Gao et al. investigate mod(mdg4), a classic trans-spliced gene in Drosophila, and report that two critical RNA sequences in the middle of the last 5′ intron, TSA and TSB, promote trans-splicing of mod(mdg4). In TSA, a 13-nt core motif is conserved across Drosophila species and is essential and sufficient for trans-splicing, which binds U1 snRNP through strong base-pairing with U1 snRNA. In TSB, a conserved secondary structure acts as an enhancer. Deletions of TSA and TSB result in developmental defects in flies.
Keywords: trans-splicing, mod(mdg4), U1 snRNP, pseudo-5′ splice site, RNA motif, Drosophila
Abstract
Unlike typical cis-splicing, trans-splicing joins exons from two separate transcripts to produce chimeric mRNA and has been detected in most eukaryotes. Trans-splicing in trypanosomes and nematodes has been characterized as a spliced leader RNA-facilitated reaction; in contrast, its mechanism in higher eukaryotes remains unclear. Here we investigate mod(mdg4), a classic trans-spliced gene in Drosophila, and report that two critical RNA sequences in the middle of the last 5′ intron, TSA and TSB, promote trans-splicing of mod(mdg4). In TSA, a 13-nucleotide (nt) core motif is conserved across Drosophila species and is essential and sufficient for trans-splicing, which binds U1 small nuclear RNP (snRNP) through strong base-pairing with U1 snRNA. In TSB, a conserved secondary structure acts as an enhancer. Deletions of TSA and TSB using the CRISPR/Cas9 system result in developmental defects in flies. Although it is not clear how the 5′ intron finds the 3′ introns, compensatory changes in U1 snRNA rescue trans-splicing of TSA mutants, demonstrating that U1 recruitment is critical to promote trans-splicing in vivo. Furthermore, TSA core-like motifs are found in many other trans-spliced Drosophila genes, including lola. These findings represent a novel mechanism of trans-splicing, in which RNA motifs in the 5′ intron are sufficient to bring separate transcripts into close proximity to promote trans-splicing.
Removal of introns by pre-mRNA splicing is one of the essential steps of RNA processing during eukaryotic gene expression and regulation (Hoskins and Moore 2012). Additionally, alternative splicing generates multiple mRNA isoforms that play key roles in development, differentiation, and diseases (Kim et al. 2008). Pre-mRNA splicing is catalyzed by a large and dynamic RNA–protein complex, the spliceosome, which contains five small nuclear RNAs (snRNAs; U1, U2, U4, U5, and U6) and >150 proteins (Will and Luhrmann 2011). Exon–exon ligation typically occurs as an intramolecular reaction (cis-splicing); however, it can occur less frequently in an intermolecular manner (trans-splicing) to generate chimeric mRNA from two different pre-mRNA molecules (Sharp 1987; Lasda and Blumenthal 2011).
Trans-splicing is widespread in lower eukaryotic trypanosomes and nematodes (Sutton and Boothroyd 1986; Zorio et al. 1994). Nearly all transcripts in Trypanosoma brucei and ∼70% of transcripts in Caenorhabditis elegans are trans-spliced with a highly conserved short spliced leader (SL), which is 39 nucleotides (nt) in trypanosomes and 22 nt in nematodes (Lasda and Blumenthal 2011). SL is trans-spliced to the 5′ end of transcripts from an ∼100-nt SL RNA by reactions similar to that of cis-splicing except that a Y-structured intermediate is formed instead of a lariat intermediate (Murphy et al. 1986; Hannon et al. 1990). SL RNA was considered to be a chimeric molecule with an exon domain (the SL) and an snRNA-like domain (Sharp 1987) in which the snRNA-like domain contains an Sm core-binding site and a 5′ splice site (5′SS) (Bruzik et al. 1988; Hannon et al. 1992). SL RNA-facilitated trans-splicing requires U2, U4, U5, and U6 snRNAs (Tschudi and Ullu 1990) but not U1 snRNA (Hannon et al. 1991). Therefore, SL RNA has been hypothesized to be assembled into SL RNP, an analog of U1 snRNP (Bruzik and Steitz 1990). However, in vitro purification using cell extracts from the parasitic nematode Ascaris lumbricoides revealed that the protein components of SL RNP are specific except for core Sm proteins (Denker et al. 2002). It has been speculated that U5 snRNA and its associated proteins may identify and align the 5′SSs in the SL RNA for the spliceosomal chemical reactions (Denker et al. 1996; Maroney et al. 1996).
Trans-splicing is found less commonly in many higher eukaryotes, including in several chloroplast and mitochondria genes in plants (Koller et al. 1987; Pereira de Souza et al. 1991), in mod(mdg4) and lola in flies (Horiuchi et al. 2003; Gabler et al. 2005), and in numerous genes in mammals (Fujieda et al. 1996; Hu et al. 2013). Recently, deep sequencing has enhanced the identification of trans-splicing events throughout the transcriptome (McManus et al. 2010; Zhang et al. 2010; Shao et al. 2012). In Drosophila, ∼80 trans-spliced genes have been identified and confirmed using interspecies hybrids, including two classic trans-spliced genes, mod(mdg4) and lola (McManus et al. 2010). The mod(mdg4) gene functions in establishing and maintaining an open chromatin conformation and therefore is being considered as an enhancer of position effect variegation (Dorn et al. 1993; Gerasimova et al. 1995; Dorn and Krauss 2003). In mod(mdg4), alternative 3′ exons from multiple loci on both DNA strands are trans-spliced to a 5′ transcript containing four common exons (Fig. 1A) and result in at least 31 isoforms (Labrador et al. 2001; Dorn and Krauss 2003; Gabler et al. 2005; McManus et al. 2010). Trans-splicing of mod(mdg4) is conserved in insects, including silkworms (Shao et al. 2012), mosquitos (Krauss and Dorn 2004), and cotton bollworms (Cai et al. 2012). The lola gene encodes a transcription factor that is involved in neuron and stem cell differentiation (Neumuller et al. 2011; Southall et al. 2014). The 5′ transcripts of lola, containing four alternative exons and four common exons, are trans-spliced to multiple 3′ exons to generate at least 80 mRNAs and 20 protein isoforms (Horiuchi et al. 2003; McManus et al. 2010).
Figure 1.
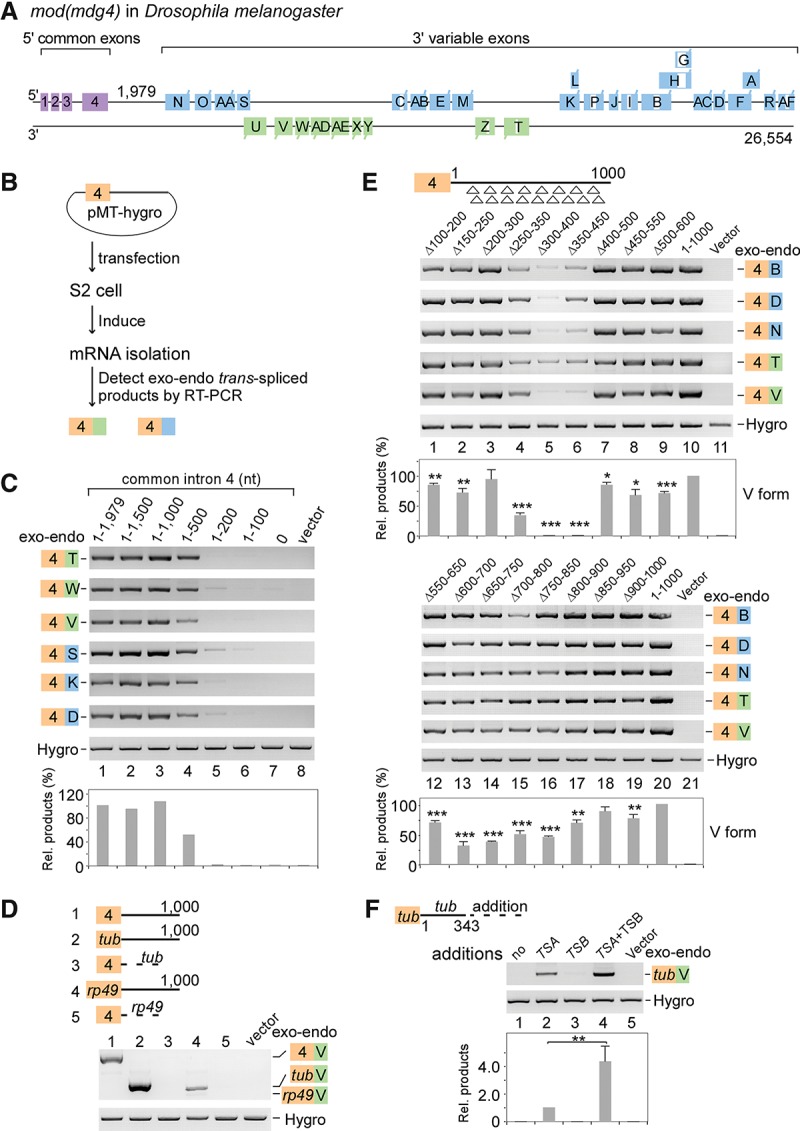
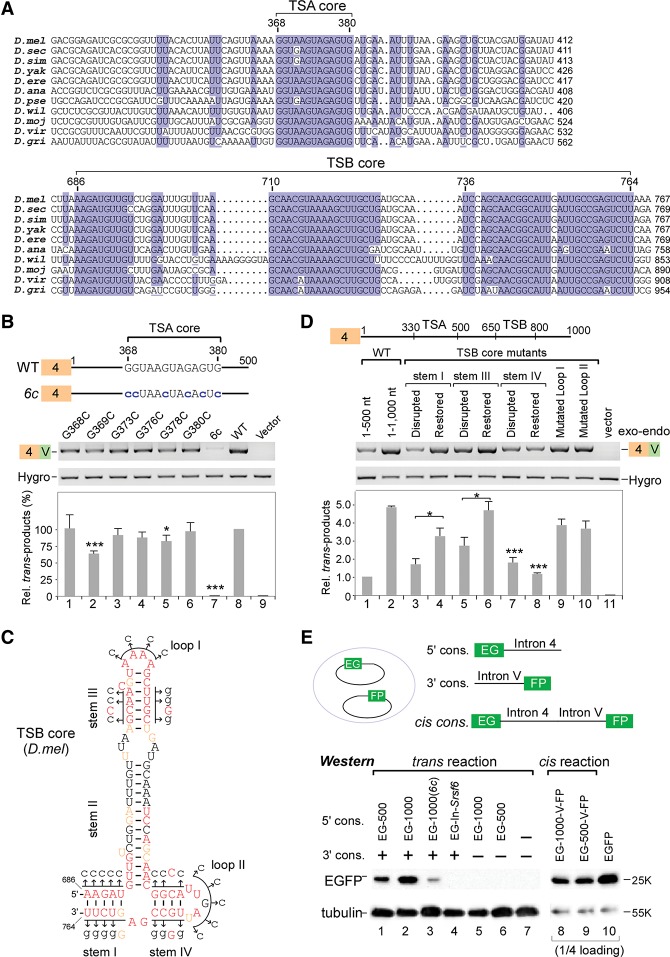
