Abstract
Exosomes are secreted vesicles of endosomal origin involved in signaling processes. We recently showed that the syndecan heparan sulfate proteoglycans control the biogenesis of exosomes through their interaction with syntenin-1 and the endosomal-sorting complex required for transport accessory component ALIX. Here we investigated the role of heparanase, the only mammalian enzyme able to cleave heparan sulfate internally, in the syndecan-syntenin-ALIX exosome biogenesis pathway. We show that heparanase stimulates the exosomal secretion of syntenin-1, syndecan and certain other exosomal cargo, such as CD63, in a concentration-dependent manner. In contrast, exosomal CD9, CD81 and flotillin-1 are not affected. Conversely, reduction of endogenous heparanase reduces the secretion of syntenin-1-containing exosomes. The ability of heparanase to stimulate exosome production depends on syntenin-1 and ALIX. Syndecans, but not glypicans, support exosome biogenesis in heparanase-exposed cells. Finally, heparanase stimulates intraluminal budding of syndecan and syntenin-1 in endosomes, depending on the syntenin-ALIX interaction. Taken together, our findings identify heparanase as a modulator of the syndecan-syntenin-ALIX pathway, fostering endosomal membrane budding and the biogenesis of exosomes by trimming the heparan sulfate chains on syndecans. In addition, our data suggest that this mechanism controls the selection of specific cargo to exosomes.
Keywords: heparan sulfate, syndecan, heparanase, syntenin, exosome biogenesis, exosomal cargo
Introduction
Exosomes are extracellular vesicles of endosomal origin, with a size ranging from 40 to 100 nm, secreted by cells. These vesicles contain various membrane and cytoplasmic components — mRNA, miRNA, lipids, activated receptors, kinases and other proteins — commonly designated as cargo. Transfer of cargo to recipient cells after the uptake of extracellular vesicles plays an important role in intercellular communication1,2,3. Although involved in many aspects of physiology and disease, the function of exosomal transfers has been studied mostly for immunological processes4,5 and tumor progression. Tumor-derived exosomes stimulate pre-metastatic niche formation6 and exosomal communication between cancer-associated fibroblasts and the primary tumor stimulates breast cancer cell motility and metastasis7.
Despite recent progress, exosome biogenesis and the mechanisms controlling sorting of specific cargo into exosomes are only partially understood at the molecular level. Intraluminal budding of the limiting membrane of multivesicular bodies (MVBs) — a type of late endosomes — creates intraluminal vesicles (ILVs) that are released in the extracellular space as exosomes upon fusion of MVBs with the cell membrane1. Several mechanisms have been suggested to control intraluminal budding. The endosomal-sorting complex required for transport (ESCRT) machinery regulates membrane budding at late endosomes8,9. Sequential activity of ESCRT-0, -I, -II and -III complexes sorts ubiquitylated membrane proteins into specific membrane domains, induces inward budding and mediates membrane abscission to form ILVs10,11. Other studies have pointed to ceramide and other lipids as mediators of exosome biogenesis by driving lateral segregation of cargo into specialized endosomal membrane regions able to bend inwards12,13.
Recently, Baietti and co-workers reported that syndecan heparan sulfate proteoglycans and their cytoplasmic adaptor syntenin-1 control the formation of exosomes. Syntenin-1 interacts directly with ALIX14, an auxiliary component of the ESCRT machinery15, through three LYPXnL motifs located in its N-terminus and with the conserved cytoplasmic domains of the syndecans, via its PDZ domains. Since ALIX binds several ESCRT proteins, syntenin-ALIX adapts syndecans and syndecan cargo to the ESCRT budding machinery at the MVBs. Heparan sulfate is essential to this mechanism of exosome formation. Syndecan cargo — most probably bound to the heparan sulfate of syndecan — is thought to cluster syndecans and create syndecan assemblies that recruit syntenin-1 and ALIX and subsequently support membrane budding. This pathway allows the recruitment of heparan sulfate-binding cargo, like FGFR1, and might select which cargo is targeted to exosomes14.
The importance of heparan sulfate for exosome production implies that heparanase might influence this process. Heparanase is the only mammalian enzyme able to cleave heparan sulfate internally, generating short fragments (of 10 to 20 residues) endowed with biological activity. Elevated heparanase expression by tumor cells correlates with increased tumor angiogenesis, tumor invasiveness and metastasis16,17,18,19. Heparanase is secreted as an inactive precursor, proheparanase, which is endocytosed after binding to LRP1 or mannose-6-phosphate receptor20. In late endosomes and lysosomes, the removal of a 6 kDa fragment from proheparanase transforms the protein into an active enzyme, an obligate heterodimer of an 8 kDa and a 50 kDa fragment21. Apart from cleaving heparan sulfate, heparanase is able to activate several signal transduction pathways independently of its enzymatic activity, resulting in altered gene expression, enhanced cell survival and cell migration22,23,24.
Here we show that heparanase functions as an important modulator of the syndecan-syntenin-ALIX pathway that supports the biogenesis of exosomes, affecting specific exosomal cargo.
Results
Exogenous proheparanase stimulates the production of exosomes that contain syntenin-1
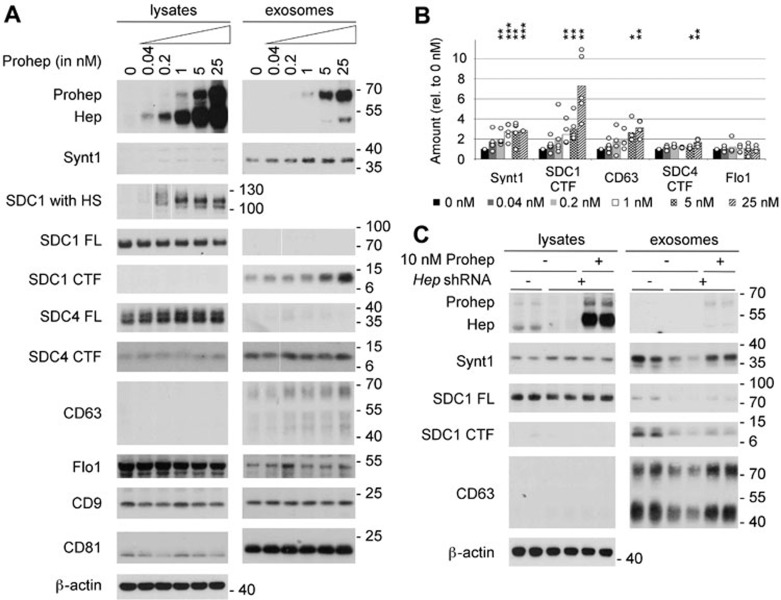
The influence of heparanase on exosome production and composition was examined by supplementing the culture medium of MCF-7 cells with increasing concentrations of purified proheparanase (0.04 to 25 nM). Proheparanase added to cells is internalized and processed into active heparanase20,21. Under the conditions used, proheparanase uptake and processing were not saturated, as documented by the increasing signals for proheparanase and mature heparanase in the lysate up to 25 nM (Figure 1A). Heparanase addition resulted in extensive trimming of the heparan sulfate on syndecan-1 (Figure 1A). By that criterion, the effect of heparanase on the heparan sulfate composition of the proteoglycans was dose-dependent, but reached a maximum at 1 nM. Exosomes were collected from equivalent amounts of culture medium, conditioned by equal numbers of cells, for equal lengths of time, as described before14. Syntenin-1, the C-terminal fragments (CTFs) of syndecan-1 and syndecan-4, CD63 and flotillin-1 were used as markers to assess exosome production and composition. Previously, syntenin-1 and the syndecans have been shown to compose exosomal cargo (with most of the syndecans transformed into CTFs upon integration in exosomes) and to be key mechanistic factors in the production of MCF-7 exosomes that contain CD63, but not of exosomes containing flotillin-114. Exosomal levels of syntenin-1 rose with increasing proheparanase concentrations in the culture medium, but reached a plateau value (3-fold increase) from 1 nM proheparanase on (Figure 1A and 1B, P < 0.01, n = 5; see the Materials and methods section for more information on the statistics used). As expected14, CTFs represented the major form of syndecan present in exosomes (Figure 1A). Note that an antibody reacting with the ectodomain might fail to detect syndecan CTFs and only document the presence of full-length forms of syndecan in exosomes25. At maximal proheparanase concentration, exosomal syndecan-1 CTF increased by ∼ 7-fold (Figure 1A and 1B, P < 0.05, n = 5). Compared to syndecan-1, the increase in syndecan-4 CTF was more modest (close to 2-fold) but significant (Figure 1A and 1B, P < 0.05, n = 4). Exosomal CD63 increased by 3-fold (Figure 1A and 1B, P < 0.05, n = 4). In contrast to the effect on exosomal syntenin-1 (cytosolic cargo), the increase of exosomal syndecan-1 and -4 CTFs and CD63 (membrane cargo binding to syntenin-1) showed no saturation at higher proheparanase concentrations. Of note, the amount of exosomal flotillin-1 did not change. The amounts of exosomal CD9 and CD81, two tetraspanins commonly used as exosomal markers, were also unaltered upon addition of heparanase (Figure 1A). At higher concentrations, some proheparanase (probably peripherally associated with exosomes) was present in the exosomal fractions (Figure 1A). These results show that addition of exogenous proheparanase specifically stimulates the production of syndecan-, CD63- and syntenin-1-containing exosomes, whereas other exosomal markers like flotillin-1, CD9 and CD81 are unaffected.
Figure 1.
