Summary
Sequencing costs are falling, but the cost of data analysis remains high, often because unforeseen problems arise, such as insufficient depth of sequencing or batch effects. Experimenting with data analysis methods during the planning phase of an experiment can reveal unanticipated problems and build valuable bioinformatics expertise in the organism or process being studied. This protocol describes using R Markdown and RStudio, user-friendly tools for statistical analysis and reproducible research in bioinformatics, to analyze and document analysis of an example RNA-Seq data set from tomato pollen undergoing chronic heat stress. Also, we show how to use Integrated Genome Browser to visualize read coverage graphs for differentially expressed genes. Applying the protocol described here and using the provided data sets represent a useful first step toward building RNA-Seq data analysis expertise in a research group.
Keywords: Integrated Genome Browser, tomato, pollen, visualization, RNA-Seq, R, differential gene expression, edgeR
1. Introduction
The term “RNA-Seq” means using sequencing platforms, typically Illumina, to produce millions of short cDNA sequences at low cost. The method resembles old-style EST library sequencing and involves similar data processing methods, but the low cost allows multiple samples to be sequenced and thus permits statistical assessment of gene expression. Because the data are sequences, rather than hybridization intensities as with microarrays, they can also elucidate aspects of gene structures, including position of introns, exons, transcription start sites, and polyadenylation sites. Using RNA-Seq, it is possible to determine not only whether a treatment or condition affects overall RNA abundance but also whether it affects splicing patterns, transcription start site use, and many other aspects of RNA transcription and posttranscriptional processing.
The RNA-Seq technique was introduced formally in the literature in 2008 with three articles published in May and July [1-3]. Of these, the best cited is Mortazavi et al. from the laboratory of Barbara Wold at Caltech [2]. This article introduced the concept of RPKM as a measure of gene expression, where RPKM refers to the number of reads per million per kilobase of transcript obtained from an expressed gene. It compared RNA-Seq to microarrays as a method for measuring gene expression and established that RNA-Seq has greater dynamic range and sensitivity than oligo-based arrays from Affymetrix, a leading brand of expression microarrays. As described in a mini-review of the paper that appeared in the same journal issue [4], this article heralded “the beginning of the end for microarrays” as a method for measuring gene expression on a whole-genome scale.
Six years later, the prediction of expression microarrays’ demise seems to be coming true, thanks to decreasing sequencing cost and increasing read lengths. In some ways, this is unfortunate, because methods for statistical analysis of expression microarrays are well developed, and there are now many excellent and well-tested software packages that implement these methods. Thanks to these tools, it is easier than ever to analyze a microarray data set. Fortunately, many of these same tools and methods are being adapted to RNA-Seq data analysis. One such tool we will introduce in this protocol is edgeR [5], which is implemented in R and is part of Bioconductor, a large collection of R libraries designed for analysis of biological data sets. EdgeR is developed and supported by groups led by Mark Robinson and Gordon Smyth, who also developed the limma (linear analysis of microarrays) package for R. One benefit of edgeR is its documentation; the User’s Guide contains many example analyses, including a case study from Arabidopsis illustrating how to detect differential expression in the face of batch effects using edgeR’s generalized linear modeling (GLM) functions. For simplicity, we will not describe GLM functions here, and instead show a simpler analysis that uses edgeR’s “classic” method for testing differential expression in a single-factor experiment. We mention the GLM functions here mainly to encourage readers to explore the User’s Guide and other materials from the edgeR authors [6, 7].
Thanks to reduced cost of sequencing and library preparation, it is now possible to conduct a well-replicated RNA-Seq study for less than a few thousand dollars. However, if unforeseen problems arise, such as insufficient sequencing depth or batch effects, the cost and time required for analysis can escalate, ultimately far exceeding that of the original experiment. Running a “mock” analysis using a well documented, published data analysis from start to finish is often a good way to learn the limitations and strengths of analysis methods, which helps to plan an experiment. Toward this end, this article describes a straightforward analysis of an RNA-Seq data set from tomato, using materials developed for the UNC Charlotte 2014 Workshop on Next-Generation Sequencing (WiNGS). For the workshop, we developed hands-on computer labs introducing experimental design, data processing, data analysis, and biological interpretation for RNA-Seq expression experiments. Here, we present a protocol for RNA-Seq data analysis taken from the workshop, focusing on data analysis using edgeR and visualization of RNA-Seq expression data using Integrated Genome Browser, a highly interactive, flexible genome visualization tool [8]. Materials presented here are freely available online at http://bitbucket.org/lorainelab/tomatopollen and http://bitly.com/rnaseq2014. Sequence data are available from the Short Read Archive under accession SRP055068.
2. Materials
2.1. RNA-Seq data sets
Descriptions of pollen samples used for RNA-Seq are available online at Bit Bucket git repository http://www.bitbucket.org/lorainelab/tomatopollen. Briefly, cDNA libraries for RNA-Seq were prepared from tomato pollen collected from plants undergoing long-term, nonlethal heat stress or from control tomato plants grown at optimal temperature. Tomato plant cultivation, application of heat stress, and collection of pollen were done in the laboratory of Nurit Firon. Pollen was collected from treatment and control groups in batches on different dates; each batch included one sample of pollen pooled from several control plants and a second sample of pollen pooled from several heat-treated plants. Libraries were prepared from RNA extracted from pollen samples and sequenced on a HiSeq instrument using paired end, 69-cycle sequencing. To prepare for the workshop, we aligned the sequence reads onto version 2.5 of the Solanum lycopersicum genome using the tophat2 program. Following this, we used the feature Counts program to generate a plain text, tab-delimited file listing the number of overlapping fragments (read pairs) for each gene in each sample. An example script (align.sh) that shows how to run these programs is available online from http://www.bitly.com/rnaseq2014 in a subfolder named Alignments.
