Abstract
Streptococcus pneumoniae is a prominent human pathogen responsible for many severe diseases and the leading cause of childhood mortality worldwide. The pneumococcus is remarkably adept at colonizing and infecting different niches in the human body, and its adaptation to dynamic host environment is a central aspect of its pathogenesis. In the last decade, increasing findings have evidenced small RNAs (sRNAs) as vital regulators in a number of important processes in bacteria. In S. pneumoniae, a small antisense RNA was first discovered in the pMV158 plasmid as a copy number regulator. More recently, genome-wide screens revealed that the pneumococcal genome also encodes multiple sRNAs, many of which have important roles in virulence while some are implicated in competence control. The knowledge of the sRNA-mediated regulation in pneumococcus remains very limited, and future research is needed for better understanding of functions and mechanisms. Here, we provide a comprehensive summary of the current knowledge on sRNAs from S. pneumoniae, focusing mainly on the trans-encoded sRNAs.
Keywords: Streptococcus pneumoniae, virulence regulation, competence, small non-coding RNAs
Introduction
Streptococcus pneumoniae, the pneumococcus, is an opportunistic pathogen responsible for a wide spectrum of human diseases, ranging from mild otitis media to more severe infections such as meningitis, sepsis, or endocarditis. It is the main etiological agent of community-acquired pneumonia, causing more deaths in young children than any other infectious disease (O’Brien et al., 2009). Pneumococcal vaccines cover only a small number of the 93 different serotypes, and the treatment of pneumococcal diseases is hampered by the emergence and spread of drug-resistant strains1. S. pneumoniae is a normal component of the human commensal flora, asymptomatically colonizing the upper respiratory tracts of children and healthy adults. Human nasopharyngeal (NS) carriage is the source of transmission from person to person and serves as the first step in pathogenesis (Bogaert et al., 2004). Transition to opportunistic pathogen often occurs after a respiratory tract infection and is triggered by unknown host and bacterial factors. Disease progression exposes S. pneumoniae to numerous environmental changes and stress conditions, and rapid adaptation is a key factor for survival and replication.
In recent years, a plethora of RNAs with regulatory functions has been discovered in many pathogenic and non-pathogenic bacteria. These small RNAs (sRNAs) accomplish a large variety of regulatory functions, and are essential elements in bacterial pathogenicity (Toledo-Arana et al., 2007; Waters and Storz, 2009; Storz et al., 2011; Bobrovskyy and Vanderpool, 2013; Caldelari et al., 2013). Often non-coding, the sRNAs can act at the level of transcription, translation or RNA degradation. The majority of them regulate pathways that sense and transfer the external signals, and adapt the cell population in response to stress and environmental changes. Some regulate replication and maintenance of plasmids and phages (Brantl, 2002) and others, such as the CRISPR RNAs, protect the core genome from foreign nucleic acids (Fineran and Charpentier, 2012). They can act through three main mechanisms: (1) by base-pairing with nucleic acids, mostly mRNA targets, having either extensive or more limited complementarity; (2) modulating the activity of proteins by mimicking other nucleic acids, or (3) acting as riboswitches, sensing physical cues or metabolites and modulating expression of downstream genes (Winkler and Breaker, 2005; Zhang et al., 2010). RNA-interacting proteins play important roles in the expression and activity of sRNAs. Nucleases have critical roles in their production, quality control, and activation, and the RNA chaperone Hfq mediates the action of many sRNAs (Vogel and Luisi, 2011; Saramago et al., 2014).
Whereas a variety of sRNAs have been identified and studied in many Gram-positive and Gram-negative bacteria, little is known about these regulators in S. pneumoniae. Recent systematic approaches have uncovered multiple chromosomal-encoded sRNAs in the pneumococcus (Livny et al., 2006, 2008; Kumar et al., 2010; Tsui et al., 2010; Acebo et al., 2012; Mann et al., 2012) but only a few have been functionally studied (Halfmann et al., 2007; Schnorpfeil et al., 2013). In this review, we outline our current knowledge on the sRNA–mediated regulation in S. pneumoniae, compiling all information available and providing a comprehensive list of the sRNAs identified and their biological functions.