Heparanase stimulates the production of syntenin-1-containing exosomes. (A) Exosome production was evaluated after overnight conditioning of MCF-7 cells with increasing concentrations of proheparanase (0.04-25 nM) and compared to that of cells not receiving proheparanase (0 nM). Exosomes were collected from equivalent amounts of culture medium, conditioned by equal numbers of cells, for equal lengths of time. For each condition both the lysate and exosomal fractions were analyzed by western blot, using cognate antibodies against heparanase, monitoring the conversion of proheparanase (Prohep) into mature heparanase (Hep) and against different exosomal markers: syntenin-1 (Synt1), syndecan-1 (SDC1), syndecan-4 (SDC4), CD63, flotillin-1 (Flo1), CD9 and CD81. Syndecan-1, which is a hybrid heparan sulfate (HS)/chondroitin sulfate proteoglycan, was analyzed using two different approaches. In one approach, the samples were digested with both heparitinase and chondroitinase ABC, removing all glycosaminoglycan chains and enabling visualization of the full-length syndecan core proteins (SDC1 FL) as sharp bands. In the other approach, the samples were digested with chondroitinase ABC only, leaving the HS on the syndecans (SDC1 with HS); comparison of 'SDC1 with HS' and 'SDC1 FL' yields information on the mass of HS on syndecans. Because of the heterogeneity in HS chain length, syndecan-1 with HS is smeared over a wide mass range in the absence of heparanase activity (and is therefore hardly visible in western blot, as illustrated by lane 1 of the lysates). With increasing heparanase activity, the HS chains on syndecan-1 are trimmed to shorter chains of more or less the same length, syndecan-1 with HS migrating as one or a few bands that are readily visualized in western blot (as illustrated by lane 6 of the lysates). Note that cell lysates contain mainly full-length syndecan core proteins; the opposite is true for exosomes, where hardly any full-length syndecan is detected and C-terminal fragments (CTFs) represent the dominant form. β-actin was used as a loading control for the lysates. Western blots are representative of five independent experiments. (B) Histogram representing the quantification of the exosomal levels of syntenin-1 (Synt1), syndecan-1 CTF (SDC1 CTF), CD63, syndecan-4 CTF (SDC4 CTF) and flotillin-1 (Flo1) in response to the addition of increasing concentrations (0 nM till 25 nM) of proheparanase. Values are relative to the exosomal levels measured in absence of exogenously added proheparanase. Bar heights represent mean values, calculated from five independent experiments. Individual data points are shown as white dots on top of the corresponding bars. *P < 0.1, **P < 0.05, ***P < 0.01 (Student's t-test, assuming normal distribution of the data points). (C) Knockdown of endogenous heparanase reduces the production of syntenin-1-containing exosomes, which can be rescued by the addition of exogenous proheparanase. Duplicate lanes show the results of two independent experiments, run side by side. B16-F10 cells are sham-transfected (−) or stably transfected with a shRNA targeting murine heparanase (+). To rescue the effects of endogenous heparanase knockdown, 10 nM human proheparanase was added to the cells. Heparanase, syntenin-1, syndecan-1 full-length (SDC1 FL), syndecan 1 CTF (SDC1 CTF) and CD63 were analyzed by western blot. Positions of molecular weight markers (in kDa) are indicated on the right of each blot. Note that (because of differences in glycosylation) the Mr of human heparanase is slightly larger than that of mouse heparanase.
The effect of heparanase on exosomal syntenin was not limited to MCF-7 cells. Proheparanase addition stimulated exosomal syntenin levels also in MDA-MB-231, MDA-MB-134 and HT1080 cells. Except for MDA-MB-134 cells, which did not express exosomal CD63, heparanase also stimulated exosomal CD63 levels (Supplementary information, Figure S1). Heparanase had little, or at best moderate effects on the total number of exosomes produced. NanoSight experiments indicated heparanase addition to MCF-7 cells increased the number of nanoparticles (with modal diameter of ∼ 120 nm) present in exosomal fractions by ∼ 30 % (Supplementary information, Figure S2). Heparanase addition had no detectable effects on exosome sedimentation. The distribution of exosomal markers between MCF-7 cell conditioned culture media materials pelleted at 10 000× (cell fragments and cell debris) and 140 000× g (exosomes) was unaffected (Supplementary information, Figure S3). Of note, unlike flotillin-1, which was present in both low- and high-speed pellets, syntenin-1, syndecan-CTFs and CD63 were exclusively detected in the 140 000× g pellets. Thus, possible trivial effects of heparanase on the physical properties of syndecan-syntenin exosomes, i.e., reducing their aggregation and increasing their partitioning in high-speed pellets during centrifugation seemed excluded. Finally, we also excluded major effects of heparanase on syntenin-exosome clearance. Recently, heparan sulfate proteoglycans in recipient cells were shown to be involved in exosome uptake or sequestration26. Thus, trimming of the heparan sulfate on cells by heparanase might reduce exosome uptake (re-internalization), leading to a net increase of exosomes in conditioned media. When incubated with conditioned media containing exosomes derived from MCF-7 cells that express eGFP-syntenin-1, MCF-7 cells, and also other types of cells (e.g., U-2 OS osteosarcoma cells), bound little of the added eGFP-syntenin-1 (i.e., did not remove significant amounts of exosomal eGFP-syntenin-1 from the culture media). Yet, confocal microscopy indicated a small fraction of the MCF-7 cells (and a somewhat larger fraction of the U-2 OS cells) bound and internalized eGFP/exosomes. Heparanase treatment did not reduce such exosomal binding (Supplementary information, Figures S4 and S5). Thus, rather than inhibiting exosome uptake, heparanase addition appears to stimulate the biogenesis of extracellular vesicles/exosomes.
Knockdown of endogenous heparanase reduces the production of syntenin-1-containing exosomes
MCF-7 cells express very little or no endogenous heparanase (Figure 1A, left top panel). Therefore, in order to study the role of endogenous heparanase, B16-F10 mouse melanoma cells were used. Stable shRNA-mediated knockdown of heparanase (Figure 1C) reduced the amount of syntenin-1 in the exosomal fractions (Figure 1C). The same was observed for exosomal CD63. Heparanase knockdown also decreased the exosomal levels of both full-length syndecan-1 and syndecan-1 CTF (Figure 1C). As in MCF-7 cells, CTFs represented the dominant form of syndecan in B16-F10 exosomes (Figure 1C).
To rule out potential artifacts due to the stable transfection and the selection process, human proheparanase was added to the growth medium of B16-F10 cells with the knockdown of endogenous heparanase (Figure 1C). Addition of 10 nM exogenous human proheparanase reversed the effect of heparanase knockdown on exosome production in B16-F10 cells. Levels of exosomal syntenin-1 and CD63 returned to the levels observed in B16-F10 cells transfected with a non-targeting shRNA (Figure 1C). The recovery of exosomal syndecan-1 (full-length and CTF), in contrast, was less complete (Figure 1C). These data indicate that endogenous levels of heparanase contribute to exosome production by cells.
The influence of heparanase on exosome production depends on cleavage of heparan sulfate
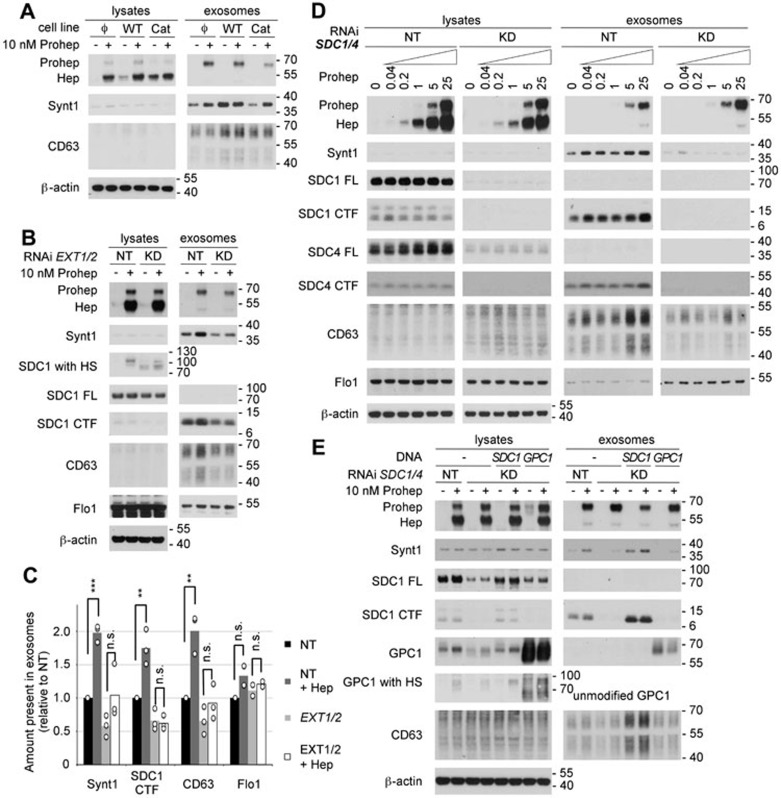
Heparanase influences a multitude of biological processes; some of these effects are directed by heparan sulfate cleavage, while others are the result of non-enzymatic signaling activity16,17,22,23,24. To distinguish between heparanase activities, we compared exosome production by stably transfected MCF-7 cells expressing either wild-type or catalytically dead human heparanase (catalytic residues mutated from glutamate to alanine, E225A and E343A). Even though MCF-7 cells stably expressing wild-type heparanase had a somewhat lower expression, at protein level, than cells expressing catalytically dead heparanase (Figure 2A), levels of exosomal syntenin-1 and CD63 in these cells were higher than in control transfected cells and in cells producing catalytically dead heparanase (Figure 2A). The levels of exosomal syntenin-1 released by cells producing catalytically dead heparanase were equal to the levels observed in exosomes from control-transfected cells. These observations indicate that the effect of heparanase on exosomes mainly depends on its enzymatic activity. While exogenously added proheparanase (10 nM) was able to increase exosomal syntenin-1 levels in control-transfected cells and in cells stably producing catalytically dead heparanase, no effect on exosomal syntenin-1 was observed in cells stably producing wild-type heparanase (Figure 2A). In the first place, these results suggest that cells having substantial heparanase activity cannot be further stimulated to produce exosomes by the addition of proheparanase, suggesting that saturation occurs at high levels of heparanase expression. Furthermore, they indicate that catalytically dead heparanase has no dominant negative effect on heparanase in exosome formation.
Figure 2.
