Abstract
Previous studies in head and neck squamous cell carcinoma (HNSCC) cell lines have revealed that the Ah receptor (AHR) plays a significant role in mediating the ‘aggressive’ phenotype of these cells, which includes enhanced inflammatory signaling (e.g. IL6) and migratory potential. Here we sought to identify putative novel targets of the AHR associated with enhanced tumor invasiveness. Global gene expression analysis identified a number of genes that are repressed upon treatment of OSC-19 or HN30 cells with an AHR antagonist. Three growth factors were targets of AHR activity; amphiregulin (AREG), epiregulin (EREG) and platelet-derived growth factor A (PDGFA) were repressed by an AHR antagonist and further examined. Quantitative PCR analysis, ELISA and siRNA-mediated knock down of AHR revealed an attenuation of basal and/or induced levels of expression of these growth factors in two HNSCC lines, following AHR antagonism. In silico analysis revealed that these growth factors possess dioxin-like response elements. Two other AHR ligands, 6-formylindolo[3,2-b]carbazole and benzo(a)pyrene also elicited similar responses. In conclusion, this study identified AREG, EREG and PDGFA as growth factor targets of AHR activity associated with metastatic phenotype of HNSCC cells, suggesting that attenuation of AHR activity may be a therapeutic strategy.
Keywords: AHR, aryl hydrocarbon receptor, growth factors, head and neck cancer, antagonist, dioxin
INTRODUCTION
The aryl hydrocarbon receptor (AHR), a ligand-activated basic helix-loop-helix/PER/ARNT/Sim transcription factor, is a xenosensor traditionally associated with xenobiotic metabolism [1]. The canonical signaling pathway typically involves unliganded AHR being tethered in a tetrameric complex composed of a dimer of heat shock protein 90, the X-associated protein 2 and a co-chaperone, p23, in the cytoplasm. Following ligand mediated activation, a conformational change in the receptor is thought to expose its nuclear localization signal. This leads the receptor to undergo translocation to the nucleus and subsequent heterodimerization with a structurally related nuclear protein, the aryl hydrocarbon nuclear translocator (ARNT), along with concomitant displacement of its cytosolic binding partners. The ligand bound AHR:ARNT complex is subsequently able to bind dioxin response elements (DREs) in the upstream region of various AHR responsive genes, often leading to their enhanced transcription [2]. A variety of exogenous and endogenous ligands of AHR have been identified, with 2,3,7,8 tetrachlorodibenzo-p-dioxin (TCDD) still being one of the most potent exogenous ligands [3].
Polycyclic aromatic hydrocarbons, halogenated aromatic hydrocarbons, polychlorinated biphenyls and several other related environmental pollutants have long been known to contain ligands for the AHR. AHR activation subsequently leads to altered metabolism, which often results in enhanced toxicity [4,5]. However, more recent studies have pointed to several ‘non-traditional’ roles for the AHR as well. These include modulation of drug metabolism [6], cholesterol metabolism [7], immune responses [8], growth and development, cellular proliferation, differentiation, apoptosis, circadian rhythm and ubiquitination of steroid hormone receptors. Several of these processes are key to cancer cell survival and tumor progression [9]. In fact, numerous studies have shown that the AHR plays a role in tumor initiation, promotion and progression [5].
We have published studies, which indicate that MCF-7 and ECC-1 cells exhibit poor inducibility of IL6 in the presence of cytokines. However, the addition of an AHR agonist and a cytokine results in a highly synergistic induction of IL6 [10–12]. The mechanism of this effect was determined to involve transcriptionally active AHR, which maintains the IL6 promoter in a de-repressed state by displacing histone deacetylase 1-containing co-repressor complexes. This in turn makes the IL6 promoter more accessible to the transcriptional machinery and subsequent acetylation of p65 [11]. A search of the literature revealed that certain tumor types exhibit relatively high IL6 production, such as head and neck tumors. This lead to the hypothesis that high constitutive IL6 expression may at least in part, be due to constitutive activation of the AHR. Further analysis has determined that AHR antagonism greatly mitigates inflammatory cytokine production, ultimately reducing the migratory and invasive phenotype of these cells [13]. It is generally believed that a high basal expression of certain inflammatory cytokines may contribute at least in part to the aggressive phenotype of certain cancers and we have established that the AHR participates in this enhanced IL6 expression. Thus, attenuation of AHR activity may constitute a viable strategy to mitigate pro-inflammatory cytokine secretion and in turn the metastatic phenotype of certain cancers.