RNAs Encoded in Extrachromosomal Elements
The first regulatory RNA discovered in pneumococcus was a plasmid-encoded bona-fide antisense RNA described by del Solar and Espinosa (1992), and its role in establishment, replication, and copy number regulation has been deeply investigated. The pMV158 is a promiscuous plasmid able to replicate in pneumococci, whose replication is initiated by the plasmid-encoded initiator protein RepB. Expression of RepB is subjected to a tight control exerted by two trans-acting plasmid elements, a transcriptional repressor protein (CopG) and an antisense RNA (RNAII; del Solar et al., 1995; Figure 1A). Both CopG and RepB are transcriptionally but not translationally coupled (López-Aguilar et al., 2013), and CopG is able to bind to their operator sequence and repress synthesis of the copG-repB mRNA (Hernández-Arriaga et al., 2009). Post-transcriptionally, the short 48-nt long antisense RNAII, whose synthesis is directed by the PctII promoter, inhibits translation of repB message by directly pairing to the region immediately upstream of its translational initiation signals (del Solar et al., 1997). Structural analyses by chemical and enzymatic probing, revealed that the RNAII consists of single stranded 5′ and 3′ tails and a hairpin, which together with the adjacent U-reach 3′ tail compose a very efficient intrinsic terminator (del Solar and Espinosa, 2001; López-Aguilar and del Solar, 2013). The most recent investigations (López-Aguilar, personal communication) demonstrated that the 5′-tail of RNAII play a critical role in the binding and translation inhibition of repB message, while the hairpin plays a secondary role. A singular binding mechanism is envisaged whereby initial pairing between complementary single stranded regions in the antisense and sense RNAs progresses upward into the corresponding hairpin to form the intermolecular duplex.
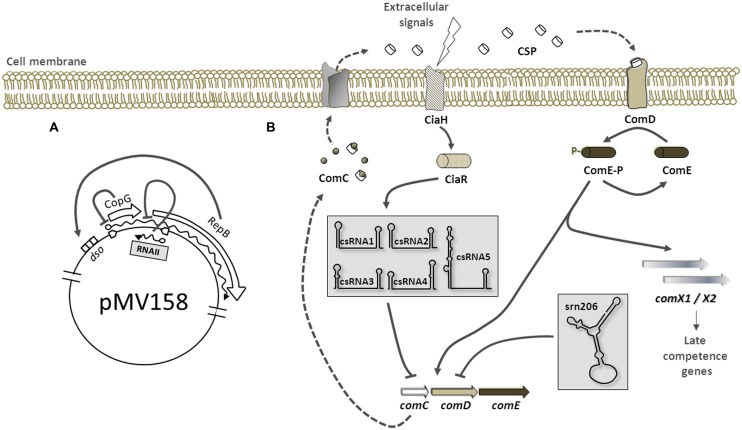
FIGURE 1.
Regulation by small RNAs (sRNAs) in pneumococci. The cytosol and the extracellular environment (upper part of the figure) are separated by the cell membrane. (A) Replication of plasmid pMV158 is initiated by RepB protein upon binding to double stranded origin (dso). RepB transcription is inhibited by CopG. The antisense RNAII represses RepB translation by base-pairing with the region immediately upstream of RepB translational initiation signals in the copG-repB message. (B) Postulated mechanism of competence regulation by sRNAs. The extracellular concentration of CSP, an exported peptide pheromone derived from precursor protein ComC, is sensed by the membrane histidine kinase ComD. Binding of CSP to ComD results in phosphorylation of ComD, which then transfers the phosphate group to the cognate response regulator ComE, thus activating transcription of early competence genes (comC, comD, comE, and comX1/X2). ComC, D and E are cotranscribed in a long mRNA. Expression of the five csRNAs (within a gray box) is activated by the CiaRH two component system. The csRNAs then associate with the SD sequence and start codon of comC inhibiting its expression through an antisense mechanism. Similarly, the srn206 (within a gray box) associates with comD message sequestering its translation initiation signals. The five csRNAs and the srn206 act together to maintain the competence switched off. Predicted secondary structure of the csRNAs (by Mfold) and the srn206 (by RNAfold) as previously published in (Halfmann et al., 2007) and (Acebo et al., 2012), respectively, is shown.
Both regulatory elements, CopG and RNAII, acts synergistically to ensure the plasmid copy number within a narrow range. The mechanism of repression by CopG has been extensively studied, as well as the RNAII mode of action, and constitutes the only regulatory RNA mechanistically studied in S. pneumoniae.
Chromosomal-Encoded sRNAs
A combination of computational predictions, transcriptome analyses, and RNA sequencing approaches has been applied to discover chromosome-encoded riboregulators in S. pneumoniae. Although the available software for sRNA identification were mainly developed on the basis of sRNA features from Gram-negative genomes, adaptation of novel programs have also enabled the discovery of a large number of sRNAs in Gram-positive bacteria. As much as 128 sRNAs were bioinformatically predicted in intergenic regions (IGRs) of the pneumococcal chromosome through two different computational approaches, using the sRNAPredict2 software and the high throughput kingdom-wide prediction tool SIPHT (Livny et al., 2006, 2008). Additionally, searching for promoters containing a consensus CiaR (response regulator)-binding sequence allowed to identify the first class of five sRNAs located within IGRs of the genome (Halfmann et al., 2007). After that, three groups reported the use of high-throughput methodologies to experimentally discover sRNAs in pneumococci. First, Kumar and coworkers found 50 putative sRNAs in the clinical isolate TIGR4 genome by high-resolution tiling microarrays (Kumar et al., 2010). Further, Acebo and coworkers identified 88 potential sRNAs by deep sequencing, 68 of which were novel candidates (Acebo et al., 2012); and, finally, Mann et al. (2012) increased the total number to 178, of which 37 were identified by at least two different methods. Differences inherent to the technique used and/or different sequence coverage may explain the moderate overlapping observed between searches. Most of these sRNAs are conserved among pneumococcal species and some (71) are also present in other bacteria, mainly in fellow streptococci (59). Interestingly, about 40% (70) appear to be unique to this human pathogen.