Two intronic RNA sequences are critical for trans-splicing of mod(mdg4). (A) Schematic of the mod(mdg4) gene locus in Drosophila melanogaster. Exons and introns from 3′ transcripts are designated according to FlyBase, and the sequence between exon 4 and the first 3′ exon N is defined as the last 5′ intron (intron 4). (B) Schematic of a trans-splicing system in Drosophila S2 cells. (C) Truncation assays of intron 4 reveal that two regions are important for trans-splicing activity. Relative trans-spliced products are averages from six isoforms that were normalized to loading controls and the full-length intron 4. (D) Replacing intron 4 of mod(mdg4) with other introns abolishes trans-splicing activity. Sequences of other genes include exon 1–intron 1 (without the 3′SS) from tubulin (tub) and rp49. (E) Trans-splicing activity of tiled 101-nt deletions in intron 4. Relative trans-spliced products of mod(mdg4) V forms were quantitated and normalized to the intact intron 4 (mean ± SEM; n = 3). For shorter tiled deletions, see Supplemental Figure S1C. (F) TSA RNA is sufficient to promote trans-splicing, while TSB RNA enhances the activity. TSA and TSB RNAs are fragments of the 330–500 nt and 650–800 nt in mod(mdg4) intron 4, respectively. Relative enhancement by TSB was normalized to TSA alone (mean ± SEM; n = 3). (Purple boxes) Chromosomal 5′ exons; (blue boxes) 3′ exons from the same DNA strand; (green boxes) 3′ exons from the opposite strand; (brown boxes) exon 4 on plasmids; (white boxes) internal cis-spliced intron in 3′ exons; (Hygro) hygromycin B. Boxes and labels are used similarly in other places unless otherwise indicated. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001.
Trans-splicing in higher eukaryotes and a few events in nematodes are not promoted by SL RNAs; we refer to these events as non-SL trans-splicing. Such non-SL trans-splicing was first observed in in vitro assays using HeLa nuclear extract, where exons from two transcripts with mutually complementary intron sequences were spliced together (Konarska et al. 1985; Solnick 1985). In the absence of complementary sequences, trans-splicing occurred at lower efficiency, which was further extensively characterized using a 5′SS-containing RNA oligo and a second RNA with a 3′SS (Konforti and Konarska 1995). Therefore, current models favor long complementary sequences bringing two separate transcripts together to promote in vivo non-SL trans-splicing. This mechanism was proposed to facilitate trans-splicing of RNAi regulatory gene ERI-6/7 in C. elegans through base-pairing in the UTRs (Fischer et al. 2008) and facilitate trans-splicing of identified genes in Giardia intestinalis and Bombyx mori through long base-pairing between introns (Kamikawa et al. 2011; Shao et al. 2012). Furthermore, this model of trans-splicing has been successfully applied to develop gene therapy strategies to replace mutated exons in several cancer cell lines (Puttaraju et al. 1999; Gruber et al. 2013).
However, it remains unclear whether all trans-splicing events in higher eukaryotes are promoted by complementary intronic sequences and how trans-splicing is promoted between transcripts that do not have obvious complementary sequences. To address these questions, we investigated trans-splicing of mod(mdg4) in Drosophila S2 cells and identified two critical RNA sequences (TSA and TSB) in the last 5′ intron of mod(mdg4). Mutagenesis analyses reveal that TSA is required and sufficient for trans-splicing and that TSB functions as an enhancer. Flies with deletions of TSA and TSB, prepared using the CRISPR/Cas9 system, exhibited defects in viability and embryonic development. Furthermore, two highly conserved core motifs across Drosophila species were found in TSA and TSB, respectively. The TSA core motif contains a pseudo-5′SS and recruits U1 snRNP through strong base-pairing with U1 snRNA to promote trans-splicing. A similar motif is also found in lola, allowing for mutual trans-splicing between lola and mod(mdg4).
Results
Identification of RNA elements required for trans-splicing of mod(mdg4)
Based on the current model that long complementary intronic sequences promote trans-splicing between two transcripts (Konarska et al. 1985; Fischer et al. 2008; Kamikawa et al. 2011), we searched the mod(mdg4) gene locus in Drosophila melanogaster (Dm) for regions of complementarity between the last 5′ intron and introns in the 3′ transcripts by BLASTN (E-value < 10); however, no obvious long base-paired regions were found. The trans-splicing of mod(mdg4), which results from one common 5′ transcript spliced to multiple 3′ transcripts, led us to hypothesize that critical RNA sequences would be located in the last common exon and intron (exon and intron 4) (Fig. 1A). To address this, plasmid-borne constructs that carry exon 4 with various lengths of intron 4 were transfected into Drosophila S2 cells; we then tested for trans-spliced products between the exogenous exon 4 and the endogenous 3′ exons of mod(mdg4) (Fig. 1B). In the presence of the full-length intron 4 (1979 nt), all tested 3′ exons were efficiently trans-spliced to the exogenous exon 4 (Fig. 1C, lane 1). When intron 4 was shortened from its 3′ end to 1500 or 1000 nt, similar levels of trans-splicing products were still detected; however, it was reduced ∼50% when intron 4 was shortened to 500 nt and was undetectable when shortened to ≤200 nt or when the 5′SS was mutated (Fig. 1C; Supplemental Fig. S1A). In addition, replacing the exogenous sequence with other exons and introns from the 5′ transcript of mod(mdg4) resulted in no detectable trans-splicing (Supplemental Fig. S1B). Similarly, intronic sequences from either Drosophila tubulin or rp49 did not generate detectable trans-spliced products (Fig. 1D). However, the presence of intron 4 was sufficient for trans-splicing of truncated exon 4 and trans-splicing of exons from tubulin or rp49 (Fig. 1D; Supplemental Fig. S1B). Taken together, these results demonstrate that the last common intron, but not other sequences, is required and sufficient for trans-splicing of mod(mdg4).
TSA RNA is sufficient for trans-splicing, whereas TSB enhances
To further define RNA sequences that facilitate trans-splicing, we performed two rounds of tiled deletions of intron 4. In the first round, deletions of nucleotides 250–350, 300–400, and 350–450 nearly abolished the trans-spliced products, and deletions of nucleotides 600–700, 650–750, 700–800, and 750–850 significantly decreased trans-splicing activity (Fig. 1E), whereas deletions of other regions did not, suggesting that two regions in intron 4 (300–500 nt and 650–800 nt) are important to the trans-splicing reaction, consistent with the above truncation assay. In the second round, the 300- to 500-nt region of intron 4 was further examined by shorter tiled deletions. Deletions of the 345- to 375-nt and 420- to 450-nt regions dramatically decreased trans-splicing activity (Supplemental Fig. S1C). Taken together, we designated the 330- to 500-nt region as “TSA RNA” and the 650- to 800-nt region as “TSB RNA.”
