Abstract
MicroRNAs (miRs, miRNAs) play central roles in gene regulation. Previously, we reported that miRNAs from somatic cell content, and handling by consumers on the degradation of miRNAs in milk; we also quantified miRNAs in dairy products. Pasteurization and homogenization caused a 63% loss of miR-200c, whereas a 67% loss observed for miR-29b was statistically significant only in skim milk. Effects of cold storage and somatic cell content were quantitatively minor (<2% loss). Heating in the microwave caused a 40% loss of miR-29b but no loss of miR-200c. The milk fat content had no effect on miRNA stability during storage and microwave heating. The concentrations of miRNAs in dairy products were considerably lower than in store-bought milk. We conclude that processing of milk by dairies and handling by consumers causes a significant loss of miRNAs.
Keywords: heating, miRNAs, milk, processing, storage
INTRODUCTION
MicroRNAs (miRs, miRNAs) are small non-coding RNAs that play essential roles in the regulation of genes at the posttranscriptional level in plants and animals.1 Mature miRNAs are about 22 nucleotides long and bind to complementary sequences in the 3′-untranslated region of mRNAs. Perfect or near perfect base pairing of the miRNAs and its target mRNAs typically results in mRNA degradation, whereas less perfect base pairing typically results in inhibition of mRNA translation.2,3 Traditionally, miRNAs have been considered endogenous regulators of genes; i.e., miRNAs synthesized by a given host regulate the expression of genes in that host. Recently, our laboratory refuted this paradigm. We provided strong evidence that (1) humans absorb biologically meaningful amounts of miRNAs from nutritionally relevant doses of cow's milk, (2) physiological concentrations of milk miRNAs affect human gene expression in vivo and in cell cultures, and (3) endogenous synthesis of miRNAs does not compensate for dietary miRNA deficiency in mice.4 Our discoveries were largely modeled on miR-29b and miR-200c but likely hold true for all miRNAs encapsulated in milk exosomes.5,6 To the best of our knowledge, our previous paper is the first to provide unambiguous evidence that miRNAs can be transferred between distinct species through dietary means. In contrast, previous claims that miRNAs from plants affect human gene expression7,8 are highly controversial and were met with skepticism by the scientific community.4,9–12 On the basis of the above observations, milk miRNAs are a novel class of bioactive food compounds as defined by the National Cancer Institute in the United States.13 The discovery that milk miRNAs are bioactive food compounds has broader implications because miRNAs play essential roles in gene regulation,2,3 cell communication,14,15 and human health.16–23
This study focused on determining the effect of milk processing, storage, somatic cell content, and handling by consumers on two miRNA, miR-29b and miR-200c, levels based on the following rationale. In bovine milk, miR-29b and miR-200c are among the most abundant miRNAs.20 miR-29b is an important regulator of bone mineralization in humans, because it increases osteoblast differentiation16 and decreases osteoclast differentiation and function.17 miR-200c decreases cancer risk by targeting the transcription factor ZEB1, which induces E-cadherin expression, thereby limiting epithelial-to-mesenchymal transition, a key event in metastasis.24,25 Also, the nucleotide sequences of miR-29b and miR-200c in bovine milk are identical to those of their human orthologues.26 Our rationale for including the somatic cell count in our analysis was to assess whether an increase in milk cells, as seen in mastitis, might be a confounder in the analysis of milk miRNAs.
In Western societies, the majority of milk is processed prior to consumption. In fact, the production and sale of raw milk dairy products is illegal in many states in the United States, and pasteurization is required.27 Moreover, while the per capita consumption of milk has declined from 236 pounds in 1982 to 195 pounds in 2012, total dairy consumption increased by 11% during the same time period.28 Therefore, we considered it worthwhile to assess the effects of processing on the miRNA content in both milk and dairy products.
Little is known about the effects of processing and storage on milk miRNA levels. In two studies, synthetic miRNAs were added to bovine milk, and their stability after exposure to harsh treatments, such as acid and RNase, was assessed and compared to the stability of endogenous miRNAs in milk.6,20 Synthetic miRNAs were rapidly degraded, whereas endogenous miRNAs were resistant to treatment. However, the harsh treatments applied in these studies are not representative of the treatments applied in commercial dairy production. In this study, we assessed the effects of pasteurization, fat content, cold storage, heating, and processing into dairy products on the content of milk miRNAs.