Among the sRNAs identified, some belonged to previously known families of Cis-acting RNAs like riboswitches or leader regions (i.e., pyr, TPP riboswitch, T-box). Others belonged to the so-called functional sRNAs, such as RNase P, the 6S RNA, or tmRNA, whose predicted biological activities and mechanism of action are based on the knowledge of sRNA orthologs in other bacterial species. Some sRNAs have been identified as BOX elements, mobile sequences exclusively found in pneumococci and closely related species, which are numerous and randomly distributed in IGRs (Knutsen et al., 2006; Croucher et al., 2011). They have the potential to form stable stem-loop structures and may affect expression levels of neighbor genes either by stabilizing mRNAs or serving as DNA-binding sites for regulatory proteins. However, the majority of the sRNAs identified could not be assigned to a functional family. Remarkably, no Cis-antisense or CRISPR RNAs have been identified so far, in what appears to be a pneumococcal singularity.
Biological Functions of Chromosomal-Encoded sRNAs
Expression of eighty out of the 178 putative sRNAs was tested by northern-blot or qRT-PCR and 70 of them were successfully validated (Halfmann et al., 2007; Kumar et al., 2010; Tsui et al., 2010; Acebo et al., 2012; Mann et al., 2012). However, knowledge about their biological function remains very limited and, with a few exceptions, no clearly defined targets or regulatory mechanisms were determined so far. Chromosome-encoded sRNAs have been mainly linked to fine-tuning metabolic processes or stress adaptation, and this often results in the lack of severe phenotypes upon deletion or overexpression. In fact, three sRNAs were deeply investigated in the D39 pneumococcal strain using deletion mutants and overexpressing strains, but no effects in common phenotypes or transcription patterns were conclusively found (Tsui et al., 2010). Nevertheless, several groups have recently succeeded in attributing diverse functions to pneumococcal sRNAs and some of them have been shown to control various aspects of virulence and participate in important regulatory networks such as competence or autolysis.
sRNAs Control Virulence
The importance of sRNAs in virulence was overlooked until recently, when the application of targeted genetic knockouts and Tn-seq transposon screening mutagenesis demonstrated that a significant portion of the pneumococcal sRNAs have important global and niche-specific roles in virulence (Mann et al., 2012). Measurements of relative fitness and competitive index of sRNA pneumococcal mutants generated by random transposon insertion in the nasopharynx, lungs, and bloodstream, rendered a comprehensive list of sRNA mutants with altered fitness. The majority showed defects in nasopharynx colonization, lung infection, or replication in bloodstream (Table 1), but a small number of mutants actually resulted in a fitness benefit in certain host sites. These data indicate that sRNAs contribute to pneumococcal pathogenesis, both for systemic infections as well as for tissue-specific tropisms. Targeted knockout mutants validated these results and demonstrated that eight of them were also attenuated in establishing invasive disease upon intranasal (IN) challenge. These attenuated mutants were deeply studied, including analysis of their ability to adhere to and invade endothelial (ET) and NS cells, as well as transcriptomic and proteomic analyses to identify putative targets (for details see Table 1).
Table 1.
Studied small RNAs (sRNAs) in pneumococci.
| sRNA name | Pathogenesis profilea | Direct target and mechanism of action | Regulatory functions |
|---|---|---|---|
| RNAII∗ | Translational repression of repB by base-pairingb | Control of plasmid replicatione | |
| F14 | Nasopharynx | ||
| F24 | Nasopharynx | ||
| F38; srn254; spd-sr17 | Nasopharynx | ||
| F51 | Nasopharynx | ||
| F52 | Nasopharynx | ||
| F63 | Nasopharynx | ||
| F64 | Nasopharynx | ||
| R16 | Nasopharynx | ||
| SN30 | Nasopharynx | ||
| SN39 | Nasopharynx | ||
| SN50 | Nasopharynx | ||
| srn142 | Nasopharynx | FMN riboswitch | |
| trn0156 | Nasopharynx | ||
| trn0760 | Nasopharynx | ||
| F66; srn502; SN27 | Nasopharynx and blood | ||
| R12; trn0830 | Nasopharynx and blood | ||
| R8 | Nasopharynx and blood | ||
| trn1025; SN46 | Nasopharynx, blood, and lungs | ||
| F41; srn277 | Nasopharynx and blood; IN Challenge | ||
| F20; srn157 | Nasopharynx and lung; IN Challenge; reduced ET and NS adhesion and invasion | ||
| F2 | Blood | ||
| F27; trn0358 | Blood | TPP riboswitch | |
| F45 | Blood | ||
| R4 | Blood | ||
| SN38 | Blood | ||
| srn279 | Blood | ||
| F5; trn0052 | Blood and lungs | ||