2.2. Software
To follow the protocol presented here, download and install a copy of the RStudio software from http://www.rstudio.org. Launch RStudio and use the Tools > Install Packages… menu to install knitr. See Note 1 on knitr. Obtain a copy of the workshop R and R Markdown code from the workshop bitbucket repository at http://www.bitbucket.org/lorainelab/tomatopollen. Either use git to clone a copy on your computer or download the repository as a “zip” file using the Download links on the site. If downloading the “zip” file, double-click the file icon to unpack it or use the “unzip” (or related) command-line utility.
3. Methods
3.1. Read counts table into RStudio
Open the Differential Expression project in RStudio. Launch RStudio and Choose File > Open Project. Navigate to the folder named Differential Expression in the unzipped tomatopollen folder. Open file Differential Expression. Rproj, the project file for the differential expression analysis code.
Open the Differential Expression Markdown file. Select the Files tab in RStudio and click the file named Differential Expression. Rmd to open it.
-
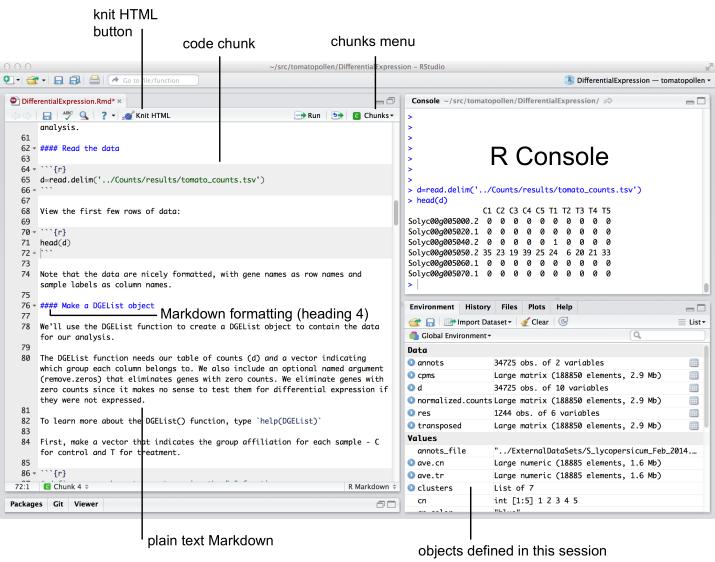
Install edgeR package if necessary. Check to see if edgeR is already installed on your system. Within the RStudio tab that displays the Differential Expression. Rmd file, locate the first “code chunk,” a part of the file RStudio recognizes as containing R code that can be executed in the R console tab (Fig. 1). Place the cursor next to the line “library(edgeR)”, the command that loads the edgeR library into the program. Run the command by clicking the Run button (top right of the tab) or by typing CNTRL-ENTER (Windows) or COMMAND-ENTER (MacOS). Doing this copies the command into the R command-line console tab and runs it. If you have not yet installed the edgeR library, you will see an error stating, “there is no package named edgeR.” In that case, install the library. Type in the R console:
>source (“http://bioconductor.org/biocLite.R”)
> biocLite(“edgeR”)
Next, load the library into the R environment. Enter:
> library(edgeR)
Load the data. Enter the command “d=read.delim(‘../Counts/results/tomato_counts.tsv’)” by typing it into the console. Alternatively, navigate to the section of the code titled “Read the Data” (around line 65) and, as before, position the cursor at that line and click Run. Observe that a new command line prompt (“>”) appears in the console, indicating that the command executed without error. Click the Environment tab and observe that a data object “d” now exists in the environment and contains 34,725 observations (obs.) of ten variables.
View the data. To view the data contained in the new data object (called a “data frame” in R), enter the next command in the file “head(d)” which prints the first six lines of data along with the header (column names) of the data frame object. Observe that there are five columns labeled C1 through C5 corresponding to control samples and five columns labeled T1 through T5 corresponding to treatment samples. The rows are labeled with tomato gene names and the values represent the number of read fragments that overlapped each gene. You can also double-click the variable name “d” in the Environment tab to open a spreadsheet view of the data within your RStudio session.
Figure 1. Example code chunk displayed in RStudio.
Embedded R code (called a “chunk”) is set apart from the rest of the Markdown document using triple back tics, followed by the letter “r” in curly braces. The Knit HTML button or the “Chunks” menu can be used to run all or part of the chunks in a Markdown document. Running “Knit HTML” Optional formatting options (such as figure width and height) can also appear between the curly braces.