MATERIALS AND METHODS
Chemicals
Guanidinium thiocyanate and ethanol were purchased for use in the NucleoSpin miRNA plasma RNA extraction kit (Macherey-Nagel, Inc., Bethlehem, PA). TRIzol was purchased from Life Technologies (Grand Island, NY).
Milk and Dairy Products
Raw, whole, 2%, and skim cow's (Bos taurus) milk was obtained from the Pennsylvania State University Creamery (University Park, PA) from separate collections in three consecutive weeks in May 2014. A diagram illustrating the preparation of whole cow's milk is presented in Figure 1. Briefly, the milk was procured from the Penn State Animal Science Department's Holsteins breed herd, using raw milk and cream routinely supplied to the Penn State Berkey Creamery, University Park, PA. The milk was stored under intermittent agitation in a 22 712.5 L raw milk silo at 2.2 °C (Feldmeier Equipment, E-015-05, Little Falls, NY). The milk contained 3.25% fat and 12.15% non-fat solids (near-infrared method, CEM, Turbo Smart5, model 907990, Matthews, NC) and was pasteurized at 75.55 °C with a 28 s holding time (APV Paraflow, serial number 20053003000302, Goldsboro, NC). The milk was homogenized at 145 bar and 60 °C (APV Gaulin homogenizer, serial number 20052410702, Lake Mills, WI) and standardized using a Westfalia separator (MSE 55-01-177, Oelde, Germany), thereby producing whole milk (3.25% fat and skim milk). The 2% fat milk was prepared by mixing 3.25% milk and skim milk. After cooling, the product was transferred to a 7200 L refrigerated storage tank at 2.2 °C (Feldmeier Equipment, E-015-05, Little Falls, NY). The product was bottled on a filling machine (Federal Manufacturing, serial number 1/12.4GL843, Milwaukee, WI) and stored in a conventional cold-milk warehouse at 3.0 °C. On our initial collection dates, milk of all fat levels were stored at 4 °C for up to 15 days, and aliquots were taken and frozen at −80 °C every other day. In a separate experiment, bovine cells were removed from raw milk by centrifugation (500g for 10 min at 4 °C) to determine whether somatic cells are a meaningful confounder when analyzing the concentrations of miRNAs in milk from healthy cows. Samples were frozen at −80 °C and shipped on dry ice to Lincoln, NE, for miRNA analysis. Samples from all fat levels of milk on day 15 were heated in the microwave for 15 s and analyzed after cooling off to room temperature. Dairy products other than milk were purchased from grocery stores in Lincoln, NE. All samples were produced and analyzed as biological repeats in triplicate.
Figure 1.
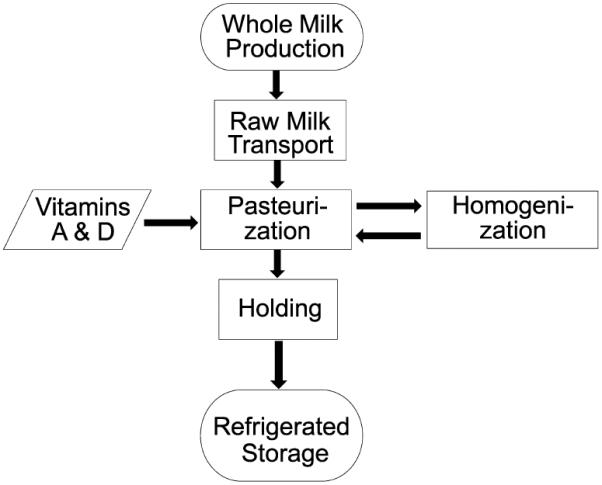
Schematic presentation of milk processing.
miRNA Analysis
Milk samples were spiked with a synthetic internal standard (25 attomoles) prior to extraction of miRNAs using miSPIKE synthetic RNA (IDT Technologies).4 Dairy products (100 mg) other than milk were extracted using TRIzol prior to the addition of the synthetic internal standard. miR-29b and miR-200c were quantified using quantitative real-time polymerase chain reaction (PCR) as described previously.4
Statistics
Analysis by Bartlett's test homogeneity suggested that variances were homogeneous.29 The paired t test was used for pairwise comparisons. One-way analysis of variance (ANOVA) and Fisher's protected least significant differences were used when comparing more than two groups. Repeated measures ANOVA was used for assessing the effects of storage time on the miRNA concentration. StatView 5.0.1 (SAS Institute, Cary, NC) was used for conducting statistical analyses. Means ± standard deviation (SD) are reported. Differences were considered statistically significant if p ≤ 0.05.