| trn0012; SN1; csRNA3 | Blood and lungs | Translational repression of comC, spr0081, spr0159, brnQ and spr1097, by base-pairingc | Competence modulation and autolysisf |
| F25; trn0332; SN11 | Blood; IN Challenge; reduced ET adhesion | ||
| F26; SN12 | Lung | Pyr regulator | |
| F29 | Lung | ||
| F47; srn368; SN24 | Lung | T-box | |
| F59; srn235; SN20 | Lung | ||
| F60; trn0485 | Lung | ||
| F62 | Lung | ||
| R14 | Lung | ||
| R6; srn400 | Lung | T-box | |
| F8; SN5; csRNA1; spd-sr56 | Lung | Translational repression of comC, spr0081, spr0159, brnQ and spr1097, by base-pairingc | Competence modulation and autolysisf,g |
| SN6; csRNA2 | Lung | Translational repression of comC, spr0081, spr0159, brnQ and spr1097, by base-pairingc | Competence modulation and autolysisf |
| SN2 | Lung | ||
| SN22 | Lung; reduced ET adhesion | ||
| SN26 | Lung | ||
| SN31 | Lung | ||
| SN32 | Lung | ||
| srn218 | Lung | ||
| trn0634 | Lung | ||
| F32; srn226; SN16; tmRNA | Lung; IN Challenge; reduced ET and NS adhesion and invasion | Trans-translationh | |
| F7; srn061; SN35; csRNA5 | Lung; IN Challenge; reduced ET adhesion | Translational repression of comC, spr0081, spr0159, brnQ and spr1097, by base-pairingc | Competence modulation and autolysisf |
| F22 | IN Challenge | ||
| F44 | IN Challenge; reduced ET adhesion | ||
| F48 | IN Challenge | ||
| srn395; SN34; 6S | Control of gene expression in stationary phaseh | ||
| srn098; SN8; RNaseP | tRNA maturationh | ||
| SN7; csRNA4 | Translational repression of comC, spr0081, spr0159, brnQ and spr1097, by base-pairingc | Competence modulation and autolysisf | |
| srn206 | Translational repression of comC, spr0081, spr0159, brnQ and spr1097, by base-pairingd | Competence modulationd |
∗Plasmid encoded antisense RNA.
aShows the contribution of sRNAs to pathogenesis on each host site as described in Mann et al. (2012). Deletion mutants with altered fitness in nasopharynx colonization, blood replication or lungs are indicated. Deletion mutants attenuated in invasive disease upon intranasal challenge (IN Challenge) or with reduced ability to adhere or to invade endothelial (ET) or nasopharyngeal (NS) cells are also indicated.
bdel Solar et al. (1997); cSchnorpfeil et al. (2013); dAcebo et al. (2012); edel Solar et al. (1995); fHalfmann et al. (2007); gTsui et al. (2010); hBiological activities predicted based on the knowledge in other bacteria.
Two sRNAs of especial interest resulted from this extensive analysis: the F20, also named as srn157, and the F32, previously identified as the tmRNA (Kumar et al., 2010; Acebo et al., 2012). Both the srn157 and tmRNA deletion mutants showed decreased adhesion/invasion of NS or ET cells, respectively, in concert with a lack of fitness and competitive index in the nasopharynx and lungs (Table 1). They also resulted in a dramatic alteration in abundance of several proteins (88 and 100, respectively), as well as in a substantial change in the gene expression profile. In case of srn157 deletion mutant, proteomic analysis indicated that proteins responsible for purine metabolism were strongly downregulated, whereas DNA synthesis and repair pathways were greatly upregulated. Thus, deletion of this sRNA had pleiotropic effects that could explain its attenuation. In the tmRNA mutant, several metabolic networks encompassing the lactose transport system and multiple PTS systems were downregulated. The tmRNA has been associated with deficiencies in stress-response and pathogenicity in other bacteria (Withey and Friedman, 2003; Okan et al., 2006, 2010; Keiler, 2008) and has a central role in the trans-translation mechanism, a RNA and protein quality control system that resolves challenges associated with stalled ribosomes on non-stop mRNAs (Giudice et al., 2014; Shimizu, 2014). This role is consistent with the strong effect in pathogenesis observed in the tmRNA mutant.
All these data provide compelling evidence that sRNAs play important roles in virulence, that their effects can arise at several levels of control, and hence these roles can be restricted to specific host tissues. However, no direct regulatory link was established yet between sRNAs and putative targets.
sRNAs Modulate Competence
Competence is a pivotal mechanism in S. pneumoniae, which regulate the expression of ∼200 genes and is involved in virulence and antibiotic resistance (Lau et al., 2001; Oggioni et al., 2004; Prudhomme et al., 2006; Kowalko and Sebert, 2008). But simultaneously, competence induction is highly stressful for the cell and needs to be tightly controlled. Thus, different layers of regulation in which proteins and regulatory RNAs act in concert to fine-tuning competence activation could be expected.