We next asked whether TSA or TSB RNA is sufficient to promote trans-splicing in S2 cells. The exogenous tubulin sequence (exon1–intron1) did not promote trans-splicing with 3′ exons of mod(mdg4); however, the addition of TSA generated a trans-spliced mRNA product between the exogenous tubulin exon and the endogenous 3′ exon of mod(mdg4) (Fig. 1F). In contrast, TSB alone did not promote trans-splicing but enhanced trans-splicing fourfold when present together with TSA (Fig. 1F). We conclude that TSA is sufficient to promote trans-splicing and functions independently of other 5′ common sequences in mod(mdg4) and that TSB functions as an enhancer element to the TSA-dependent trans-splicing reaction.
Conserved motifs in TSA and TSB across Drosophila species are critical
To identify functional motifs in TSA and TSB, we aligned intron 4 sequences from homologous mod(mdg4) genes in other available Drosophila genomes and found two highly conserved regions (Supplemental Fig. S2). The first conserved region is a nearly invariant 13-nt RNA motif (GGUA/GAGUAGAGUG) located in the previously described TSA; the second forms highly conserved secondary structures in TSB and varies from 79 to 95 nt long across Drosophila species (Fig. 2A; Supplemental Fig. S3). Therefore, we designate these two conserved regions as the “TSA core” and “TSB core.”
Figure 2.
Mutations in the highly conserved RNA motifs of intron 4 significantly decrease trans-splicing activity. (A) TSA and TSB contain highly conserved core motifs across Drosophila species. For sequence alignment of the full-length intron 4, see Supplemental Figure S2. (B) Mutations in the TSA core motif significantly decrease trans-splicing activity. Relative trans-spliced products were normalized to loading controls and the wild-type (WT) intron 4. Mean ± SEM; n = 3; (*) P < 0.05; (***) P < 0.001. (C) Highly conserved secondary structure of TSB core RNA from D. melanogaster. (Red) 100% conservation; (brown) >75% conservation; (black) <75% conservation. Mutated sites used in D are indicated by arrows here. For structures of TSB cores in other Drosophila species, see Supplemental Figure S3. (D) Secondary structures of stems I and III in the TSB core are required for enhancing trans-splicing activity. Mutated sites are indicated in C. Relative activities were normalized to loading controls and the wild-type intron 4 (1–500 nt). Mean ± SEM; n = 3. (E) Validation of trans-splicing at the protein level. The coding sequence (CDS) of EGFP is split into two halves (EG and FP) followed by intronic sequences; trans-spliced products were detected by Western blot using anti-EGFP antibody. (EG-500) Exon EG with 1–500 nt of intron 4; (EG-1000) exon EG with 1–1000 nt of intron 4; (6c) all six Gs are mutated to Cs in the TSA core; (In-Srsf6) 1–548 nt of Srsf6 intron 4. Cis-splicing constructs were used as controls.
We generated a series of mutations to investigate the function of the TSA and TSB cores. In the TSA core, six single G-to-C mutations exhibited various reductions of trans-splicing activity in which the G369C mutant provided the strongest inhibition (Fig. 2B). Importantly, when all six guanosines were mutated to cytosines (6c), trans-splicing activity was almost totally abolished (Fig. 2B). Moreover, the TSA core alone or with additional short sequences was sufficient to promote trans-splicing between the exogenous tubulin exon and endogenous 3′ exons of mod(mdg4) (Supplemental Fig. S1D). These results demonstrate that the highly conserved TSA core is a critical element for trans-splicing and suggest that a new mechanism promotes trans-splicing in Drosophila.
The 79-nt-long Dm-TSB core contains 48 nt that are invariant in other Drosophila species. Using the mfold server (Zuker 2003), TSB core sequences were predicted to form stable conserved secondary structures containing four stems and two loops (Fig. 2C; Supplemental Fig. S3). The most notable difference between TSB core secondary structures across Drosophila species is the length of stem II. To address the contribution of these structures in the TSB core, we generated mutations at conserved nucleotides to disrupt base-pairing in stems, restore base-pairing by compensatory mutations, and change sequences in loops (Fig. 2C). When base-pairing in stem I or III was disrupted (nucleotides in one chain were mutated to cytosines), the enhancement by TSB RNA was significantly reduced from 4.9-fold by the wild-type sequence to 1.7-fold and 2.7-fold by stem I and III mutants, respectively (Fig. 2D). Importantly, the reduced enhancement was partially rescued by compensatory mutations that restore base-pairing in stem I or III (Fig. 2D, lanes 3–6). This indicates that the structures of stems I and III are critical for TSB's enhancement to trans-splicing activity. Disruption of stem IV base-pairing significantly reduced the enhancement, but compensatory mutations of stem IV could not rescue the impairment (Fig. 2D, lanes 7,8), implying the importance of the primary sequence rather than its structure; alternatively, the restoration construct did not fold as predicted. Last, trans-splicing activity was not significantly altered when the loop I or II sequence was mutated (Fig. 2D, lanes 9,10). These results demonstrated that the secondary structures of the TSB core are critical to enhance the trans-splicing activity of mod(mdg4).
To verify the effect of these findings on protein expression and confirm that the chimeric mRNAs detected above were not artificially generated by homology-driven template switching during RT–PCR, we split the EGFP coding sequence (CDS) into two halves and separately fused them with intronic sequences from mod(mdg4) on two plasmids (Fig. 2E, top). The upstream exon (EG) was followed by various intron 4 sequences, while the downstream exon (FP) was fused with 3′ intron V. Thus, EGFP can be expressed only when the two exons are trans-spliced. Consistent with the above mRNA analyses, EGFP was detected when either 1–1000 nt (TSA+TSB) or 1–500 nt (TSA only) of intron 4 were included. The intron 4 sequence containing both TSA and TSB provided higher expression of EGFP, and the TSA core mutant EG-1000(6c) significantly decreased EGFP expression, whereas replacement of intron 4 with an intron from the Drosophila Srsf6 gene did not produce EGFP (Fig. 2E, lanes 1–4). These results again argue that the TSA core motif is the critical element in trans-splicing. In addition, cis-splicing constructs were tested and showed more efficient EGFP expression (Fig. 2E, lanes 8,9).
Lack of a 5′SS in 3′ transcripts of mod(mdg4) determines alternative trans-splicing
The mod(mdg4) gene has at least 31 identified 3′ exons, which are located in several regions transcribed from both DNA strands (Yu et al. 2014). Using the splice site-finding software Human Splicing Finder (HSF) (Desmet et al. 2009), we found no strong 5′SS sequences in introns between the alternatively trans-spliced exons except the identified internal cis-spliced introns (Supplemental Fig. S4). One possible explanation is that the presence of a 5′SS would promote cis-splicing between 3′ exons and inhibit its trans-splicing to the 5′ exon. To test this, we cloned 3′ intron–exon sequences of mod(mdg4) and analyzed their trans-spliced products with endogenous exon 4 in S2 cells (Fig. 3A). Two 3′ exons, U and V with their own introns or the introns of tubulin or rp49, were trans-spliced to the endogenous exon 4 in the absence of a 5′SS (Fig. 3A, top); however, trans-splicing was abolished and resulted in high levels of cis-splicing when a 5′SS (CAAG/GUAAGU) was added (Fig. 3A, bottom). This demonstrates that the presence of a 5′SS in the 3′ intron inhibits trans-splicing. Furthermore, in the absence of a strong 5′SS, trans-splicing still occurred at high levels when 3′ introns and exons from mod(mdg4) were replaced by control genes (Fig. 3B), suggesting that the primary sequence of the 3′ introns and exons is not an important factor for trans-splicing of mod(mdg4).
Figure 3.
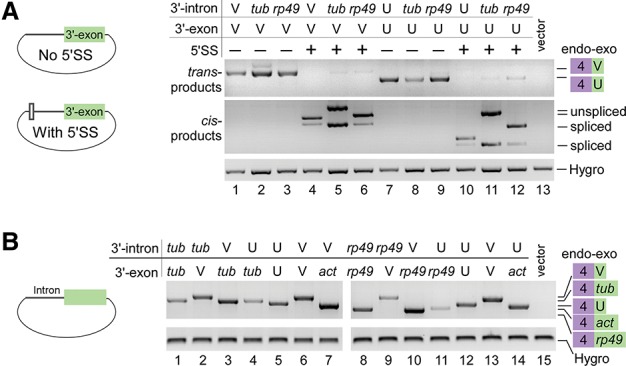
Lack of a 5′SS in the 3′ transcripts is required for trans-splicing of mod(mdg4). (A) An additional 5′SS abolishes trans-splicing and results in self-cis-splicing. (B) Replacing sequences of the 3′ intron and exons of mod(mdg4) did not obviously affect trans-splicing activity. (tub) Exon 1 and/or intron 1 (no 5′SS) of tubulin; (act) exon 1 and/or intron 1 (no 5′SS) of actin; (endo–exo) trans-spliced products between the endogenous 5′ exon 4 and exogenous 3′ exons.