RESULTS
Pasteurization and homogenization of raw milk resulted in a 63 ± 28% decrease of miR-200c in whole milk; effects were similar for 2% fat milk and skim milk (Figure 2A). The effect was pronounced for miR-29b, for which a significant decrease (67 ±less 18%) was observed only in skim milk (Figure 2B). Cold storage of milk did not affect the concentration of mir-29b and miR-200c in whole milk, 2% milk, and skim milk up to 15 days; 2% fat milk is shown as a representative example in Figure 3. Somatic cells are not meaningful confounds regarding the analysis of miRNAs in milk from healthy cows. When somatic cells were removed from raw milk by centrifugation and analyzed for miRNA content, the cellular miRNAs were found to contribute less than 2% of the total miRNAs present in raw milk before centrifugation: 1.1 ± 0.9% for miR-29b and 0.14 ± 0.08% for miR-200c.
Figure 2.
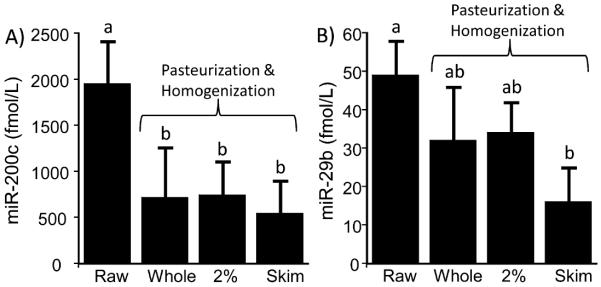
Loss of (A) miR-200c and (B) miR-29b during milk pasteurization and homogenization of milk with different fat contents. (a and b) Significantly different (n = 3 biological replicates; p < 0.05).
Figure 3.
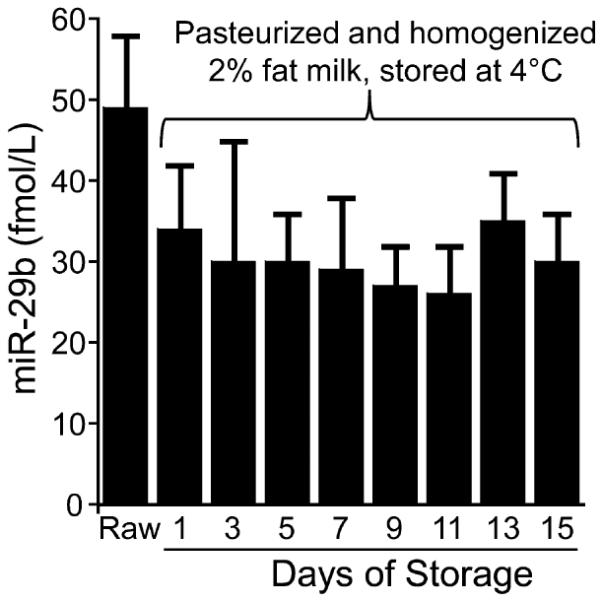
Storage at 4 °C did not affect the concentrations of miR-29b in pasteurized and homogenized 2% fat milk (n = 3 biological replicates; p > 0.05).
Processing in the household has the potential to cause a considerable loss of some miRNAs in milk. For example, the concentration of miR-29b decreased by 40 ± 28% when processed milk was heated in the microwave and cooled to room temperature compared to milk before heating (Figure 4). In contrast, when milk was heated in the microwave, the concentration of miR-200c was not statistically different compared to unheated controls. The concentrations of miRNAs varied considerably among the dairy products tested (Table 1) but were generally lower than the concentrations in pasteurized whole milk (compare to Figure 2). Fresco queso dip was a notable exception and contained higher concentrations of miRNAs than those observed in milk.
Figure 4.