Current treatment options for most cancers typically involve surgery followed by radiotherapy or chemotherapy. A combination of chemoradiotherapy, rather than individual therapies, has been found to yield better survival rates, especially for cases of locally advanced or recurrent head and neck squamous cell carcinomas (HNSCCs) [14]. However, despite these aggressive therapies, at least 50% of patients with locally advanced HNSCCs develop either locoregional or distant relapses within 2 years of treatment, demanding interdisciplinary, novel treatment approaches that would be curative rather than merely palliative [14–16]. HNSCCs constitute the 8th most common cancer in the US with a median overall survival rate of less than a year for recurrent or metastatic HNSCCs, despite the availability of third generation chemotherapeutic drugs and targeted therapy [15]. Therefore, identification of new targets for therapeutic intervention should constitute an effective and novel approach for use in ‘combination’ treatments with existing therapies.
In this report we examined whole genome expression profiles of two highly metastatic head and neck tumor cell lines, OSC-19 and HN30, following treatment with either TCDD or the AHR antagonist, CH223191 [17], to determine additional putative targets of AHR. Regulation of AHR activity may constitute a novel strategy to reduce the highly metastatic and malignant phenotype of these cells. Growth factors have been documented as participants in the aggressive phenotype of cancer. Along with cytokines, they have been shown to play a prominent role in dictating tumor cell malignancy [18,19]. Therefore, one therapeutic approach for cancers that exhibit aberrant growth factor signaling, would target repression of growth factor secretion and their subsequent downstream signaling pathways. Given that previous studies from this laboratory have already documented that HNSCC cells possess relatively high basal levels of inflammatory cytokines, which likely contribute to their malignant phenotype, the study presented here establishes that the AHR also plays a role in driving the expression of several key growth factors in these aggressive carcinoma cell lines.
MATERIALS AND METHODS
Chemicals
2,3,7,8 tetrachlorodibenzo-p-dioxin (TCDD) was a gift from Dr. Stephen Safe, Texas A&M University. CH223191 and 6-formylindolo[3, 2-b]carbazole (FICZ) was purchased from ChemBridge Corporation (San Diego, CA) and Enzo Life Sciences (Farmington, NY), respectively.
Cell culture and treatments
OSC-19 and HN30 head and neck squamous cell carcinoma (HNSCC) cell lines were kindly provided by Dr. Jeffrey Meyers (MD Anderson Cancer Center) and J. Silvo Gutkind (NIH), respectively. OSC-19 cells were cultured in α MEM media supplemented with 10% fetal bovine serum (FBS, Hyclone Labs), 1% sodium pyruvate and 1000 units/ml penicillin and 0.1mg/ml streptomycin (Sigma Aldrich, St. Louis, MO). While HN30 cells were cultured in high glucose 1:1 DMEM:F12 (Sigma Aldrich, St. Louis, MO) supplemented with 10% fetal bovine serum (Hyclone Labs) and 1000 units/ml penicillin and 0.1mg/ml streptomycin.
Both cell lines were subjected to one of the following treatment regimes: (a) vehicle (DMSO), (b) TCDD (10 nM), (c) CH223191 (500 nM) or (d) pre-treatment with CH223191 (500 nM) for 2 h followed by post-treatment with TCDD (500 nM). In general, all treatments were carried out in serum-free media supplemented with 5 mg/ml bovine serum albumin (BSA). All treatments were for 24 h, and CH223191 was added every 12 h unless otherwise mentioned. To examine the effect of other prototypic AHR ligands on the expression of putative AHR targets, both OSC-19 and HN30 cells were also treated with FICZ, or benzo(a)pyrene for 24 h, in addition to treatments (a), (b) (c) and (d). To show the effect of an inflammatory signal, such as IL1B, in the absence or presence of the AHR ligand, TCDD, IL1B was added at a concentration of 10 ng/ml and incubated for 24 h. Therefore these studies utilized treatments (a), (b) (c) and (d) in addition to pre-treatment with CH223191 (500 nM) for 2 h followed by post-treatment with IL1B (10 ng/ml) alone, treatment with TCDD (500 nM) and IL1B (10 ng/ml) in combination as well as pre-treatment with CH223191 (500 nM) for 2 h, followed by post-treatment with TCDD (500 nM) and IL1B (10 ng/ml).
