Abstract
Background
Alternaria is considered one of the most common saprophytic fungal genera on the planet. It is comprised of many species that exhibit a necrotrophic phytopathogenic lifestyle. Several species are clinically associated with allergic respiratory disorders although rarely found to cause invasive infections in humans. Finally, Alternaria spp. are among the most well known producers of diverse fungal secondary metabolites, especially toxins.
Description
We have recently sequenced and annotated the genomes of 25 Alternaria spp. including but not limited to many necrotrophic plant pathogens such as A. brassicicola (a pathogen of Brassicaceous crops like cabbage and canola) and A. solani (a major pathogen of Solanaceous plants like potato and tomato), and several saprophytes that cause allergy in human such as A. alternata isolates. These genomes were annotated and compared. Multiple genetic differences were found in the context of plant and human pathogenicity, notably the pro-inflammatory potential of A. alternata. The Alternaria genomes database was built to provide a public platform to access the whole genome sequences, genome annotations, and comparative genomics data of these species. Genome annotation and comparison were performed using a pipeline that integrated multiple computational and comparative genomics tools. Alternaria genome sequences together with their annotation and comparison data were ported to Ensembl database schemas using a self-developed tool (EnsImport). Collectively, data are currently hosted using a customized installation of the Ensembl genome browser platform.
Conclusion
Recent efforts in fungal genome sequencing have facilitated the studies of the molecular basis of fungal pathogenicity as a whole system. The Alternaria genomes database provides a comprehensive resource of genomics and comparative data of an important saprophytic and plant/human pathogenic fungal genus. The database will be updated regularly with new genomes when they become available. The Alternaria genomes database is freely available for non-profit use at http://alternaria.vbi.vt.edu.
Keywords: Database, Alternaria, Fungal genome, Sequence, Annotation, Comparative genomics, Plant pathogen, Allergy, Saprophyte
Background
Alternaria species are a major cause of necrotrophic diseases of plants and some of the most common fungi encountered by humans. There are several noteworthy examples of Alternaria spp. as major plant pathogens including but not limited to, A. brassicicola and A. solani. A. brassicicola causes black spot disease (also called dark leaf spot) on virtually every important cultivated Brassica spp. [1-3]. Black spot disease is of worldwide economic importance. For example, black spot can be a devastating foliar and seed-borne disease resulting in severe yield reductions in crops such as cabbage, broccoli, canola and rapeseed [4-6]. A. solani is the causal agent of early blight disease of several major Solanaceous crops including potato and tomato. Early blight caused by A. solani is considered one of the most destructive diseases of potatoes and tomatoes in the world [7,8].
Alternaria spp. are among the most well known producers of diverse secondary metabolites, especially toxins [9]. Over 70 small molecule compounds have been reported from Alternaria [9]. Some of these metabolites are potent mycotoxins (e.g. alternariol, alternariol methyl ether, tenuazonic acid, etc.) with mutagenic and teratagenic properties, and have been linked to certain forms of cancer [10]. The occurrence of potentially harmful Alternaria metabolites in food and food products is becoming an increasing environmental concern [11]. Other toxins are host specific or non-host specific phytotoxins and are important virulence factors during plant pathogenesis. To date many of the genes responsible for the production of these specialized metabolites are unknown although recently the genes responsible for production of the HDAC inhibitor depudecin in A. brassicicola was elucidated as well as the toxin Alternariol and Alternariol methyl ether in A. alternata [12-14]. Annotated genome sequence information was critical for these discoveries.
In addition to harboring many important plant pathogenic species, Alternaria spores are one of the most common and potent indoor and outdoor sources of airborne allergens. Epidemiological studies from a variety of locations worldwide indicate that Alternaria sensitivity is closely linked with the development of atopic asthma and up to 70% of mold-allergic patients have skin test reactivity to Alternaria [15-17]. Alternaria sensitivity has been shown to not only be a risk factor for asthma, but can also directly lead to the development of severe and potentially fatal asthma often more than any other fungus [15-19]. Although some research has been performed on the physiological and molecular identification of Alternaria allergens only three major and several minor allergenic proteins have been described [20]. The biological role of these allergens and other secreted fungal products in the development of allergy and asthma is very poorly understood. Thus there is clearly a need to elucidate the role of Alternaria immunoreactive proteins and other molecules such as secondary/specialized metabolites in the development of allergic diseases from both diagnostic and immunotherapeutic perspectives.
In this article, we introduce the Alternaria genomes database that provides tools to browse and visualize genome sequences, genome annotations, whole genome alignments, and homologous data of the fungal genus Alternaria.