The effect of heparanase on exosome production depends on the modification of the heparan sulfate (HS) on syndecans. (A) The role of heparanase enzymatic activity on exosome production was investigated by comparing MCF-7 cells stably expressing wild-type heparanase (WT), catalytically dead heparanase (Cat) or empty vector (Φ) in western blot, in the absence (−) or presence (+) of 10 nM exogenously added proheparanase. (B) The importance of HS was analyzed by treating MCF-7 cells with RNAi targeting EXT1 and EXT2 (KD). Non-targeting RNAi (NT) was used as a control. Cells were challenged with 10 nM proheparanase (+) or left untreated (−). Heparanase activity, reducing the HS on syndecan, was apparent from the migration of chondroitinase ABC-treated syndecan-1 present in cell lysates (SDC1 with HS). EXT1 and EXT2 knockdown leads to the appearance of syndecan-1 that is not substituted with HS (a band running slightly > 70 kDa, after chondroitinase ABC digestion only), not detectable in cells treated with non-targeting RNAi, where all syndecan is substituted with HS (and is larger than SDC FL). A small amount of the syndecan-1 still carried HS and was affected by heparanase addition (yielding a band slightly > 100 kDa, after chondroitinase ABC digestion only), indicating an incomplete knockdown of EXT1 and EXT2. Western blots are representative of three independent experiments. (C) Quantification of the effect of EXT1 and EXT2 knockdown. Histograms representing the exosomal levels of syntenin-1, syndecan-1 CTF, CD63 and flotillin-1 in the different conditions tested (non-targeting RNAi, black bars; non-targeting RNAi and heparanase, dark gray bars; EXT1 and EXT2 RNAi, light gray bars; EXT1 and EXT2 RNAi and heparanase, white bars). Values are relative to the exosomal levels measured in cells treated with non-targeting RNAi and in the absence of exogenously added proheparanase. Bar heights represent mean values, calculated from three independent experiments. Individual data points are shown as white dots on top of the corresponding bars. **P < 0.05, ***P < 0.01, n.s., not significant (Student's t-test, assuming normal distribution of the data points). (D) Likewise, the role of syndecans was investigated by treating MCF-7 cells with RNAi targeting syndecan-1 and -4 (KD). Non-targeting RNAi (NT) was used as a control. Cells were treated with increasing concentrations of proheparanase (0 to 25 nM) and both the lysates and exosomal fractions were analyzed. Western blots are representative of three independent experiments. Note that lysate and exosome samples derived from NT-and KD-treated cells, separated by a blank space in Figure 2D, were run in the same gel, but not side by side, and that the band intensities in each row are directly comparable. (E) The ability of glypican-1 to rescue the effect of heparanase on exosome production in absence of syndecans was investigated by knocking down both syndecan-1 and -4 (KD) and overexpressing glypican-1 (GPC1) in the same cells. Glypican-1, when deglycanated by heparitinase and chondroitinase ABC treatment (GPC1) or when unmodified (because of overexpression) has a mass of ∼ 60 kDa. Treatment with chondroitinase ABC only reveals 'GPC1 with HS' of a mass of ∼ 90 kDa in heparanase-exposed cells, indicating the HS on glypican-1 is trimmed the same way as the HS on syndecans. Yet, extra glypican fails to rescue the exosomal accumulations of syntenin and CD63. Rescue by expression of mouse syndecan-1 (SDC1) was used as a positive control. Cells treated with non-targeting RNAi (NT) served as reference. Molecular weight markers (in kDa) are indicated on the right of each blot. Western blots are representative of two independent experiments. Note that MCF-7 cells use mainly Man-6-P receptors for heparanase internalization, and that neither the knock down of EXT1 and EXT2 nor the knock down of syndecans has an effect on the uptake of proheparanase and its conversion into mature active form.
The need for heparanase enzymatic activity suggests a role for heparan sulfate in the mechanism through which heparanase stimulates exosome production. To test this hypothesis heparan sulfate polymerases EXT1 and EXT2, catalyzing the elongation of the heparan sulfate precursor27, were knocked down, reducing heparan sulfate production. Combined knockdown of EXT1 and 2 reduced the amount of heparan sulfate on syndecan, as evident from western blots, showing a downward band shift of syndecan-1 after enzymatic removal of its chondroitin sulfate while preserving the heparan sulfate (SDC1 with HS). However, a small amount of the syndecan-1 was still substituted with heparan sulfate (and modified by added heparanase), indicating an incomplete knockdown of EXT1 and 2 (Figure 2B). Upon knockdown of EXT1 and 2, the levels of syntenin-1, syndecan-1 CTF and CD63 in exosomes were reduced approximately by half (Figure 2B and 2C). Moreover, effects of exogenously added heparanase (10 nM) on exosomal syntenin-1, CD63 and syndecan-1 CTF were nonsignificant (Figure 2B and 2C). The small, nonsignificant effect of exogenous heparanase was in line with the evidence for a large but incomplete EXT 1 and 2 knockdown. Cells treated with non-targeting RNAi, in contrast, were responsive to heparanase (Figure 2B and 2C, P < 0.05, n = 3). Of note, exosomal flotillin-1 was unaffected by EXT 1 and 2 knockdown, and unaffected by the addition of exogenous proheparanase (Figure 2B and 2C). Thus, in the absence of substrate (i.e., heparan sulfate) wild-type heparanase has little or no effect on exosomes. Together, these data suggest an important role for heparanase-modified heparan sulfate in exosome production.
Syndecans play a key role in the effect of heparanase on exosome production
Previous work identified syndecans (binding syntenin-1) among the key mechanistic components involved in the biogenesis of exosomes containing syntenin-114. In combination with the data shown above, this suggests a mechanistic role for heparanase-modified syndecan in (heparanase-mediated) exosome production. MCF-7 cells express syndecan-1 and syndecan-4. Therefore, the effect of a combined knockdown was investigated. Absence of full-length syndecan-1 and strong reduction of full-length syndecan-4 in cell lysates (compared to MCF-7 cells treated with non-targeting RNAi) demonstrated effective knockdown of both syndecans (Figure 2D). Of note, syndecan-1 and -4 knockdown did not substantially influence heparanase uptake and processing, as judged from the amounts of proheparanase and mature protein in the cell lysates (Figure 2D). As expected, after knockdown of syndecan-1 and -4, syndecan-1 CTF and syndecan-4 CTF were depleted from the exosomal fractions, and, in contrast to cells treated with non-targeting RNAi, addition of proheparanase had no detectable effect on syndecan-1 and -4 CTF accumulation in exosomes (Figure 2D). Both exosomal syntenin-1 and CD63 decreased upon combined syndecan-1 and -4 knockdown. More importantly, the amount of syntenin-1 and CD63 left in the exosomal fraction upon combined syndecan knockdown did not increase upon the addition of exogenous proheparanase (Figure 2D). These results clearly demonstrate that heparanase acts through syndecans, to exert its effect on exosomes. As observed before (Figures 1A, 2B and 2C), levels of exosomal flotillin-1 did not respond to the addition of proheparanase (Figure 2D), confirming that flotillin-1 represents a heparanase-insensitive cargo. Of note, the level of proheparanase present in the exosomal fractions was unaffected by EXT1/2 knockdown (Figure 2B) and syndecan-1/4 knockdown (Figure 2D). These observations indicate that this proheparanase is probably not directly associated with syntenin-1 exosomes.
The combined knockdown of syndecan-1 and -4 removes most of the heparan sulfate present at the surface of MCF-7 cells. Glypicans (1 to 6) constitute the second major group of membrane heparan sulfate proteoglycans. Unlike syndecans, glypicans do not have a transmembrane domain, but are linked to the membrane by a glycosylphosphatidylinositol anchor and, therefore, are unable to bind syntenin-1 directly. To confirm that heparanase effects on exosome production depend on syndecans, and not on cell surface heparan sulfate per se, we tested whether glypican-1 can compensate for the loss of syndecan-1 and -4, using mouse syndecan-1 (resistant to the RNAi targeting endogenous syndecan-1) as a control. As described above, exosomal syntenin-1, syndecan-1 CTF and CD63 were reduced upon the knockdown of syndecan-1 and -4 (Figure 2E) and the effect of exogenous proheparanase (10 nM) on the exosomal levels of these markers was less potent than in cells treated with non-targeting RNAi (Figure 2E). Transient expression of mouse syndecan-1 rescued the effect of heparanase on exosomal syntenin-1 and CD63. In contrast, transient expression of glypican-1, although heparan sulfate-modified — and therefore restoring heparan sulfate expression in cells with syndecan knockdown — was unable to rescue the effect of heparanase on exosomal syntenin-1 and CD63 (Figure 2D). This suggests heparanase specifically depends on syndecans to exert its effect on exosomes.
Heparanase influences the biogenesis of vesicles of endosomal origin, enhancing syndecan endocytosis and intraluminal budding.