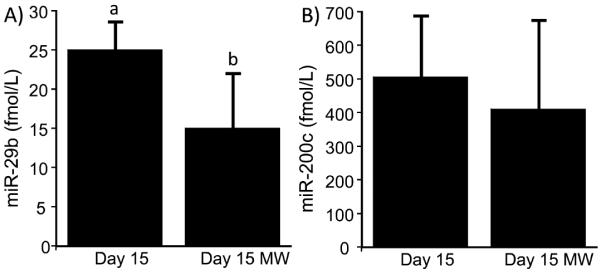
Loss of (A) miR-29b and (B) miR-200c during heating of whole milk in the microwave after 15 days of storage at 4 °C. Abbreviation: MW, microwaved. (a and b) Significantly different (n = 3 biological replicates; p < 0.05).
Table 1.
Concentration of miRNAs, miR-29b and miR-200c, in Dairy Productsa
| miRNA |
||
|---|---|---|
| product | miR-29b (fmol/kg) | miR-200c (fmol/kg) |
| Best Choice yogurt | 0.9 ± 0.10 | 37.6 ± 2.8 |
| Fresco queso dip | 36.1 ± 5.5 | 1029.8 ± 478.6 |
| Greek yogurt | 14.2 ± 3.9 | 462.3 ± 126.9 |
| half and half | 3.0 ± 0.17 | 513.3 ± 159.2 |
| heavy whip cream | 2.6 ± 1.3 | 342.0 ± 132.9 |
| parmesan cheese | 4.9 ± 1.9 | 232.0 ± 64.5 |
| Upstate Farm yogurt | 2.4 ± 1.0 | 216.9 ± 93.8 |
Data are means ± SD; n = 3.
DISCUSSION
In a recent paper, we reported the importance of milk miRNAs for gene regulation in humans.4 That report has major implications for the roles of milk and possibly other dairy products in human health. Cow's milk contains meaningful quantities of 245 miRNAs,20,30 and 71.4% of these miRNAs are predicted to target about 11 000 human transcripts (unpublished observations). In addition to the roles of miR-29b and miR-200c in bone health and cancer prevention,16,17,24,25 respectively, miRNAs have been implicated in various aspects of human health and disease, including hypertension, insulin resistance and diabetes, hyperlipidemia and atherosclerosis, reproduction, immune function, and Crohn's disease.18,20,21,31–33
We propose that milk has a meaningful effect on human health, mediated by miRNA-dependent gene regulation. The potential importance of dietary milk miRNA intake is supported by data suggesting that (1) Americans consume large quantities of milk and dairy products,28 (2) a large proportion of milk miRNAs is encapsulated in extracellular vesicles, thereby providing protection against degradation5,6 and a pathway for cellular uptake by endocytosis,34,35 and (3) milk miRNAs are resistant against degradation during storage (this study).
Our previous studies of milk miRNAs in humans and mice were conducted using 1% fat milk from the grocery store.4 On the basis of this study, the content of miRNAs is about 2 times higher in unprocessed milk compared to pasteurized, store- bought milk. Note that we have no intent recommending the consumption of raw cow's milk by humans, because of food safety concerns associated with raw milk. We observed that a loss of milk miRNAs occurred only during pasteurization, homogenization, and processing to dairy products other than milk. This observation is consistent with previous studies of milk miRNAs. For example, endogenous miRNAs were not degraded when milk was exposed to harsh treatments, such as low pH or treatment with RNase.6,20 It is reasonable to propose that encapsulation of miRNAs in extracellular vesicles5 prevents miRNA degradation based on the following lines of evidence: (1) When synthetic miRNAs are added to milk and subjected to low pH or RNase treatment, the miRNAs are rapidly degraded.6,20 (2) When exosome membranes in milk were disrupted by sonication for preparing miRNA-depleted mouse diets in previous studies, miR-29b was rapidly degraded to concentrations below the detection limit.4 Presumably, degradation was due to milk RNases gaining access to miRNAs released from exosomes. (3) When milk was fermented to produce yogurt, miRNA concentrations decreased to levels much lower than in milk (this study). We speculate that the decrease was due to the lysis of exosomes during fermentation and the large amounts of RNases produced by microbes. (4) When milk was homogenized, miRNA concentrations decreased by on average 50% (this study). We speculate that the decrease was caused by a disruption of exosome mebranes by shear forces applied during homogenization. Collectively, our studies suggest that milk and perhaps dairy products have the potential to contribute to the miRNA body pool in humans.
