resuspended in 0.5 ml PBS solution with 50 μg/ml propidium iodide and 20 μg/ml RNase A for 30 min at 37 °C. Samples were then analyzed for their DNA content by FACSCalibur (Becton Dickinson).
Apoptosis analysis
For apoptosis analysis, HepG2 cells were plated in 6-well plates (2 × 105 cells per well). Twelve hours later, the cells were transfected with miR-27b or scrambled miRNA control. Two days after transfection, cells were treated with 1 μM doxorubicin for 24 h and then cultured in the absence of doxorubicin for another 24 h. Apoptotic cells were detected by Annexin V-FITC Apoptosis Detection Kit (Sigma) and analyzed by FACSCalibur.
RNA extraction and q-PCR
Total RNA was extracted using Trizol reagent (Invitrogen). 1 μg RNA was reversely transcribed into cDNA with M-MLV Reverse Transcriptase (Promega). q-PCR was performed with SYBR Premix Ex Taq (TaKaRa) on an ABI 7500 fast real-time PCR system (Applied Biosystems). Primer sequences will be provided upon request. Quantification of mature miRNAs was performed with a stem-loop real-time PCR.
Antibodies
Antibodies targeting the following epitopes were used for western blot assays: p53 (Cell Signaling, no 2524, 1:1 000), p53 phosphorylated at Ser15 (Cell Signaling, no 9284, 1:1 500), caspase-3 (Cell Signaling, no 9662, 1:1 000), cleaved caspase-3 (Cell Signaling, no 9664, 1:1 000), CYP1B1 (Abcam, ab33586, 1:2 000), CCNG1 (Santa Cruz, sc-320, 1:1 500) and β-Actin (Sigma, A2228, 1:6 000).
Vector constructs
The open reading frames of the CCNG1 or CYP1B1 gene were amplified and cloned into a lentiviral pCDH vector, the corresponding primers are listed in Supplementary information, Table S11. For the luciferase reporter construct containing the wild-type 3′-UTRs, single-strand oligos and their complementary strands were annealed to generate a ∼100 bp double-strand DNA fragment containing the miR-27b recognition sequences in the 3′-UTRs of CCNG1 or CYP1B1. Of note, the four alternative transcripts of CCNG1 harbor the miR-27b binding sites in their 3′-UTRs. For sequence-deleted 3′-UTRs, oligos without the miR-27b recognition sequences were annealed. Both wild-type and seed sequence-deleted 3′-UTRs of CCNG1, or CYP1B1 were cloned downstream of a cytomegalovirus (CMV) promoter-driven firefly luciferase cassette in a pcDNA3.0 vector. The corresponding oligos are listed in Supplementary information, Table S11.
Lentivirus production and transfection
pCDH-CCNG1 or pCDH-CYP1B1 was co-transfected with the packaging plasmid psPAX2 and the envelope plasmid pMD2.G into HEK-293FT cells using Calcium phosphate transfection. Virus particles were harvested 48 h after transfection. HepG2 cells were infected with virus particles plus 4 μg/ml Polybrene (Sigma).
Oligonucleotide transfection
miR-27b mimics and scrambled miRNA mimics used for in vitro studies were purchased from Dharmacon. In vivo Ready miR-27b mimics and scrambled miRNA control used for in vivo experiment were purchased from Ambion. siRNAs of CCNG1, CYP1B1, and scrambled siRNA control were purchased from Ribobio. Oligonucleotide transfection was performed with Lipofectamine RNAiMAX Reagent (Invitrogen).
Luciferase assay
HepG2 cells were cultured in 12-well plates (1 × 105 cells per well) and co-transfected with 50 nM miR-27b mimics or scrambled miRNA control, 0.25 μg firefly luciferase reporter containing the 3′-UTR and 50 ng pRL-CMV Renilla luciferase reporter. After 48 h, the luciferase activity was measured with the dual luciferase reporter assay system (Promega).