TSA and TSB are required for fly development
Benefiting from the recently developed CRISPR/Cas9 system (Ren et al. 2013), we performed two fly experiments to investigate in vivo functions of TSA and TSB.
We generated flies with deletions of TSA (TSA−/−, Δ340–487 nt) or TSB (TSB−/−, Δ646–799 nt) (Fig. 4A). Neither TSA−/− nor TSB−/− adults showed visible phenotypes; however, only 44.6% of TSA−/− and 33.5% of TSB−/− embryos hatched, showing a semilethal phenotype (Fig. 4B, lanes 2,3). Since the embryonic development of TSB−/− flies unexpectedly exhibited more defects than TSA−/− flies, we further crossed deletion flies with wild-type flies in both directions and investigated the heterozygous embryos. The hatching rate of TSB+/− was similar to that of the wild type when the paternal fly was TSB−/− but was significantly reduced when the maternal fly was TSB−/−. In contrast, hatching rates of TSA+/− heterozygotes were significantly affected in crosses of both directions (Fig. 4B, lanes 4–7). Moreover, RT–PCR analyses revealed that all tested mod(mdg4) isoforms were obviously decreased in the adult internal reproductive organs from TSA−/− flies and exhibited strong accumulation of unspliced pre-mRNA, whereas trans-spliced isoforms in TSB−/− fly samples exhibited subtle and inconsistent changes depending on the isoforms (Fig. 4C). These results reveal that deletion of TSA significantly reduces trans-spliced isoforms of mod(mdg4) in vivo and affects survival of the progeny of both male and female flies, whereas deletion of TSB does not obviously affect the survival of the progeny of male flies, but a serious reduction in the survival of female flies was observed.
Figure 4.
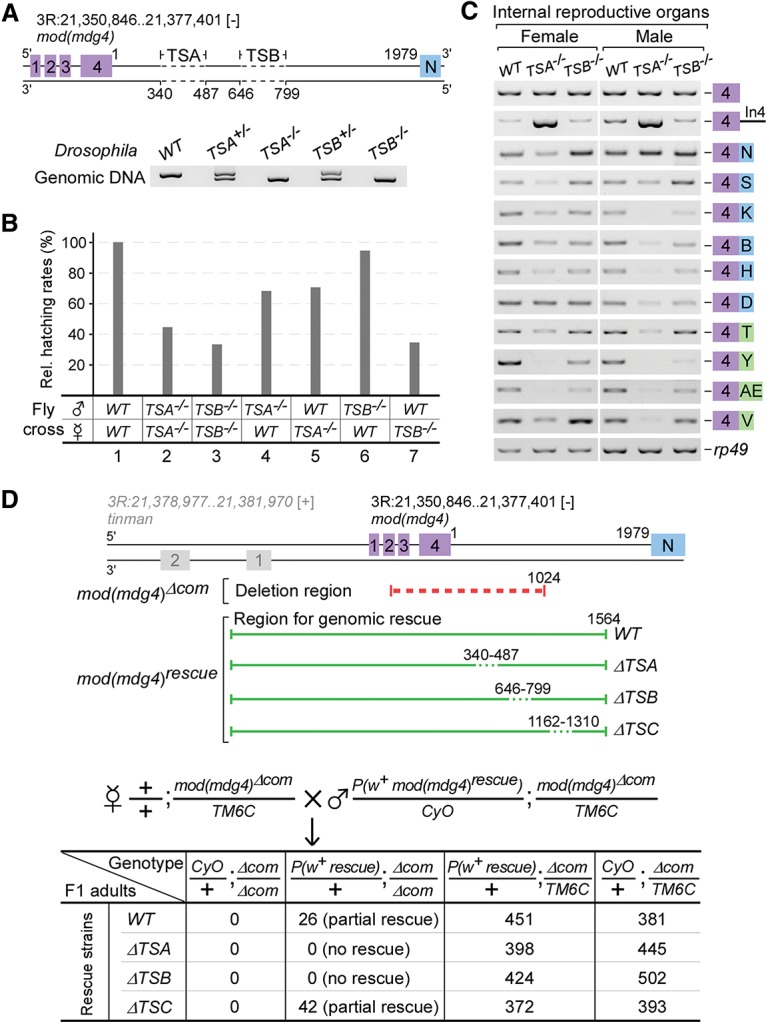
In vivo deletions of TSA and TSB result in development and viability defects in flies. (A) Schematic of in vivo deletions of TSA and TSB induced using the CRISPR/Cas9 system. Homozygous and heterozygous flies were confirmed by genomic PCR and sequencing. (B) Deletions of TSA and TSB result in semilethal phenotypes. Hatching rates were determined based on 300 embryos from each strain and normalized to the wild-type (WT) flies. (C) Deletion of TSA significantly decreases in vivo trans-spliced mod(mdg4) isoforms. (D) TSA and TSB are both required for the genomic rescue of lethality caused by deletion of the 5′ common region of mod(mdg4). F1 adults were counted based on their genotypes. (Red dashed lines) Deleted common region; (green lines) regions for genomic rescue (in which deleted elements are indicated by green dashed lines).
It has been reported that a transgenic fragment containing the entire 5′ transcribed region of mod(mdg4) could partially rescue fly lethality caused by transposon insertion at the common region of mod(mdg4) (Buchner et al. 2000). To address whether the presence of TSA or TSB is required for the rescue, we first generated a deletion allele, mod(mdg4)Δcom, in which the 5′ common region of the locus was deleted using the CRISPR/Cas9 system, causing recessive lethality; we then generated transgenic flies with various 5′ common regions of mod(mdg4) inserted at the attP40 site on chromosome II for genomic rescue crosses (Fig. 4D, top). The wild-type and ΔTSC (deletion of a control region in intron 4) transgenic alleles could partially rescue the lethality of mod(mdg4)Δcom; however, the transgenic allele without the TSA or TSB sequence (ΔTSA or ΔTSB allele) could not (Fig. 4D, bottom), demonstrating that TSA and TSB are critical elements for trans-splicing of mod(mdg4) in vivo.
TSA RNA associates with U1 snRNP components
The TSA core motif contains a notable 5′SS-like sequence (GGUA/GAGUAGAGUG), and Dm-TSA RNA could form a secondary structure similar to SL RNAs from trypanosome and nematode (Supplemental Fig. S5A), raising the possibility that TSA could function like SL RNA to facilitate trans-splicing in Drosophila. However, constructs in which the TSA sequence was replaced with SL RNAs (SL from T. brucei or SL1 and SL2 from C. elegans) promoted little trans-splicing, which was not further decreased by mutations in either the 5′SSs or Sm-binding sites (Supplemental Fig. S5B), indicating that TSA RNA in Drosophila is not an analog of SL RNAs in lower eukaryotes.
To gain insights into how TSA and TSB facilitate trans-splicing, we identified associated factors by affinity purification of in vitro transcribed RNA with MS2 RNA-binding sequences. After incubation with S2 cell lysate, RNA-associated proteins were purified and identified by mass spectrometry (Fig. 5A). In comparison with TSB and the control (MS2 RNA-binding sequence only) RNAs, TSA RNA specifically purified components of U1 snRNP, including U1-70K, U1A, SmB, D1, D2, D3, E, and G, in which peptides from U1-70K were the most abundant (Fig. 5B; Supplemental Table S1). In contrast, TSB RNA specifically associated with several identified proteins but no U1 proteins (Supplemental Table S1). To confirm the TSA RNA and U1 snRNP interaction, we analyzed associated proteins by Western blot. Consistent with the mass spectrometry data, wild-type TSA RNA purified both U1-70K and SmD2, but the TSA-6c mutant, the control, and TSB RNA did not (Fig. 5C). Similarly, TSA RNA specifically pulled down U1 snRNA, whereas the TSA-6c mutant and TSB RNA did not (Fig. 5D). These results demonstrate that both RNA and protein components of U1 snRNP are specifically associated with TSA in vitro.
Figure 5.