The first chromosomal-encoded sRNAs described in pneumococcus are part of the regulon of the two-component system CiaRH (Halfmann et al., 2007). These five csRNAs (cia-dependent small RNAs), designated from 1 to 5, are non-coding RNAs between 87 and 151 nt-long with a high degree of similarity to each other. Their predicted secondary structure consists of a stem-loop at the 5′-end and an unpaired region followed by a terminator stem-loop. Sequences complementary to the Shine-Dalgarno and the start codon AUG in the unpaired region suggested that csRNAs may control initiation of translation. CiaRH two-component system has been implicated in β-lactam susceptibility, autolysis, bacteriocin production, competence, and virulence, and some of these functions appear to be mediated by the csRNAs. For instance, stationary-phase autolysis was affected by csRNA4 and csRNA5 (Halfmann et al., 2007), and csRNA5 was defective in lung infectivity (Mann et al., 2012). But one of the most apparent phenotypes associated with CiaRH is blocking of spontaneous competence upon CiaRH activation (Guenzi et al., 1994; Müller et al., 2011). On this regard, csRNA1 was first shown to act negatively on competence development (Tsui et al., 2010), reversing the natural competence induction phenotype of the ΔCiaRH mutant, but no direct link between this csRNA and competence genes as targets could be established. Recently, csRNA target prediction analysis and evaluation by translational fusions identified six genes, which were all downregulated by the csRNAs acting additively (Schnorpfeil et al., 2013). One of these target genes was comC, encoding the precursor of the competence stimulating peptide CSP. Hyperactivation of CiaRH in the absence of csRNAs did not block competence development and partial disruption of comC complementarity to the csRNAs greatly diminished csRNA-mediated repression and relieved competence from CiaRH-dependent control. Therefore, CiaRH competence control is mediated by csRNAs, which block production of CSP precursor thereby inhibiting competence development (Figure 1B). Interestingly, CiaRH also controls production of the serine protease HtrA, which acts negatively on competence by degradation of CSP (Cassone et al., 2012). Which negative CiaRH-dependent control mechanism prevails, csRNA- or HtrA-mediated, depends strongly on growth conditions.
In addition to these five redundant csRNAs, another non-coding RNA, the srn206, has been suggested to participate in competence control. The srn206 is a highly structured 120-nt long RNA that was predicted to associate to the translation initiation region of comD mRNA (Acebo et al., 2012). ComD is the histidine kinase of the ComDE two-component system that, upon induction by CSP, allows the entrance into a competent state of pneumococcal cells (Claverys et al., 2009; Johnston et al., 2014). Binding of CSP to ComD protein results in a phosphorylation cascade that finally activates the transcription of competence genes. Target prediction analysis suggested that srn206 could regulate ComD levels by sequestering its start codon and ribosome-binding site, thereby preventing activation of competence (Figure 1B). In fact, overexpression of srn206 significantly reduced the transformation efficiency of pneumococcal cells in response to exogenous CSP (Acebo et al., 2012). Nevertheless, no direct link between srn206 and ComD translational repression could be established so far and more experiments are required to uncover the precise role of srn206 in competence.
Therefore, although more investigation is required, current data suggests that different pneumococcal sRNAs may participate in competence modulation acting at distinct levels of the competence cascade, resembling the quorum sensing circuits described in other bacteria (Bejerano-Sagie and Xavier, 2007).
Concluding Remarks and Perspectives
As shown above, numerous sRNAs have been identified in S. pneumoniae, but the understanding of sRNA-mediated regulation is largely insufficient and identification of targets and modes of action is still missing. Nevertheless, important aspects have been uncovered. For instance, the use of a multi-organ Tn-seq approach revealed that many sRNAs display global roles in discrete host tissues during disease, and provided a comprehensive list of sRNAs playing distinct roles in pathogenesis in the nasopharynx, the lung or the bloodstream. The analysis of sRNA contribution to pneumococcal pathogenesis in different host sites may provide a framework for future investigations to elucidate the precise function of these sRNAs. Moreover, a regulatory circuit including the concerted action of proteins and regulatory RNAs appears to control activation of competence in pneumococci. In this circuit, the five redundant csRNAs and the srn206 act together, contributing to the maintenance of the competence on–off switch. Interestingly, all streptococcal genomes harbor from two to six csRNAs genes and their expression was validated for some of them (Marx et al., 2010). Therefore, the study of pneumococcal regulatory RNAs may uncover novel sRNA functions in other streptococcal species.
Trans-encoded antisense RNAs often require the action the RNA chaperone Hfq (Vogel and Luisi, 2011). This protein is present in 50% of all sequenced bacterial species, and a few species like Bacillus anthracis encode even two. In Gram-negative bacteria, Hfq is essential for activity and/or stability of most trans-encoded sRNAs. However, in Gram-positives its role is still controversial, and examples of Hfq-dependent antisense regulation have been reported only in Listeria monocytogenes (Nielsen et al., 2010) and Clostridium difficile (Boudry et al., 2014). As other streptococci, S. pneumoniae lacks an homolog of Hfq, and future research is needed to know whether pneumococcal trans-acting sRNAs require RNA chaperones to function or whether they have evolved fundamentally different mechanisms of action. Identification of a pneumococcal Hfq-like protein might be helpful to detect additional sRNAs and to identify targets.
