Abstract
An important prerequisite for computational neuroanatomy is the spatial normalization of the data. Despite its importance for the success of the subsequent statistical analysis, image alignment is dealt with from the perspective of image matching, while its influence on the group analysis is neglected. The choice of the template, the registration algorithm as well as the registration parameters, all confound group differences and impact the outcome of the analysis. In order to limit their influence, we perform multiple registrations by varying these parameters, resulting in multiple instances for each sample. In order to harness the high dimensionality of the data and emphasize the group differences, we propose a supervised dimensionality reduction technique that takes into account the organization of the data. This is achieved by solving a supervised dictionary learning problem for block-sparse signals. Structured sparsity allows the grouping of instances across different independent samples, while label supervision allows for discriminative dictionaries. The block structure of dictionaries allows constructing multiple classifiers that treat each dictionary block as a basis of a subspace that spans a separate band of information. We formulate this problem as a convex optimization problem with a geometric programming (GP) component. Promising results that demonstrate the potential of the proposed approach are shown for an MR image dataset of Autism subjects.
1 Introduction
Computational Anatomy (CA) employs statistical methods in order to analyze and model anatomical structures across individuals. Typical CA approaches include Voxel Based Analysis (VBA) [2] and high dimensional pattern-classification [7]. These are complementary techniques and suffer from different limitations.
On the one hand, VBA employs mass univariate linear statistical tests on voxel values in order to identify regional individual differences. The simplicity of the statistical models limits its ability to capture multivariate relationships. On the other hand, high dimensional pattern-classification is able to recover multivariate relationships that characterize group differences while accurately classifying individuals. Nonetheless, it requires a dimensionality reduction step in order to cope with the challenges due to the high dimensional small sample size data that are typical in medical imaging.
A common assumption behind all CA techniques is that the data are optimally brought in correspondence through a registration process. However, the optimality of the spatial normalization is evaluated through measures that usually reflect the intensity agreement of the voxels. While these criteria are relevant in the case of image matching, they are potentially insufficient, or even irrelevant, in the case of group analysis. As a consequence, the choice of registration parameters (i.e., regularization weight) may act as confounding factor for the subsequent statistical analysis.
Motivated by this observation, we propose a novel approach for computational anatomy that is insensitive to the pre-processing step (i.e., registration). In order to limit the influence of registration parameters, we expand the space where the statistical analysis takes place by performing multiple registrations for varying degrees of regularization. In this space, group analysis is performed by simultaneous clustering and classification. The proposed approach is formulated as supervised dictionary learning [9, 3] endowed with a block-sparsity inducing norm [5, 6] in order to harness the underlying structure of the data and enhance the discriminative power of the estimated elements.
The remainder of this paper is divided into 3 sections. Section 2 details the proposed method. Section 3 presents empirical results obtained using an Autism Spectrum Disorder dataset. Section 4 provides a brief discussion of the implications of this work and possible applications in the context of computational anatomy.
2 Method
Motivation
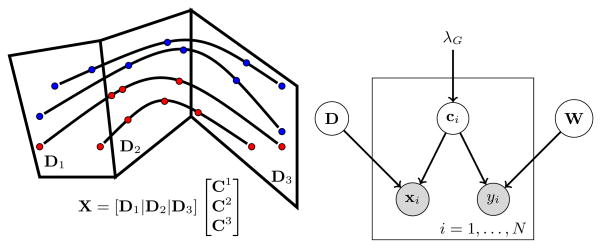
Suppose that the data set comprises of multiple instances per sample where each instance potentially conveys a new band of information. Then our goal is to accurately model the entire dataset in the span of a dictionary such that different blocks of the dictionary capture the information contained in these bands. Furthermore, we want the dictionary, and specifically, each band of the dictionary to be discriminative with respect to the observed labels. The visualization of this goal is illustrated in figure 1.
Fig. 1.
Left: A graphical depiction of subspace clustering for multiple instance data. Black curves indicate the domain of instances for an independent subject and red points indicate the sampled instances. Dictionary blocks D1, D2, D3 indicate the basis of the subspaces where clusters of data reside and where separate classifiers discriminate. Right: The graphical model of supervised block sparse dictionary learning. Gray denotes observed variable. D is the dictionary, ci are loading coefficients, W are the discriminative parameters, (xi, yi) is the sample/label pair, λG is the hyperparameter that controls block sparsity
Supervised Block Sparse Dictionary Learning
To introduce the formulation, first we define the variables. Let X ∈ ℝd×n denote the spatially normalized data stored in a tall matrix. Let y ∈ ℝn denote the labels. The aim is to find an appropriate lower dimensional representation C ∈ ℝp×n where each row corresponds to loading coefficients for the bases of the lower dimensional space. The bases can also be called the atoms of a dictionary D ∈ ℝd×p that we aim to learn. Furthermore, we want the dictionary to have sub-blocks D = [D1|…|Dnb] such that the corresponding loading coefficients can discriminate the labels y using a different discriminative model {w1, …, wnb} for each block.