Some uncertainties remain to be addressed in future studies. For example, this study was modeled on the basis of miR-29b and miR-200c; however, there is a possibility that distinct miRNAs may be differentially metabolized.8,36 Another layer of uncertainty is the possible effects of feeding regimens, season, and breed on the miRNA content in milk. Moreover, while this study suggests that somatic cells in milk from healthy cows do not contribute meaningful amounts to the total miRNA content in milk, it is possible that the increased somatic cell count in milk from cows suffering from mastitis37 may cause an artificial increase in milk miRNA concentrations. Our previous studies suggest that plasma miRNA concentrations decrease by 61% in mice fed a milk miRNA-depleted diet for 4 weeks. This observation is consistent with milk miRNAs contributing meaningful quantities to the miRNA body pool but does not necessarily establish the essentiality of dietary miRNA intake. Clearly, this is an uncertainty that will need to be addressed in future studies. Finally, it is conceivable that miRNAs from foods other than milk also contribute toward the total body pool of miRNAs.
There is precedent for degradation of milk compounds during processing. Examples include the thermal and non thermal degradation of modified peptides38 and DNA encapsulated in silica.39 The immediate relevance of these previous publications for the stability of miRNAs in milk is uncertain.
ACKNOWLEDGMENTS
The authors thank Deanna Jane Bartos and Abigail Sido for help with milk sample collection and processing.
Funding Support was provided by National Institutes of Health (NIH) Grant P20GM104320, National Research Initiative Grant 2014-06605, the Gerber Foundation (to Janos Zempleni), the National Research Initiative Grant 2009-55200-05197 from the United States Department of Agriculture (USDA) National Institute for Food and Agriculture, and startup funds from the College of Agricultural Sciences, Penn State University (to Jairam Vanamala).
ABBREVIATIONS USED
- miR
microRNA
- miRNA
microRNA
Footnotes
The authors declare no competing financial interest.
REFERENCES
- (1).Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nat. Rev. Mol. Cell Biol. 2013;14:475–488. doi: 10.1038/nrm3611. [DOI] [PubMed] [Google Scholar]
- (2).Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J, Di Padova F, Lin SC, Gram H, Han J. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- (3).Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336:237–240. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Baier SR, Nguyen C, Xie F, Wood JR, Zempleni J. MicroRNAs are absorbed in biologically meaningful amounts from nutritionally relevant doses of cow's milk and affect gene expression in peripheral blood mononuclear cells, HEK-293 kidney cell cultures, and mouse livers. J. Nutr. 2014;144:1495–1500. doi: 10.3945/jn.114.196436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Zhou Q, Li M, Wang X, Li Q, Wang T, Zhu Q, Zhou X, Wang X, Gao X, Li X. Immune-related microRNAs are abundant in breast milk exosomes. Int. J. Biol. Sci. 2012;8:118–123. doi: 10.7150/ijbs.8.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Kosaka N, Izumi H, Sekine K, Ochiya T. microRNA as a new immune-regulatory agent in breast milk. Silence. 2010;1:7. doi: 10.1186/1758-907X-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Zhang L, Hou D, Chen X, Li D, Zhu L, Zhang Y, Li J, Bian Z, Liang X, Cai X, Yin Y, Wang C, Zhang T, Zhu D, Zhang D, Xu J, Chen Q, Ba Y, Liu J, Wang Q, Chen J, Wang J, Wang M, Zhang Q, Zhang J, Zen K, Zhang CY. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: Evidence of cross-kingdom regulation by microRNA. Cell Res. 2012;22:107–126. doi: 10.1038/cr.2011.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Zhou Z, Li X, Liu J, Dong L, Chen Q, Liu J, Kong H, Zhang Q, Qi X, Hou D, Zhang L, Zhang G, Liu Y, Zhang Y, Li J, Wang J, Chen X, Wang H, Zhang J, Chen H, Zen K, Zhang CY. Honeysuckle-encoded atypical microRNA2911 directly targets influenza A viruses. Cell Res. 2015;25:39–49. doi: 10.1038/cr.2014.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Snow JW, Hale AE, Isaacs SK, Baggish AL, Chan SY. Ineffective delivery of diet-derived microRNAs to recipient animal organisms. RNA Biol. 2013;10:1107–1116. doi: 10.4161/rna.24909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Dickinson B, Zhang Y, Petrick JS, Heck G, Ivashuta S, Marshall WS. Lack of detectable oral bioavailability of plant microRNAs after feeding in mice. Nat. Biotechnol. 2013;31:965–967. doi: 10.1038/nbt.2737. [DOI] [PubMed] [Google Scholar]
- (11).Chen X, Zen K, Zhang CY. Reply to lack of detectable oral bioavailability of plant microRNAs after feeding in mice. Nat. Biotechnol. 2013;31:967–969. doi: 10.1038/nbt.2741. [DOI] [PubMed] [Google Scholar]
- (12).Wang K, Li H, Yuan Y, Etheridge A, Zhou Y, Huang D, Wilmes P, Galas D. The complex exogenous RNA spectra in human plasma: An interface with human gut biota? PLoS One. 2012;7:e51009. doi: 10.1371/journal.pone.0051009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).National Cancer Institute (NCI) [accessed July 6, 2014];NCI Dictionary of Cancer Terms. http://www.cancer.gov/dictionary?cdrid=703278.
