Abstract
We used whole-exome sequencing to identify variants other than APOE associated with the rate of hippocampal atrophy in amnestic mild cognitive impairment. An in-silico predicted missense variant in REST (rs3796529) was found exclusively in subjects with slow hippocampal volume loss and validated using unbiased whole-brain analysis and meta-analysis across 5 independent cohorts. REST is a master regulator of neurogenesis and neuronal differentiation that has not been previously implicated in Alzheimer’s disease. These findings nominate REST and its functional pathways as protective and illustrate the potential of combining next-generation sequencing with neuroimaging to discover novel disease mechanisms and potential therapeutic targets.
Late-onset Alzheimer’s disease (LOAD) is a progressive neurodegenerative condition for which there is presently no disease-modifying treatment.1 With the heritability of LOAD estimated to be as high as 80%, a better understanding of genetic susceptibility factors has the potential to advance strategies for early detection and treatment.2 Recent large-scale genome-wide association studies have identified and confirmed approximately 21 LOAD-associated genes in addition to APOE, where the ε4 allele is the best established and most significant genetic risk factor.3 However, only about 50% of LOAD heritability is accounted for by all of the known LOAD susceptibility genes including APOE, leaving a substantial proportion of the heritability remaining to be identified.4
Most genetic studies in LOAD have focused on identifying variants associated with case–control status. Genetic influences on quantitative intermediate phenotypes such as rate of brain tissue loss and subsequent cognitive decline remain understudied despite enhanced statistical power and mechanistic explanatory contributions. 5 Recently, using a novel strategy combining an extreme-trait design with whole-exome sequencing (WES) followed by neuroimaging genetics in a larger sample, we performed the first genome-wide analysis to identify functional exonic single nucleotide variants (SNVs) associated with rate of hippocampal neurodegeneration. 6 Our prior study identified 2 functional nonsynonymous SNVs other than the well-described APOE ε4 allele where the minor alleles were associated with more rapid hippocampal volume loss in participants with APOE ε3/ε3 genotypes. In the present study, we adapted our previous approach to discover protective functional missense SNVs associated with a slower atrophy rate of hippocampal volume in subjects with mild cognitive impairment (MCI), an early stage of the LOAD continuum. To discover variants independent of APOE ε4, we again focused our analyses on APOE ε3/ε3 subjects.
Subjects and Methods
Subjects
All individuals included in these analyses were participants in the Alzheimer’s Disease Neuroimaging Initiative Phase 1 (ADNI-1) and its subsequent extensions (ADNI-GO/2), Add- NeuroMed, Indiana Memory and Aging Study (IMAS), and Multi Institutional Research on Alzheimer Genetic Epidemiology studies (MIRAGE; see the reference6 for details). To reduce the possibility of spurious findings due to population stratification effects, we selected only non-Hispanic Caucasian participants from each cohort (ADNI-1, ADNI-GO/2, IMAS, Add- NeuroMed, and MIRAGE) using the same method, that is, selecting only subjects who clustered with CEU (Utah residents with Northern and Western European ancestry from the CEPH [Centre de’Etude du Polymorphism Humain] collection)+TSI (Toscani in Italia) populations using HapMap 3 genotype data and multidimensional scaling analysis after performing standard quality control procedures for genetic markers and subjects.