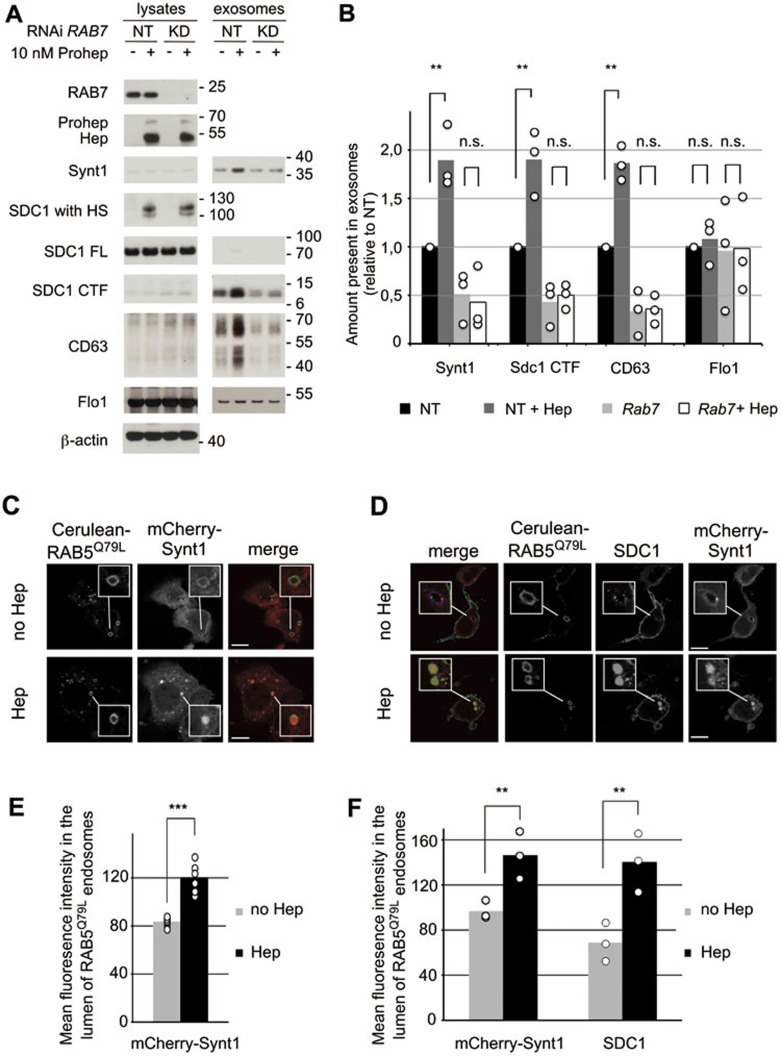
RAB7 regulates several steps in endosome trafficking, including the trafficking downstream of MVB biogenesis28. In MCF-7 cells, RAB7 knockdown results in large MVBs, filled with ILVs, and reduces the accumulation of exosomes into the extracellular space14. As such, RAB7 knockdown allows for investigating whether the vesicles under study are of endosomal origin. RAB7 knockdown did not influence heparanase uptake, processing and activity (Figure 3A). However, RAB7 knockdown abolished the heparanase-mediated increases in exosomal syntenin-1, syndecan-1 CTF and CD63, whereas levels of exosomal flotillin-1 were unaffected (Figure 3A and 3B). Thus, heparanase affects the production of vesicles that are of endosomal origin, the operational definition of exosomes. Of note, consistent with the notion that heparanase affects endosomal pathways, exogenous addition of heparanase or providing the cells with heparanase activity stimulated the endocytosis of syndecan-1 and syndecan-4 in MCF-7 cells (Supplementary information, Figure S6).
Figure 3.
Heparanase influences the biogenesis of vesicles of endosomal origin, enhancing intraluminal budding. (A) To investigate whether the extracellular vesicles affected by heparanase were of endosomal origin, RAB7 was knocked down (RAB7 RNAi) in MCF-7 cells. Non-targeting RNAi (−) served as a control. Cells were left untreated (−) or treated with proheparanase (10 nM). In both experiments, heparanase activity was evaluated using the migration pattern of chondroitinase ABC-, but not heparitinase-treated syndecan-1 present in cell lysates (SDC1 with HS). Molecular weight markers (in kDa) are indicated on the right of each blot. Western blots are representative of three independent experiments. (B) Quantification of the effect of RAB7 knockdown. Histograms representing the exosomal levels of syntenin-1, syndecan-1 CTF and CD63 in the different conditions tested (non-targeting RNAi, black bars; non-targeting RNAi with heparanase, dark gray bars; RAB7 knockdown, light gray bars; RAB7 knockdown with heparanase, white bars). Values are relative to the levels (intensities of the signals) measured in exosomes derived from cells treated with non-targeting RNAi in the absence of proheparanase. Bar heights represent mean values, calculated from three independent experiments. Individual data points are shown as white dots on top of the corresponding bars. **P < 0.05, n.s., not significant (Student's t-test, assuming normal distribution of the data points). (C) Confocal micrographs of MCF-7 cells co-transfected with mCherry-syntenin-1 (red in merge) and Cerulean-RAB5Q79L (green in merge). Note the presence of mCherry-syntenin-1, a cytosolic protein, inside the Cerulean-RAB5Q79L endosomes upon heparanase treatment (50 nM). (D) Confocal micrographs of MCF-7 cells co-transfected with mCherry-syntenin-1 (red in merge), Cerulean-RAB5Q79L (blue in merge) and syndecan-1 (green in merge), scoring the accumulations (budding) of mCherry-syntenin-1 and of syndecan-1 cytoplasmic domain inside vacuoles outlined by Cerulean-RAB5Q79L. (E) Quantification of intraluminal budding of mCherry-syntenin-1, as in Figure 3C, by measuring the fluorescence of mCherry-syntenin-1 in the lumen of RAB5Q79L-positive endosomes, corrected for the size of the RAB5Q79L-positive endosomes (mean gray value per pixel). Bar heights represent mean values calculated from six independent experiments, scoring at least 30 cells per experiment. Individual data points (mean luminal fluorescence intensity per individual experiment) are shown as white dots on top of the corresponding bars. ***P < 0.01 (Student's t-test, assuming normal distribution of the data points). (F) Quantification of intraluminal budding of mCherry-syntenin-1 and syndecan-1 cytoplasmic domain, as in Figure 3D, by measuring the fluorescence intensity of mCherry-syntenin-1 and syndecan-1 cytoplasmic domain in the lumen of RAB5Q79L-positive endosomes. Bar heights represent mean values calculated from three independent experiments, scoring at least 30 cells per experiment. Individual data points are shown as white dots on top of the corresponding bars. **P < 0.05 (Student's t-test, assuming normal distribution of the data points).
The syndecan-syntenin-ALIX exosome pathway affects intraluminal budding at the limiting membrane of endosomes13,14, a process generating MVBs and thereby critical for the formation of exosomes. To test whether heparanase affects intraluminal budding (of endosomal syndecan and associated syntenin) we took advantage of the Q79L mutant of RAB5, which leads to enlarged endosomes resembling MVBs. As such this mutant allows for studying intraluminal budding at the resolution of light microscopy29. We transiently co-expressed Cerulean-RAB5Q79L with mCherry-syntenin-1 and, using confocal microscopy, investigated the effect of heparanase on the concentration of mCherry-syntenin-1 in large endosomes outlined by Cerulean-RAB5Q79L (Figure 3C). We found that upon addition of exogenous heparanase the mean fluorescence intensity of mCherry-syntenin-1 in the lumen of Cerulean-RAB5Q79L endosomes increased by 43% (Figure 3C and 3E, P < 0.01, n = 6). Similar observations were made when Cerulean-RAB5Q79L, mCherry-syntenin-1 and syndecan-1 were co-expressed in MCF-7 cells. The mean fluorescence intensity of mCherry-syntenin-1 and (immunofluorescent signals for) syndecan-1 (cytoplasmic domain) inside the lumen of Cerulean-RAB5Q79L-positive endosomes increased by 51% (Figure 3D and 3F, P < 0.05, n = 3) and 103% (Figure 3D and 3F, P < 0.05, n = 3), respectively. Together, these data underpin the notion that exogenous heparanase impacts intraluminal budding and therefore exosome biogenesis.
Heparanase acts through syntenin-1, ALIX and the syntenin-ALIX interaction to stimulate intraluminal budding
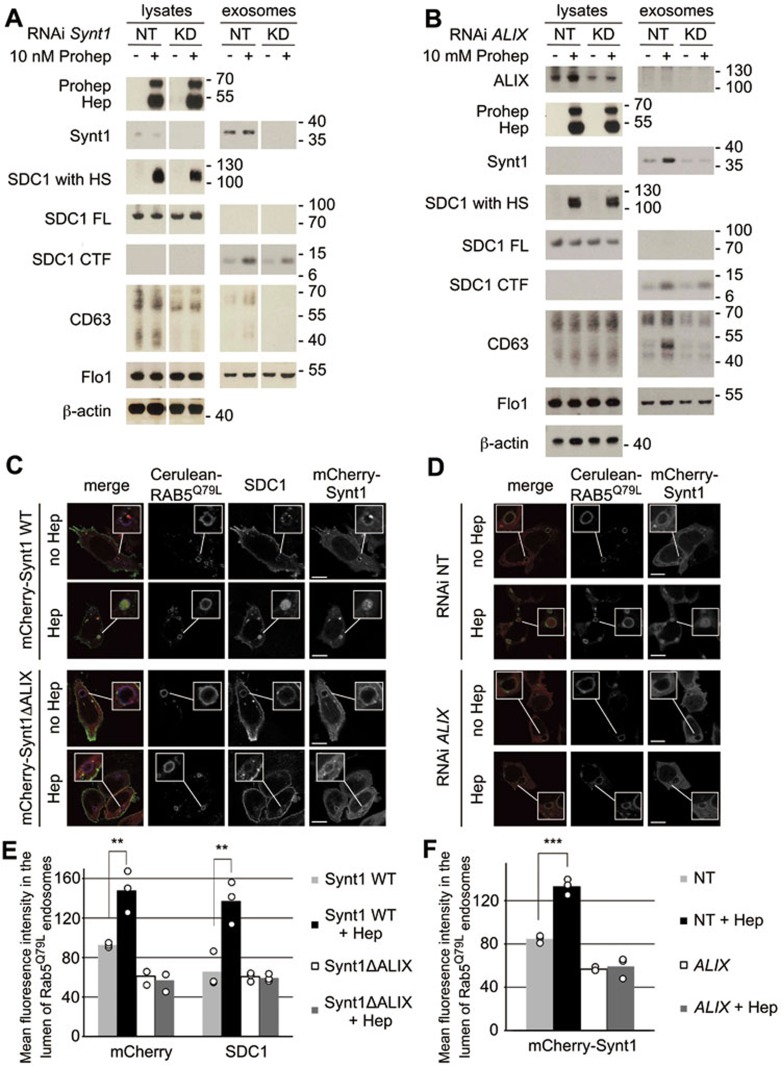
Along with syndecans, syntenin-1 and ALIX are key mechanistic components in the production of exosomes, influencing intraluminal budding into MVBs14. To investigate the roles of syntenin-1 and ALIX in the effect of heparanase on exosome production, syntenin-1 (Figure 4A) and ALIX (Figure 4B) were knocked down in MCF-7 cells, in the presence or absence of exogenously added proheparanase. Reducing the syntenin-1 or ALIX expression levels in cells had no significant influence on the uptake and processing of proheparanase, and on the trimming of syndecan by heparanase, as judged from the amount of internalized heparanase and the migration of heparan sulfate-modified syndecan-1, respectively (Figure 4A and 4B). Knockdown of syntenin-1 reduced the effect of heparanase on exosomal levels of syndecan-1 CTF and CD63 (Figure 4A). ALIX knockdown also markedly reduced the stimulatory effect of heparanase on exosomal syntenin-1, CD63 and syndecan-1 CTF (Figure 4B). Exosomal levels of flotillin-1 were unaffected by syntenin or ALIX knockdown or the addition of exogenous proheparanase (Figure 4A and 4B). These data indicate that heparanase exerts its effects on exosomes through syntenin-1 and ALIX.