Content and construction
The Alternaria genomes database houses genome sequences, genome annotation and genome comparison data from 25 species, including saprophytes, necrotrophic plant pathogens and species associated with human diseases like allergic airway disorders (Table 1). These genomes were analyzed using a pipeline that incorporated multiple computational and comparative genomics tools. Genomes (i.e. genomic sequences, in the form of contigs or supercontigs) were assembled from Sanger or next-generation sequencing reads and then used as the input for the pipeline. These sequences were analyzed through multiple annotation modules, including repetitive sequence annotation, gene prediction, protein function and domain structure annotation. Comparative genomics analyses were also performed including whole genome alignment and homology analysis.
Table 1.
Description of the sequenced Alternaria genomes
| Species name | Strain codes | Additional information | Sequencing technologies | Genome sequence size (Mb) | Contigs/super-contigs | Contigs/super-contigs N50(kb) | Predicted genes (#) |
|---|---|---|---|---|---|---|---|
| A. alternata | ATCC 66891, EGS 34–016, BMP 0269 | Allergic diseases of human, leaf spot, rots of plants | 454 | 33.2 | 499 | 300 | 11635 |
| A. alternata | ATCC 11680, BMP 0238, IHEM 4706 | Allergic diseases of human, leaf spot, rots of plants (possibly A. tenuissima) | 454 | 33.8 | 797 | 450 | 12323 |
| A. brassicicola | ATCC 96836, EGS 42–002, BMP 1950 | Blackspot of brassica | Sanger | 29.6 | 4039/838 | 18/ 2400 | 10514 |
| A. alternata | ATCC 66982, EGS 34–039, BMP 0270 | Allergic disease of human, leaf spot, rots of plants | Illumina | 33.5 | 393 | 757 | 12290 |
| A. arborescens | ATCC 204491, EGS 39–128, BMP 0308 | Stem canker of tomato | Illumina | 34.0 | 1332 | 624 | 14741 |
| A. citriarbusti | EGS 46–140, BMP 2343, SH-MIL-8 s | Brown/black spot of citrus | Illumina | 34.1 | 2273 | 48 | 12606 |
| A. destruens | ATCC 204363, EGS 46–069, BMP 0317 | Infecting and suppressing dodder (weed) | Illumina | 41.8 | 31070 | 3 | 14814 |
| A. fragariae | BMP 3062, NAF-8 | Black spot disease of strawberry | Illumina | 33.2 | 1027 | 78 | 12272 |
| A. gaisen | EGS 90–0512, BMP 2338 | Black spot, ring spot disease of pear | Illumina | 34.6 | 7485 | 10 | 13902 |
| A. tangelonis | EGS 45–080, BMP 2327, BC2-RLR-1 s | Leaf spot of citrus | Illumina | 34.0 | 2459 | 37 | 12639 |
| A. longipes | EGS 30–033, BMP 0313 | Black/brown leaf spot of tobacco | Illumina | 36.3 | 3412 | 137 | 13219 |
| A. mali | BMP 3064, IFO8984 | Leaf ring spot of apple | Illumina | 34.7 | 2682 | 35 | 12715 |
| A. mali | BMP 3063, M-71 | Leaf ring spot of apple | Illumina | 34.1 | 4439 | 21 | 12727 |
| A. turkisafria | BMP 3436, SH-MIL-20s | Leaf spot of citrus | Illumina | 34.0 | 2347 | 33 | 12739 |
| A. tenuissima | ATCC 96828, EGS 34–015, BMP 0304 | Leaf spot of plants | Illumina | 33.5 | 676 | 662 | 12276 |
| A. limoniasperae | EGS 44–159, BMP 2335 | Leaf spot of citrus | Illumina | 35.1 | 2796 | 50 | 12966 |
| A. carthami | BMP 1963, CBS 635.80 | Leaf spot and blight of safflower | Illumina | 34.5 | 9340 | 72 | 12071 |
| A. capsici | ATCC MYA-998, EGS 45–075, BMP 0180 | Leaf spot of solanaceae (pepper) | Illumina | 34.0 | 13743 | 31 | 11487 |
| A. crassa | BMP 0172, ACR1 | Leaf spot of solanaceae | Illumina | 35.0 | 12126 | 54 | 11663 |
| A. dauci | ATCC 36613, BMP 0167 | Leaf blight of carrots | Illumina | 32.1 | 12030 | 13 | 11981 |
| A. macrospora | BMP 1949, CH3 | Leaf spot of cotton | Illumina | 31.7 | 3153 | 37 | 11961 |
| A. porri | BMP 0178, Z6B | Purple blotch, leaf blight and bulb rot of Allium (onion) | Illumina | 31.2 | 16767 | 9 | 12232 |
| A. solani | BMP 0185 | Early blight of potatoes and tomatoes | Illumina | 32.9 | 5613 | 144 | 11726 |
| A. tagetica | EGS 44–044, BMP 0179 | Leaf spot of marigold | Illumina | 35.1 | 16372 | 72 | 11999 |
| A. tomatophila | BMP 2032, CBS 109156 | Leaf spot of tomato | Illumina | 34.1 | 10185 | 22 | 12601 |
Genome sequencing and assembly
Alternaria genomes were sequenced using various sequencing technologies including whole genome shotgun method with Sanger sequencing, GS-FLX 454, and Illumina HiSeq (Table 1). Genomes were assembled from sequencing reads using PCAP [21] (for Sanger sequencing), Newbler [22] (for GS-FLX 454), and Velvet [23] (for Illumina HiSeq). The physical map of A. brassicicola was constructed by generating fingerprints from the CSU-K35 A. brassicicola BAC library that were then used to scaffold the genome (Dang et al., unpublished).