WES Analysis
To implement the extreme-trait design for WES, 8 matched pairs of non-Hispanic Caucasian male participants were selected by on the basis of rapid versus slow hippocampal volume change (annualized percentage of change [APC]) on magnetic resonance imaging (MRI) over 2 years. Participants had a diagnosis of amnestic MCI at the baseline visit and APOE ε3/ε3 genotype. The 8 pairs were approximately matched on age, educational level, and handedness. One member of each pair had a relatively rapid loss in hippocampal volume over the first 2 years of the study (rapid group) and the other member of each pair had a stable or relatively slow rate of hippocampal volume loss (slow group; p<0.001). The rapid group had an average 2-year APC of −4.99% (range=−3.11 to −7.15), whereas the slow group had an average 2-year APC of 0.16% (range=1.59 to −1.47). WES was performed on blood-derived genomic DNA samples using Agilent’s SureSelect Human All Exon 50Mb kit (Agilent Technologies, Santa Clara, CA) and a HiSeq2000 (Illumina, San Diego, CA; paired-end 105bp reads). Short-read sequences in the target region were mapped to the National Center for Biotechnology Information reference human genome (build 37.64) and analyzed using our previously established WES pipeline.6 Among all exonic variants identified by WES, we specifically focused on identification of variants carried only in the slow group, that is, variants in coding regions in which >4 of 8 subjects in the slow group had at least 1 alternative allele, but where all 8 subjects in the rapid group had the same alleles at the locus as the reference human genome.
Image Processing and Imaging Genetics Analysis
Automated MRI analysis procedures were detailed in previous studies.7–10 We performed imaging genetics analyses including multivariate analyses of cortical thickness and gray matter (GM) density using longitudinal and cross-sectional imaging phenotypes to investigate further the association in the 315 remaining ADNI-1 APOE ε3/ε3 participants after removing those included in the WES. Age at baseline, gender, years of education, and total intracranial volume were used as covariates. Left and right hemispheres are significantly correlated with each other.11 The 9 p-values in the Table were combined into a global chi-square test statistic using Fisher’s method.12 To account for correlation among the p-values, we computed the p-value of this global test statistic using N=100,000 permutations. Since the smallest of the 9 p-values has an unadjusted p-value less than or equal to the global test statistic pvalue, the multiple comparison corrected p-value of the smallest p-value is equal to the protecting global test statistic p-value by the logic of Fisher’s protected least significant difference procedure. Meta-analysis of the remaining cohorts was then performed to validate the association with right hippocampal volume at baseline using Stouffer’s weighted z score.
TABLE.
Association Results (p-values) of Quantitative Trait Analysis Using a Dominant Model of rs3796529 in the Remaining Alzheimer’s Disease Neuroimaging Initiative Phase 1 Participants
| Trait | All APOE ε3/ε3, n=315
|
APOE ε3/ε3 MCI, n=135
|
||||
|---|---|---|---|---|---|---|
| β | 95% CI | p | β | 95% CI | p | |
| Volume | ||||||
|
| ||||||
| Right hippocampus | 163.5 | 28.6, 298.4 | 0.0182 | 209.4 | 6.7, 412.1 | 0.0451 |
|
| ||||||
| Left hippocampus | 110.6 | −14.8, 236.1 | 0.0850 | 165.6 | −15.3, 346.5 | 0.0751 |
|
| ||||||
| Mean hippocampus | 137.1 | 13.3, 260.8 | 0.0307 | 187.5 | 4.3, 370.6 | 0.0470 |
|
| ||||||
| Slope | ||||||
|
| ||||||
| Right hippocampus | 10.5 | −35.9, 56.9 | 0.6568 | −12.4 | −89.3, 64.5 | 0.7524 |
|
| ||||||
| Left hippocampus | 36.3 | −6.9, 79.5 | 0.1010 | 26.1 | −41.5, 93.7 | 0.4515 |
|
| ||||||
| Mean hippocampus | 21.8 | −18.0, 61.7 | 0.2838 | 3.5 | −56.9, 63.8 | 0.9110 |
|
| ||||||
| APC | ||||||
|
| ||||||
| Right hippocampus | −0.06 | −0.99, 0.86 | 0.8964 | −0.19 | −1.86, 1.49 | 0.8268 |
|
| ||||||
| Left hippocampus | 0.22 | −0.89, 1.32 | 0.7013 | −0.67 | −2.65, 1.30 | 0.5046 |
|
| ||||||
| Mean hippocampus | 0.07 | −0.82, 0.96 | 0.8725 | −0.43 | −2.04, 1.19 | 0.6047 |
APC=annualized percentage of change; CI=confidence interval; MCI=mild cognitive impairment.