3.2. Exploratory data analysis
The next step in an RNA-Seq data analysis is to do exploratory data analysis, which means: visualize and summarize aspects of the data in order to build familiarity with the data, determine overall quality of the data, and identify problems that could complicate further analysis. One common problem is sample label switching; sometimes investigators switch treatment or control samples. Another common problem is batch effects; e.g., the time and date of sample collection can introduce bias into the data. The edgeR, limma, and other expression analysis packages have methods that can detect differentially expressed genes in the face of batch effects, and so it is important to notice any problems when launching an analysis.
Create DGEList object. Run the next command in the file (group=c(‘C’,‘C’,‘C’,‘C’,‘C’,‘T’,‘T’,‘T’,‘T’,‘T’)) which defines vector of character values indicating the sample types (“C” for control and “T” for treatment) in the data frame. Run the next command (dge=DGEList(d,group=group,remove.zeros=TRUE)), which creates the DGEList object named dge from the group vector group and the data frame object d that was created in the preceding section. To eliminate genes with zero counts, use the remove.zeros option (remove.zeros=TRUE in the command line above). The DGEList object is just a container for data already loaded into the environment; the edgeR library methods are designed for operations and analyses on DGEList objects, which is why we need to create one before proceeding to the next steps.
Observe read counts. Run the code in the section titled “Take a moment to look at the DGEList object.” Enter the name of the dge variable into the Console to observe its contents; this causes a summary of object contents to be printed in abbreviated form. The “sample” component lists the number of reads obtained from each library; there were some libraries with small numbers of reads (T2 had the least) and other libraries contained more. This highlights the need to normalize the data to account for sequencing depth before proceeding with differential expression analysis. Read counts indicate the sum of column values listing reads per gene; reads that mapped outside gene regions were not counted.
Normalize expression data stored in the DGEList object. Position the cursor next to the line dge=calc Norm Factors(dge) and run the command. This command applies the calc Norm Factors function to the dge object; it both returns a copy of the dge object while also changing it. That is, it adds new information to the object, which is merely a container for data. In this case, it updated the normalization factors that were stored in the samples component of dge. See Note 2 for discussion of normalization of RNA-Seq expression data.
Review normalization factors. Enter the name of the object (dge) into the console to view its contents as before. Now, the norm.factors column contains new values. Observe there is an approximate relationship between the normalization factors and the number of counts per library, but this relationship is not perfectly linear.
-
Create an MDS plot. Enter the next commands, which show a plot summarizing the variation between samples. Use the Run button or enter the following commands into the R console to run the code and make the plot:
> cn.color=‘blue’
> tr.color=‘brown’
> main=‘MDS Plot for Count Data’
> colors=c(rep(cn.color,5),rep(tr.color,5))
> plotMDS(dge,main=main,labels=colnames(dge$counts),col=colors,las=1)
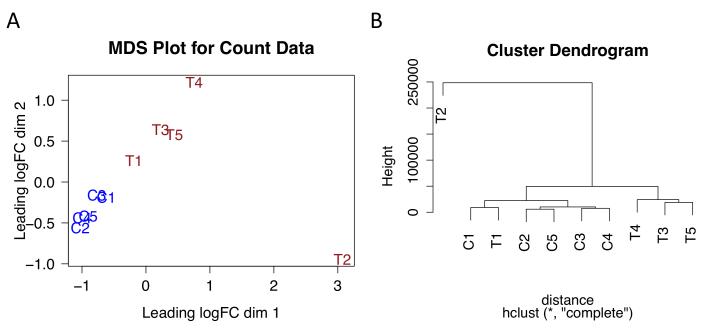
Examine the plot. (See Fig. 2A.) This plot is a multidimensional scaling (MDS) plot. Observe that the treated (T) and control (C) samples are separated along dimension 2 (the y axis), except for sample T2. Sample T2 is separate from the other treatment samples, occupying the right lower corner, and therefore appears to be an outlier. See Note 3 for discussion of MDS plots.
-
View hierarchical clustering plot. Run the code in the next code chunk titled Hierarchical Clustering or enter the following commands into the R console:
> normalized.counts=cpm(dge)
> transposed=t(normalized.counts)
> distance=dist(transposed)
> clusters=hclust(distance)
> plot(clusters)
Examine the plot. This plot shows a clustering dendrogram where samples that are most similar occupy closer positions in the tree, while samples that are less similar are separated by larger numbers of branch points (Fig. 2B). As before, sample T2 occupies a position different from all the others. Also, samples C1 and T1 form a cluster, indicating they are more similar to each other than to the other control or treatment samples. This plot suggests there may be batch effects affecting the first set of samples (C1 and T1) but the other samples are not affected, and also T2 is an outlier. Based on this, it may be wise to repeat our subsequent analysis steps leaving out T2 and/or C1 and T1. Alternatively, we could include a term to cancel out batch effects in our linear modeling in EdgeR. To start, however, we will stick with a simple comparison between treatment and control samples.
Figure 2. MDS and clustering plots showing relationships between sample types.
(A) Multidimensional scaling (MDS) plot. Distance between sample labels indicates similarity. (B) Cluster dendrogram. Number of branches separating samples indicates similarity. Treatment samples are named T1 through T5. Control samples are named C1 through C5.
3.3. Differential expression analysis
Differential expression analysis means identifying genes with RNA levels that were different across experimental groups. Here, we assess RNA levels as the number of reads that overlap the entire gene region. To identify differentially expressed genes, we first estimate variance and then use the variance estimates to determine if the treatment causes a significant change in gene expression.