Lactate Dehydrogenase (LDH) assay
Cytotoxicity of CH223191 in media and lysates of OSC-19 and HN30 cells treated for 24 h, was measured using an LDH based in vitro toxicology assay kit (Sigma Aldrich, St. Louis, MO) as per manufacturer’s instructions.
Microarray analysis
Total RNA, >200 nucleotides long, from OSC-19 and HN30 cells subjected to treatment regimes (a), (b) and (c), was isolated using the RNeasy mini kit (Qiagen, Valencia, CA) according to manufacturer’s instructions. RNA integrity was determined using the NanoDrop 2000 (Thermo Scientific, Wilmington, DE) and 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). RNA (260/280 >2.0 and 260/230 1.5–2.0 as determined by the Nanodrop and RIN ≥ 9.0 as determined by the Bioanalyzer) was used for further analysis. Subsequent generation of cDNA, labeling and generation of in vitro transcribed cRNA, fragmentation of cRNA, hybridization, washing and staining of the arrays (Human Genome 133 Plus 2.0 arrays, Affymetrix, Santa Clara, CA) was carried out according to the manufacturer’s instructions (Affymetrix, Santa Clara, CA) at the Pennsylvania State University microarray core facility, University Park, PA. Data, following normalization, was subsequently analyzed using ArrayStar (DNAStar, Madison, WI) and Ingenuity Pathway Analysis (Ingenuity Systems, Redwood City, CA).
Gene expression
Following analysis of microarray data, select gene targets considered altered were analyzed by quantitative real-time PCR (qPCR) using a Biorad MyiQ single color real time PCR detection system equipped with MyiQ software v1.0.410 (Biorad laboratories, Hercules, CA). One microgram total RNA, extracted from OSC-19 and HN30 cells subject to the treatment regimes (a), (b), (c) and (d), was reverse transcribed to cDNA (ABI high capacity cDNA archive kit, Applied Biosystems, Carlsbad, CA), diluted 1:10 and used for the quantitation of gene expression changes using the appropriate primers and Perfecta SYBR Green supermix (Quanta Biosciences, Gaithersburg, MD) according to manufacturer’s protocol. The sequence of the primers used is listed in Supplementary Table S1. Data was analyzed using the ‘standard curve’ method where the starting quantity of transcripts of the gene of interest was normalized to that of a housekeeping gene, following comparison to a standard curve [20].
ELISA assay
OSC-19 and HN30 cells were subject to treatment regimes (a), (b), (c) and (d) for a total of 36 h, media was collected, centrifuged at 140 g for 5 min., aliquoted and stored at −20°C until further use. The remaining cells were washed once with 1X phosphate buffered saline (PBS, pH 7.4), lysed in RIPA buffer (10 mM TrisHCl, pH 8.0, 1 mM EDTA, 0.5 mM EGTA, 140 mM NaCl, 1% TritonX-100, 0.1% sodium deoxycholate, 0.1% SDS and 300 mM sodium molybdate) containing 1X proteosomal inhibitor cocktail (EMD Millipore, Billerica, MA), centrifuged at 12000g for 15 min at 4°C to remove any cellular debris, aliquoted and stored at −20°C until further use. CH223191 was dosed at 9 h intervals. Amphiregulin (AREG), epiregulin (EREG), fibroblast growth factor 2 (FGF2) and platelet derived growth factor A (PDGFA) were quantified using ELISA kits for the respective proteins according to manufacturer’s instructions (AREG and PDGFA- Abcam, Cambridge, MA, FGF2-Biolegend, San Diego, CA and EREG-Uscn Life Science Inc., Wuhan, China).