In a graphical sense, the problem of supervised dictionary learning can be described as minimizing a joint energy with 5 variables 1) D: the dictionary, 2) C: the loading coefficients, 3) W: the discriminative model 4) X: observed signal and 5) y: observed labels. The dependencies of these variables is the following: D and C generate X and C and W discriminate y. The graphical model can be seen in figure 1.
While the graphical model provides the dependency structure between the pertinent random variables, it is necessary to define a measure of goodness of fit for potential choices of these random variables in order to formulate an optimization program to solve for the hidden variables D, C, W. If we let
denote the ℓth block of the loading coefficients for sample i, we can define the joint energy of these variables as the following term
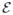 :
:
| (1) |
The terms above can be as explained as such:
is the data reconstruction error.
is the place holder for the hinge loss function
is a joint term for enforcing block sparsity and minimizing classification loss
λG and λD are generative and discriminative penalty parameters, respectively.
The main novelty in our work is the term . This term diverges from the typical sparse dictionary learning regularization term λ||ci||1 in 3 ways:
is the mixed L2/1 norm of the coefficients broken up into nb blocks. This term enforces sparsity in block selection rather than atom selection which adds additional structure. It implicitly clusters data along subspaces that are spanned by the atoms in each block [5][6].
If the basis block ℓ is not used to represent subject then εi,ℓ is penalized less in the objective. εi,ℓ is the margin violation term for the subpace spanned by dictionary block ℓ. This way the corresponding subspace specific hyperplane wℓ is not affected by this sample.
If εi,ℓ is large then the block-sparsity of is penalized more: objective aims to not represent sample i from the subspace spanned by dictionary block ℓ because it is poorly discriminated there.
Given this energy function for each sample, the overall objective that we aim to minimize for a given dataset is:
| (2) |
This is a difficult optimization to perform since like most dictionary learning and clustering tasks; the objective is not convex, specifically with respect to D, C. In addition, the term constitutes a geometric programming (GP) form. Nevertheless, the formulation is block-convex in C and D, W, b, ε and we may perform an iterative procedure to obtain a local minimum. The iterative optimization procedure is described in Algorithm 1.
The overview of the algorithm is as follows: The input samples X, labels Y and hyperparameters that set number of blocks in the dictionary (nb), the number of atoms per block (na) and penalty terms for block sparsity λG, discrimination loss penalty λD and model complexity λM are initially specified. All model parameters W and b are set to 0, initially. The initial dictionary D is set to a random Gaussian matrix with normalized columns. Then iteratively {C} and {D, W, b, ε} are optimized. The gradients for each variable are omitted due to space limitations.
Once D, W, b are trained, during test time, an unobserved sample is classified using the following rule:
| (5) |
In words, the predicted label y* is one which the minimum energy stated in equation (1) can be obtained after optimizing for the loading coefficients.
Since the energy
 can be interpreted as the negative joint log probability of the graphical model, our framework readily handles the multiple instance data in the following sense:
can be interpreted as the negative joint log probability of the graphical model, our framework readily handles the multiple instance data in the following sense:
Algorithm 1.
Block sparse supervised dictionary learning (BS-SDL)
| Input: X ℝd×n, Y ∈ {−1, +1}n (training signals); na (number of atoms per block); nb (number of blocks); λG, λD, λM (parameters) | ||||
| Output: D ∈ ℝd×(nbna)(dictionary); w1, …, wnb ∈ ℝna, b1, …, bnb ∈ ℝ (parameters) | ||||
| Initialization: Set D to a random Gaussian matrix with normalized columns. Set w1, …, wnb, b1, …, bnb to zero. Set εi,ℓ = 1 for i = 1, …, n ℓ = 1, …, nb | ||||
| Loop: Repeat until convergence (or a fixed number of iterations) | ||||
• Supervised block-sparse coding:
D, W, b, ε, optimize w.r.t. C
|
| (6) |
where {xj} denotes the set of multiple instances belonging to the test subject.
3 Experiments and Results
Data
We validated our framework using data from a dataset comprising of Autism Spectrum Disorder (ASD) patients and controls. Autism dataset images were T1-weighted MR scans from a 3T scanner, acquired sagitally using volumetric 3D MPRAGE with 0.8 mm × 0.8 mm in plane resolution and 0.9 mm thick sagittal slices. The ASD dataset included 206 subjects (105 patients/101 controls) representing both sexes and no age matching.