Patient analysis
For patient analysis, all the data in Figures 2D-2F, 4H-4J, and 7 were retrieved from TCGA data portal (cancergenome.nih.gov) updated by the end of November 30, 2014. Information for these patients contains miRNA expression, mRNA expression, SNP, DNA methylation data, and clinical information. All these data were used in level three. For detailed information of gene copy number variation analysis, promoter methylation analysis, patient stratification and survival analysis, see Supplementary information, Data S1.
Statistical analysis
Statistical calculation was performed in GraphPad Prism 5 software. All dose-response curve experiments were analyzed by F-test. A two-tailed P < 0.05 was taken to indicate statistical significance. For analyzing the survival of patients, two-tailed log-rank test was applied. All the other statistical comparisons between experimental groups were analyzed by two-tailed Student's t-test. Data are presented as mean ± SEM.
Acknowledgments
We are grateful to Drs Bakiri Latifa, Mofang Liu, Ruimin Huang, Erwei Song, Hai Jiang, Hongbin Ji, and Lihua Min for critical comments on the manuscript. This study was funded by the National Natural Science Foundation of China (31225016, 81170418 and 81125016), the Ministry of Science and Technology of China (2014CB910601, 2011ZX09307-302-01, 2011CB966304 and 2011CB910204), the Science and Technology Commission of Shanghai Municipality (12JC1409500 and 14XD1404200), and the Natural Science Foundation of Jiangsu Province (BK20131084).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Identification of miRNAs that enhance drug sensitivity by functional screen.
miR-27b is the master miRNA for enhancing drug sensitivity in liver cancers.
Expression levels of drug sensitizing miRNAs and miR-23b∼27b∼24-1 gene cluster members in liver cancers.
The combinational therapy of miR-27b and doxorubicin shows synergistic therapeutic effect on liver cancers and kidney cancers in vivo.
miR-27b enhances the drug response by activating p53.
miR-27b directly targets CCNG1 to activate p53 activity.
The miR-27b-CCNG1-p53 mechanism accounted for the regulation of sensitivity to sorafenib.
Knockdown of CCNG1 did not confer drug sensitivity in p53 mutant cell lines.
CYP1B1 is post-transcriptionally regulated by miR-27b.
miR-27b promotes drug sensitivity by additionally reducing CYP1B1 levels.
The establishment of the CYP1B1 gene signature.
miR-27b promotes good drug responses in patients with wild-type p53 or high CYP1B1 levels.
miRNAs promoting HepG2 cells sensitive or resistant to doxorubicin in the high-throughput screen
miRNAs increasing or decreasing HepG2 cell numbers in the high-throughput screen
Pathways regulated by each miRNA in the miRNA-pathway network
Information of 49 paired liver cancers and 66 paired kidney cancers
The original data of subcutaneously transplanted tumors
Fold change of genes upon forced miR-27b expression
Fold change of genes upon forced miR-27b expression
p53 status of different cancer cell lines
CYP1B1 signature genes
Information of 29 liver cancer patients and 60 kidney cancer patients with drug treatment
Primers for vector construction
Supplemenal Methods
References
- Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer. 2013;13:714–726. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- Corrie PG. Cytotoxic chemotherapy: clinical aspects. Medicine. 2008;36:24–28. [Google Scholar]
- Llovet JM, Ricci S, Mazzaferro V, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- Escudier B, Eisen T, Stadler WM, et al. Sorafenib in advanced clear-cell renal-cell carcinoma. N Engl J Med. 2007;356:125–134. doi: 10.1056/NEJMoa060655. [DOI] [PubMed] [Google Scholar]
- Chen PJ, Furuse J, Han KH, et al. Issues and controversies of hepatocellular carcinoma-targeted therapy clinical trials in Asia: experts' opinion. Liver Int. 2010;30:1427–1438. doi: 10.1111/j.1478-3231.2010.02292.x. [DOI] [PubMed] [Google Scholar]