Genome annotation
Genome annotation was performed using a custom pipeline (Figure 1). Assembled genomes were first scanned for repetitive sequences (both transposable elements and simple repeats) using multiple tools including REPET [24], RepeatScout [25], RepeatModeler and RepeatMasker (http://www.repeatmasker.org). Protein-coding gene prediction was then carried out using JIGSAW [26] that combined gene models discovered by various de novo and homology-based gene prediction tools including Genewise [27], FgeneSH (http://softberry.com), AUGUSTUS [28], Genemark-ES [29], and GeneID [30]. We also generated RNA-Seq data for A. alternata ATCC 66981 which were aligned to the genome using TopHat [31] with Bowtie [32], and de novo transcripts were constructed using Cufflinks [33]. These data were used internally to evaluate gene predictions. Predicted genes were then conceptually translated to protein sequences that served as the input for most of the functional annotation tasks. Non-coding genes were also annotated using tRNAScan-SE [34] and RNAmmer [35].
Figure 1.
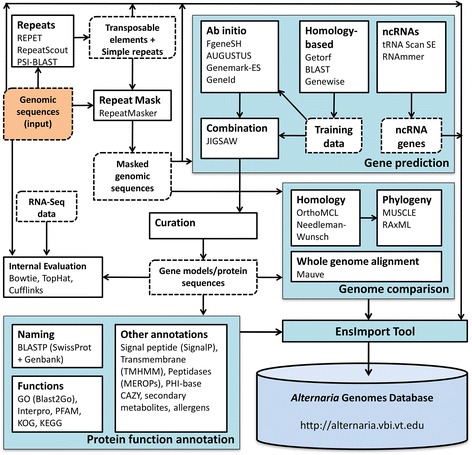
Alternaria genome annotation pipeline.
Various computational functional annotations were performed on the conceptual protein sequences. The proteins were first searched against Genbank [36] and SwissProt [37] using BLAST to identify known proteins with similar sequences. The name/description of the known proteins was then transferred to the predicted proteins following the standard operating procedure (SOP) developed for fungi by the Broad Institute [38]. Protein domain and family annotation was performed using the Interpro database [39] and PFAM [40]. Gene ontology annotation was performed using Blast2GO [41] and Interpro.
Various fungal-related and additional annotations were also carried out using the pipeline. Signal peptides were predicted using SignalP [42], WoLF-Psort [43], and Phobius [44]. Transmembrane proteins were predicted using TMHMM [45]. Pathogenicity-related gene candidates were identified via multiple annotation data including BLAST search against PHI-base [46]. Carbohydrate Active Enzymes were identified according to the CAZY database [47] and dbCAN [48]. Potential allergens were identified using BLAST based homology searches and Allerdictor [49]. Proteases were annotated using the batched BLAST search tool from the MEROPS database [50]. Secondary metabolites were identified using SMURF [51].
Genome comparison
Multiple genome comparison tasks were performed that utilized the genome sequences as well as the predicted genes/proteins from multiple species. Whole genome pairwise alignment was performed using Mauve progressive alignment software [52,53]. Orthologs and paralogs were identified using bidirectional best BLAST hits and Markov clustering via OrthoMCL [54].
Porting data to Ensembl database schema
Annotation and comparison data of Alternaria genomes are presented via the popular Ensembl genome browser platform [55] that was customized and installed at the Virginia Bioinformatics Institute. Outputs from the genome annotation pipeline as well as outputs from comparative genomics analyses were processed and converted to Ensembl compatible MySQL databases (both core and compara databases) using EnsImport, a custom suite of scripts we developed in Perl. EnsImport supports multiple standard file formats such as FASTA, AGP, GFF3 and XMFA, and outputs from widely-used tools such as BLAST, Interpro, RepeatMasker, OrthoMCL and Blast2GO.
Utility and discussion
Using Ensembl genome browser platform, the Alternaria genomes database provides a rich set of user-friendly tools to browse and visualize sequences, annotation, and comparison data. Data export and search features are also available. Detailed instructions on how to use the Ensembl browser are available on the ‘Help & Documentation’ section of the database. Here we only describe the most relevant features in the context of the Alternaria genomes project.