Estimate dispersion (variance). Execute the next two commands “dge=estimate Common Disp(dge)” and “dge=estimate Tagwise Disp(dge)”, which add estimates of variance to the “dge” object. These steps are required before performing the next step, which uses within-group estimates of variance to determine if a gene’s expression has changed due to the treatment. See Note 4 for more discussion on variance estimation.
Test differential expression. Here, we will employ the simplest method for testing differential gene expression in edgeR, which utilizes a form of Fisher’s exact test, which we can use because the samples were normalized in the previous steps. Move the cursor to the code chunk in the section titled “Gene Expression Analysis” and run the command “dex=exact Test (dge, pair=c(“C”,“T”),dispersion=“tagwise”)”, which creates a DGEExact differential expression results object. The object contains the log (base 2) fold-change and p value result for each gene.
-
Add false discovery rate (FDR) to DGEExact object. When we write the results to a spreadsheet, we will include FDR and not p value. Either execute the next two lines in the file starting in the section titled “Multiple Hypothesis testing correction” or type into the R console:
> fdrvalues=p.adjust(dex$table$PValue, method=‘BH’)
> dex$table$fdr=fdrvalues
-
Evaluate possible FDR cutoffs. Use the “decide Tests DGE” and “summary” functions as shown in the section titled “Picking an FDR cutoff” to determine how many genes were called as differentially expressed (DE) at different FDR cutoffs. Enter:
> summary(decideTestsDGE(dex,p=0.05))
> summary(decideTestsDGE(dex,p=0.01))
> summary(decideTestsDGE(dex,p=0.005))
-
Select a cutoff. Observe that the previous commands print tables listing the number of genes that were not changed (0), upregulated (1), and downregulated (-1) using FDR cutoffs given by the “p” option. Use the results from this to determine an acceptable false discovery rate for the experiment. At FDR of 0.005, there were around 700 downregulated DE genes and around 550 upregulated genes. At FDR 0.005, which means we expect that around 5 in 1,000 DE genes are false discoveries, we can expect fewer than ten false discoveries in our list of DE genes. To use FDR of 0.005 as the threshold for deciding differential expression, enter:
>cutoff=0.005
-
Visualize differential gene expression. Move the cursor to the code chunk in the section titled “Get an overview of the DE genes in the data set” and run the next commands to create a plot summarizing differential gene expression analysis results. Alternatively, type the following commands into the R console:
> de = decideTestsDGE(dex, p = cutoff, adjust = “BH”)
> detags = rownames(dex)[as.logical(de)]
> plotSmear(dex, de.tags = detags)
> abline(h = c(-1, 1), col = “blue”)
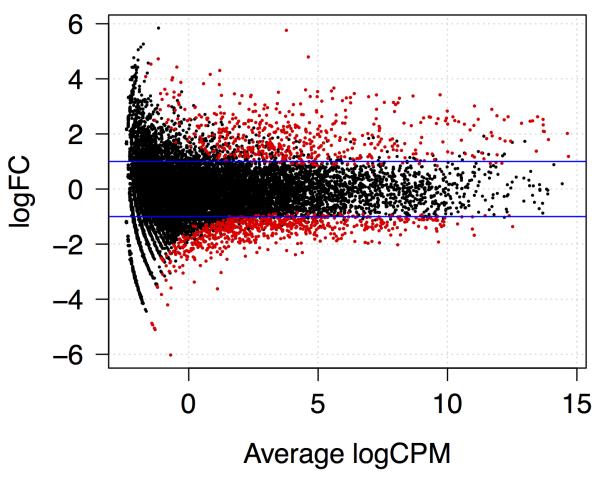
Examine the plot. Observe how the plot shows the relationship between overall expression level measured in CPM (counts per million) on the x axis and log2 fold-change (FC) on the y axis (Fig. 3). Red points indicate genes found to be significantly upregulated or downregulated by the treatment. Observe that the treatment changed expression of many genes, and for most, the fold-change was greater than two.
-
Writing results files. Move the cursor to the next code chunk and run it. To save time, select Run Current Chunk under the Chunks menu at the upper right of the tab. Alternatively, enter the following commands into the R console:
> cpms=cpm(dge$counts) > cn=grep(‘C’,colnames(cpms))
> tr=grep(‘T’,colnames(cpms))
> ave.cn=apply(cpms[,cn],1,mean)
> ave.tr=apply(cpms[,tr],1,mean)
> res=data.frame(gene=row.names(dex$table),- fdr=dex$table$fdr,
- logFC=dex$table$logFC,
- Cn=ave.cn,
- Tr=ave.tr)
> de=res$fdr<=cutoff
> res=res[de,]
> annots_file=‘../ExternalDataSets/S_lycopersicum_Feb_2014.bed’
> annots=read.delim(annots_file,sep=‘\t’,header=F)[,13:14]
> names(annots)=c(‘gene’,‘description’)
> res=merge(res,annots,by.x=‘gene’,by.y=‘gene’)
> res=res[order(res$fdr),]
> res=res[,c(‘fdr’,‘logFC’,‘Cn’,‘Tr’,‘gene’,‘description’)]
> out_file=‘results/tomatoDE.tsv’
> write.table(res,file=out_file,row.names=F,sep=‘\t’,quote=F)
> out_file=‘results/forLycoCyc.tsv’
> write.table(res[,c(‘gene’,‘logFC’)],file=out_file,quote=FALSE, sep=‘\t’,col.names=FALSE,row.names=FALSE)
Review results file. Now, the data file tomatoDE.tsv ready to be opened in Excel or other spreadsheet program has been saved to the results subfolder. Also, a second file named “forLycoCyc.tsv” has been generated; this file is designed for visualization in the LycoCyc “Omics” viewer, which enables identification of differentially expressed enzymes annotated to metabolic pathways in the LycoCyc database. See Note 5 for information about LycoCyc.) Open “tomatoDE.tsv” in Excel or other spreadsheet program. When prompted, indicate that the file contains tab-separated values (tsv). If using Excel, click the row label for the second line of text to select the entire row and choose Window > Freeze Panes. Use the vertical scrollbar to move the data display up and down; note that the first row, which contains column headings, remains in place, making it easier to review the data. See Note 6 for descriptions of data in the file.