Results
The average coverage of each base in the target regions was ~40×, and 89,400 SNVs, of which 5,941 (6.6%) were not found in the dbSNP database (dbSNP 137), were identified within the target regions and received a Phred-based quality score of ≥30. For the quality of variant calls, the observed transition-to-transversion ratio for the variants in the coding region was 3.14, and genotypes determined by sequencing and the Illumina 610-Quad array were 99.4% concordant. Of 89,400 SNVs, there were 50,396 exonic, 945 splicing, and 29,236 intronic variants. A total of 25,144 nonsynonymous and 25,234 synonymous SNVs were found in the protein-coding regions.
To identify functional exonic variants associated with atrophy rate of hippocampal volume in APOE ε3/ ε3 MCI participants, further analysis focused on 25,144 nonsynonymous SNVs and 945 SNVs within the regions of the splice sites. After determining the minor allele frequency of variants in the slow and rapid groups, we identified variants carried only by the slow group. We identified 56 nonsynonymous SNVs that were found exclusively in at least 4 of 8 subjects in the slow atrophy group, but not in any of the 8 subjects in the rapid atrophy group. Among these SNVs, the variant that accounted for the greatest group difference (present in 5 subjects in the slow group but not in any of the 8 subjects in the rapid group) and was predicted in-silico as a “functional” missense variant (ANNOVAR 2012)13 was rs3796529 (REST; RE1-silencing transcription factor). rs3796529 has a minor allele frequency >15% in the European American population from the National Heart, Lung, and Blood Institute Exome Sequencing Project Database.
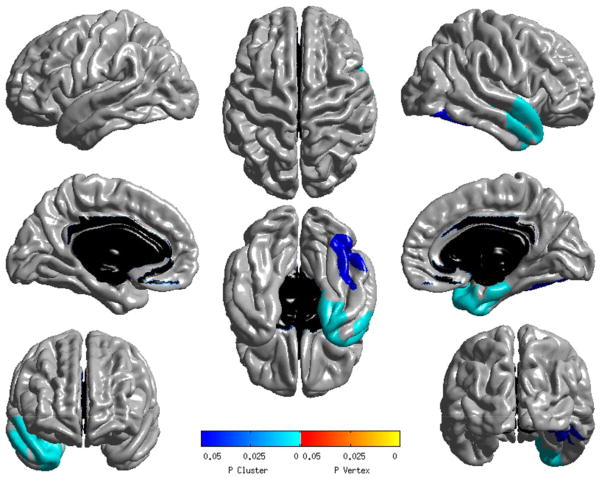
We investigated further the association of rs3796529 by conducting a quantitative trait analysis of hippocampal volume and unbiased whole-brain analyses of cortical thickness and GM density using a dominant model in the remaining 315 ADNI-1 APOE ε3/ε3 participants. Combined analysis of the 9 imaging phenotypes using Fisher’s method showed that at least one measure and rs3796529 are marginally associated in participants with APOE ε3/ε3 (p=0.061). In particular, rs3796529 showed a marginal association with right hippocampal volume in all participants with APOE ε3/ε3 (corrected p=0.061) as well as in MCI participants with APOE ε3/ε3 (see Table). APOE ε3/ε3 participants with minor alleles of rs3796529 had larger hippocampal volumes. The Figure displays the results of the main dominant effect of rs3796529 (TT, TC>CC; minor allele: T) using surface-based analysis of baseline MRI scans.14 Highly significant clusters associated with rs3796529 were found in the right temporal cortical region, where individuals carrying at least one minor allele showed greater mean cortical thickness compared with the participants carrying no minor allele. No significant cortical regions were associated with rate of cortical thickness loss (slope) over 2 years for rs3796529. The voxel-wise analysis results of the association between rs3796529 and baseline GM density in a dominant model showed weak associations in the hippocampus (p<0.05, uncorrected, data not shown), which were similar to those obtained from the cortical thickness analyses. Subjects carrying at least one minor allele showed larger mean GM density compared with the participants carrying no minor allele. A suggestive cluster from the dominant effect on rate of GM density loss over 2 years was found in the bilateral hippocampal region (p<0.05, uncorrected, data not shown), and participants carrying no minor allele showed more rapid GM density loss over 2 years compared with the participants carrying at least one minor allele.