- Park JW, Amarapurkar D, Chao Y, et al. Consensus recommendations and review by an International Expert Panel on Interventions in Hepatocellular Carcinoma (EPOIHCC) Liver Int. 2013;33:327–337. doi: 10.1111/liv.12083. [DOI] [PubMed] [Google Scholar]
- Nanus DM, Garino A, Milowsky MI, Larkin M, Dutcher JP. Active chemotherapy for sarcomatoid and rapidly progressing renal cell carcinoma. Cancer. 2004;101:1545–1551. doi: 10.1002/cncr.20541. [DOI] [PubMed] [Google Scholar]
- Qin S. Guidelines on the diagnosis and treatment of primary liver cancer (2011 edition) Chinese Clinl Oncol. 2012;1 doi: 10.3978/j.issn.2304-3865.2012.07.01. [DOI] [PubMed] [Google Scholar]
- Tacar O, Sriamornsak P, Dass CR. Doxorubicin: an update on anticancer molecular action, toxicity and novel drug delivery systems. J Pharm Pharmacol. 2013;65:157–170. doi: 10.1111/j.2042-7158.2012.01567.x. [DOI] [PubMed] [Google Scholar]
- Longley DB, Johnston PG. Molecular mechanisms of drug resistance. J Pathol. 2005;205:275–292. doi: 10.1002/path.1706. [DOI] [PubMed] [Google Scholar]
- Huang SD, Holzel M, Knijnenburg T, et al. MED12 controls the response to multiple cancer drugs through regulation of TGF-beta receptor signaling. Cell. 2012;151:937–950. doi: 10.1016/j.cell.2012.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap TA, Omlin A, de Bono JS. Development of therapeutic combinations targeting major cancer signaling pathways. J Clin Oncol. 2013;31:1592–1605. doi: 10.1200/JCO.2011.37.6418. [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Bader AG. miR-34 — a microRNA replacement therapy is headed to the clinic. Front Genet. 2012;3:120. doi: 10.3389/fgene.2012.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013;12:847–865. doi: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garofalo M, Croce CM. MicroRNAs as therapeutic targets in chemoresistance. Drug Resist Updat. 2013;16:47–59. doi: 10.1016/j.drup.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garofalo M, Di Leva G, Romano G, et al. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell. 2009;16:498–509. doi: 10.1016/j.ccr.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Fornari F, Milazzo M, Chieco P, et al. MiR-199a-3p regulates mTOR and c-Met to influence the doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. 2010;70:5184–5193. doi: 10.1158/0008-5472.CAN-10-0145. [DOI] [PubMed] [Google Scholar]
- Ding J, Huang SL, Wang Y, et al. Genome-wide screening reveals that miR-195 targets the TNF-α/NF-κB pathway by down-regulating IκB kinase alpha and TAB3 in hepatocellular carcinoma. Hepatology. 2013;58:654–666. doi: 10.1002/hep.26378. [DOI] [PubMed] [Google Scholar]
- Knasmuller S, Mersch-Sundermann V, Kevekordes S, et al. Use of human-derived liver cell lines for the detection of environmental and dietary genotoxicants; current state of knowledge. Toxicology. 2004;198:315–328. doi: 10.1016/j.tox.2004.02.008. [DOI] [PubMed] [Google Scholar]
- Jia D, Dong R, Jing Y, et al. Exome sequencing of hepatoblastoma reveals novel mutations and cancer genes in the Wnt pathway and ubiquitin ligase complex. Hepatology. 2014;60:1686–1696. doi: 10.1002/hep.27243. [DOI] [PubMed] [Google Scholar]
- Birmingham A, Selfors LM, Forster T, et al. Statistical methods for analysis of high-throughput RNA interference screens. Nat Methods. 2009;6:569–575. doi: 10.1038/nmeth.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra A, Mishra L, Li SL. Technologies for deriving primary tumor cells for use in personalized cancer therapy. Trends Biotechnol. 2013;31:347–354. doi: 10.1016/j.tibtech.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell P, Villanueva A, Llovet JM. Molecular targeted therapies in hepatocellular carcinoma: from pre-clinical models to clinical trials. J Hepatol. 2008;49:1–5. doi: 10.1016/j.jhep.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin L, Andersen JN, Futreal PA. Cancer genomics: from discovery science to personalized medicine. Nat Med. 2011;17:297–303. doi: 10.1038/nm.2323. [DOI] [PubMed] [Google Scholar]