Figure 3. Plot showing relationship between average expression and fold-change.
Horizontal lines indicate fold-change of two. Red indicates genes called as differentially expressed at FDR of 0.005 or smaller.
3.4. Gene Ontology enrichment analysis
The Gene Ontology (GO) is a controlled vocabulary of terms that classify gene products by biological process, molecular function, or cellular localization. Using GO term enrichment analysis, we can identify entire categories or families of genes that are differentially regulated due to a treatment in either a microarray or an RNA-Seq experiment. By enrichment, we mean: a higher than expected percentage of the genes annotated to a term are differentially expressed. For example, if 15% of genes annotated to GO molecular function term “calcium ion binding” (GO:0005509) are differentially expressed, but only 5% of genes overall are differentially expressed, this suggests that a process or function related to calcium signaling may be perturbed due the treatment being tested. Thus, the key to useful GO term enrichment analysis is to identify when these percentages (15% versus 5%) are significantly different. Toward this end, most tools for GO enrichment use variations on Fisher’s exact test for contingency tables to identify significant GO terms. However, for RNA-Seq analysis, it may be misleading to use Fisher’s test because of size bias among genes in some GO categories; that is, larger genes are more likely to be sampled in RNA-Seq than smaller ones, making them more likely to be detected in differential expression analysis, and genes in the same GO category (especially the molecular function) are likely to have similar sequence and thus similar sizes [7]. In microarray expression analysis, because the method of detection is independent of transcript size, no such bias exists, and so tools designed for GO enrichment analysis of array results may not work as well for RNA-Seq. For RNA-Seq data, it is probably better to use methods that take transcript size into account when testing for enrichment. So, for the WiNGS workshop, we introduced the GOSeq library [9] that takes size of transcript into account when identifying enriched GO categories.
Open Gene Ontology project. Use the File > Open menu in RStudio to open the Gene Ontology. Rproj file in the Gene Ontology directory. Under the Files tab, select the file named TomatoGO.Rmd.
-
Install GOSeq. If you are running the code for the first time, you will need to install GOSeq. Find the section in TomatoGO.Rmd titled “Load GOSeq library.” Delete the “#” comment characters and run the following commands:
> source(“http://bioconductor.org/biocLite.R”)
> biocLite(“goseq”)
Restore the comment characters (#) to avoid reinstalling GOSeq. See Note 7 on installing GOSeq.
Knit HTML. Rather than run the code chunk by chunk, click the Knit HTML button, which will run all the code in the file and make an HTML report (see Fig. 1).
Open the HTML file. Click the file named TomatoGO.html in the Files menu and select “View in Web Browser.” Read the report, making note of how the code from the code chunks in the Markdown file is executed and the results added to the HTML report.
Use Excel (or other spreadsheet viewer program) to open and explore results files. Three tab-separated, plain text files list GO categories enriched among genes that were upregulated (GO_up.tsv), downregulated (GO_down.tsv), or both (GO_all_de.tsv). The file genesInSigGO.tsv reports genes, log2 fold-change, descriptions, and GO categories for differentially expressed tomato genes in enriched categories.
Test how changing FDR thresholds for differential expression affects GO term enrichment results. For this, reopen the Differential Expression project. Select File > Open Project in New Window to open the Differential Expression project without closing the GOSeq project. Open the R Markdown file named Differential Expression. Rmd. Use the RStudio editor to change FDR setting by editing the line “cutoff=0.005”. Click Knit HTML or select Chunks > Run All to rerun all the code in the Differential Expression. Rmd file. After rerunning the file, there should now be a new version of tomato_DE.tsv in Differential Expression/results. Return to the GOseq project and rerun the TomatoGO.Rmd by clicking Knit HTML or selecting Chunks > Run All. Look again at the output file to find out how the results changed when the threshold for DE genes was raised or lowered.
3.5. Visualizing tomato RNA-Seq data in Integrated Genome Browser
Integrated Genome Browser is an open source, freely available genome browser that enables fast exploration of sequence alignments and coverage graphs from high-throughput sequencing experiments [8]. Unlike other tools (e.g., IGV from the Broad Institute), users can click to select (and count) items in the display, search the Web or run BLAST searches on genes, and even view/copy gene and read sequence data, which is useful when designing PCR primers to validate results. IGB has far too many features to describe here; to find out more, visit the IGB Web site at http://www.bioviz.org or use the Help menu in IGB to view the User’s Guide or contact the IGB support team.