Figure 4.
Heparanase acts through syntenin-1, ALIX and the syntenin-ALIX interaction to stimulate intraluminal budding. (A) The role of syntenin-1 and (B) the role of ALIX in the effect of heparanase on exosomes were investigated by the knockdown of syntenin-1 and ALIX (using Synt1 RNAi and ALIX RNAi, respectively) in MCF-7 cells. Non-targeting RNAi (NT) served as a control. Cells were left untreated (−) or treated with proheparanase (10 nM). In both the experiments, heparanase activity was evaluated using the migration pattern of chondroitinase ABC-, but not heparitinase-treated syndecan-1 present in cell lysates (SDC1 with HS). Molecular weight markers (in kDa) are indicated on the right of each blot. Western blots are representative of two independent experiments. Note that lysate and exosome samples derived from NT and KD cells, separated by a blank space in A, were run in the same gel, but not side by side, and that the band intensities in each row are directly comparable. (C) Confocal micrographs of MCF-7 cells co-transfected with wild-type mCherry-syntenin-1 (mCherry-Synt1 WT) or with mCherry-syntenin-1 harboring mutant LYP sequences and therefore defective in ALIX-binding (mCherry-Synt1-ΔALIX; red in merge), Cerulean-RAB5Q79L (blue in merge) and syndecan-1 (green in merge). Intraluminal budding of wild-type and mutant mCherry-syntenin-1 and of syndecan-1 cytoplasmic domain was scored in the presence (Hep) or absence (no Hep) of heparanase. (D) Confocal micrographs of MCF-7 cells co-transfected with mCherry-syntenin-1 (red in merge) and Cerulean-RAB5Q79L (blue in merge). In addition, the cells were transfected with RNAi targeting ALIX (ALIX RNAi) or with non-targeting RNAi (NT RNAi). Intraluminal budding of mCherry-syntenin-1 was evaluated in the presence (Hep) or absence (no Hep) of heparanase. (E) Quantification of intraluminal budding of mCherry-syntenin-1 and syndecan-1 cytoplasmic domain, as in C and (F) quantitation of intraluminal budding of mCherry-syntenin-1, as in D, measuring the mean fluorescence intensity of mCherry-syntenin-1 and syndecan-1 in the lumen of RAB5Q79L-positive endosomes (mean gray value per pixel). Bar heights represent mean values calculated from three independent experiments, scoring at least 30 cells per experiment. Individual data points are shown as white dots on top of the corresponding bars. **P < 0.05, ***P < 0.01 (Student's t-test, assuming normal distribution of the data points).
We also investigated the importance of ALIX for heparanase-stimulated intraluminal budding. ALIX was knocked down in MCF-7 cells co-expressing Cerulean-RAB5Q79L and mCherry-syntenin-1, and intraluminal budding of mCherry-syntenin-1 was measured in the presence or absence of exogenously added proheparanase and compared to the budding in cells treated with non-targeting RNAi, used as control (Figure 4D). As observed before (Figure 3C and 3E), in cells treated with non-targeting RNAi heparanase stimulated the intraluminal fluorescence intensity of mCherry-syntenin-1 by roughly 50% (Figure 4D and 4F, P < 0.01, n = 3). ALIX knockdown reduced the basal level of intraluminal budding of mCherry-syntenin-1 by 30% (P < 0.01, n = 3). Moreover, ALIX knockdown abolished the effect of heparanase on intraluminal budding of mCherry-syntenin-1 (Figure 4D and 4F).
Finally, we tested whether syntenin-ALIX interaction was required for the effect of heparanase. Syntenin-1 and ALIX interact directly through three LYPXnL motifs in the N-terminal domain of syntenin-1, which is essential in the syndecan-syntenin-ALIX exosomal pathway14. To this end, MCF-7 cells were co-transfected with Cerulean-RAB5Q79L, syndecan-1 and either wild-type mCherry-syntenin-1 or the mCherry-syntenin-1-ΔALIX mutant. In this mutant, the three LYPXnL motifs have been changed to LAAXnL, abolishing the interaction between syntenin-1 and ALIX14. Intraluminal budding of mCherry-syntenin-1 and syndecan-1 was studied in the presence or absence of exogenously added proheparanase (Figure 4C). As observed before (Figure 3D and 3F), heparanase augmented the concentration (i.e., fluorescence intensity) of wild-type mCherry-syntenin-1 and syndecan-1 in the lumen of RAB5Q79L-positive endosomes by ∼ 50 % and 100 %, respectively (Figure 4C and 4E, P < 0.05, n = 3). Compared to the levels of intraluminal fluorescence measured in cells expressing wild-type syntenin-1, basal levels of intraluminal budding of mCherry-syntenin-1-ΔALIX (i.e., budding in the absence of heparanase) were lower by 35% (P < 0.01, n = 3, Figure 4C and 4E). Of note, heparanase did not stimulate the intraluminal budding of mCherry-syntenin-1-ΔALIX. Moreover, in the presence of mCherry-syntenin-1-ΔALIX, heparanase was not able to stimulate intraluminal budding of syndecan-1 (Figure 4C and 4E). These experiments clearly demonstrate that heparanase depends on syntenin-1, ALIX and the direct interaction of syntenin-1 with ALIX to exert its effect on exosome biogenesis, i.e., acts as a modulator of the syndecan-syntenin-ALIX pathway.
Discussion
Previously, we have shown that the syndecan-syntenin-ALIX axis controls the biogenesis of syntenin-1-exosomes14. ALIX, through interaction with the cytoplasmic adaptor syntenin-1, adapts syndecans and syndecan cargo to the ESCRT budding machinery at the level of endosomal membranes. Heparan sulfate is essential to this mechanism of exosome formation. Syndecan cargo — most probably bound to the heparan sulfate of syndecan — is thought to cluster syndecans and create syndecan assemblies that recruit syntenin-1 and ALIX and subsequently support membrane budding14. Recently, Thompson and colleagues reported that heparanase stimulates the release of extracellular vesicles/exosomes in a concentration-dependent fashion and alters the composition of these extracellular vesicles25.
Our study shows heparanase exerts its effect on exosomes through the syndecan-syntenin-ALIX pathway. First, heparanase-mediated stimulation of exosome production depends on the syndecans (but not glypicans; Figure 2D and 2E). Second, syntenin-1, ALIX and the interaction of syntenin with ALIX control the effect of heparanase (Figure 4). Third, as shown by the RAB7 knockdown (Figure 3A and 3B), the extracellular vesicles that are affected by heparanase are of endosomal origin, proving these vesicles are exosomes, an issue not addressed in the Thompson study25. Last, heparanase stimulates the endocytosis of syndecan and stimulates the accumulation of syndecan and syntenin-1 in the lumen of endosomes (Figure 3C to 3F). At present, it is not clear whether heparanase stimulates the number of ILVs/exosomes made, which are possibly ILVs/exosomes that belong to a specific category, or stimulates the loading of ILVs/exosomes with specific cargo or both. Heparanase appears to only slightly stimulate the total number of exosomes produced (Supplementary information, Figure S2). However, if syntenin-1-exosomes constitute a minor subpopulation of the vesicles made by non-stimulated cells, such (small) difference might still correspond to a large increase in syntenin-exosome numbers, whereas other subpopulations are unaffected by heparanase. Possible additional effects of heparanase on exosome aggregation (Supplementary information, Figure S3) or exosome uptake (Supplementary information, Figures S4 and S5) have been excluded or contribute minimally to the changes in exosome composition. Contrary to the findings of a recent study30, knockdowns of syndecans or syntenin-1 in MCF-7 cells have no effect on heparanase uptake or processing (Figures 2D and 4A). We found that proheparanase uptake by MCF-7 cells largely depends on mannose-6-phosphate receptor, which possibly explains this difference (Roucourt et al., unpublished data). Thus, heparanase requires syndecans and syntenin-1 as effectors, rather than for its own routing to endosomes and activation.
The effects of heparanase on exosome production depend on heparan sulfate (Figure 2B), and more specifically on the cleavage of heparan sulfate (Figure 2A). The need for heparanase enzymatic activity is also in line with the observations of Thompson et al.25. Importantly, heparanase enzymatic activity does not remove all heparan sulfate from proteoglycans. Comparison of the migration patterns of the chondroitinase-treated heparanase-exposed syndecan-1 with that of chondroitinase- and heparitinase-treated syndecan-1 reveals that the former has a mass that is roughly 20 kDa larger, suggesting that sizeable heparan sulfate chains remain attached to the proteoglycan core proteins, even in the presence of high levels of heparanase. Although the nature of the remaining heparan sulfate is unknown, its presence on syndecans seems to be essential for exosome formation. Of note, the complete removal of heparan sulfate with heparitinase abrogates syntenin-1-exosome production14. Glypican, providing an alternative source of heparan sulfate and substrate for heparanase, cannot substitute for syndecans (Figure 2E). Taken together, all elements show that heparanase is an important modulator of the syndecan-syntenin-ALIX exosome biogenesis pathway by cleaving the heparan sulfate chains on the syndecans, yielding syndecans with trimmed heparan sulfate, rather than short free heparan sulfate fragments directly influencing exosomal secretion.