The breadth of pneumococcal species and clinical isolates is an important issue for pneumococcal pathogenesis. They often differed in aspects such as invasiveness or antibiotic resistance, and comparison of their sRNA repertoire may help to elucidate their biological activity. Furthermore, the use of sRNAs as diagnostic tools and platforms for the development of antimicrobial therapies has long been suggested as an important outcome of sRNA studies. Clearly there are many exciting frontiers and unanswered questions in research on bacterial sRNAs and it is likely that important insights will come from breakthroughs in methodology. Understanding the ways that bacteria respond to and influence communities and how they survive such diverse environments will benefit from further studies of sRNAs.
Acknowledgments
This work was supported by the Fondo de Investigación Sanitaria (FIS; PI11/00656) and CIBER Enfermedades Respiratorias (an initiative of the Instituto de Salud Carlos III). PA was the recipient of a contract funded by the FIS project PI11/00656 and JW was the recipient of grant from the Inov Contacto C18 program, attributed by the Agência para o Investimento e Comércio Externo de Portugal with Portuguese and European funds.
Footnotes
References
- Acebo P., Martin-Galiano A. J., Navarro S., Zaballos A., Amblar M. (2012). Identification of 88 regulatory small RNAs in the TIGR4 strain of the human pathogen Streptococcus pneumoniae. RNA 18 530–546 10.1261/rna.027359.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejerano-Sagie M., Xavier K. B. (2007). The role of small RNAs in quorum sensing. Curr. Opin. Microbiol. 10 189–198 10.1016/j.mib.2007.03.009 [DOI] [PubMed] [Google Scholar]
- Bobrovskyy M., Vanderpool C. K. (2013). Regulation of bacterial metabolism by small RNAs using diverse mechanisms. Annu. Rev. Genet. 47 209–232 10.1146/annurev-genet-111212-133445 [DOI] [PubMed] [Google Scholar]
- Bogaert D., De Groot R., Hermans P. W. M. (2004). Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect. Dis. 4 144–154 10.1016/S1473-3099(04)00938-7 [DOI] [PubMed] [Google Scholar]
- Boudry P., Gracia C., Monot M., Caillet J., Saujet L., Hajnsdorf E., et al. (2014). Pleiotropic role of the RNA chaperone protein Hfq in the human pathogen Clostridium difficile. J. Bacteriol. 196 3234–3248 10.1128/JB.01923-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brantl S. (2002). Antisense RNAs in plasmids: control of replication and maintenance. Plasmid 48 165–173 10.1016/S0147-619X(02)00108-7 [DOI] [PubMed] [Google Scholar]
- Caldelari I., Chao Y., Romby P., Vogel J. (2013). RNA-mediated regulation in pathogenic bacteria. Cold Spring Harb. Perspect. Med. 3 a010298 10.1101/cshperspect.a010298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassone M., Gagne A. L., Spruce L. A., Seeholzer S. H., Sebert M. E. (2012). The HtrA protease from Streptococcus pneumoniae digests both denatured proteins and the competence-stimulating peptide. J. Biol. Chem. 287 38449–38459 10.1074/jbc.M112.391482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverys J. P., Martin B., Polard P. (2009). The genetic transformation machinery: composition, localization, and mechanism. FEMS Microbiol. Rev. 33 643–656 10.1111/j.1574-6976.2009.00164.x [DOI] [PubMed] [Google Scholar]
- Croucher N. J., Vernikos G. S., Parkhill J., Bentley S. D. (2011). Identification, variation and transcription of pneumococcal repeat sequences. BMC Genomics 12:120 10.1186/1471-2164-12-120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Solar G., Acebo P., Espinosa M. (1995). Replication control of plasmid pLS1: efficient regulation of plasmid copy number is exerted by the combined action of two plasmid components, CopG and RNA II. Mol. Microbiol. 18 913–924 10.1111/j.1365-2958.1995.18050913.x [DOI] [PubMed] [Google Scholar]
- del Solar G., Acebo P., Espinosa M. (1997). Replication control of plasmid pLS1: the antisense RNA II and the compact rnaII region are involved in translational regulation of the initiator RepB synthesis. Mol. Microbiol. 23 95–108 10.1046/j.1365-2958.1997.1981561.x [DOI] [PubMed] [Google Scholar]
- del Solar G., Espinosa M. (1992). The copy number of plasmid pLS1 is regulated by two trans-acting plasmid products: the antisense RNA II and the repressor protein, RepA. Mol. Microbiol. 6 83–94 10.1111/j.1365-2958.1992.tb00840.x [DOI] [PubMed] [Google Scholar]