Even when statistical methods are robust and well understood, it is important to visualize the data in order to check that the methods are working as expected. That is, one should always “sanity check” one’s differential expression analysis by looking at a few examples to make sure that no code or logic errors have contaminated the data analysis thus far. Even better, viewing the data in a graphical format, such as in IGB, can trigger unanticipated insights and suggest new experiments. Here, we explain how to view and work with RNA-Seq coverage graphs and also how to investigate the function of individual genes using the Google and BLAST search features.
Start Integrated Genome Browser. If running IGB for the first time, visit http://www.bioviz.org/igb and follow the instructions to download and launch IGB. If IGB is already installed, it should be available as a shortcut icon on your computer’s desktop. If so, double-click the shortcut icon to launch IGB.
Open the tomato genome. On startup, IGB displays a start screen with a carousel of images representing the latest genome assemblies for human, mouse, blueberry, rice, Arabidopsis, fruit fly, and many other species important for research or agriculture. Starting with IGB 8.2, you will also see an image of tomato (near the blueberry image) linking to the latest tomato assembly. For IGB 8.1 or earlier versions, use the Current Genome tab on the right side of the IGB window to select species Solanum lycopersicum and then the Feb. 2014 (SL2.50) genome assembly, the most recent publicly available genome assembly for tomato.
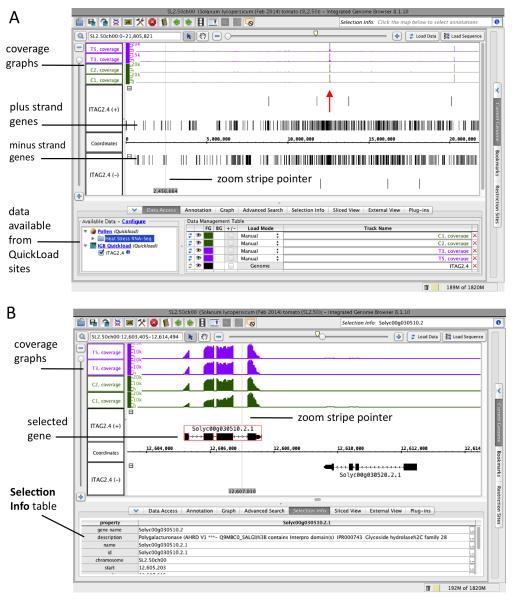
Observe that the tomato gene models load automatically into two tracks labeled ITAG2.4 (+) and ITAG2.4 (−) corresponding to the plus (forward) and minus (reverse) strands of the chromosome. (Fig. 4A). When you first choose a genome, the reference gene models associated with that genome version automatically load from the IGBQuickLoad.org site into IGB. For tomato, the reference gene models are from the ITAG 2.4 genome annotations, originally harvested from the Sol Genomics Web site. By default, IGB separates gene models into two tracks, one for each strand. To make more space for visualizing the RNA-Seq data, you can merge the strand tracks into a single track. To merge the tracks, select the Annotation tab and select the +/− “combined” box in the Labels and Strand section.
Load data sets from pollen RNA-Seq. In the Data Access tab (lower left of the IGB screen) note there is an area labeled Available Data (Fig. 4A). The Pollen data source includes read alignments, junction features from a Loraine lab program called Find Junctions, and coverage graphs representing the number of reads that overlap base positions in the genome. See Note 8 for more information about how the files were made and for instructions on how to load your own files into the viewer. To load RNA-Seq pollen coverage graphs, select Pollen > Heat Stress RNA-Seq > SM > Graph. (SM are data sets from reads that mapped to one location, and MM are data sets from reads that mapped multiple times.) Select the checkboxes next to T3, T5, C2, and C1 samples; these libraries were sequenced to roughly the same depth. For charts showing sequencing depth, see the HTML file named “AssessingCounts.html” in the “Counts” of your downloaded tomato pollen folder.
Load coverage graph data. Click the Load Data button at the top right of the IGB window to load coverage graphs data into the main IGB display. The y axis for each graph indicates the lower and upper range values for any base position in the current view; when IGB first loads a coverage graph, it sets the y axis scale to encompass the largest and smallest y values from the graph.
Adjust scale of coverage graphs. Values in the coverage graphs range from 0 (no expression) to more than 20,000. Select the Graph tab and click the Select All button to select all visible graphs. Use the sliders in the sections labeled Height and Y Axis Scale to change graph height and the scale of values being shown. To adjust the scale for a single graph, click in the IGB main window to deselect the graphs and then click a track label next to the graph you want to adjust. Note there is a region near the center of chromosome SL2.50ch00 (ITAG2.4 name) with nearly 20,000 reads.
Zoom in on a region of high expression. Click the highly expressed region between 10,000,000 and 15,000,000 on SL2.50ch00, near the position indicated by the arrow in Fig. 4A. Observe that when you click a location in the IGB display, a thin gray line with a base position label appears in the location you clicked. This is the IGB zoom stripe pointer and it focuses zooming on the selected position. Observe that once you’ve selected a location by clicking it, you can drag the horizontal zoomer (top right slider) to the left or right to zoom in or out. During zooming, the zoom stripe remains in one location while the display stretches or contracts around it. This is an important difference between IGB and other genome browsers that makes navigation through multiple scales much easier. Also, you can zoom in on a region quickly by click-dragging the arrow tool over the coordinates axis.