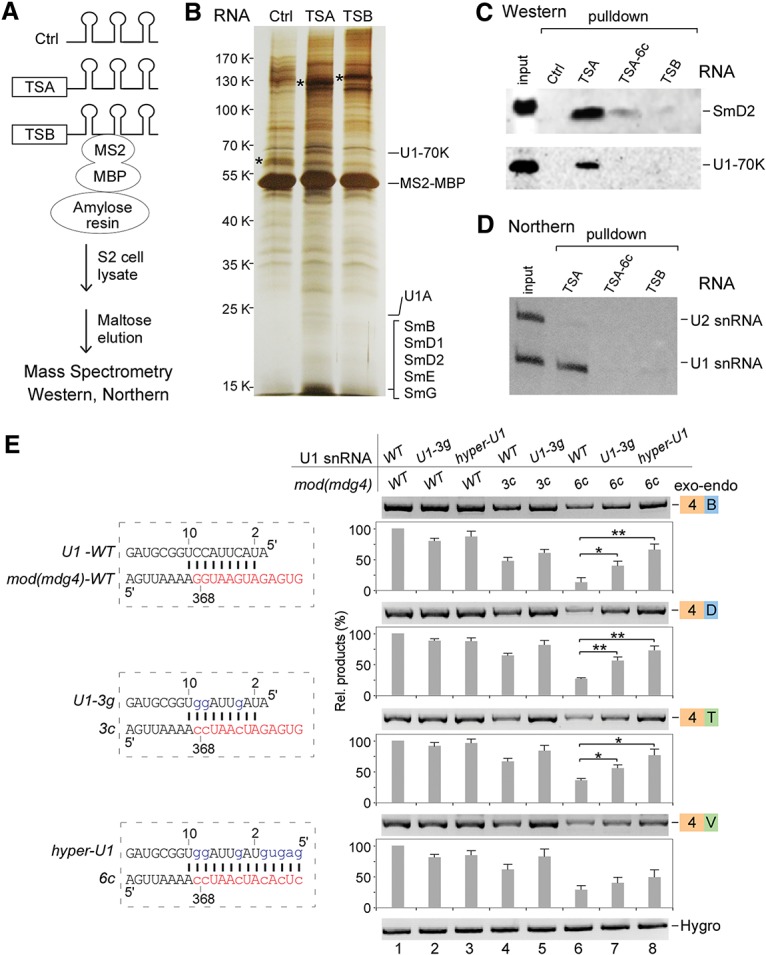
The TSA core motif binds with U1 snRNP to facilitate trans-splicing of mod(mdg4). (A) Schematic of RNA affinity purification. (Ctrl) 6xMS2-RNA-binding sites only. (B) Silver staining of affinity-purified proteins. Analyzed by mass spectrometry, TSA and TSB specifically associated proteins are listed in Supplemental Table S1. Proteins that specifically associated with TSA are indicated. (*) Visualized RNAs. (C) Western blot analyses of TSA RNA-associated U1 snRNP proteins. (TSA-6c) 6Gs-to-6Cs mutations in the TSA core motif. (D) Northern blot analysis revealed that U1 snRNA is specifically associated with TSA. (E) Compensatory changes in U1 snRNA partially rescue the decreased trans-splicing activities of the TSA core motif mutations. The wild-type (WT) TSA core motif forms 9 base pairs (bp) with the 5′ end of U1 snRNA. mod(mdg4) mutants 3c and 6c disrupt this base-pairing, but U1-3g and hyper-U1 mutants will restore or enhance the base-pairing. Relative trans-spliced products were normalized to loading controls and the wild-type constructs. Mean ± SEM; n = 3; (*) P < 0.05; (**) P < 0.01.
The TSA core contains a pseudo-5′SS and forms 9 base pairs (bp) with the 5′ end of U1 snRNA (Fig. 5E, left), and mutations in the TSA core significantly reduced trans-splicing activity. Therefore, we hypothesized that base-pairing between the TSA core and U1 snRNA is a critical interaction for trans-splicing of mod(mdg4). To test this, we constructed U1 snRNA mutants that restore (U1-3g) or enhance (hyper-U1) base-pairing with TSA core mutants and predicted that such compensatory mutations of U1 snRNA would rescue the decreased trans-splicing activity of TSA core mutants in S2 cells. Consistent with the previous assays, both the TSA core mutants 3c and 6c decreased trans-splicing of mod(mdg4) (Fig. 5E, lanes 4,6). However, additional expression of U1 snRNA with compensatory mutations, U1-3g and hyper-U1, significantly rescued the decreased trans-splicing of the 3c and 6c mutants (Fig. 5E, lanes 6–8), demonstrating that the TSA core motif recruits U1 snRNP through base-pairing with the 5′ end of U1 snRNA to promote trans-splicing.
lola has a TSA core-like motif and is mutually trans-spliced with mod(mdg4)
Trans-splicing of lola gene transcripts occurs between the last 5′ exon (exon 8) and alternative 3′ exons (Fig. 6A; Horiuchi et al. 2003; McManus et al. 2010). Aligning available intron 8 sequences of lola from five Drosophila species, we found nine conserved regions (Supplemental Fig. S6) in which one region (2191CAGGUA/GAGUAUGCU) was similar to the TSA core in mod(mdg4) containing a 5′SS with strong base-pairing (12 bp) to the 5′ end of U1 snRNA (Fig. 6B). Deletion of a fragment containing this conserved region (Δ2150–2250 nt) dramatically decreased trans-spliced products of lola in S2 cells (Fig. 6C), implying the existence of a TSA core-like motif in lola. Considering similar gene structures (Figs. 1A, 6A), we hypothesized that trans-splicing may occur between lola and mod(mdg4), that the 5′ exons of lola could be trans-spliced to the 3′ exons of mod(mdg4), and vice versa. Indeed, we detected multiple lola-mod(mdg4) or mod(mdg4)-lola chimeric mRNAs that were generated by trans-splicing between endogenous and exogenous transcripts [i.e., chromosomal exon 4 of mod(mdg4) efficiently trans-spliced to the chromosomal or plasmid-borne 3′ exons AA and G of lola] (Fig. 6D). Meanwhile, mod(mdg4)-lola chimeric trans-splicing activity was significantly reduced when using the mod(mdg4) 6c mutant (Fig. 6D, lane 3), demonstrating that trans-splicing of lola and mod(mdg4) requires the TSA core motif and shares a mechanism similar to trans-splicing within the mod(mdg4) gene locus.
Figure 6.
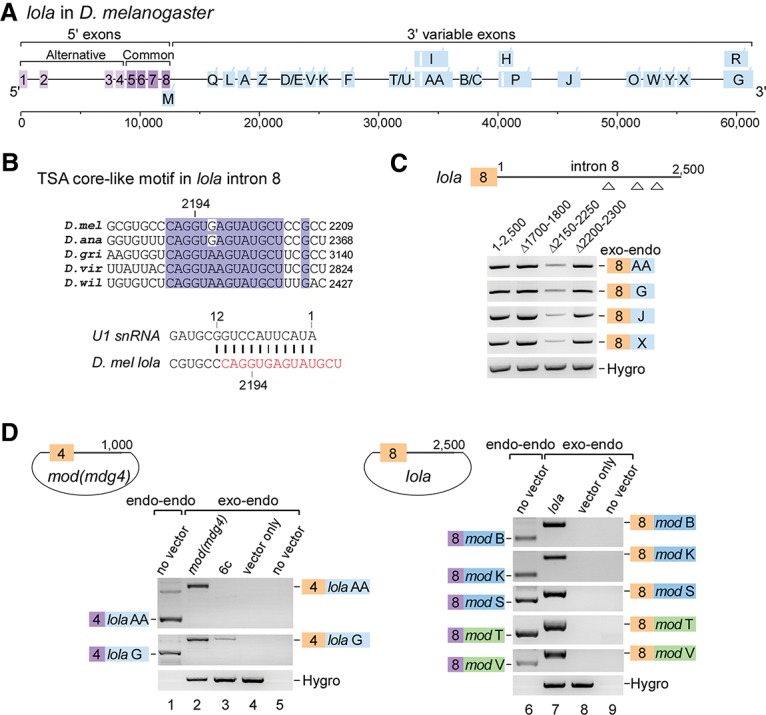
lola is mutually trans-spliced with mod(mdg4). (A) Schematic of the lola gene locus in D. melanogaster. All 3′ exons are designated according to isoforms in FlyBase. (Light purple boxes) 5′ alternative exons; (dark purple boxes) common exons; (blue boxes) 3′ alternative exons; (white boxes) internal cis-introns. (B) The conserved TSA core-like motif in the last 5′ intron of lola, which forms 12 bp with U1 snRNA. For sequence alignment of the full-length intron 8 from available Drosophila species, see Supplemental Figure S6. (C) Deletion of a region containing the TSA core-like motif in lola significantly decreases trans-splicing activity. (D) The lola and mod(mdg4) genes are mutually trans-spliced. Both chromosomal and plasmid-borne exon 4s of mod(mdg4) are trans-spliced to the chromosomal 3′ exons of lola (left) and vice versa (right). Trans-spliced products formed between chromosomal exons (endo–endo) and between plasmid-borne and chromosomal exons (exo–endo) are indicated. Boxes in colors are used similarly to in other figures.
Discussion
Trans-splicing increases genome complexity by generating chimeric mRNAs through intermolecular splicing reactions between two distinct pre-mRNA transcripts, which can be transcribed from distant genomic loci (Lasda and Blumenthal 2011). It has been reported that trans-splicing undergoes two transesterification steps similar to those the cis-splicing does (Yu et al. 1993; MacMorris et al. 2007) except leaving a Y-structured intron rather than a lariat intron (Murphy et al. 1986; Kamikawa et al. 2011). One of the key questions in trans-splicing is how those two nascent transcripts are brought into close proximity to allow subsequent catalysis of splicing. Several mechanisms have been proposed for trans-splicing events in higher eukaryotes, such as close cellular transcription sites, SR protein interactions, and complementary intronic sequences (Konarska et al. 1985; Bruzik 1996; Furuyama and Bruzik 2002; Gruber et al. 2013). In this study, through an effective trans-splicing detection system in Drosophila S2 cells and fly strains with precise deletions generated using the CRISPR/Cas9 system, two critical RNA motifs were successfully identified in the last common intron of the 5′ transcript of mod(mdg4). We demonstrate that one of the conserved motifs, the TSA core, strongly binds to U1 snRNP, providing a novel mechanism to bring separate transcripts together and promote trans-splicing in higher eukaryotes.