Genome region view
For each species, users can access and visualize a genomic region along with annotated functional and non-functional elements such as repetitive elements, predicted protein-coding gene models, and RNA coding gene models (Figure 2). A genomic region can be a whole (or part of) a contig or supercontig. Zooming functionality allows for intuitively scaling region views based on location. Each type of element (functional and non-functional) is displayed in a separate track using a unique color. Users can click on an individual element (e.g. repeats, genes, transcripts) to open a popup menu to access available annotation. The tracks can be displayed or hidden using the display configuration tool.
Figure 2.
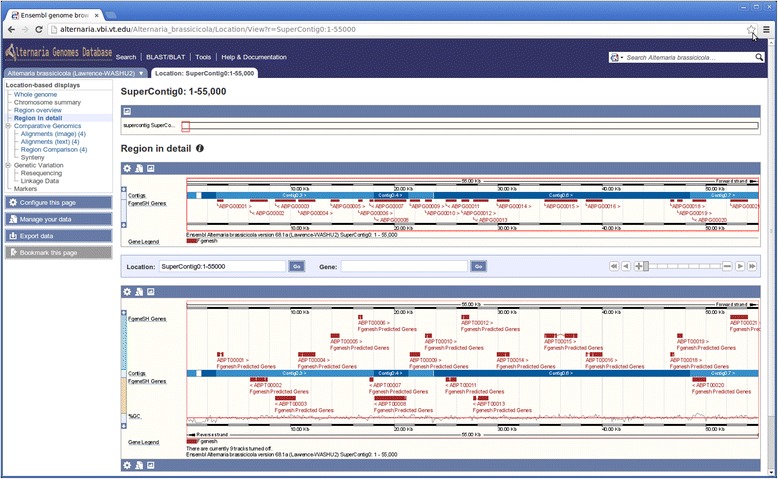
A screenshot of the Alternaria genomes database that shows a region of an A. brassicicola supercontig along with the predicted genes and transcripts.
Annotation view
The majority of functional annotation data in the database is for protein coding genes. For each gene/protein, extensive annotations include gene structure and sequence, gene description, location, protein domain architectures (e.g. Interpro, PFAM), gene ontology assignments, signal peptides, transmembrane structures and other annotation data (Figure 3). These annotation data are available and presented in multiple tightly linked web interfaces in the browser.
Figure 3.
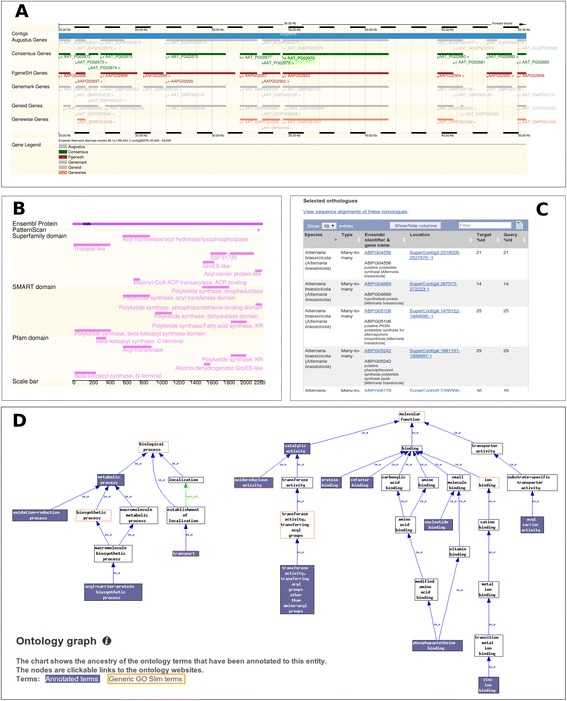
Examples of annotation and comparison views for an Alternaria alternata polyketide synthase gene (AAT_PG02879). (A) Contig view of the gene, (B) Domain annotation, (C) Orthologous genes in other Alternaria genomes, (D) Gene ontology annotation.
Comparative genomics view
The comparative browsing feature of Ensembl platform allows for conveniently viewing and visualizing comparative genomics data side-by-side with annotation data. Aligned regions between two genomes identified via whole genome pairwise alignments are displayed together with functional and non-functional elements such as repetitive elements and gene models (Figure 4). This feature allows for easy investigation of the conserved genomic regions between multiple genomes. Whole genome alignments can be visualized using graphical representation as well as displayed in text formats such as FASTA and ClustalW. Orthologs and paralogs of a gene can be easily retrieved in a table that contains links to access protein alignments and related annotation data (Figure 3C).
Figure 4.