FIGURE.
Association of whole-exome sequencing–identified variant rs3796529 in a larger sample of 315 Alzheimer’s Disease Neuroimaging Initiative Phase 1 subjects by surface-based analysis (SurfStat) of cortical thickness at baseline. Statistical maps of SurfStat were thresholded using random field theory (RFT) correction with a corrected significance level of 0.05. Left hemisphere is shown on the left and P is the RFT-corrected p-value.
Meta-analysis of the 4 remaining cohorts yielded a one-sided Stouffer weighted z score of 1.53, indicating a marginal association of rs3796529 with right hippocampal volume at baseline (p=0.063). Using Fisher’s method to combine this result with the ADNI-1 corrected p-value of 0.061 yielded a combined p-value of 0.025. Thus, based on the evidence from the 5 independent cohorts, we found an association between right hippocampal volume and the REST gene at a significance level of 0.025.
Discussion
This is the first study to show that rs3796529 (REST) is associated with baseline hippocampal volume and rate of hippocampal GM density loss. Our results suggested that APOE ε3/ε3 individuals carrying at least one minor allele of rs3796529 had larger hippocampal volumes and slower GM density loss compared with the participants carrying no minor allele. Thus, the minor allele of rs3796529 confers a protective effect on hippocampal morphology.
The missense variant rs3796529 is located within exon 4 of REST on chromosome 4. REST is a master negative transcriptional regulator of adult hippocampal neurogenesis that recruits chromatin-modifying enzymes, indicating a potential epigenetic role.15–17 REST also plays a vital role in neuronal differentiation and modulates gene expression patterns related to fundamental neuronal functions including ion channels and synaptic plasticity.18 Dysregulation of REST has been implicated in the pathogenesis of several diseases such as Huntington disease and Down syndrome.18,19 However, this gene has not previously been associated with Alzheimer’s disease (AD). This new association of a gene involved in neurogenesis and neuronal differentiation and function suggests that its expression might underlie protection against neurodegenerative processes in AD.
Although mitigated somewhat by the extreme-trait design, a limitation of the present report is that with only 16 WES samples it is not possible to reach significance after Bonferroni correction for any variant. Therefore, we set our initial arbitrary a priori threshold for variants of interest at ≥4 of 8 subjects in the slow atrophy group, but not in any of the 8 subjects in the rapid atrophy group.
In conclusion, our data further suggest that the minor allele of rs3796529 in the REST gene on chromosome 4q12 may be protective for rate of hippocampal volume loss and GM density in participants with the common APOE ε3/ε3 genotype. These findings warrant further investigation in independent replication, analysis of maternal parental history, and functional genomic characterization to determine whether REST may constitute a viable therapeutic target. At a broader level, this study demonstrates the potential of next-generation sequencing combined with quantitative imaging phenotypes for discovery of disease mechanisms and novel candidate therapeutic targets.