We obtained multiple lines of evidence to support that the intronic TSA core motif plays a fundamental role in trans-splicing of mod(mdg4). First, this 13-nt RNA motif is one of the few highly conserved regions in the nearly 2-kb length of mod(mdg4) intron 4s in all Drosophila species (Supplemental Fig. S2), and similar sequences are also observed in mod(mdg4) homologous genes in other insects (data not shown), showing a significant evolutionary importance of this motif. Second, TSA RNA is required and sufficient to promote trans-splicing in our trans-splicing detection system. Containing a pseudo-5′SS sequence, the TSA core motif binds U1 snRNP through strong base-pairing with the 5′ end of U1 snRNA. This association can be disrupted by mutations within this sequence, which significantly decreased trans-splicing activity, and trans-splicing activity was restored by compensatory mutations in U1 snRNA. Last, in vivo deletion of a region containing the TSA core results in developmental defects of flies. The second conserved motif found in intron 4 of mod(mdg4), the TSB core, has a conserved secondary structure and associates with chromatin proteins and splicing factors to enhance trans-splicing of mod(mdg4).
Therefore, we propose a model in which two intronic RNA sequences, TSA and TSB, are critical to promote trans-splicing of mod(mdg4) (Fig. 7). First, U1 snRNP binds to TSA RNA in the last 5′ intron of mod(mdg4) through strong base-pairing between the conserved TSA core and U1 snRNA. Second, TSB RNA forms a highly conserved secondary structure that is required for enhancing the trans-splicing activity of mod(mdg4). Recruitment of spliceosomal factors by TSA and TSB RNAs might introduce a bridging interaction between the 5′ and 3′ trans-spliced introns; however, it remains unclear how this happens. TSB association with chromatin proteins implies that TSB-specific enhancement of trans-splicing could be regulated by transcription and could explain the observed regulation of trans-spliced mod(mdg4) isoform abundance by Brahma, the ATPase subunit of the SWI/SNF chromatin remodeling complex, and the local density of RNA polymerase II (Yu et al. 2014).
Figure 7.
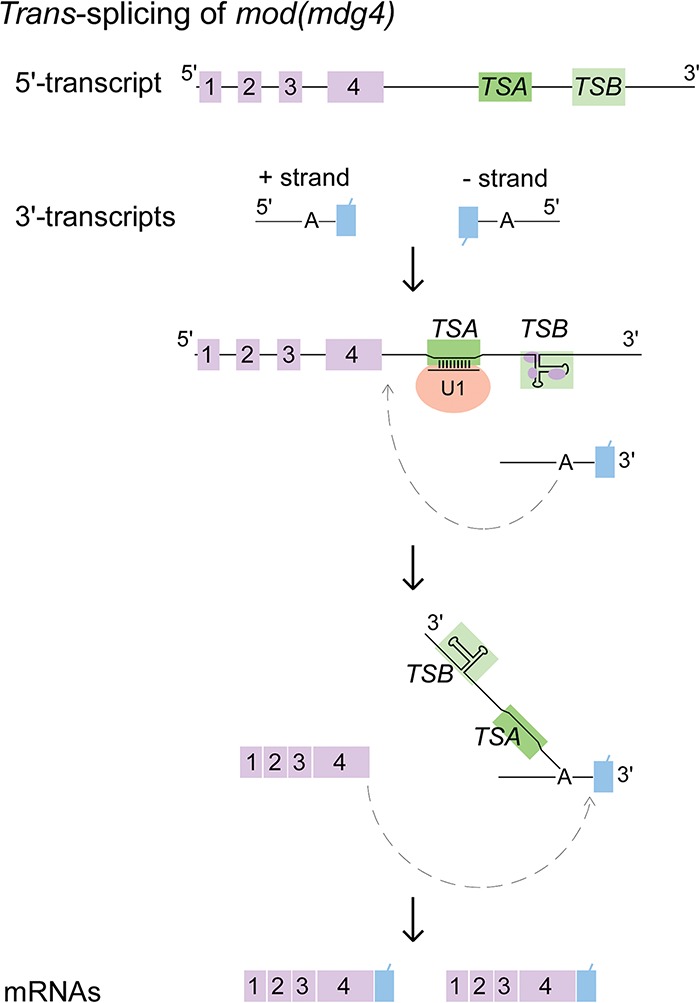
Trans-splicing of mod(mdg4) is facilitated by two intronic RNA motifs. Highly conserved TSA and TSB RNAs located in the last 5′ intron are critical for trans-splicing of mod(mdg4). TSA RNA is sufficient to promote trans-splicing in Drosophila, which binds U1 snRNP through strong base-pairing between its highly conserved 13-nt TSA core motif and the 5′ end of U1 snRNA. TSB RNA forms a highly conserved secondary structure and enhances the trans-splicing activity of mod(mdg4). Including the formation of a Y-structured intermediate, the two-step chemical reactions of trans-splicing are indicated.
Introns often contain many pseudo-5′SSs (Zhang et al. 2005); however, spliceosomal A complex formation on these sites is inefficient due to unproductive U2 snRNP recruitment (Dhir et al. 2010). There are 10 and 16 pseudo-5′SSs predicted with high scores in the last 5′ introns of mod(mdg4) and lola, respectively (Supplemental Fig. S7). Why is the TSA core motif so important for trans-splicing of these two genes? First, we found that among those pseudo-5′SS-containing sequences, the TSA core motif in each intron forms the strongest base-pairing with U1 snRNA: 9 bp in mod(mdg4) and 12 bp in lola (Supplemental Fig. S7). Second, TSA and TSB core motifs are highly conserved across Drosophila species, whereas other pseudo-5′SS sequences are not (Supplemental Figs. S2, S6). In addition, deletion of the TSA core significantly, but not totally, decreased trans-splicing activity in both S2 cells and flies, suggesting that other pseudo-5′SS sequences might function as backup and inefficiently facilitate trans-splicing in the absence of the optimal sequence.
Additionally, as another determinant of trans-splicing, the absence of a strong 5′SS in the trans-spliced 3′ introns is necessary to avoid competition from cis-splicing. We found that adding a 5′SS to the alternative 3′ transcripts of mod(mdg4) resulted in internal cis-splicing and abolished trans-splicing (Fig. 3A). Consistent with this, trans-spliced genes in Drosophila have been classified into three categories: genes with multiple 3′-terminal exons, genes with multiple first exons, or genes with very large introns (McManus et al. 2010), where the lack of strong splice sites is common.
After recruitment of spliceosomal factors by TSA and TSB, there are several possible models to explain the follow-up trans-splicing of mod(mdg4). Binding of U1 snRNP to nascent transcripts prevents shortening and the use of cryptic polyadenylation sites (telescripting) (Kaida et al. 2010). The presence of a strong pseudo-5′SS in intron 4 of mod(mdg4) could increase the abundance of mod(mdg4) transcripts containing intron 4 available for trans-splicing. However, this is unlikely to be the mechanism, as there are no significant changes in transcript levels when introns either contain a mutated TSA core motif or completely lack the sequence (Fig. 4C). In addition, recursive cis-splicing has been well characterized in Drosophila and is often used for excising long introns in which the key sequence is a ratcheting point (Y)nNCAG/GUAAGU, similar to the TSA core motif (Burnette et al. 2005; Venables et al. 2012). However, our data do not support a recursive splicing model for mod(mdg4). First, if the trans-splicing reaction occurs between the TSA core motif and downstream 3′SS, the newly generated 3′SS of D. melanogaster is a suboptimal 3′SS (AAG/) and could be an even worse in other Drosophila species, such as in Drosophila virilis (GGG/) (Fig. 2A). Second, compensatory mutations in U1 snRNA rescue the decreased trans-splicing activity of TSA core motif mutants in which the 5′SS is totally mutated (Fig. 5E), and recursive splicing cannot occur.
Trans-splicing of mod(mdg4) and trans-splicing of lola share a similar mechanism, both using a conserved intronic TSA core motif to promote the reaction (Fig. 6). In order to verify whether this is a common mechanism of trans-splicing in Drosophila, we searched the last 5′ introns of other trans-splicing events in Drosophila (McManus et al. 2010) and found that 16 of the 126 introns have pseudo-5′SS-containing sequences, which form ≥9 bp with the 5′ end of U1 snRNA (i.e., TSA core-like motifs) (Supplemental Table S2). Under such strict searching criteria, this result implies that the TSA core-mediated mechanism of trans-splicing could represent a general mechanism for a class of trans-splicing events in Drosophila.