- (14).Turchinovich A, Samatov TR, Tonevitsky AG, Burwinkel B. Circulating miRNAs: Cell—cell communication function? Front. Genet. 2013;4:119. doi: 10.3389/fgene.2013.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Xu L, Yang BF, Ai J. MicroRNA transport: A new way in cell communication. J. Cell. Physiol. 2013;228:1713–1719. doi: 10.1002/jcp.24344. [DOI] [PubMed] [Google Scholar]
- (16).Li Z, Hassan MQ, Jafferji M, Aqeilan RI, Garzon R, Croce CM, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J. Biol. Chem. 2009;284:15676–15684. doi: 10.1074/jbc.M809787200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Rossi M, Pitari MR, Amodio N, Di Martino MT, Conforti F, Leone E, Botta C, Paolino FM, Del Giudice T, Iuliano E, Caraglia M, Ferrarini M, Giordano A, Tagliaferri P, Tassone P. miR-29b negatively regulates human osteoclastic cell differentiation and function: Implications for the treatment of multiple myeloma-related bone disease. J. Cell. Physiol. 2013;228:1506–1515. doi: 10.1002/jcp.24306. [DOI] [PubMed] [Google Scholar]
- (18).Nothnick WB. The role of micro-RNAs in the female reproductive tract. Reproduction. 2012;143:559–576. doi: 10.1530/REP-11-0240. [DOI] [PubMed] [Google Scholar]
- (19).Abu-Halima M, Hammadeh M, Schmitt J, Leidinger P, Keller A, Meese E, Backes C. Altered microRNA expression profiles of human spermatozoa in patients with different spermatogenic impairments. Fertil. Steril. 2013;99:1249–1255. doi: 10.1016/j.fertnstert.2012.11.054. [DOI] [PubMed] [Google Scholar]
- (20).Izumi H, Kosaka N, Shimizu T, Sekine K, Ochiya T, Takase M. Bovine milk contains microRNA and messenger RNA that are stable under degradative conditions. J. Dairy Sci. 2012;95:4831–4841. doi: 10.3168/jds.2012-5489. [DOI] [PubMed] [Google Scholar]
- (21).Arnold CN, Pirie E, Dosenovic P, McInerney GM, Xia Y, Wang N, Li X, Siggs OM, Karlsson Hedestam GB, Beutler B. A forward genetic screen reveals roles for Nfkbid, Zeb1, and Ruvbl2 in humoral immunity. Proc. Natl. Acad. Sci. U. S. A. 2012;109:12286–12293. doi: 10.1073/pnas.1209134109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Nijhuis A, Biancheri P, Lewis A, Bishop CL, Giuffrida P, Chan C, Feakins R, Poulsom R, Di Sabatino A, Corazza GR, MacDonald TT, Lindsay JO, Silver AR. In Crohn's disease fibrosis-reduced expression of the miR-29 family enhances collagen expression in intestinal fibroblasts. Clin. Sci. 2014;127:341–350. doi: 10.1042/CS20140048. [DOI] [PubMed] [Google Scholar]
- (23).Amodio N, Di Martino MT, Foresta U, Leone E, Lionetti M, Leotta M, Gulla AM, Pitari MR, Conforti F, Rossi M, Agosti V, Fulciniti M, Misso G, Morabito F, Ferrarini M, Neri A, Caraglia M, Munshi NC, Anderson KC, Tagliaferri P, Tassone P. miR-29b sensitizes multiple myeloma cells to bortezomib-induced apoptosis through the activation of a feedback loop with the transcription factor Sp1. Cell Death Dis. 2012;3:e436. doi: 10.1038/cddis.2012.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J. Biol. Chem. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9(6):582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Langer AJ, Ayers T, Grass J, Lynch M, Angulo FJ, Mahon BE. Nonpasteurized dairy products, disease outbreaks, and state laws—United States, 1993–2006. Emerging Infect. Dis. 2012;18:385–391. doi: 10.3201/eid1803.111370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).United States Department of Agriculture Economic Research Service [accessed Sept 25, 2013];Dairy Data. http://www.ers.usda.gov/data-products/dairy-data.aspx.