Pre-processing
The acquired images were all bias corrected, skull stripped and had cerebellum removed. The resulting brain images were then segmented into white matter (WM), gray matter (GM) and ventricle (VN). The images were then registered to a template brain using registration algorithm in [10] using varying smoothness penalty terms from λ = 0 to λ = 1 to obtain 11 instances per independent subject. The resulting deformation field and the segmentation results were used to obtain WM, GM, VN tissue density maps [4], which were then used for the subsequent analysis. In order to reduce computational burden, the dimensionality of the data was reduced to 1000 (which amounts to ≈ 95% of the energy) by the random matrix projections method described in [8].
Parameter Selection
As in all generative models, an important issue is hyper parameter selection. In our model, we have 5 hyperparameters: 1) λG: generative penalty, 2) λD: discriminative loss penalty, 3) λM : discriminative model complexity penalty, 4) nb: number of blocks/clusters, 5) na: number of atoms per block.
Cross validating to select all of these parameters is a daunting task, therefore we limited our cross validation to smaller set of values for each hyperparameter obtained by the following heuristics: Since the number of instances per each sample was 11, it could be assumed that there are at most 11 possible bands of information. We limited the number of blocks to be half of this value: nb ∈ {1, 2, 3, 4, 5}. We limited the number of atoms per block to na ∈ {10, 20, 40, 80} to control overfitting as suggested in [3]. We fixed the λM = 1. λD and λG were cross validated from possible values of {0, 1, 2, 8}. A robust parameter combination that we found among these values was λG = 2, λD = 8. All nb ≥ 3 achieved a plateau performance given na ≥ 20. We used nb = 3 and na = 20.
Classification Results
We compared the classification performance of our method with both linear support vector machine (lin-SVM) and Gaussian kernel support vector machine (rbf-SVM). The input data for these methods was both single instances (denoted by sing.) obtained by choosing the best registration parameter by cross validation and also multiple instances (denoted by mult.). The multiple instance data was classified using two protocols. The first approach is a variant of multiple instance SVM [1] that was adapted for neuroimaging datasets in [11] (denoted by ensemble-MI-SVM). The second approach consists of naively concatenating all instances of a subject in a single vector (denoted by naive-concat.) and using these to train the lin-SVM and rbf-SVM. All of these methods underwent cross validation to determine the best kernel width and hinge loss penalty. All classification was done by separating the data in two parts and training on the half of data and testing on the other half. The splits were randomly permuted 100 times to obtain a confidence interval. Our method is denoted as block sparse supervised dictionary learning (BS-SDL). The results are given in Table 1.
Table 1.
Mean area under the ROC curve (AUC) and standard deviation for 100 permutations of 50% training and 50% testing
| Method | Mean AUC | St.Dev |
|---|---|---|
| BS-SDL | 0.7131 | 0.0410 |
| sing.lin-SVM | 0.6819 | 0.0509 |
| sing.rbf-SVM | 0.7200 | 0.0492 |
| mult.lin-SVM.naive-concat. | 0.6762 | 0.0527 |
| mult.rbf-SVM.naive-concat. | 0.6744 | 0.0529 |
| mult.ensemble-MI-SVM | 0.6735 | 0.0516 |
As it can be seen, our method performs comparably well to some of the state of art methods for both single instance learning and multiple instance learning. Although rbf-SVM has a slightly higher mean AUC than our method, a t-test has shown that the difference is not significant. In addition, the contribution of our work is that BS-SDL discriminates as well as rbf-SVM in a dimension much lower (60) than the input dimension of rbf-SVM (1000). This shows that our model effectively performs feature selection. Furthermore, the block dictionaries learned shed insight into how the data clusters.
Clustering Results
Although we did not have clinical labels to assess the quality of clustering, inspecting the loading coefficients learned for our training samples demonstrates that enforcing structured sparsity indeed does cluster the data in some way. In figure 2, the block sparsity of the loading coefficients C can be observed along with some of the atoms that comprise the corresponding dictionary blocks. We intend to do further analysis in future work to assess the interpretation of this clustering action.
Fig. 2.