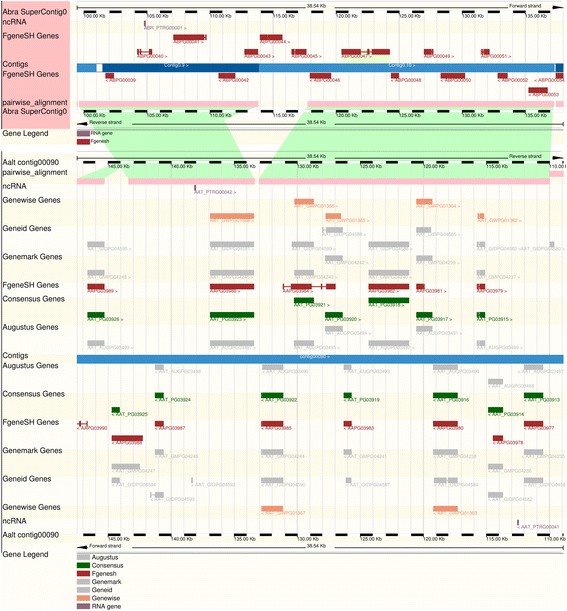
An example of a syntenic region between Alternaria brassicicola and A. alternata . The aligned blocks (in pink) between genomic sequences are connected by green bands.
Database search
Users may query the database using sequence alignment search (e.g. BLAST) and text search. The built-in search feature of the Ensembl platform allows for BLAST searches against genomic sequences, predicted transcript and protein sequences (Figure 5). Full text search for gene names is also available as a built-in feature in Esembl platform. However, for newly sequenced species, a large portion of the predicted genes are not named or annotated with highly reliable descriptions. In such cases, information on the hits with known proteins or protein families and domains can be used to explore the functions of the genes. Therefore, we implemented a more comprehensive search module that allows for full text search within annotation from multiple sources including BLAST and Interpro hits and incorporated this module in the Alternaria genomes database (Figure 5).
Figure 5.
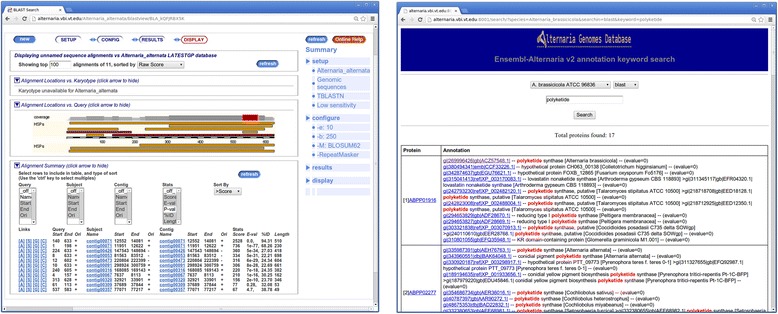
Search features of Alternaria genomes database that allows for sequence alignment search using BLAST (left) and Interepro and BLAST hit description search (right).
Data export
Ensembl built-in functionality allows for exporting multiple types of data to various formats. Raw sequence and annotation data can be easily exported in multiple formats such as FASTA and GFF via available tools in Ensembl. A button to access data export feature is located on the left pane in the interface of the database. It is also possible to export the graphical visualization of multiple types of annotation and comparison data to multiple image formats that are suitable for publication or further editing.
Conclusion
Over the past few years, efforts in sequencing fungal genomes have facilitated the studies of the molecular basis of fungal pathogenicity as a whole system [56-59]. The Alternaria genomes database provides a comprehensive resource of genomics and comparative genomics data of an important plant and human pathogenic fungal genus Alternaria. In addition, the database may prove useful for discovery of genes encoding industrial enzymes, antibiotics, and other molecules with utility in medicine and agriculture.
These genome annotation and comparison data have recently facilitated several large-scale functional genomics studies that resulted in the discovery of many new genes that contribute to virulence especially secondary metabolite genes, mitogen-activated protein (MAP) kinases, and transcription factors in A. brassicicola [13,14,60-68]. Alternaria genome annotation and comparison data have also enabled comprehensive comparative studies of Alternaria genomes in the context of plant and human pathogenicity [69] (several other manuscripts are under preparation).
The use of the familiar Ensembl browser platform makes browsing and visualizing Alternaria genome annotation and comparison data convenient. As we continue our efforts in Alternaria genome sequencing and analysis, we will update this database as new genomes and relevant annotation data become available.
Availability and requirements
The Alternaria genomes database is freely available for non-commercial use at http://alternaria.vbi.vt.edu.
Acknowledgements
The authors wish to acknowledge William Spooner for helpful discussions regarding Ensembl installation. This work was supported by the grants NSF DEB-0918298 and NIFA 2004-35600-15030 to CL. This article is dedicated to the late Dr. Dennis L. Knudson.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
HD and CL conceived the experiments and analyses. HD carried out all of the bioinformatics analyses. HD, BP, TP and CL wrote the manuscript, which was reviewed by all authors. All authors read and approved the final manuscript.
Contributor Information
Ha X Dang, Email: hd@vt.edu.
Barry Pryor, bmpryor@email.arizona.edu.
Tobin Peever, Email: tpeever@wsu.edu.
Christopher B Lawrence, Email: cblawren@vt.edu.