- del Solar G., Espinosa M. (2001). In vitro analysis of the terminator T(II) of the inhibitor antisense rna II gene from plasmid pMV158. Plasmid 45 75–87 10.1006/plas.2000.1503 [DOI] [PubMed] [Google Scholar]
- Fineran P. C., Charpentier E. (2012). Memory of viral infections by CRISPR-Cas adaptive immune systems: acquisition of new information. Virology 434 202–209 10.1016/j.virol.2012.10.003 [DOI] [PubMed] [Google Scholar]
- Giudice E., Macé K., Gillet R. (2014). Trans-translation exposed: understanding the structures and functions of tmRNA-SmpB. Front. Microbiol. 5:113 10.3389/fmicb.2014.00113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenzi E., Gasc A. M., Sicard M. A., Hakenbeck R. (1994). A two-component signal-transducing system is involved in competence and penicillin susceptibility in laboratory mutants of Streptococcus pneumoniae. Mol. Microbiol. 12 505–515 10.1111/j.1365-2958.1994.tb01038.x [DOI] [PubMed] [Google Scholar]
- Halfmann A., Kovacs M., Hakenbeck R., Bruckner R. (2007). Identification of the genes directly controlled by the response regulator CiaR in Streptococcus pneumoniae: five out of 15 promoters drive expression of small non-coding RNAs. Mol. Microbiol. 66 110–126 10.1111/j.1365-2958.2007.05900.x [DOI] [PubMed] [Google Scholar]
- Hernández-Arriaga A. M., Rubio-Lepe T. S., Espinosa M., del Solar G. (2009). Repressor CopG prevents access of RNA polymerase to promoter and actively dissociates open complexes. Nucleic Acids Res. 37 4799–4811 10.1093/nar/gkp503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston C., Campo N., Bergé M. J., Polard P., Claverys J.-P. (2014). Streptococcus pneumoniae, le transformiste. Trends Microbiol. 22 113–119 10.1016/j.tim.2014.01.002 [DOI] [PubMed] [Google Scholar]
- Keiler K. C. (2008). Biology of trans-translation. Annu. Rev. Microbiol. 62 133–151 10.1146/annurev.micro.62.081307.162948 [DOI] [PubMed] [Google Scholar]
- Knutsen E., Johnsborg O., Quentin Y., Claverys J.-P., Håvarstein L. S. (2006). BOX elements modulate gene expression in Streptococcus pneumoniae: impact on the fine-tuning of competence development. J. Bacteriol. 188 8307–8312 10.1128/JB.00850-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalko J. E., Sebert M. E. (2008). The Streptococcus pneumoniae competence regulatory system influences respiratory tract colonization. Infect. Immun. 76 3131–3140 10.1128/IAI.01696-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R., Shah P., Swiatlo E., Burgess S. C., Lawrence M. L., Nanduri B. (2010). Identification of novel non-coding small RNAs from Streptococcus pneumoniae TIGR4 using high-resolution genome tiling arrays. BMC Genomics 11:350 10.1186/1471-2164-11-350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau G. W., Haataja S., Lonetto M., Kensit S. E., Marra A., Bryant A. P., et al. (2001). A functional genomic analysis of type 3 Streptococcus pneumoniae virulence. Mol. Microbiol. 40 555–571 10.1046/j.1365-2958.2001.02335.x [DOI] [PubMed] [Google Scholar]
- Livny J., Brencic A., Lory S., Waldor M. K. (2006). Identification of 17 Pseudomonas aeruginosa sRNAs and prediction of sRNA-encoding genes in 10 diverse pathogens using the bioinformatic tool sRNAPredict2. Nucleic Acids Res. 34 3484–3493 10.1093/nar/gkl453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livny J., Teonadi H., Livny M., Waldor M. K. (2008). High-throughput, kingdom-wide prediction and annotation of bacterial non-coding RNAs. PLoS ONE 3:e3197 10.1371/journal.pone.0003197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Aguilar C., del Solar G. (2013). Probing the sequence and structure of in vitro synthesized antisense and target RNAs from the replication control system of plasmid pMV158. Plasmid 70 94–103 10.1016/j.plasmid.2013.02.005 [DOI] [PubMed] [Google Scholar]
- López-Aguilar C., Ruiz-Masó J. A., Rubio-Lepe T. S., Sanz M., del Solar G. (2013). Translation initiation of the replication initiator repB gene of promiscuous plasmid pMV158 is led by an extended non-SD sequence. Plasmid 70 69–77 10.1016/j.plasmid.2013.01.011 [DOI] [PubMed] [Google Scholar]