- (29).Abacus Concepts, Inc. StatView. Abacus Concepts, Inc.; Berkeley, CA: 1996. [Google Scholar]
- (30).Chen X, Gao C, Li H, Huang L, Sun Q, Dong Y, Tian C, Gao S, Dong H, Guan D, Hu X, Zhao S, Li L, Zhu L, Yan Q, Zhang J, Zen K, Zhang CY. Identification and characterization of microRNAs in raw milk during different periods of lactation, commercial fluid, and powdered milk products. Cell Res. 2010;20:1128–1137. doi: 10.1038/cr.2010.80. [DOI] [PubMed] [Google Scholar]
- (31).Li S, Zhu J, Zhang W, Chen Y, Zhang K, Popescu LM, Ma X, Lau WB, Rong R, Yu X, Wang B, Li Y, Xiao C, Zhang M, Wang S, Yu L, Chen AF, Yang X, Cai J. Signature microRNA expression profile of essential hypertension and its novel link to human cytomegalovirus infection. Circulation. 2011;124:175–184. doi: 10.1161/CIRCULATIONAHA.110.012237. [DOI] [PubMed] [Google Scholar]
- (32).Fernandez-Valverde SL, Taft RJ, Mattick JS. MicroRNAs in β-cell biology, insulin resistance, diabetes and its complications. Diabetes. 2011;60(7):1825–1831. doi: 10.2337/db11-0171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Soh J, Iqbal J, Queiroz J, Fernandez-Hernando C, Hussain MM. MicroRNA-30c reduces hyperlipidemia and atherosclerosis in mice by decreasing lipid synthesis and lipoprotein secretion. Nat. Med. 2013;19:892–900. doi: 10.1038/nm.3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Feng D, Zhao WL, Ye YY, Bai XC, Liu RQ, Chang LF, Zhou Q, Sui SF. Cellular internalization of exosomes occurs through phagocytosis. Traffic. 2010;11:675–687. doi: 10.1111/j.1600-0854.2010.01041.x. [DOI] [PubMed] [Google Scholar]
- (35).Escrevente C, Keller S, Altevogt P, Costa J. Interaction and uptake of exosomes by ovarian cancer cells. BMC Cancer. 2011;11:108. doi: 10.1186/1471-2407-11-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Squadrito ML, Baer C, Burdet F, Maderna C, Gilfillan GD, Lyle R, Ibberson M, De Palma M. Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell Rep. 2014;8:1432–1446. doi: 10.1016/j.celrep.2014.07.035. [DOI] [PubMed] [Google Scholar]
- (37).Dohoo IR, Meek AH. Somatic cell counts in bovine milk. Can. Vet. J. 1982;23:119–125. [PMC free article] [PubMed] [Google Scholar]
- (38).Meltretter J, Wust J, Pischetsrieder M. Modified peptides as indicators for thermal and nonthermal reactions in processed milk. J. Agric. Food Chem. 2014;62:10903–10915. doi: 10.1021/jf503664y. [DOI] [PubMed] [Google Scholar]
- (39).Bloch MS, Paunescu D, Stoessel PR, Mora CA, Stark WJ, Grass RN. Labeling milk along its production chain with DNA encapsulated in silica. J. Agric. Food Chem. 2014;62:10615–10620. doi: 10.1021/jf503413f. [DOI] [PubMed] [Google Scholar]