Acknowledgments
Data collection and sharing for this project was funded by the ADNI (NIH grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging and the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Alzheimer’s Association, Alzheimer’s Drug Discovery Foundation, BioClinica, Biogen Idec, Bristol-Myers Squibb, Eisai, Elan Pharmaceuticals, Eli Lilly, F. Hoffmann-La Roche and its affiliated company Genentech, GE Healthcare, Innogenetics, IXICO, Janssen Alzheimer Immunotherapy Research and Development, Johnson & Johnson Pharmaceutical Research and Development, Medpace, Merck, Meso Scale Diagnostics, NeuroRx Research, Novartis Pharmaceuticals, Pfizer, Piramal Imaging, Servier, Synarc, and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (FNIH; www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Samples from the National Cell Repository for AD, which receives government support under a cooperative agreement grant (U24 AG21886) awarded by the National Institute on Aging (NIA), were used in this study. Additional support for data analysis was provided by NIA AG041232, National Library of Medicine (NLM) 4R00 LM011384-02, NIA R01 AG19771, P30 AG10133, National Cancer Institute R01 CA101318, NLM R01 LM011360, R01 AG09029, R01 AG25259, P30 AG13846, IIS-1117335, RC2 AG036535, and U01 AG032984 from the NIH, the FNIH, grant #87884 from the Indiana Economic Development Corporation, and the NIH National Institute of Neurological Disorders and Stroke (R01NS059873). The AddNeuroMed study was funded by the European Union as part of the FP6 InnoMed program. S.J.F., A.S., and S.L. were supported by the Biomedical Research Centre for Mental Health at South London and Maudsley National Health Service (NHS) Foundation Trust and Institute of Psychiatry, King’s College London, the Biomedical Research Unit for Dementia at South London and Maudsley NHS Foundation Trust and Institute of Psychiatry, King’s College London, and Alzheimer’s Research UK. The National Institute of Health Research provided infrastructure funding for the Biomedical Research Unit at Maudsley.
Footnotes
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
Authorship
All authors contributed substantively to this work. K.N., S.K., S.L.R., L.S., M.H.I., M.J.H., and A.J.S. were involved in study conception and design. K.N., J.J.C., H.L., V.K.R., S.S., Y.L., M.H.I., and A.J.S. were involved in data organization, WES analysis, and statistical analyses. P.S.A., R.C.P., R.C.G., C.R.J., M.W.W., and A.J.S. were involved in coordination and data collection and processing for ADNI. B.C.M., M.R.F., B.G., and A.J.S. were involved in coordination and data collection and processing for IMAS. S.J.F., S.L., A.S., P.M., B.V., M.T., I.K., and H.S. were involved in coordination and data collection and processing for Add- NeuroMed. C.T.B., K.L.L., and L.A.F. were involved in coordination and data collection and processing for MIRAGE. K.N. and A.J.S. drafted the report and prepared the figure and table. All authors were involved in reviewing and editing of the manuscript.
Potential Conflicts of Interest
P.S.A.: personal fees, NeuroPhage, Elan Corporation, Wyeth, Eisai, Schering-Plough, Bristol-Myers Squibb, Eli Lilly, Merck, Roche, Amgen, Genentech, Abbott, Pfizer, Novartis, Bayer, Astellas, Dainippon, Biomarin, Solvay, Otsuka, Daiichi, AstraZeneca, Janssen, Medivation, Ichor, Toyama, Lundbeck, Biogen Idec, iPerian, Probiodrug, Somaxon, Biotie, Anavex and Kyowa Hakko Kirin Pharma; grants, Lilly, Janssen, Baxter, NIA, FNIH. R.C.P.: personal fees, Pfizer, Janssen Alzheimer Immunotherapy, GE Healthcare, Roche, Merck, Genentech. C.R.J.: consultancy, Janssen Research & Development; grants, NIH; research funding, Alexander Family Alzheimer’s Disease Research Professorship of the Mayo Foundation. M.W.W.: scientific advisory board, Pfizer, Eli Lilly; editorial advisory board, Alzheimer’s & Dementia, Magnetic Resonance Imaging; travel expenses, Eli Lilly, Decision Resources, Kenes International, ADRC, UCSD, ADRC, UCSD, Sanofi-Aventis Group, CHU (University Hospital, Toulouse, France), AC Immune, New York Academy of Sciences, National Brain Research Center (India, for Johns Hopkins Medicine, USA); consultancy, Eli Lilly, Alzheimer’s Drug Discovery Foundation, Avid Radiopharmaceuticals, Clearview Healthcare Partners, Perceptive Informatics, Smartfish, Decision Resources, Araclon Biotech, Synarc, Merck, Genentech. A.J.S.: consultancy, Eli Lilly, Siemens Healthcare; investigator initiated grant, Siemens Healthcare.