Use move tools to reposition. To reposition the display to the left or right, either click the green arrows in the IGB toolbar or change the cursor to the move tool (hand icon) and click-drag the display. Alternatively, change back to selection tool (arrow icon) and click-drag the mouse into left or right side of the IGB window to scroll.
Zoom in on the highly expressed gene (Solyc00g030510.2) and select it. Change the cursor back to the selection tool (arrow) by clicking the arrow button on the toolbar. Select the gene (Fig. 4B) by clicking its label or one of the thin lines representing introns. (Blocks represent exons; thicker blocks mark translated exons.) When an item is selected, a red outline appears around it. Observe that the Selection Info text area in the upper right of the display shows the name of the selected item. Click the Selection Info tab (lower tab, Figure 4B) or click the “i” information button next to the Selection Info box (upper right) to view text annotations associated with selected gene. Observe that this gene is annotated as encoding polygalacturonase and does not appear to be upregulated or downregulated by the treatment.
View coverage graphs for a differentially expressed gene. Open the tab-separated (tsv) file named tomatoDE.tsv created in the Differential Expression results folder in a previous section. The file is already sorted, so that the most significantly changed genes appear at the top of the file. Observe that the gene with the smallest FDR is Solyc05g055400.2, annotated as encoding a cytochrome P450 gene. In the top left text area of IGB, type (or copy and paste) the name of the gene into the text box and click the magnifying class to search for the gene. Observe that if the gene is found, IGB zooms and scrolls to the location on chromosome 5 where this gene is located. Use the horizontal slider to zoom all the way out and click the Load Data button to load coverage graphs for the entire chromosome. Observe that all the data load into the tracks, but the graphs are on different scales due to differences in sequencing depth.
Use the Graph tab to configure the graphs. Click the Graph tab and click the Select All button to select all graphs. Next, select Percentile in the section labeled Y Axis Scale. Enter 50 (for 50th percentile) in the Max text box and type ENTER. This ensures that within each track, the maximum value of the y axis will be the 50th percentile value of that track. This scaling by percentile is a primitive form of normalization, allowing you to compare relative height and shape of graphs between tracks.
Return to Solyc05g055400.2. As before, enter the gene name Solyc05g055400.2 in the text box (top left). Click the magnifying glass search icon or type ENTER to find the gene. Zoom out to view several genes in the vicinity of Solyc05g055400.2 (Fig. 5A). Observe that some genes appear to have equivalent expression in all samples. Make note of them and then search for these genes in the spreadsheet, which lists DE genes only. For example, look for Solyc05g055420.2, which is near to Solyc05g055400.2 and is not DE. Observe that the coverage graphs above Solyc05g055420.2 are about the same height, while the coverage graphs above Solyc05g055400.2 are of different heights (Fig. 5A). Observe also that the treatment coverage graphs (T3 and T5) are of slightly different heights. This indicates that not only did the treatment induce expression of Solyc05g055400.2, it also increased variability of expression of Solyc05g055400.2. Recall that in the MDS plot (see Fig. 2A), the treatment (T1 through T5) samples were farther apart than the control samples. This increase in variability that goes along with increased expression seems to be a general feature of RNA-Seq and array expression experiments we have analyzed; control samples, no matter what the experiment, seem to be more uniform than treatment samples (Loraine, unpublished observations).
Investigate the annotations for Solyc05g055400.2. Right-click the gene model and observe that a menu appears with options to search the Web (Fig. 5B). Select BLASTP option to search the annotated protein sequencing against the non-redundant protein database at NCBI. The top hits are from tomato and have 100% identity with the query sequence; these are from the same gene model that was translated and entered into the “nr” database. Observe that the other best matching proteins are from other plant species and mostly are annotated as “hypothetical” and “predicted” cytochrome P450, family 77, subfamily A proteins.
Make an image for slides or publication. Select File > Export Image to save an SVG (vector graphics), PNG, or JPEG format image showing the entire IGB frame or just the data display area with or without the track labels. Use the image quality settings to select image dimensions and DPI (Fig. 5C).
Figure 4. Integrated Genome Browser showing pollen data sets and gene models.
(A) Whole chromosome view with coverage graphs and gene models from the ITAG2.4 annotations release. Coverage graphs are from treatment (T3, T5) and control (C1, C2) samples. The arrow indicates the location of the most highly expressed gene on the chromosome. (B) Zoomed-in view of the highly expressed gene indicated in (A). The gene encodes a putative polygalacturonase.
Figure 5. Zooming in on a differentially expressed gene.
(A) Coverage graphs for a differentially expressed (DE) gene. For comparison, a nearby gene that is not differentially expressed is also shown. (B) Right-click context menu. Right-clicking a gene model triggers options to do a BLAST search, view the sequence in a new window, or search Google. (C) Image export. Selecting File > Export Image saves an image file with the current view within IGB.
Acknowledgements
The example data set was from the Workshop in Next-Generation Sequencing (WiNGS), which was cosponsored by the NSF Research Coordination Network on Integrative Pollen Biology (award 0955431), the NSF Plant Genome Research Program (award 1238051), and the Department of Bioinformatics and Genomics at UNC Charlotte. NIH R01 grant number 21737838 supports development of the IGB software.