- Benson AB, Abrams TA, Ben-Josef E, et al. NCCN clinical practice guidelines in oncology: hepatobiliary cancers. J Natl Compr Canc Netw. 2009;7:350–391. doi: 10.6004/jnccn.2009.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin SB, Ohm JE. Epigenetic gene silencing in cancer — a mechanism for early oncogenic pathway addiction. Nat Rev Cancer. 2006;6:107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- Szakacs G, Paterson JK, Ludwig JA, Booth-Genthe C, Gottesman MM. Targeting multidrug resistance in cancer. Nat Rev Drug Discov. 2006;5:219–234. doi: 10.1038/nrd1984. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. human microRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Li HY, Jensen MR, et al. Cyclin G recruits PP2A to dephosphorylate Mdm2. Mol Cell. 2002;9:761–771. doi: 10.1016/s1097-2765(02)00504-x. [DOI] [PubMed] [Google Scholar]
- Jensen MR, Factor VM, Fantozzi A, Helin K, Huh CG, Thorgeirsson SS. Reduced hepatic tumor incidence in cyclin G1-deficient mice. Hepatology. 2003;37:862–870. doi: 10.1053/jhep.2003.50137. [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y, Nakajima M, Takagi S, Taniya T, Yokoi T. MicroRNA regulates the expression of human cytochrome P4501B1. Cancer Res. 2006;66:9090–9098. doi: 10.1158/0008-5472.CAN-06-1403. [DOI] [PubMed] [Google Scholar]
- Rochat B, Morsman JM, Murray GI, Figg WD, Mcleod HL. Human CYP1B1 and anticancer agent metabolism: Mechanism for tumor-specific drug inactivation. J Pharmacol Exp Ther. 2001;296:537–541. [PubMed] [Google Scholar]
- Marquardt JU, Thorgeirsson SS. SnapShot: Hepatocellular carcinoma. Cancer Cell. 2014;25:550.e1. doi: 10.1016/j.ccr.2014.04.002. [DOI] [PubMed] [Google Scholar]
- Sotiriou C, Piccart MJ. Opinion — taking gene-expression profiling to the clinic: when will molecular signatures become relevant to patient care. Nat Rev Cancer. 2007;7:545–553. doi: 10.1038/nrc2173. [DOI] [PubMed] [Google Scholar]
- Miller LD, Smeds J, George J, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci USA. 2005;102:13550–13555. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gry M, Rimini R, Stromberg S, et al. Correlations between RNA and protein expression profiles in 23 human cell lines. BMC Genomics. 2009;10:365. doi: 10.1186/1471-2164-10-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M, Oksvold P, Fagerberg L, et al. Towards a knowledge-based human protein atlas. Nat Biotechnol. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stransky N, et al. The Cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- Cheng AL, Kang YK, Chen Z, et al. Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: a phase III randomised, double-blind, placebo-controlled trial. Lancet Oncol. 2009;10:25–34. doi: 10.1016/S1470-2045(08)70285-7. [DOI] [PubMed] [Google Scholar]
- Hatziapostolou M, Polytarchou C, Aggelidou E, et al. An HNF4alpha-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell. 2011;147:1233–1247. doi: 10.1016/j.cell.2011.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermeking H. MicroRNAs in the p53 network: micromanagement of tumour suppression. Nat Rev Cancer. 2012;12:613–626. doi: 10.1038/nrc3318. [DOI] [PubMed] [Google Scholar]
- Rukov JL, Shomron N. MicroRNA pharmacogenomics: post-transcriptional regulation of drug response. Trends Mol Med. 2011;17:412–423. doi: 10.1016/j.molmed.2011.04.003. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasinski AL, Slack FJ. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11:849–864. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobrero AF, Maurel J, Fehrenbacher L, et al. EPIC: phase III trial of cetuximab plus irinotecan after fluoropyrimidine and oxaliplatin failure in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:2311–2319. doi: 10.1200/JCO.2007.13.1193. [DOI] [PubMed] [Google Scholar]