‘Promoter Scan’ for the presence of DRE-like elements
AREG, EREG, FGF2 and PDGFA were scanned for the presence of DRE-like consensus sequences (G/T N T/G CGTG A/C) in all possible orientations. 2500 bp upstream of the transcription start site was scanned using SCOPE v 2.1.0 [21]. The March 2006 (NCBII36/hg18) assembly of the Human genome was used by SCOPE for the analysis.
Gene silencing
Repression of AHR expression was carried out using Dharmacon small interfering RNA (siRNA) [control oligo D-001810-0X and AHR oligo J-004990-07]. Cells (2 × 106) along with either control oligo or AHR oligo (final concentration 1.4 µmol/L) in 100 µl nucleofection solution (272 mM sucrose +1 mM magnesium chloride) were electroporated using the Amaxa Nucleofector 2b device (Lonza, Allendale, NJ) as per manufacturer’s protocol using the manufacturer’s preset T-016 program.
Statistical analysis
All treatments were carried out in triplicate. Analysis was performed using Excel (Microsoft Corporation, Redmond, WA) and GraphPad Prism 5 v5.03 graphing and statistical software (GraphPad Software Inc., San Diego, CA). Data was analyzed by one way analysis of variance (ANOVA), Tukey’s multiple comparison or student-t tests. P values < 0.05 were considered statistically significant (* p<0.05; ** p<0.01; *** p<0.001).
RESULTS
Global gene expression analyses identify putative novel growth factor targets of AHR activity
We have previously shown that HNSCC cell lines have elevated inflammatory signaling [11]. For this study we utilized two well-characterized HNSCC cell lines. OSC-19 cells were chosen because these HNSCC cells are a model for head and neck cancer invasion and metastasis in orthotropic nude mice [22], while NH30 cells have been used extensively to study various aspects of head and neck cancer [23,24]. Gene expression analysis was performed with RNA isolated from OSC-19 and HN30 cells treated with either TCDD or CH223191. The latter compound has exhibited potent AHR antagonist activity, has not been reported to exhibit any other biological activities and the appropriate concentrations to use in cell culture experiments have been determined [17]. A comparison of TCDD versus CH223191 treatment revealed 264 and 256 genes altered, respectively. A total of 58 probesets, representing ~ 54 genes, were altered in both OSC-19 and HN30 cell lines. Of these 58 probe sets, 41 were up-regulated by comparison of control versus TCDD treated cells and significantly down-regulated by CH223191 treatment, relative to TCDD (Table 1). Further analysis by network mapping of these commonly altered probesets, using Ingenuity Pathway analysis (IPA, Redwood City, CA), revealed ~24 altered gene targets, with 7 sharing no connectivity to the rest of the network (Figure S1 and S2). All of these probesets constituted genes whose expression was significantly down-regulated by CH223191 relative to cells treated with TCDD. A subset of 24 genes was validated by qPCR and ANGPLT4, CTNNA1, HAS3, MMP1 and RUNX2 were all altered by modulation of AHR activity (Figure S3). However, these genes were not further examined.
Table 1.