References
- 1.Querfurth HW, LaFerla FM. Alzheimer’s disease. N Engl J Med. 2010;362:329–344. doi: 10.1056/NEJMra0909142. [DOI] [PubMed] [Google Scholar]
- 2.Gatz M, Reynolds CA, Fratiglioni L, et al. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry. 2006;63:168–174. doi: 10.1001/archpsyc.63.2.168. [DOI] [PubMed] [Google Scholar]
- 3.Lambert JC, Ibrahim-Verbaas CA, Harold D, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamboh MI, Demirci FY, Wang X, et al. Genome-wide associationstudy of Alzheimer’s disease. Transl Psychiatry. 2012;2:e117. doi: 10.1038/tp.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sherva R, Tripodis Y, Bennett DA, et al. Genome-wide association study of the rate of cognitive decline in Alzheimer’s disease. Alzheimer Dement. 2014;10:45–52. doi: 10.1016/j.jalz.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nho K, Corneveaux JJ, Kim S, et al. Whole-exome sequencing and imaging genetics identify functional variants for rate of change in hippocampal volume in mild cognitive impairment. Mol Psychiatry. 2013;18:781–787. doi: 10.1038/mp.2013.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashburner J, Friston KJ. Voxel-based morphometry—the methods. Neuroimage. 2000;11:805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- 8.Risacher SL, Shen L, West JD, et al. Longitudinal MRI atrophy biomarkers: relationship to conversion in the ADNI cohort. Neurobiol Aging. 2010;31:1401–1418. doi: 10.1016/j.neurobiolaging.2010.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis. II: Inflation, flattening, and a surface-based coordinate system. Neuroimage. 1999;9:195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- 10.Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage. 1999;9:179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- 11.Hasboun D, Chantome M, Zouaoui A, et al. MR determination of hippocampal volume: comparison of three methods. AJNR Am J Neuroradiol. 1996;17:1091–1098. [PMC free article] [PubMed] [Google Scholar]
- 12.Zaykin DV, Zhivotovsky LA, Westfall PH, Weir BS. Truncated product method for combining P-values. Genet Epidemiol. 2002;22:170–185. doi: 10.1002/gepi.0042. [DOI] [PubMed] [Google Scholar]
- 13.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Worsley KJ, Taylor JE, Tomaiuolo F, Lerch J. Unified univariate and multivariate random field theory. Neuroimage. 2004;23(suppl 1):S189–S195. doi: 10.1016/j.neuroimage.2004.07.026. [DOI] [PubMed] [Google Scholar]
- 15.Gao Z, Ure K, Ding P, et al. The master negative regulator REST/ NRSF controls adult neurogenesis by restraining the neurogenic program in quiescent stem cells. J Neurosci. 2011;31:9772–9786. doi: 10.1523/JNEUROSCI.1604-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noh KM, Hwang JY, Follenzi A, et al. Repressor element-1 silencing transcription factor (REST)-dependent epigenetic remodeling is critical to ischemia-induced neuronal death. Proc Natl Acad Sci U S A. 2012;109:E962–E971. doi: 10.1073/pnas.1121568109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu M, Zheng L, Han B, et al. REST regulates DYRK1A transcription in a negative feedback loop. J Biol Chem. 2011;286:10755–10763. doi: 10.1074/jbc.M110.174540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bahn S, Mimmack M, Ryan M, et al. Neuronal target genes of the neuron-restrictive silencer factor in neurospheres derived from fetuses with Down’s syndrome: a gene expression study. Lancet. 2002;359:310–315. doi: 10.1016/S0140-6736(02)07497-4. [DOI] [PubMed] [Google Scholar]
- 19.Buckley NJ, Johnson R, Zuccato C, et al. The role of REST in transcriptional and epigenetic dysregulation in Huntington’s disease. Neurobiol Dis. 2010;39:28–39. doi: 10.1016/j.nbd.2010.02.003. [DOI] [PubMed] [Google Scholar]