4. Notes
Written by Yihui Xie while a graduate student in statistics at Iowa State University, knitr is a optional add-in library for R that enables writing statistical reports that contain R code embedded in code “chunks” or inserted in-line with plain text. Reports can be written using LaTeX (typesetting language) or the much simpler and easier to learn R Markdown language. When you “knit” a document using knitr, the embedded code is executed and a document is produced that contains nicely formatted text along with the output of the R code, including figures. In this way, you can create easy-to-reproduce reports and experiment with how different parameters (such as a p value cutoff) affect analysis results.
Some libraries had more reads than others and so counts per gene are not directly comparable; we have to normalize. However, there is another aspect of normalization that we need to take into account, which is that a treatment (like heat) may greatly increase in the expression of a subset of genes, thus “consuming” counts that might have otherwise come from less highly expressed genes and making those genes appear downregulated when actually they were not. For a deeper discussion, see [7].
MDS (multidimensional scaling plots) enable identification of larger trends or biases in a data set. Typically, biological replicates from the same group cluster together, but sometimes samples from the same batch cluster together, indicating possible batch effects. In this experiment, sample T2 clusters far away from the other treatment samples, which are separated as a group in the y dimension from the control samples.
To start, we need to estimate dispersion, which reflects the degree to which variation in expression depends on expression level. We need this to model gene expression and test whether gene expression has changed due to the treatment. The details of how this works are explained in the edgeR user’s guide. In a nutshell, the basic idea is that we use the negative binomial model to estimate a dispersion parameter for each gene, which edgeR calls “the degree of inter-library variability” for that particular gene.
The file named “forLycoCyc.tsv” is a tab separated designed for upload into the LycoCyc Cellular Overview viewer, which shows a diagram of metabolic pathways annotated in the LycoCyc metabolic pathways database and can be used to visualize differentially expressed metabolic pathway genes. The “forLycoCyc.tsv” file contains two columns: gene names in the first column and log2 fold-change in the second column. To use the Cellular Overview to visualize expression data, visit the Cellular Overview Web site (http://solcyc.solgenomics.net/overviewsWeb/celOv.shtml) and select Overlay Experimental Data > Upload Data from File from the OPERATIONS menu. A new window will appear. Next, click the Browse button and select file forLycoCyc.tsv. Enter “1” in the text box labeled “Data column(s) to use.” Click the Submit button to upload the file and add expression data to the Cellular Overview, which uses data from the second column in the file to color-code pathway steps according to whether genes associated with this steps were upregulated or downregulated.
The file named “tomatoDE.tsv” contains four columns and many rows of data – one for each differentially expressed gene. The column with heading “fdr” indicates false discovery rate for differential expression. Column named “logFC” indicates the log2 of the fold-change between the treated and untreated samples. Negative values indicate genes that were downregulated and positive values indicate genes that were upregulated. Columns named “Cn” and “Tr” indicate the average, normalized counts per million (CPM) for the gene in the control and treatment samples. The column named “gene” contains the gene id for each gene and the column named “description” contains annotation text from the Sol Genomics Web site. These are also the same gene descriptions that appear in IGB.
You may not be able to install GOSeq without first upgrading your R installation. For information on how to install (or upgrade) R on your platform, visit the R project Web site at http://r-project.org.
Files available shown in the IGB Available Data list were made by aligning RNA-Seq reads onto the tomato genome with tophat2, which created BAM alignment files and TopHat junction (TH) feature files. Reads were then further processed using bedtools to generate coverage graphs and Find Junctions (FJ) to create junction features. Reads were also separated into two groups: “MM” for reads that mapped to multiple locations and “SM” for reads that mapped to one location. For more information, click the “Pollen” hyperlink in the Available Data Sets section of the Data Access Panel. Clicking the hyperlink will open a Web browser showing the Web site where the data files are stored; documentation on the site also describes how files were created. Note also that IGB can also open local files. To open your own files, first select a genome or use File > Open Genome to open a genome descriptor file (genome.txt) or fasta file with genomic sequence. Then use the File > Open menu to open a local file or click-drag the file from your desktop into the IGB display area. For more information, see the IGB User’s Guide or contact the IGB support team. Links to the User’s Guide and IGB support are available from the IGB Help menu.
5. References
- [1].Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Nature methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- [3].Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Shendure J. Nature methods. 2008;5:585–587. doi: 10.1038/nmeth0708-585. [DOI] [PubMed] [Google Scholar]
- [5].Robinson MD, McCarthy DJ, Smyth GK. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Nikolayeva O, Robinson MD. Methods in molecular biology. 2014;1150:45–79. doi: 10.1007/978-1-4939-0512-6_3. [DOI] [PubMed] [Google Scholar]
- [7].Oshlack A, Wakefield MJ. Biol Direct. 2009;4:14. doi: 10.1186/1745-6150-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Nicol JW, Helt GA, Blanchard SG, Jr., Raja A, Loraine AE. Bioinformatics. 2009;25:2730–2731. doi: 10.1093/bioinformatics/btp472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Young MD, Wakefield MJ, Smyth GK, Oshlack A. Genome Biol. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]