- Geyer CE, Forster J, Lindquist D, et al. Lapatinib plus capecitabine for HER2-positive advanced breast cancer. N Engl J Med. 2006;355:2733–2743. doi: 10.1056/NEJMoa064320. [DOI] [PubMed] [Google Scholar]
- Sun TM, Du JZ, Yao YD, et al. Simultaneous delivery of siRNA and paclitaxel via a ”two-in-one” micelleplex promotes synergistic tumor suppression. ACS Nano. 2011;5:1483–1494. doi: 10.1021/nn103349h. [DOI] [PubMed] [Google Scholar]
- Ohtsuka T, Jensen MR, Kim HG, Kim KT, Lee SW. The negative role of cyclin G in ATM-dependent p53 activation. Oncogene. 2004;23:5405–5408. doi: 10.1038/sj.onc.1207693. [DOI] [PubMed] [Google Scholar]
- Sato Y, Yoshizato T, Shiraishi Y, et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet. 2013;45:860–867. doi: 10.1038/ng.2699. [DOI] [PubMed] [Google Scholar]
- Roe JS, Kim H, Lee SM, Kim ST, Cho EJ, Youn HD. p53 stabilization and transactivation by a von Hippel-Lindau protein. Mol Cell. 2006;22:395–405. doi: 10.1016/j.molcel.2006.04.006. [DOI] [PubMed] [Google Scholar]
- McFadyen MC, Melvin WT, Murray GI. Cytochrome P450 enzymes: novel options for cancer therapeutics. Mol Cancer Ther. 2004;3:363–371. [PubMed] [Google Scholar]
- McFadyen MC, Murray GI. Cytochrome P450 1B1: a novel anticancer therapeutic target. Future Oncol. 2005;1:259–263. doi: 10.1517/14796694.1.2.259. [DOI] [PubMed] [Google Scholar]
- Offer SM, Butterfield GL, Jerde CR, Fossum CC, Wegner NJ, Diasio RB. microRNAs miR-27a and miR-27b directly regulate liver dihydropyrimidine dehydrogenase expression through two conserved binding sites. Mol Cancer Ther. 2014;13:742–751. doi: 10.1158/1535-7163.MCT-13-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Lazikani B, Banerji U, Workman P. Combinatorial drug therapy for cancer in the post-genomic era. Nat Biotechnol. 2012;30:679–691. doi: 10.1038/nbt.2284. [DOI] [PubMed] [Google Scholar]
- Sharma SV, Haber DA, Settleman J. Cell line-based platforms to evaluate the therapeutic efficacy of candidate anticancer agents. Nat Rev Cancer. 2010;10:241–253. doi: 10.1038/nrc2820. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Identification of miRNAs that enhance drug sensitivity by functional screen.
miR-27b is the master miRNA for enhancing drug sensitivity in liver cancers.
Expression levels of drug sensitizing miRNAs and miR-23b∼27b∼24-1 gene cluster members in liver cancers.
The combinational therapy of miR-27b and doxorubicin shows synergistic therapeutic effect on liver cancers and kidney cancers in vivo.
miR-27b enhances the drug response by activating p53.
miR-27b directly targets CCNG1 to activate p53 activity.
The miR-27b-CCNG1-p53 mechanism accounted for the regulation of sensitivity to sorafenib.
Knockdown of CCNG1 did not confer drug sensitivity in p53 mutant cell lines.
CYP1B1 is post-transcriptionally regulated by miR-27b.
miR-27b promotes drug sensitivity by additionally reducing CYP1B1 levels.
The establishment of the CYP1B1 gene signature.
miR-27b promotes good drug responses in patients with wild-type p53 or high CYP1B1 levels.
miRNAs promoting HepG2 cells sensitive or resistant to doxorubicin in the high-throughput screen
miRNAs increasing or decreasing HepG2 cell numbers in the high-throughput screen
Pathways regulated by each miRNA in the miRNA-pathway network
Information of 49 paired liver cancers and 66 paired kidney cancers
The original data of subcutaneously transplanted tumors
Fold change of genes upon forced miR-27b expression
Fold change of genes upon forced miR-27b expression
p53 status of different cancer cell lines
CYP1B1 signature genes
Information of 29 liver cancer patients and 60 kidney cancer patients with drug treatment
Primers for vector construction
Supplemenal Methods