Materials and methods
Strains and plasmids
For plasmids used in Drosophila S2 cells, sequences from mod(mdg4), lola, or other genes in D. melanogaster were inserted into modified pMT/V5-His B vector (Invitrogen) with hygromycin B and P copia promoter as described (Yang et al. 2013). Plasmids were transfected into S2 cells by Effectene transfection reagent (Qiagen) and induced by 0.5 mM CuSO4 at 12 h after transfection; cells were collected 24 h later for isolation of RNA and proteins. The sequence of U1 snRNA, including a ±1-kb region, was amplified from genomic DNA using primers listed in Supplemental Table S3. The CDS of EGFP was amplified from vector pEGFP-N1 (Invitrogen) and split after nucleotide G489.
RT–PCR and sequence analyses
Total RNA was extracted using Trizol (Invitrogen), and reverse transcription was performed by a first strand cDNA synthesis kit (RevertAid, Thermo) using either oligo(dT) or specific primers. Trans-spliced products were amplified by Ex Taq DNA polymerase (TaKaRa), confirmed by Sanger sequencing, and quantified using a Tanon-2500 gel analysis system. All sequences from the Drosophila species were downloaded from FlyBase and aligned using DNAman version 4.0 (Lynnon BioSoft). Potential splice sites were identified according to scores obtained by the online software HSF (Desmet et al. 2009).
RNA affinity purification and mass spectrometry
Along with 6xMS2 RNA-binding sites (Zhou et al. 2002), studied RNAs were in vitro transcribed by T7 RNA polymerase (Promega) and gel-purified. MS2-MBP protein was expressed in Escherichia coli and purified by Superdex 200 10/300 GL column (GE) (Tange et al. 2005). Ninety picomoles of RNA was mixed with 270 pmol of MS2-MBP protein for 20 min on ice in 50 μL of buffer 1 (20 mM Tris-Cl at pH 7.4, 100 mM KCl, 10 mM MgCl2, 1% Triton X-100, 0.5 mM DTT, 50 µg/mL yeast tRNA, 5 mM creatine phosphate, 0.5 mM ATP, 200 U/mL RNase inhibitor, protease inhibitors). The mixture was then applied to amylose resin (New England Biolabs) and tumbled with 0.1% BSA to reduce background before adding 1.2 mL of lysate from 2 × 107 S2 cells (Spain et al. 2010) in buffer 1. After six washes with buffer 1, bead-bound proteins were eluted with buffer 2 (12 mM maltose, 20 mM Tris-Cl at pH 7.9, 100 mM KCl) and separated on SDS-PAGE. Obtained proteins were then analyzed using a Q-Exactive hybrid quadrupole orbitrap mass spectrometer (Thermo Scientific) with a Dionex UltiMate 3000 RSLCnano system. Peptide identification and protein assembly were performed on a Thermo Proteome Discoverer 1.4.1 platform. For each tandem mass spectrometry data set, a single search was performed against the corresponding UniProtKB/Swiss-Prot database using the SEQUEST and percolator algorithms.
Western and Northern analyses
Antiserum against Drosophila U1-70K was generated by immunizing rabbits with the peptide DGKKIDSKRVLVDV (amino acids 164–177). Anti-SNRPD2 (SmD2) antibody was purchased from Abcam (ab155030). Western blots of anti-EGFP and anti-tubulin were probed using mAb 7G9 (Abmart) and mAb DM1A (Sigma), respectively. Northern analysis was performed by transferring RNAs from 8 M urea-PAGE gel to Hybond N membrane (Amersham). Membranes were then probed by DIG-labeled (Roche) antisense DNAs complementary to the full-length U snRNAs in D. melanogaster.
Fly strains and mutagenesis
Culture and crosses of D. melanogaster were carried out on standard medium and at standard temperature. Mutagenesis of the Drosophila mod(mdg4) gene were performed using the recently developed CRISPR/Cas9 system (Ren et al. 2013). Briefly, target sequences of two guide RNAs (sgRNA) (listed in Supplemental Table S3) were selected for each genomic deletion. Each pair of sgRNA plasmids was coinjected into embryos of transgenic line nanos-Cas9(attP40) by Core Facility of Drosophila Resource and Technique, Shanghai Institute for Biological Sciences. G0 flies were then crossed with the TM3/TM6 balancer strain (Bloomington no. 5906); subsequently, G1 male flies were crossed with the same balancer strain followed by genomic DNA screening for deletion strains. Obtained flies were then backcrossed at least five generations into a controlled uniform homogeneous genetic background (Bloomington no. 5905) to eliminate potential off-target events.
To generate constructs for genomic rescue of lethality caused by the deletion of mod(mdg4) common region, a 6690-bp wild-type fragment containing a sequence from part of the upstream gene tinman to nucleotide 1564 of mod(mdg4) intron 4 was amplified and cloned into the pUAST(attB) vector. Three parallel deletion constructs—ΔTSA (340–487 nt), ΔTSB (646–799 nt), and a control, ΔTSC (1162–1310 nt)—were constructed by overlapping PCR based on the above wild-type plasmid. Genomic rescue constructs were microinjected and incorporated into the attP40 site on chromosome II. Obtained flies were then applied in crosses to test their ability to rescue the lethality of the homozygous mod(mdg4)Δcom strain. For each cross, total offspring from six parallel vials were counted based on their genotypes.
Supplementary Material
Acknowledgments
We thank C. Query and B. Kosmyna at Albert Einstein College of Medicine and L. Rabinow at Centre de Neurosciences de Université Paris Sud XI for critical reading and discussion of the manuscript, H. Cheng at Shanghai Institutes for Biological Sciences for pMS2-MBP plasmid, and other members in the Xu laboratory for data entries and discussions. This work was supported by grants from National Natural Science Foundation of China (NSFC) (31270842 and 31472045) and National Basic Research Program of China (2012CB114101) to Y.-Z.X., and NSFC (31400653) to W.S.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.258863.115.
References
- Bruzik JP. 1996. Splicing glue: a role for SR proteins in trans splicing? Microb Pathog 21: 149–155. [DOI] [PubMed] [Google Scholar]
- Bruzik JP, Steitz JA. 1990. Spliced leader RNA sequences can substitute for the essential 5′ end of U1 RNA during splicing in a mammalian in vitro system. Cell 62: 889–899. [DOI] [PubMed] [Google Scholar]
- Bruzik JP, Van Doren K, Hirsh D, Steitz JA. 1988. Trans splicing involves a novel form of small nuclear ribonucleoprotein particles. Nature 335: 559–562. [DOI] [PubMed] [Google Scholar]
- Buchner K, Roth P, Schotta G, Krauss V, Saumweber H, Reuter G, Dorn R. 2000. Genetic and molecular complexity of the position effect variegation modifier mod(mdg4) in Drosophila. Genetics 155: 141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette JM, Miyamoto-Sato E, Schaub MA, Conklin J, Lopez AJ. 2005. Subdivision of large introns in Drosophila by recursive splicing at nonexonic elements. Genetics 170: 661–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai MJ, Liu W, He HJ, Wang JX, Zhao XF. 2012. Mod(mdg4) participates in hormonally regulated midgut programmed cell death during metamorphosis. Apoptosis 17: 1327–1339. [DOI] [PubMed] [Google Scholar]
- Denker JA, Maroney PA, Yu YT, Kanost RA, Nilsen TW. 1996. Multiple requirements for nematode spliced leader RNP function in trans-splicing. RNA 2: 746–755. [PMC free article] [PubMed] [Google Scholar]
- Denker JA, Zuckerman DM, Maroney PA, Nilsen TW. 2002. New components of the spliced leader RNP required for nematode trans-splicing. Nature 417: 667–670. [DOI] [PubMed] [Google Scholar]
- Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C. 2009. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res 37: e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhir A, Buratti E, van Santen MA, Luhrmann R, Baralle FE. 2010. The intronic splicing code: multiple factors involved in ATM pseudoexon definition. EMBO J 29: 749–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn R, Krauss V. 2003. The modifier of mdg4 locus in Drosophila: functional complexity is resolved by trans splicing. Genetica 117: 165–177. [DOI] [PubMed] [Google Scholar]
- Dorn R, Krauss V, Reuter G, Saumweber H. 1993. The enhancer of position-effect variegation of Drosophila, E(var)3-93D, codes for a chromatin protein containing a conserved domain common to several transcriptional regulators. Proc Natl Acad Sci 90: 11376–11380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer SE, Butler MD, Pan Q, Ruvkun G. 2008. Trans-splicing in C. elegans generates the negative RNAi regulator ERI-6/7. Nature 455: 491–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujieda S, Lin YQ, Saxon A, Zhang K. 1996. Multiple types of chimeric germ-line Ig heavy chain transcripts in human B cells: evidence for trans-splicing of human Ig RNA. J Immunol 157: 3450–3459. [PubMed] [Google Scholar]