Exactly how heparanase-cleaved heparan sulfate linked to syndecans facilitates exosome formation is not clear yet. Figure 5 models the potential mechanisms of action. Central to the model is the concept that syntenin is recruited to membranes by clustered 'bait', engaging the tandem PDZ domains of syntenin31. One possible explanation is that the newly generated heparan sulfate ends are permissive for syndecan clustering or render syndecan clustering more efficient. Compared to the original non-reducing ends of non-cleaved heparan sulfate, these new ends are likely to be enriched in sugars carrying one or more sulfates32,33, increasing the propensity of chain-ends to be occupied by ligand. Such change in charge distribution over the chain might stimulate ligand-induced clustering of heparan sulfate chains, and as a result syndecan clustering. Alternatively, shorter heparan sulfate chains or a reduction of the total negative charge on syndecan might diminish steric hindrance or negative charge repulsion, respectively. Whichever mechanism involved, all would facilitate syndecans approaching each other and the clustering of syndecans, which has been shown to support exosome production14, probably by facilitating the membrane-recruitment of syntenin-ALIX-ESCRT complexes. Thus, in the absence or in the relative shortage of heparanase activity, the levels of syndecan that can recruit syntenin-1 might be limiting. In addition, proheparanase and heparanase activity accelerate syndecan internalization, possibly enhancing the amount of endosomal syndecan (Supplementary information, Figure S6). Increased levels of endosomal syndecan and endosomal syndecan processing might contribute to syndecan clustering (as described above) and enhance the membrane-recruitment of syntenin-ALIX-ESCRT complexes.
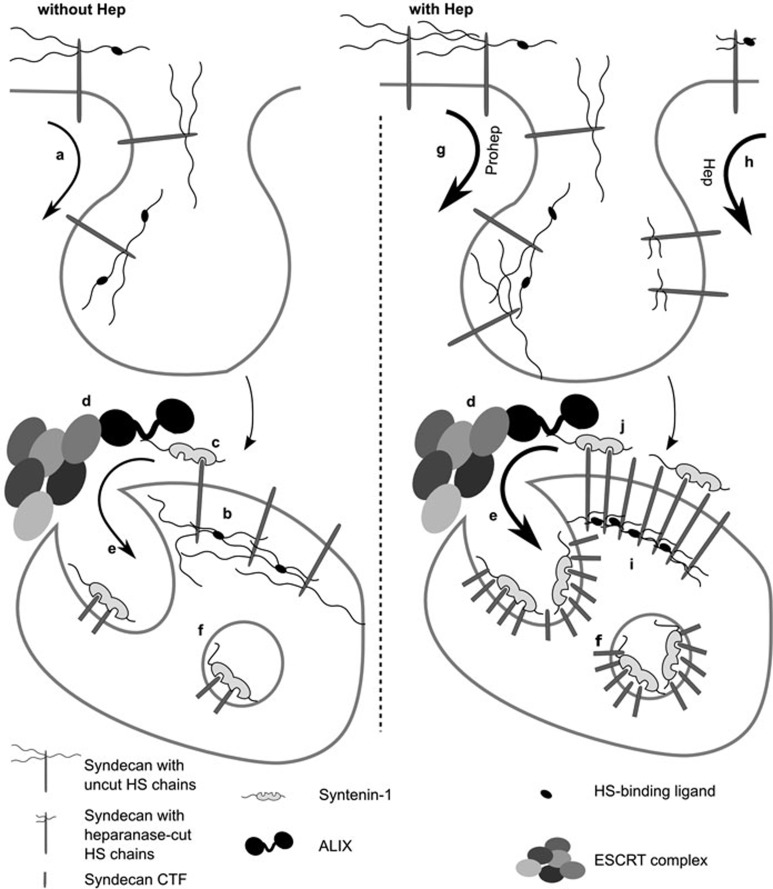
Figure 5.
Schematic representation of the potential mechanism of action of heparanase in exosome biogenesis. (a) Syndecans are continuously internalized by cells. (b) Endocytosed syndecans are clustered by the ligands binding to their heparan sulfate (HS) chains, a process that might already be initiated at the cell surface. (c) These syndecan assemblies subsequently recruit syntenin, (d) ALIX and the ESCRT machinery. (e) As a result, syndecan and cargo bound to syndecan is recruited to membrane buds protruding into the lumen of MVBs. At this stage, the extracellular portion of the syndecans is cleaved by endosomal protease, generating syndecan CTFs. (f) The ESCRT machinery pinches off these buds and as such liberates intraluminal vesicles, loaded with syntenin and syndecan CTFs, into the lumen of MVBs. (g) As a ligand of HS, proheparanase might directly stimulate the clustering and subsequent endocytosis of syndecans at the cell membrane. (h) In addition, and more importantly, heparanase activity trims the HS chains on syndecan, possibly regulating the engagements and further stimulating the internalization of syndecans. Both the mechanisms lead to a higher concentration of endosomal syndecan. (i) Trimming the HS chains on syndecans might also stimulate clustering of syndecans at the limiting membrane of MVBs. Enhancement of the syndecan engagements, reduction of syndecan self-repulsion and effects on the dimensions and spacing of syndecans in syndecan-ligand complexes, by the trimming of the HS on syndecan, are several possible and non-mutually exclusive mechanisms by which heparanase may foster syndecan clustering. (j) Enhanced clustering of syndecans increases the recruitment and binding of syntenin-1 to endosomal syndecan assemblies and ultimately may cause the binding of syntenin-1 to larger associations of syndecans, increasing the syndecan-syntenin stoichiometry in syndecan-syntenin complexes and exosomes. Note that the diagram makes no statement on the accumulation of ALIX and ESCRTs in the intraluminal vesicles and exosomes, which were not investigated here.
Of note, whereas the effect of heparanase on exosomal syntenin-1 plateaus, the amounts of exosomal cargo that composes the direct 'bait' for the PDZ domains of syntenin (i.e., syndecan CTF and CD63) continue to increase with increasing heparanase concentrations (Figure 1A and 1B). These findings correspond to previous observations that the ratio of 'bait' or cargo to syntenin in exosomes is not constant14. Thus, while intraluminal budding/exosome formation appears to be triggered by the organization of syntenin bait (i.e., syndecan-CTF) in structures of higher order, syntenin may adapt only to a part of the bait present in these organizations. This situation would be similar to the incorporation of viral proteins in budding membranes, where, due to lateral associations between coat proteins and late domains also functioning in trans, not all individual copies of the GAG-proteins that end up in the viral coats need to be provided with direct links to the budding machineries34. Similarly, a sizeable fraction, but not all of the syntenin-dependent cargo that ends up in exosomes might need to be directly linked to syntenin. Thus, where syntenin may become limiting at some point (and exosome numbers might stagnate), heparanase might still increase the 'lateral association' or clustering of syndecans, further stimulating the incorporation of syndecans and syndecan-associated proteins in exosomes. Differences in the magnitude of the heparanase effects on recruitment of specific syndecan CTFs to exosomes (i.e., up to 7-fold stimulations for syndecan-1 CTF compared to the rather modest influence on syndecan-4 CTF), then suggest at least subtle differences between the syndecan family members, i.e., differential localization, trafficking, heparan sulfate composition, access of heparanase to syndecans or syndecan-cargo associations.
Apart from the role of heparanase in the syndecan-syntenin-ALIX exosome biogenesis pathway, our data reveal that heparanase stimulates the targeting of specific cargo to exosomes. While exosomal syntenin-1, syndecan CTFs and CD63 increase with increasing heparanase levels, exosomal flotillin-1 remains unaffected, even at high concentrations of heparanase (Figure 1A and 1B). This would be in line with our previous observations of flotillin-1 not being the cargo of syntenin-1 exosomes14. Of note, heparanase was reported to stimulate exosomal accumulation of flotillin-1 in CAG human myeloma cells25. While remaining unexplained, differences in the selectivity of heparanase effects might reflect the complexity of exosome biogenesis pathways and the differences in direct and possibly also indirect effects of heparanase on these different pathways in different cell lines and tissues. For example, heparanase-activated signaling might ultimately also impact on ubiquitin-regulated budding mechanisms. Yet, similarly to flotillin-1, exosomal levels of CD9 and CD81, two tetraspanins commonly used as exosomal markers, are also not affected by heparanase. The specificity of the heparanase effect in MCF-7 cells suggests that different mechanisms for selecting exosomal cargo exist at the level of the MVBs. Even more, it underpins the hypothesis of different exosomal populations formed through specific biogenesis pathways, one of which would be the syndecan-syntenin-ALIX pathway. Most probably, specific cargo is selected through the interaction with heparan sulfate and heparanase-trimmed heparan sulfate. Heparanase-stimulated exosomal accumulation of hepatocyte growth factor and VEGF25, two heparan sulfate-binding growth factors, is suggestive of such a mechanism. On the other hand, cargo like CD63 might be recruited to exosomes through the interaction of its C-terminal PDZ-binding motif with the PDZ domains of syntenin-114. Further research will have to clarify to what extent the different components of the syndecan-syntenin-ALIX pathway participate in the selection of specific exosomal cargo and whether this specificity is reflected in the existence of distinctive exosomal subpopulations or merely reflects the different pathways taking part in the biogenesis and variable loading of the same exosomes.
Taken together, the above findings identify heparanase as a fundamental modulator of exosome biogenesis via the syndecan-syntenin-ALIX pathway, by cleaving and 'activating' the heparan sulfate chains of the syndecans. It will be of particular interest to find out the mechanisms governing this activation, and to delineate the influence of heparanase, i.e., the heparanase-activated syndecan-syntenin-ALIX machinery, on the overall composition of exosomal cargo, and more specifically on the exosomal incorporation of (phosphorylated) receptors, growth factors and morphogens, many of which interact with heparan sulfate and depend on heparan sulfate for activity. Intercellular communication via specific exosomal cargo (Wnt11 and c-Met, respectively) has been shown to regulate crucial processes like cancer cell motility, the onset of metastasis7 and premetastatic niche formation6. Intriguingly, the molecular players and pathways involved are strongly influenced by heparan sulfate, arguing for a pivotal role of the syndecan-syntenin-ALIX machinery and its modulators, like heparanase, in physiological processes linked to exosomes and the transfers of exosomal cargo.