References
- 1.Neergaard P. Danish species of Alternaria and Stemphylium. 1945:560 pp.
- 2.Sigareva MA, Earle ED. Camalexin induction in intertribal somatic hybrids between Camelina sativa and rapid-cycling Brassica oleracea. Theor Appl Genet. 1999;98:164–70. doi: 10.1007/s001220051053. [DOI] [Google Scholar]
- 3.Westman AL, Kresovich S, Dickson MH. Regional variation in Brassica nigra and other weedy crucifers for disease reaction to Alternaria brassicicola and Xanthomonas campestris pv. campestris. Euphytica. 1999;106:253–9. doi: 10.1023/A:1003544025146. [DOI] [Google Scholar]
- 4.Daebeler FD, Riedel A, Riedel V. Wiss Z Wilhelm Pieck-Univ Rostock Naturwissenschaftliche Reiche. 1986;35:52.
- 5.MacKinnon SL, Keifer P, Ayer WA. Components from the phytotoxic extract of Alternaria brassicicola, a black spot pathogen of canola. Phytochemistry. 1999;51:215–21. doi: 10.1016/S0031-9422(98)00732-8. [DOI] [Google Scholar]
- 6.Pedras MSC, Chumala PB, Jin W, Islam MS, Hauck DW. The phytopathogenic fungus Alternaria brassicicola: Phytotoxin production and phytoalexin elicitation. Phytochemistry. 2009;70:394–402. doi: 10.1016/j.phytochem.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 7.Chaerani R, Voorrips RE. Tomato early blight (Alternaria solani): the pathogen, genetics, and breeding for resistance. J Gen Plant Pathol. 2006;72:335–47. doi: 10.1007/s10327-006-0299-3. [DOI] [Google Scholar]
- 8.Olanya OM, Honeycutt CW, Larkin RP, Griffin TS, He Z, Halloran JM. The effect of cropping systems and irrigation management on development of potato early blight. J Gen Plant Pathol. 2009;75:267–75. doi: 10.1007/s10327-009-0175-z. [DOI] [Google Scholar]
- 9.Montemurro N, Visconti A. Alternaria metabolites-chemical and biological data. Alternaria Biol Plant Dis Metab. 1992:449-57.
- 10.Liu GT, Qian YZ, Zhang P, Dong ZM, Shi ZY, Zhen YZ, et al. Relationships between Alternaria alternata and oesophageal cancer. IARC Sci Publ. 1991:258-62. [PubMed]
- 11.Bottalico A, Logrieco A. Toxigenic Alternaria species of economic importance. Mycotoxins Agric Food Saf. 1998;65:108. [Google Scholar]
- 12.Kwon HJ, Owa T, Hassig CA, Shimada J, Schreiber SL. Depudecin induces morphological reversion of transformed fibroblasts via the inhibition of histone deacetylase. Proc Natl Acad Sci U S A. 1998;95(7):3356–61. doi: 10.1073/pnas.95.7.3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saha D, Fetzner R, Burkhardt B, Podlech J, Metzler M, Dang H, et al. Identification of a Polyketide Synthase Required for Alternariol (AOH) and Alternariol-9-Methyl Ether (AME) Formation in Alternaria alternata. PLoS One. 2012;7:e40564. doi: 10.1371/journal.pone.0040564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wight WD, Kim K-H, Lawrence CB, Walton JD. Biosynthesis and role in virulence of the histone deacetylase inhibitor depudecin from Alternaria brassicicola. Mol Plant Microbe Interact. 2009;22:1258–67. doi: 10.1094/MPMI-22-10-1258. [DOI] [PubMed] [Google Scholar]
- 15.Gergen PJ, Turkeltaub PC. The association of individual allergen reactivity with respiratory disease in a national sample: Data from the second National Health and Nutrition Examination Survey, 1976–1980 (NHANES II) J Allergy Clin Immunol. 1992;90(4, Part 1):579–88. doi: 10.1016/0091-6749(92)90130-T. [DOI] [PubMed] [Google Scholar]
- 16.Halonen M, Stern DA, Wright AL, Taussig LM, Martinez FD. Alternaria as a major allergen for asthma in children raised in a desert environment. Am J Respir Crit Care Med. 1997;155:1356–61. doi: 10.1164/ajrccm.155.4.9105079. [DOI] [PubMed] [Google Scholar]
- 17.Salo PM, Arbes SJ, Jr, Sever M, Jaramillo R, Cohn RD, London SJ, et al. Exposure to Alternaria alternata in US homes is associated with asthma symptoms. J Allergy Clin Immunol. 2006;118:892–8. doi: 10.1016/j.jaci.2006.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Hollaren MT, Yunginger JW, Offord KP, Somers MJ, O’Connell EJ, Ballard DJ, et al. Exposure to an aeroallergen as a possible precipitating factor in respiratory arrest in young patients with asthma. N Engl J Med. 1991;324:359–63. doi: 10.1056/NEJM199102073240602. [DOI] [PubMed] [Google Scholar]
- 19.Andersson M, Downs S, Mitakakis T, Leuppi J, Marks G. Natural exposure to Alternaria spores induces allergic rhinitis symptoms in sensitized children. Pediatr Allergy Immunol Off Publ Eur Soc Pediatr Allergy Immunol. 2003;14:100–5. doi: 10.1034/j.1399-3038.2003.00031.x. [DOI] [PubMed] [Google Scholar]
- 20.Bush RK, Prochnau JJ. Alternaria-induced asthma. J Allergy Clin Immunol. 2004;113:227–34. doi: 10.1016/j.jaci.2003.11.023. [DOI] [PubMed] [Google Scholar]