Gene expression values of genes up-regulated following treatment with TCDD and significantly down-regulation by the AHR antagonist, CH223191, relative to cells treated with TCDD alone
| Gene | DMSO vs. TCDDa |
DMSO vs. CHa |
TCDD vs. CHa |
DMSO vs. TCDDb |
DMSO vs. CHb |
TCDD vs. CHb |
|---|---|---|---|---|---|---|
| Cytokines | ||||||
| IL1B | 1.97c | −1.70 | −3.34 | 1.70 | −1.20 | −1.95 |
| IL6 | 2.22 | −1.68 | −3.74 | 3.08 | −1.14 | −3.52 |
| Growth factors | ||||||
| PDGFA | 15.06 | −1.38 | −14.06 | 2.26 | −1.16 | −2.38 |
| FGF2 | 1.74 | −1.26 | −2.19 | 1.24 | −1.43 | −1.77 |
| EREG | 2.99 | −1.68 | −5.02 | 1.60 | −1.49 | −2.38 |
| Enzymes | ||||||
| AGPAT9 | 2.55 | −1.28 | −3.28 | 3.03 | −1.74 | −5.28 |
| CYP1A1 | 36.89 | −41.78 | −1541.33 | 28.50 | −5.34 | −152.21 |
| CYP1B1 | 31.53 | −8.12 | −256.01 | 10.56 | −2.48 | −26.17 |
| GDA | 3.48 | −1.55 | −5.23 | 2.56 | −1.81 | −4.65 |
| PTGS2 | 3.01 | −2.18 | −6.57 | 3.57 | −1.60 | −5.17 |
| HAS3 | 2.87 | −1.68 | −4.82 | 2.50 | −1.58 | −3.94 |
| Transporters | ||||||
| ABCG2 | 2.18 | −1.39 | −3.03 | 3.32 | −1.41 | −4.68 |
| SLC22A4 | 2.90 | −1.35 | −3.90 | 2.31 | −1.03 | −2.38 |
| SLC3A2 | 1.91 | −1.27 | −2.42 | 1.92 | −1.25 | −2.39 |
| Transcription factors | ||||||
| FOSL1 | 2.95 | −1.92 | −5.66 | 3.02 | −1.53 | −4.64 |
| RUNX2 | 3.52 | −1.33 | −4.68 | 3.31 | −1.12 | −3.71 |
| Others | ||||||
| GPR110 | 1.87 | −1.36 | −2.55 | 2.23 | −1.01 | −2.25 |
| AHRR | 5.32 | −2.24 | −11.89 | 5.96 | −1.12 | −6.69 |
| ANXA10 | 1.91 | −1.55 | −2.96 | 2.99 | −1.23 | −3.69 |
| SERPINB2 | 7.00 | −2.52 | −17.67 | 89.40 | −3.83 | −342.32 |
| TIPARP | 2.83 | −1.27 | −3.58 | 4.52 | −1.36 | −6.14 |
| CTNNA1 | 1.31 | −1.10 | −1.44 | 1.04 | −1.04 | −1.08 |
| MMP1 | 1.79 | 1.03 | −1.73 | 2.33 | −1.38 | −3.22 |
| ANGPTL4 | 1.20 | −1.42 | −1.70 | 4.63 | −1.54 | −7.12 |
OSC-19 cells;
HN-30;