- Mann B., van Opijnen T., Wang J., Obert C., Wang Y.-D., Carter R., et al. (2012). Control of virulence by small RNAs in Streptococcus pneumoniae. PLoS Pathog. 8:e1002788 10.1371/journal.ppat.1002788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx P., Nuhn M., Kovacs M., Hakenbeck R., Bruckner R. (2010). Identification of genes for small non-coding RNAs that belong to the regulon of the two-component regulatory system CiaRH in Streptococcus. BMC Genomics 11:661 10.1186/1471-2164-11-661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M., Marx P., Hakenbeck R., Brückner R. (2011). Effect of new alleles of the histidine kinase gene ciaH on the activity of the response regulator CiaR in Streptococcus pneumoniae R6. Microbiol. Read. Engl. 157 3104–3112 10.1099/mic.0.053157-0 [DOI] [PubMed] [Google Scholar]
- Nielsen J. S., Lei L. K., Ebersbach T., Olsen A. S., Klitgaard J. K., Valentin-Hansen P., et al. (2010). Defining a role for Hfq in Gram-positive bacteria: evidence for Hfq-dependent antisense regulation in Listeria monocytogenes. Nucleic Acids Res. 38 907–919 10.1093/nar/gkp1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien K. L., Wolfson L. J., Watt J. P., Henkle E., Deloria-Knoll M., McCall N., et al. (2009). Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 374 893–902 10.1016/S0140-6736(09)61204-6 [DOI] [PubMed] [Google Scholar]
- Oggioni M. R., Iannelli F., Ricci S., Chiavolini D., Parigi R., Trappetti C., et al. (2004). Antibacterial activity of a competence-stimulating peptide in experimental sepsis caused by Streptococcus pneumoniae. Antimicrob. Agents Chemother. 48 4725–4732 10.1128/AAC.48.12.4725-4732.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okan N. A., Bliska J. B., Karzai A. W. (2006). A Role for the SmpB-SsrA system in Yersinia pseudotuberculosis pathogenesis. PLoS Pathog. 2:e6 10.1371/journal.ppat.0020006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okan N. A., Mena P., Benach J. L., Bliska J. B., Karzai A. W. (2010). The smpB-ssrA mutant of Yersinia pestis functions as a live attenuated vaccine to protect mice against pulmonary plague infection. Infect. Immun. 78 1284–1293 10.1128/IAI.00976-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudhomme M., Attaiech L., Sanchez G., Martin B., Claverys J.-P. (2006). Antibiotic stress induces genetic transformability in the human pathogen Streptococcus pneumoniae. Science 313 89–92 10.1126/science.1127912 [DOI] [PubMed] [Google Scholar]
- Saramago M., Bárria C., Dos Santos R. F., Silva I. J., Pobre V., Domingues S., et al. (2014). The role of RNases in the regulation of small RNAs. Curr. Opin. Microbiol. 18 105–115 10.1016/j.mib.2014.02.009 [DOI] [PubMed] [Google Scholar]
- Schnorpfeil A., Kranz M., Kovács M., Kirsch C., Gartmann J., Brunner I., et al. (2013). Target evaluation of the non-coding csRNAs reveals a link of the two-component regulatory system CiaRH to competence control in Streptococcus pneumoniae R6. Mol. Microbiol. 89 334–349 10.1111/mmi.12277 [DOI] [PubMed] [Google Scholar]
- Shimizu Y. (2014). Biochemical aspects of bacterial strategies for handling the incomplete translation processes. Front. Microbiol. 5:170 10.3389/fmicb.2014.00170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz G., Vogel J., Wassarman K. M. (2011). Regulation by small RNAs in bacteria: expanding frontiers. Mol. Cell 43 880–891 10.1016/j.molcel.2011.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Arana A., Repoila F., Cossart P. (2007). Small non-coding RNAs controlling pathogenesis. Curr. Opin. Microbiol. 10 182–188 10.1016/j.mib.2007.03.004 [DOI] [PubMed] [Google Scholar]
- Tsui H. C., Mukherjee D., Ray V. A., Sham L. T., Feig A. L., Winkler M. E. (2010). Identification and characterization of non-coding small RNAs in Streptococcus pneumoniae serotype 2 strain D39. J. Bacteriol. 192 264–279 10.1128/JB.01204-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J., Luisi B. F. (2011). Hfq and its constellation of RNA. Nat. Rev. Microbiol. 9 578–589 10.1038/nrmicro2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters L. S., Storz G. (2009). Regulatory RNAs in bacteria. Cell 136 615–628 10.1016/j.cell.2009.01.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler W. C., Breaker R. R. (2005). Regulation of bacterial gene expression by riboswitches. Annu. Rev. Microbiol. 59 487–517 10.1146/annurev.micro.59.030804.121336 [DOI] [PubMed] [Google Scholar]
- Withey J. H., Friedman D. I. (2003). A salvage pathway for protein structures: tmRNA and trans-translation. Annu. Rev. Microbiol. 57 101–123 10.1146/annurev.micro.57.030502.090945 [DOI] [PubMed] [Google Scholar]
- Zhang J., Lau M. W., Ferré-D’Amaré A. R. (2010). Ribozymes and riboswitches: modulation of RNA function by small molecules. Biochemistry 49 9123–9131 10.1021/bi1012645 [DOI] [PMC free article] [PubMed] [Google Scholar]