- 21.Huang X, Wang J, Aluru S, Yang S-P, Hillier L. PCAP: a whole-genome assembly program. Genome Res. 2003;13:2164–70. doi: 10.1101/gr.1390403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–80. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–9. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flutre T, Duprat E, Feuillet C, Quesneville H. Considering transposable element diversification in de novo annotation approaches. PLoS One. 2011;6:e16526. doi: 10.1371/journal.pone.0016526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Price AL, Jones NC, Pevzner PA. De novo identification of repeat families in large genomes. Bioinformatics. 2005;21:i351–8. doi: 10.1093/bioinformatics/bti1018. [DOI] [PubMed] [Google Scholar]
- 26.Allen JE, Salzberg SL. JIGSAW: integration of multiple sources of evidence for gene prediction. Bioinformatics. 2005;21:3596–603. doi: 10.1093/bioinformatics/bti609. [DOI] [PubMed] [Google Scholar]
- 27.Birney E, Clamp M, Durbin R. GeneWise and Genomewise. Genome Res. 2004;14:988–95. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stanke M, Waack S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics. 2003;19(suppl 2):ii215–25. doi: 10.1093/bioinformatics/btg1080. [DOI] [PubMed] [Google Scholar]
- 29.Ter-Hovhannisyan V, Lomsadze A, Chernoff YO, Borodovsky M. Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Res. 2008;18:1979–90. doi: 10.1101/gr.081612.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blanco E, Parra G, Guigó R. Using geneid to identify genes. Curr Protoc Bioinforma Ed Board Andreas Baxevanis Al. 2007;Chapter 4:Unit 4.3. [DOI] [PubMed]
- 31.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinforma Oxf Engl. 2009;25:1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roberts A, Pimentel H, Trapnell C, Pachter L. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics. 2011;27:2325–9. doi: 10.1093/bioinformatics/btr355. [DOI] [PubMed] [Google Scholar]
- 34.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–64. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lagesen K, Hallin P, Rødland EA, Staerfeldt H-H, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–8. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benson DA, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2013;42:D32–7. doi: 10.1093/nar/gkt1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Consortium UP. Update on activities at the Universal Protein Resource (UniProt) in 2013. Nucleic Acids Res. 2013;41(Database issue):D43–7. doi: 10.1093/nar/gks1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haas BJ, Zeng Q, Pearson MD, Cuomo CA, Wortman JR. Approaches to fungal genome annotation. Mycology. 2011;2:118–41. doi: 10.1080/21501203.2011.606851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, Bateman A, et al. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 2012;40(Database issue):D306–12. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, et al. Pfam: the protein families database. Nucleic Acids Res. 2013;42:D222–30. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, et al. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36:3420–35. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–95. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 43.Horton P, Park K-J, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, et al. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35(Web Server issue):W585–7. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Käll L, Krogh A, Sonnhammer ELL. Advantages of combined transmembrane topology and signal peptide prediction--the Phobius web server. Nucleic Acids Res. 2007;35(Web Server issue):W429–32. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J Mol Biol. 2001;305:567–80. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 46.Winnenburg R, Urban M, Beacham A, Baldwin TK, Holland S, Lindeberg M, et al. PHI-base update: additions to the pathogen host interaction database. Nucleic Acids Res. 2008;36(Database issue):D572–6. doi: 10.1093/nar/gkm858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The carbohydrate-active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 2009;37(Database issue):D233–8. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yin Y, Mao X, Yang J, Chen X, Mao F, Xu Y. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012;40(Web Server issue):W445–51. doi: 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dang HX, Lawrence CB. Allerdictor: fast allergen prediction using text classification techniques. Bioinforma Oxf Engl. 2014. [DOI] [PMC free article] [PubMed]