Materials and Methods
Expression vectors and reagents
For gain-of-function and rescue experiments in cells, cDNA (encoding full-length non-tagged human syntenin-1 and human glypican-1) was cloned in pcDNA3.1/Zeo (Invitrogen)35,36. Constructs were verified by sequencing. pcDNA3.1 encoding full length mouse syndecan 1 was a kind gift from Prof Alan Rapraeger (University of Wisconsin, USA). Empty vector was used as control. pmCherry-syntenin-1, pmCherry-syntenin-1-ΔALIX (Y5A, P6A, Y46A, P47A, Y50A, P51A), pcDNA3.1/Zeo-syndecan-1 and pCerulean-RAB5Q79L were described previously13,14. Lentiviral vector pKLO.1puro carrying a shRNA targeting the mouse heparanase 3′UTR (TRCN0000098110, obtained from BCCM/LMBP collection, Ghent University, Belgium) was used to knockdown mouse heparanase. pKLO.1 with non-targeting shRNA (SHC002) served as the control. For stable heparanase expression, cDNA encoding wild-type or catalytically dead (E225A and E343A) human heparanase were cloned in pMSCVpuro retroviral vector (Clontech). Empty vector served as control. All RNAi targeting human sequences (ON-Target plus for syntenin-1MQ-008270-00, syndecan 1 J-010621-11-0020, syndecan 4 J-003706-08-0020, EXT1 L-011030-00, EXT2 L-011031-00 and RAB7 L-010388-00) and the non-targeting control RNAi (siSTABLE) were purchased from Dharmacon. Rabbit polyclonal antibodies against human (Rb1) and mouse syntenin-1 (Rb96), ALIX (Rb67), and mouse monoclonal antibodies against human glypican-1 (S1), human/mouse syndecan-1/3 intracellular domain (2E9) and human syndecan-4 ectodomain (8G3) were described previously14,31,37,38,39,40. Other antibodies were from commercial sources and were used as recommended by the manufacturer: antibodies against RAB7 (H-50) and β-actin (AC-15) were from Santa Cruz; antibody against the intracellular domain of syndecan 4 was from Abnova; antibodies against human CD63 (MEM-259) was from Abcam; antibody against HA (16B12) was from Covance; anti-flotillin-1 was from BD Biosciences. A home-made HA-tagged Nanobody, binding heparanase, in combination with anti-HA antibody, was used to detect human and mouse (pro)heparanase. Recombinant proheparanase was purchased from the Protein Service Facility of VIB.
Cells and transfection
MCF-7 (human mammary ductal carcinoma) cells were obtained from ATCC. B16-F10 (mouse melanoma) cells were a kind gift from Prof Anton Roebroeck (KU Leuven, Belgium) Cells were routinely grown in DMEM/F12 (1:1) medium (Life Technologies) supplemented with 10% fetal bovine serum (Life Technologies). For exosome production, cells were cultured in DMEM/F12 (1:1) with HEPES, pH 7.3, supplemented with 0.1% BSA (Sigma-Aldrich). For RNAi experiments (throughout the manuscript RNAi refers to the use of siRNA for transient knockdown), cells were transfected at 30% confluency with 100 nM RNAi using Oligofectamine (Invitrogen) according to the protocol provided by the manufacturer. Cells (and exosomes produced during the 16 h of culture) were collected for analysis 48 h later. For transient expressions (in absence of RNAi treatment), the cells were transfected the day after plating with 1 μg DNA per 85-mm (56 cm2) dish using X-tremeGENE 9 (Roche) according to instructions provided by the manufacturer. In case cells were transfected with both RNAi and DNA, RNAi was transfected first (using Oligofectamine), 1 day after plating the cells, according to the instructions of the manufacturer. The next day (2 days after plating the cells), cells were transiently transfected as described above. Cells and exosomes were collected 48 h later. For creating cell lines stably expressing wild-type or catalytically dead heparanase (E225A and E343A), the φNX-ampho retroviral packaging cell line (gift from Prof Garry Nolan, Stanford University, USA) was transfected at a density of 50% with pMSCVpuro encoding wild-type or catalytically dead heparanase, or pMSCVpuro empty as control, using X-tremeGENE 9 (Roche). Retroviral particles were collected after 48 h, filtered (Millex-HV 0.45-μm PVDF filters, Millipore) and transferred on MCF-7 cells. This procedure was repeated the day after and the cells were selected for transfection with 800 μg/ml hygromycin (Roche). The procedure to create B16-F10 cells with stable knockdown of heparanase was similar, except for the use of the MISSION lentivrial packaging mix (Sigma-Aldrich) in combination with pKLO.1 vectors described above and 800 ng/ml puromycin (Invivogen) selection.
Exosomes and total cell lysates
For comparative analyses, in gain- and loss-of-function studies, exosomes were collected from equivalent amounts of culture medium, conditioned by equivalent amounts of cells, for equivalent lengths of time, as described before14. In brief, when reaching 70% of confluency, the cell layers were rinsed and refreshed with DMEM/F12 with HEPES pH 7.3 supplemented with 0.1% BSA. The same procedure was repeated 8 h later and (where indicated) proheparanase was added to the medium. The conditioned media containing the exosomes were collected 16 h later, pooling media from two corresponding 85-mm culture dishes. Exosomes were isolated from these media by three sequential centrifugation steps at 4 °C: 10 min at 800× g, to remove cells; 30 min at 10 000× g, to remove cell debris; and 3 h at 140 000× g using a swinging bucket SW41 Ti rotor with a k factor of 265 (Beckman Coulter), to pellet exosomes, followed by one rinse (suspension in PBS/centrifugation at 140 000× g). Exosomal pellets were re-suspended in lysis buffer (PBS supplemented with 1% NP40, 1 mM EDTA, 5 μg/ml leupeptin, 1 μg/ml pepstatin and 1 mM phenylmethylsulphonyl fluoride). Corresponding cell layers were washed in cold PBS, and scraped on ice in lysis buffer. Lysates from two corresponding 85-mm culture dishes were pooled, and cleared by centrifugation at 16 000× g, for 30 min at 4 °C. Cell lysates were normalized by protein content, and corresponding exosome preparations were adjusted accordingly. None of the DNA or RNAi transfections significantly affected cell survival or growth, requiring major (> 10%) adjustments of the samples.
Western blot
To allow detection of full-length syndecan core proteins, the samples were digested with (0.006 U/ml) heparitinase (Amsbio) and (0.166 U/ml) chondroitinase ABC (Amsbio) for 3 h at 37 °C in heparitinase buffer (0.1% Triton, 0.1 M NaCl, 1 mM CaCl2, 50 mM 6-aminohexanoic acid and 50 mM HEPES, at pH 7). Samples were fractionated in 4%-12% gradient gels (NuPAGE Novex Bis-Tris gels, Invitrogen), electro-transferred to Hybond-C extra 0.45-μm nitrocellulose membrane (GE Healthcare Life Sciences) and incubated with the indicated antibodies. Signals were visualized with enhanced chemiluminescence detection reagent (GE Healthcare Life Sciences) and were quantified by densitometric scanning using ImageJ Software (National Institutes of Health).
Fluorescence microscopy
For microscopic analysis, cells were plated on non-coated eight-well chamber slides (Nalge Nunc International). 20 000 cells were plated per well. The next morning (16 h after plating) medium was changed to DMEM/F12 with HEPES, pH 7.3, supplemented with 0.1% BSA and the cells were transfected with pmCherry-syntenin-1, pmCherry-syntenin-1-ΔALIX, pCerulean-RAB5Q79L and pcDNA3.1-Zeo-syndecan-1, in the combinations indicated, using XtremeGene 9 (Roche) as described by the manufacturer. Then, 8 h later (24 h after plating), the cells were transferred to DMEM/F12 with HEPES, pH 7.3, supplemented with 0.1% BSA, without or with 50 nM heparanase. After overnight incubation (40 h after plating) the cells were fixed with PBS-4% paraformaldehyde and mounted with Citifluor. In case cells were transfected with both RNAi and DNA, RNAi was transfected while plating the cells, using RNAiMAX (Life Technologies) as described by the manufacturer. The next morning (24 h after plating) the cells were transfected with DNA and further treated as described above.
For the immunocytochemical detection of the syndecan-1 cytoplasmic domain, after fixation, cells were permeabilized with 0.1% Triton X-100 in PBS, followed by incubation in PBS-glycine 0.1 M and then blocked with PBS-1% BSA containing 10% goat serum. Primary antibodies (25 μg/ml of mAb 2E9) and Alexa Fluor 488-conjugated secondary goat anti-mouse antibodies (diluted 1 in 1 000) were diluted in blocking buffer and applied successively to cells before mounting with Citifluor.
Images were obtained using a confocal Olympus Fluoview 1 000 instrument (Olympus, Pennsylvania-Allentown, USA). The intensity of the mCherry-syntenin-1 or Alexa Fluor-488 signals in the lumen of RAB5Q79L-positive endosomes, i.e., total fluorescence corrected for the size of the lumen of the RAB5Q79L-positive endosomes (the sum of the gray values of all the pixels in the selection divided by the number of pixels or mean grey value per pixel), was quantified using ImageJ.
Statistical analysis
Throughout the manuscript, Student's t-test is used for assessing statistical significance of observed differences. The Student's t-test assumes a normal distribution of the data. Because of the limited number of data points per condition (maximally 5) normal distribution of the data points could not be formally examined. For all statistical analyses we are assuming normal distribution of the data points.