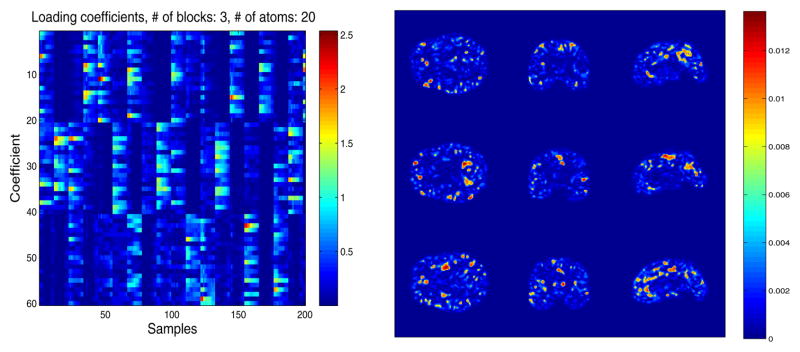
Left: The learned block sparse loading coefficients C, Right: Most utilized gray matter atoms for each block (rows) of the learned dictionary D. Note: Images are absolute values for visual clarity
4 Discussion
In this work, we have presented a novel dictionary learning method that is both discriminative and has clustering capabilities. We achieved this by promoting formation of independent blocks within the learned dictionary that only operate on a subset of data. In addition, adaptively penalizing the block sparsity of the loading coefficients by the classification loss allows the formation of discriminative dictionary blocks. We used this methodology to address an important issue in computational anatomy: the selection of pre-processing parameters. In particular, our method allows to circumvent the potentially biased pre-processing parameter selection step by being able to operate on multiple instances generated by pre-processing the data under various parameter settings. It is of importance to note that although we demonstrate the effectiveness of our model using instances obtained by registrations at varying levels of smoothness, our framework is able to handle multiple instances generated by other processes such as segmentation, selection of varying registration templates or even selection of different registration algorithms. Another advantage of our work is that it is not limited to only multiple instanced or single instanced data as it can readily perform classification on both. In the future, we would like validate our method on additional datasets where heterogeneity is more evident to assess our clustering performance. In conclusion, perhaps the most significant strength of our model is that it is competitive in terms in classification performance with respect to some state of the art methods. Furthermore, we achieve this performance using far fewer input dimensions due to dictionary construction.
References
- 1.Andrews S, Tsochantaridis I, Hofmann T. Support vector machines for multiple-instance learning. Advances in Neural Information Processing Systems. 2003:561–568. [Google Scholar]
- 2.Ashburner J, Friston KJ. Voxel-based morphometry - the methods. Neuroimage. 2000;11(6):805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- 3.Batmanghelich NK, Taskar B, Davatzikos C. Generative-discriminative basis learning for medical imaging. IEEE Transactions on Medical Imaging. 2012;31(1):51–69. doi: 10.1109/TMI.2011.2162961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davatzikos C, Genc A, Xu D, Resnick SM. Voxel-based morphometry using the ravens maps: methods and validation using simulated longitudinal atrophy. NeuroImage. 2001;14(6):1361–1369. doi: 10.1006/nimg.2001.0937. [DOI] [PubMed] [Google Scholar]
- 5.Eldar YC, Kuppinger P, Bolcskei H. Block-sparse signals: Uncertainty relations and efficient recovery. IEEE Transactions on Signal Processing. 2010;58(6):3042–3054. [Google Scholar]
- 6.Elhamifar E, Vidal R. Sparse subspace clustering: Algorithm, theory, and applications. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2013;35(11):2765–2781. doi: 10.1109/TPAMI.2013.57. [DOI] [PubMed] [Google Scholar]
- 7.Fan Y, Shen D, Gur RC, Gur RE, Davatzikos C. Compare: classification of morphological patterns using adaptive regional elements. IEEE Transactions on Medical Imaging. 2007;26(1):93–105. doi: 10.1109/TMI.2006.886812. [DOI] [PubMed] [Google Scholar]
- 8.Halko N, Martinsson PG, Tropp JA. Finding structure with randomness: Probabilistic algorithms for constructing approximate matrix decompositions. SIAM Review. 2011;53(2):217–288. [Google Scholar]
- 9.Mairal J, Bach F, Ponce J. Task-driven dictionary learning. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2012;34(4) doi: 10.1109/TPAMI.2011.156. [DOI] [PubMed] [Google Scholar]
- 10.Ou Y, Sotiras A, Paragios N, Davatzikos C. Dramms: Deformable registration via attribute matching and mutual-saliency weighting. Medical Image Analysis. 2011;15(4):622–639. doi: 10.1016/j.media.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Varol E, Gaonkar B, Davatzikos C. Classifying medical images using morphological appearance manifolds. 2013 IEEE 10th International Symposium on Biomedical Imaging (ISBI); IEEE; 2013. pp. 744–747. [DOI] [PMC free article] [PubMed] [Google Scholar]