- 50.Rawlings ND, Barrett AJ, Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2012;40:D343–50. doi: 10.1093/nar/gkr987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khaldi N, Seifuddin FT, Turner G, Haft D, Nierman WC, Wolfe KH, et al. SMURF: genomic mapping of fungal secondary metabolite clusters. Fungal Genet Biol FG B. 2010;47:736–41. doi: 10.1016/j.fgb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Darling ACE, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li L, Stoeckert CJ, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–89. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, et al. Ensembl 2012. Nucleic Acids Res. 2011;40:D84–90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, Arroyo J, et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature. 2005;438:1151–6. doi: 10.1038/nature04332. [DOI] [PubMed] [Google Scholar]
- 57.Hane JK, Lowe RGT, Solomon PS, Tan K-C, Schoch CL, Spatafora JW, et al. Dothideomycete–plant interactions illuminated by genome sequencing and EST analysis of the wheat pathogen stagonospora nodorum. Plant Cell Online. 2007;19:3347–68. doi: 10.1105/tpc.107.052829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mabey Gilsenan J, Cooley J, Bowyer P. CADRE: the Central Aspergillus Data REpository 2012. Nucleic Acids Res. 2012;40(Database issue):D660–6. doi: 10.1093/nar/gkr971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cerqueira GC, Arnaud MB, Inglis DO, Skrzypek MS, Binkley G, Simison M, et al. The Aspergillus Genome Database: multispecies curation and incorporation of RNA-Seq data to improve structural gene annotations. Nucleic Acids Res. 2013;42:D705–10. doi: 10.1093/nar/gkt1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cho Y, Davis JW, Kim K-H, Wang J, Sun Q-H, Cramer RA, Jr, et al. A high throughput targeted gene disruption method for Alternaria brassicicola functional genomics using linear minimal element (LME) constructs. Mol Plant-Microbe Interact MPMI. 2006;19:7–15. doi: 10.1094/MPMI-19-0007. [DOI] [PubMed] [Google Scholar]
- 61.Kim K-H, Cho Y, LA Rota M, Cramer RA, Jr, Lawrence CB. Functional analysis of the Alternaria brassicicola non-ribosomal peptide synthetase gene AbNPS2 reveals a role in conidial cell wall construction. Mol Plant Pathol. 2007;8:23–39. doi: 10.1111/j.1364-3703.2006.00366.x. [DOI] [PubMed] [Google Scholar]
- 62.Cho Y, Cramer RA, Jr, Kim K-H, Davis J, Mitchell TK, Figuli P, et al. The Fus3/Kss1 MAP kinase homolog Amk1 regulates the expression of genes encoding hydrolytic enzymes in Alternaria brassicicola. Fungal Genet Biol. 2007;44:543–53. doi: 10.1016/j.fgb.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 63.Craven KD, Vélëz H, Cho Y, Lawrence CB, Mitchell TK. Anastomosis is required for virulence of the fungal necrotroph Alternaria brassicicola. Eukaryot Cell. 2008;7:675–83. doi: 10.1128/EC.00423-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cho Y, Kim K-H, La Rota M, Scott D, Santopietro G, Callihan M, et al. Identification of novel virulence factors associated with signal transduction pathways in Alternaria brassicicola. Mol Microbiol. 2009;72:1316–33. doi: 10.1111/j.1365-2958.2009.06689.x. [DOI] [PubMed] [Google Scholar]
- 65.Kim K-H, Willger SD, Park S-W, Puttikamonkul S, Grahl N, Cho Y, et al. TmpL, a transmembrane protein required for intracellular redox homeostasis and virulence in a plant and an animal fungal pathogen. PLoS Pathog. 2009;5:e1000653. doi: 10.1371/journal.ppat.1000653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cho Y, Srivastava A, Ohm RA, Lawrence CB, Wang K-H, Grigoriev IV, et al. Transcription factor Amr1 induces melanin biosynthesis and suppresses virulence in Alternaria brassicicola. PLoS Pathog. 2012;8:e1002974. doi: 10.1371/journal.ppat.1002974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Srivastava A, Ohm RA, Oxiles L, Brooks F, Lawrence CB, Grigoriev IV, et al. A zinc-finger-family transcription factor, AbVf19, is required for the induction of a gene subset important for virulence in Alternaria brassicicola. Mol Plant-Microbe Interact MPMI. 2012;25:443–52. doi: 10.1094/MPMI-10-11-0275. [DOI] [PubMed] [Google Scholar]
- 68.Srivastava A, Cho IK, Cho Y. The Bdtf1 gene in Alternaria brassicicola is important in detoxifying brassinin and maintaining virulence on Brassica species. Mol Plant-Microbe Interact MPMI. 2013;26:1429–40. doi: 10.1094/MPMI-07-13-0186-R. [DOI] [PubMed] [Google Scholar]
- 69.Hu J, Chen C, Peever T, Dang H, Lawrence C, Mitchell T. Genomic characterization of the conditionally dispensable chromosome in Alternaria arborescens provides evidence for horizontal gene transfer. BMC Genomics. 2012;13:171. doi: 10.1186/1471-2164-13-171. [DOI] [PMC free article] [PubMed] [Google Scholar]