Acknowledgments
We thank Dr Baietti MF for her feedback on this study and for critical comments during the redaction of this manuscript, Dr Granjeaud S for advice on statistical analysis and the Imaging Core Facility of the KU Leuven for access to confocal microscopes. This work was supported by the Fund for Scientific Research-Flanders (FWO, G.0683.09N), the Concerted Actions Program of the KU Leuven (GOA/12/016 and GOA/2006/13), the VIB, the Belgian Federation against Cancer (Stichting tegen Kanker, SCIE2006-36 and 214-2008) and the Interuniversity Attraction Poles of the Prime Ministers Services (IUAP). The TRC 1/1.5 human and mouse shRNA libraries of BCCM/LMBP (Ghent University, Belgium) were obtained through structural funding for medium-scale research infrastructure (HERCULES, AUGE/09/040).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Heparanase stimulates exosome production in cell lines other than MCF-7.
Heparanase effects on exosome number and size.
Heparanase effects on exosome sedimentation.
Heparanase effects on syntenin-1-exosome binding and uptake.
Heparanase does not influence syntenin-exosome binding and uptake.
Heparanase stimulates the endocytosis of syndecan-1 and syndecan-4.
References
- Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- Skog J, Wurdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo G, Nijman HW, Stoorvogel W, et al. B lymphocytes secrete antigen-presenting vesicles. J Exp Med. 1996;183:1161–1172. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thery C, Duban L, Segura E, Veron P, Lantz O, Amigorena S. Indirect activation of naive CD4+ T cells by dendritic cell-derived exosomes. Nat Immunol. 2002;3:1156–1162. doi: 10.1038/ni854. [DOI] [PubMed] [Google Scholar]
- Peinado H, Aleckovic M, Lavotshkin S, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luga V, Zhang L, Viloria-Petit AM, et al. Exosomes mediate stromal mobilization of autocrine Wnt-PCP signaling in breast cancer cell migration. Cell. 2012;151:1542–1556. doi: 10.1016/j.cell.2012.11.024. [DOI] [PubMed] [Google Scholar]
- Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev Cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- Wollert T, Hurley JH. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature. 2010;464:864–869. doi: 10.1038/nature08849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH, Hanson PI. Membrane budding and scission by the ESCRT machinery: it's all in the neck. Nat Rev Mol Cell Biol. 2010;11:556–566. doi: 10.1038/nrm2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trajkovic K, Hsu C, Chiantia S, et al. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- Ghossoub R, Lembo F, Rubio A, et al. Syntenin-ALIX exosome biogenesis and budding into multivesicular bodies are controlled by ARF6 and PLD2. Nat Commun. 2014;5:3477. doi: 10.1038/ncomms4477. [DOI] [PubMed] [Google Scholar]
- Baietti MF, Zhang Z, Mortier E, et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol. 2012;14:677–685. doi: 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- Hurley JH, Odorizzi G. Get on the exosome bus with ALIX. Nat Cell Biol. 2012;14:654–655. doi: 10.1038/ncb2530. [DOI] [PubMed] [Google Scholar]
- Vlodavsky I, Friedmann Y, Elkin M, et al. Mammalian heparanase: gene cloning, expression and function in tumor progression and metastasis. Nat Med. 1999;5:793–802. doi: 10.1038/10518. [DOI] [PubMed] [Google Scholar]
- Kelly T, Miao HQ, Yang Y, et al. High heparanase activity in multiple myeloma is associated with elevated microvessel density. Cancer Res. 2003;63:8749–8756. [PubMed] [Google Scholar]
- Edovitsky E, Elkin M, Zcharia E, Peretz T, Vlodavsky I. Heparanase gene silencing, tumor invasiveness, angiogenesis, and metastasis. J Natl Cancer Inst. 2004;96:1219–1230. doi: 10.1093/jnci/djh230. [DOI] [PubMed] [Google Scholar]
- Ilan N, Elkin M, Vlodavsky I. Regulation, function and clinical significance of heparanase in cancer metastasis and angiogenesis. Int J Biochem Cell Biol. 2006;38:2018–2039. doi: 10.1016/j.biocel.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Vreys V, Delande N, Zhang Z, et al. Cellular uptake of mammalian heparanase precursor involves low density lipoprotein receptor-related proteins, mannose 6-phosphate receptors, and heparan sulfate proteoglycans. J Biol Chem. 2005;280:33141–33148. doi: 10.1074/jbc.M503007200. [DOI] [PubMed] [Google Scholar]
- Zetser A, Levy-Adam F, Kaplan V, et al. Processing and activation of latent heparanase occurs in lysosomes. J Cell Sci. 2004;117:2249–2258. doi: 10.1242/jcs.01068. [DOI] [PubMed] [Google Scholar]
- Gingis-Velitski S, Zetser A, Flugelman MY, Vlodavsky I, Ilan N. Heparanase induces endothelial cell migration via protein kinase B/Akt activation. J Biol Chem. 2004;279:23536–23541. doi: 10.1074/jbc.M400554200. [DOI] [PubMed] [Google Scholar]
- Zetser A, Bashenko Y, Edovitsky E, Levy-Adam F, Vlodavsky I, Ilan N. Heparanase induces vascular endothelial growth factor expression: correlation with p38 phosphorylation levels and Src activation. Cancer Res. 2006;66:1455–1463. doi: 10.1158/0008-5472.CAN-05-1811. [DOI] [PubMed] [Google Scholar]
- Fux L, Feibish N, Cohen-Kaplan V, et al. Structure-function approach identifies a COOH-terminal domain that mediates heparanase signaling. Cancer Res. 2009;69:1758–1767. doi: 10.1158/0008-5472.CAN-08-1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CA, Purushothaman A, Ramani VC, Vlodavsky I, Sanderson RD. Heparanase regulates secretion, composition, and function of tumor cell-derived exosomes. J Biol Chem. 2013;288:10093–10099. doi: 10.1074/jbc.C112.444562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson HC, Svensson KJ, van Kuppevelt TH, Li JP, Belting M. Cancer cell exosomes depend on cell-surface heparan sulfate proteoglycans for their internalization and functional activity. Proc Natl Acad Sci USA. 2013;110:17380–17385. doi: 10.1073/pnas.1304266110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrazin S, Lamanna WC, Esko JD. Heparan sulfate proteoglycans. Cold Spring Harb Perspect Biol. 2011;3pii:a004952. doi: 10.1101/cshperspect.a004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlandingham PA, Ceresa BP. Rab7 regulates late endocytic trafficking downstream of multivesicular body biogenesis and cargo sequestration. J Biol Chem. 2009;284:12110–12124. doi: 10.1074/jbc.M809277200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg C, Bremnes B, Mehlum A, et al. FYVE and coiled-coil domains determine the specific localisation of Hrs to early endosomes. J Cell Sci. 2001;114:2255–2263. doi: 10.1242/jcs.114.12.2255. [DOI] [PubMed] [Google Scholar]
- Shteingauz A, Ilan N, Vlodavsky I. Processing of heparanase is mediated by syndecan-1 cytoplasmic domain and involves syntenin and alpha-actinin. Cell Mol Life Sci. 2014;71:4457–4470. doi: 10.1007/s00018-014-1629-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Tomatis D, Rosas M, et al. Characterization of syntenin, a syndecan-binding PDZ protein, as a component of cell adhesion sites and microfilaments. Mol Biol Cell. 2001;12:339–350. doi: 10.1091/mbc.12.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Shriver Z, Venkataraman G, El Shabrawi Y, Sasisekharan R. Tumor cell surface heparan sulfate as cryptic promoters or inhibitors of tumor growth and metastasis. Proc Natl Acad Sci USA. 2002;99:568–573. doi: 10.1073/pnas.012578299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson SB, Liu J. Multi-faceted substrate specificity of heparanase. Matrix Biol. 2013;32:223–227. doi: 10.1016/j.matbio.2013.02.006. [DOI] [PubMed] [Google Scholar]
- Wills JW, Cameron CE, Wilson CB, Xiang Y, Bennett RP, Leis J. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J Virol. 1994;68:6605–6618. doi: 10.1128/jvi.68.10.6605-6618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David G, Lories V, Decock B, Marynen P, Cassiman JJ, Van den Berghe H. Molecular cloning of a phosphatidylinositol-anchored membrane heparan sulfate proteoglycan from human lung fibroblasts. J Cell Biol. 1990;111:3165–3176. doi: 10.1083/jcb.111.6.3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grootjans JJ, Zimmermann P, Reekmans G, et al. Syntenin, a PDZ protein that binds syndecan cytoplasmic domains. Proc Natl Acad Sci USA. 1997;94:13683–13688. doi: 10.1073/pnas.94.25.13683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David G, van der Schueren B, Marynen P, Cassiman JJ, van den Berghe H. Molecular cloning of amphiglycan, a novel integral membrane heparan sulfate proteoglycan expressed by epithelial and fibroblastic cells. J Cell Biol. 1992;118:961–969. doi: 10.1083/jcb.118.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boeck H, Lories V, David G, Cassiman JJ, van den Berghe H. Identification of a 64 kDa heparan sulphate proteoglycan core protein from human lung fibroblast plasma membranes with a monoclonal antibody. Biochem J. 1987;247:765–771. doi: 10.1042/bj2470765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lories V, Cassiman JJ, Van den Berghe H, David G. Multiple distinct membrane heparan sulfate proteoglycans in human lung fibroblasts. J Biol Chem. 1989;264:7009–7016. [PubMed] [Google Scholar]
- Zimmermann P, Meerschaert K, Reekmans G, et al. PIP(2)-PDZ domain binding controls the association of syntenin with the plasma membrane. Mol Cell. 2002;9:1215–1225. doi: 10.1016/s1097-2765(02)00549-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Heparanase stimulates exosome production in cell lines other than MCF-7.
Heparanase effects on exosome number and size.
Heparanase effects on exosome sedimentation.
Heparanase effects on syntenin-1-exosome binding and uptake.
Heparanase does not influence syntenin-exosome binding and uptake.
Heparanase stimulates the endocytosis of syndecan-1 and syndecan-4.