- Furuyama S, Bruzik JP. 2002. Multiple roles for SR proteins in trans splicing. Mol Cell Biol 22: 5337–5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabler M, Volkmar M, Weinlich S, Herbst A, Dobberthien P, Sklarss S, Fanti L, Pimpinelli S, Kress H, Reuter G, et al. 2005. Trans-splicing of the mod(mdg4) complex locus is conserved between the distantly related species Drosophila melanogaster and D. virilis. Genetics 169: 723–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimova TI, Gdula DA, Gerasimov DV, Simonova O, Corces VG. 1995. A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell 82: 587–597. [DOI] [PubMed] [Google Scholar]
- Gruber C, Koller U, Murauer EM, Hainzl S, Huttner C, Kocher T, South AP, Hintner H, Bauer JW. 2013. The design and optimization of RNA trans-splicing molecules for skin cancer therapy. Mol Oncol 7: 1056–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon GJ, Maroney PA, Denker JA, Nilsen TW. 1990. Trans splicing of nematode pre-messenger RNA in vitro. Cell 61: 1247–1255. [DOI] [PubMed] [Google Scholar]
- Hannon GJ, Maroney PA, Nilsen TW. 1991. U small nuclear ribonucleoprotein requirements for nematode cis- and trans-splicing in vitro. J Biol Chem 266: 22792–22795. [PubMed] [Google Scholar]
- Hannon GJ, Maroney PA, Yu YT, Hannon GE, Nilsen TW. 1992. Interaction of U6 snRNA with a sequence required for function of the nematode SL RNA in trans-splicing. Science 258: 1775–1780. [DOI] [PubMed] [Google Scholar]
- Horiuchi T, Giniger E, Aigaki T. 2003. Alternative trans-splicing of constant and variable exons of a Drosophila axon guidance gene, lola. Genes Dev 17: 2496–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins AA, Moore MJ. 2012. The spliceosome: a flexible, reversible macromolecular machine. Trends Biochem Sci 37: 179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu GJ, Chen J, Zhao XN, Xu JJ, Guo DQ, Lu M, Zhu M, Xiong Y, Li Q, Chang CC, et al. 2013. Production of ACAT1 56-kDa isoform in human cells via trans-splicing involving the ampicillin resistance gene. Cell Res 23: 1007–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, Dreyfuss G. 2010. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature 468: 664–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamikawa R, Inagaki Y, Tokoro M, Roger AJ, Hashimoto T. 2011. Split introns in the genome of Giardia intestinalis are excised by spliceosome-mediated trans-splicing. Curr Biol 21: 311–315. [DOI] [PubMed] [Google Scholar]
- Kim E, Goren A, Ast G. 2008. Alternative splicing and disease. RNA Biol 5: 17–19. [DOI] [PubMed] [Google Scholar]
- Koller B, Fromm H, Galun E, Edelman M. 1987. Evidence for in vivo trans splicing of pre-mRNAs in tobacco chloroplasts. Cell 48: 111–119. [DOI] [PubMed] [Google Scholar]
- Konarska MM, Padgett RA, Sharp PA. 1985. Trans splicing of mRNA precursors in vitro. Cell 42: 165–171. [DOI] [PubMed] [Google Scholar]
- Konforti BB, Konarska MM. 1995. A short 5′ splice site RNA oligo can participate in both steps of splicing in mammalian extracts. RNA 1: 815–827. [PMC free article] [PubMed] [Google Scholar]
- Krauss V, Dorn R. 2004. Evolution of the trans-splicing Drosophila locus mod(mdg4) in several species of Diptera and Lepidoptera. Gene 331: 165–176. [DOI] [PubMed] [Google Scholar]
- Labrador M, Mongelard F, Plata-Rengifo P, Baxter EM, Corces VG, Gerasimova TI. 2001. Protein encoding by both DNA strands. Nature 409: 1000. [DOI] [PubMed] [Google Scholar]
- Lasda EL, Blumenthal T. 2011. Trans-splicing. Wiley Interdiscip Rev RNA 2: 417–434. [DOI] [PubMed] [Google Scholar]
- MacMorris M, Kumar M, Lasda E, Larsen A, Kraemer B, Blumenthal T. 2007. A novel family of C. elegans snRNPs contains proteins associated with trans-splicing. RNA 13: 511–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroney PA, Yu YT, Jankowska M, Nilsen TW. 1996. Direct analysis of nematode cis- and trans-spliceosomes: a functional role for U5 snRNA in spliced leader addition trans-splicing and the identification of novel Sm snRNPs. RNA 2: 735–745. [PMC free article] [PubMed] [Google Scholar]
- McManus CJ, Duff MO, Eipper-Mains J, Graveley BR. 2010. Global analysis of trans-splicing in Drosophila. Proc Natl Acad Sci 107: 12975–12979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy WJ, Watkins KP, Agabian N. 1986. Identification of a novel Y branch structure as an intermediate in trypanosome mRNA processing: evidence for trans splicing. Cell 47: 517–525. [DOI] [PubMed] [Google Scholar]
- Neumuller RA, Richter C, Fischer A, Novatchkova M, Neumuller KG, Knoblich JA. 2011. Genome-wide analysis of self-renewal in Drosophila neural stem cells by transgenic RNAi. Cell Stem Cell 8: 580–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira de Souza A, Jubier MF, Delcher E, Lancelin D, Lejeune B. 1991. A trans-splicing model for the expression of the tripartite nad5 gene in wheat and maize mitochondria. Plant Cell 3: 1363–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puttaraju M, Jamison SF, Mansfield SG, Garcia-Blanco MA, Mitchell LG. 1999. Spliceosome-mediated RNA trans-splicing as a tool for gene therapy. Nat Biotechnol 17: 246–252. [DOI] [PubMed] [Google Scholar]
- Ren X, Sun J, Housden BE, Hu Y, Roesel C, Lin S, Liu LP, Yang Z, Mao D, Sun L, et al. 2013. Optimized gene editing technology for Drosophila melanogaster using germ line-specific Cas9. Proc Natl Acad Sci 110: 19012–19017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao W, Zhao QY, Wang XY, Xu XY, Tang Q, Li M, Li X, Xu YZ. 2012. Alternative splicing and trans-splicing events revealed by analysis of the Bombyx mori transcriptome. RNA 18: 1395–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PA. 1987. Trans splicing: variation on a familiar theme? Cell 50: 147–148. [DOI] [PubMed] [Google Scholar]
- Solnick D. 1985. Trans splicing of mRNA precursors. Cell 42: 157–164. [DOI] [PubMed] [Google Scholar]
- Southall TD, Davidson CM, Miller C, Carr A, Brand AH. 2014. Dedifferentiation of neurons precedes tumor formation in Lola mutants. Dev Cell 28: 685–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spain MM, Caruso JA, Swaminathan A, Pile LA. 2010. Drosophila SIN3 isoforms interact with distinct proteins and have unique biological functions. J Biol Chem 285: 27457–27467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton RE, Boothroyd JC. 1986. Evidence for trans splicing in trypanosomes. Cell 47: 527–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tange TO, Shibuya T, Jurica MS, Moore MJ. 2005. Biochemical analysis of the EJC reveals two new factors and a stable tetrameric protein core. RNA 11: 1869–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschudi C, Ullu E. 1990. Destruction of U2, U4, or U6 small nuclear RNA blocks trans splicing in trypanosome cells. Cell 61: 459–466. [DOI] [PubMed] [Google Scholar]
- Venables JP, Tazi J, Juge F. 2012. Regulated functional alternative splicing in Drosophila. Nucleic Acids Res 40: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Luhrmann R. 2011. Spliceosome structure and function. Cold Spring Harb Perspect Biol 3: a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Wang XY, Zhang ZM, Pu J, Fan YJ, Zhou J, Query CC, Xu YZ. 2013. Splicing proofreading at 5′ splice sites by ATPase Prp28p. Nucleic Acids Res 41: 4660–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YT, Maroney PA, Nilsen TW. 1993. Functional reconstitution of U6 snRNA in nematode cis- and trans-splicing: U6 can serve as both a branch acceptor and a 5′ exon. Cell 75: 1049–1059. [DOI] [PubMed] [Google Scholar]
- Yu S, Waldholm J, Bohm S, Visa N. 2014. Brahma regulates a specific trans-splicing event at the mod(mdg4) locus of Drosophila melanogaster. RNA Biol 11: 134–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XH, Leslie CS, Chasin LA. 2005. Computational searches for splicing signals. Methods 37: 292–305. [DOI] [PubMed] [Google Scholar]
- Zhang G, Guo G, Hu X, Zhang Y, Li Q, Li R, Zhuang R, Lu Z, He Z, Fang X, et al. 2010. Deep RNA sequencing at single base-pair resolution reveals high complexity of the rice transcriptome. Genome Res 20: 646–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Sim J, Griffith J, Reed R. 2002. Purification and electron microscopic visualization of functional human spliceosomes. Proc Natl Acad Sci 99: 12203–12207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorio DA, Cheng NN, Blumenthal T, Spieth J. 1994. Operons as a common form of chromosomal organization in C. elegans. Nature 372: 270–272. [DOI] [PubMed] [Google Scholar]
- Zuker M. 2003. Mfold Web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.