Numbers represent fold change with a ‘−‘ sign indicating down-regulation
Expression of multiple growth factors was mitigated by the AHR antagonist, CH223191
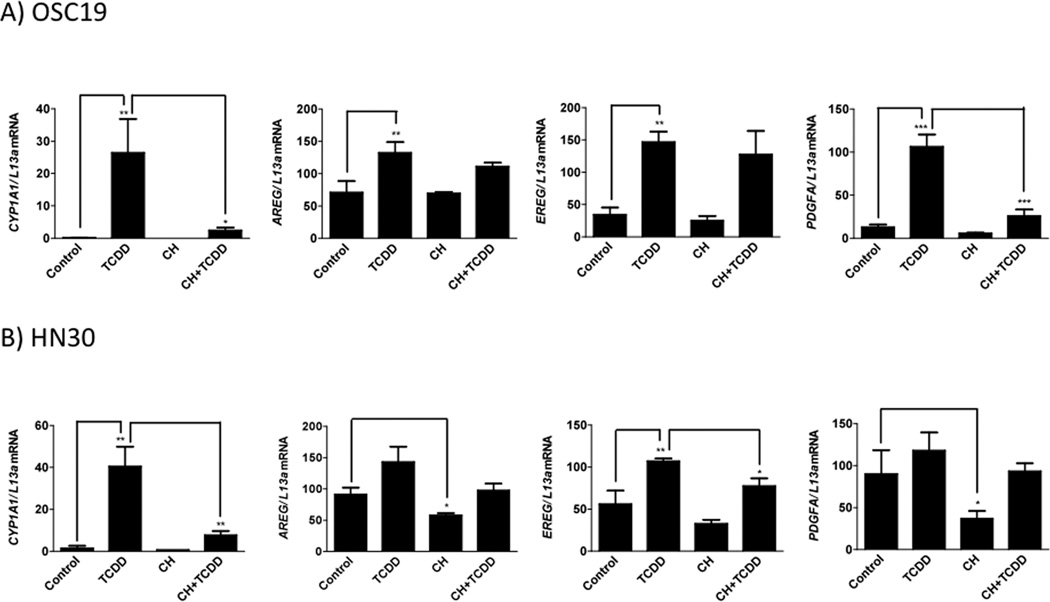
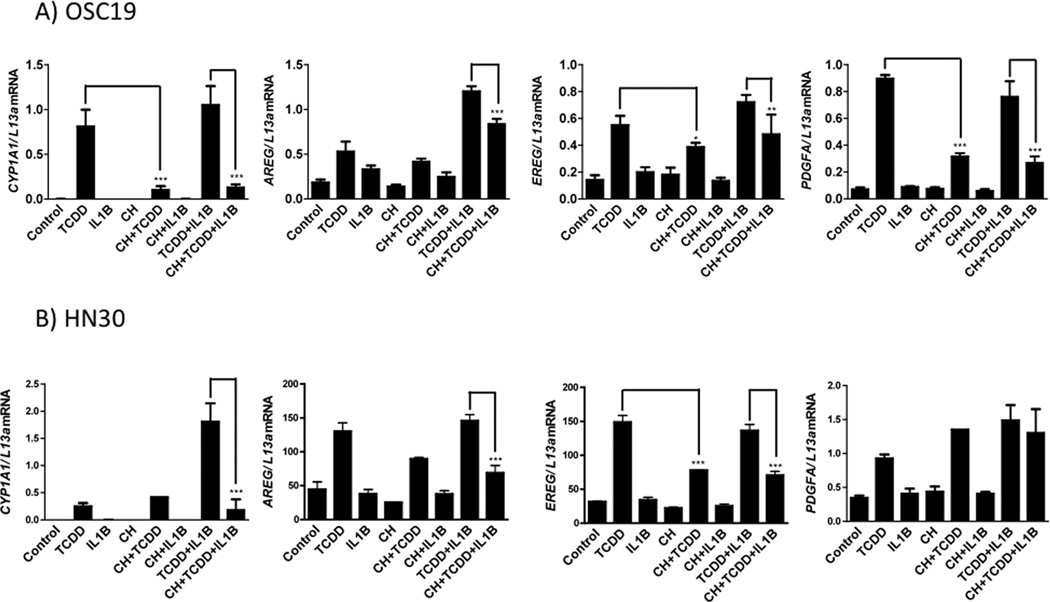
Epiregulin (EREG), fibroblast growth factor 2 (FGF2) and platelet derived growth factor A (PDGFA) were altered growth factors identified within the microarray dataset. These growth factors exhibited an enhanced expression upon TCDD treatment and subsequent down-regulation following the addition of CH223191 in the presence or absence of TCDD. Though absent from the network, AREG was considered on the basis of other ongoing studies in our laboratory. Next, the expression profiling data was validated using qRT-PCR. The growth factors examined exhibited an attenuation of both basal and induced gene expression in both OSC-19 and HN30 cells (Figure 1). For example, the basal gene expression of AREG and PDGFA was significantly mitigated in HN30 cells upon CH223191 treatment. Furthermore, TCDD-induced expression of PDGFA and EREG was significantly decreased by CH223191 in OSC-19 and HN30, respectively. The expression of the AHR target gene CYP1A1 was determined to assess ligand-mediated modulation of AHR activity. FGF2 was not induced upon TCDD treatment in HN30 cells and also failed to display a significant attenuation of basal or induced expression, in both OSC-19 and HN30 cells (Figure S4). Overall, these results confirm that the expression of EREG, AREG and PDGFA are significantly modulated by the level of AHR activity in HNSCC cell lines.
Figure 1.
Gene expression analysis of putative AHR target genes and CYP1A1. OSC-19 (A) and HN30 (B) cells were treated with either TCDD (10 nM), CH223191 (500 nM) or both for 24 h. RNA isolated and mRNA levels determined by qRT-PCR
Protein levels of multiple growth factors were attenuated by CH223191 exposure
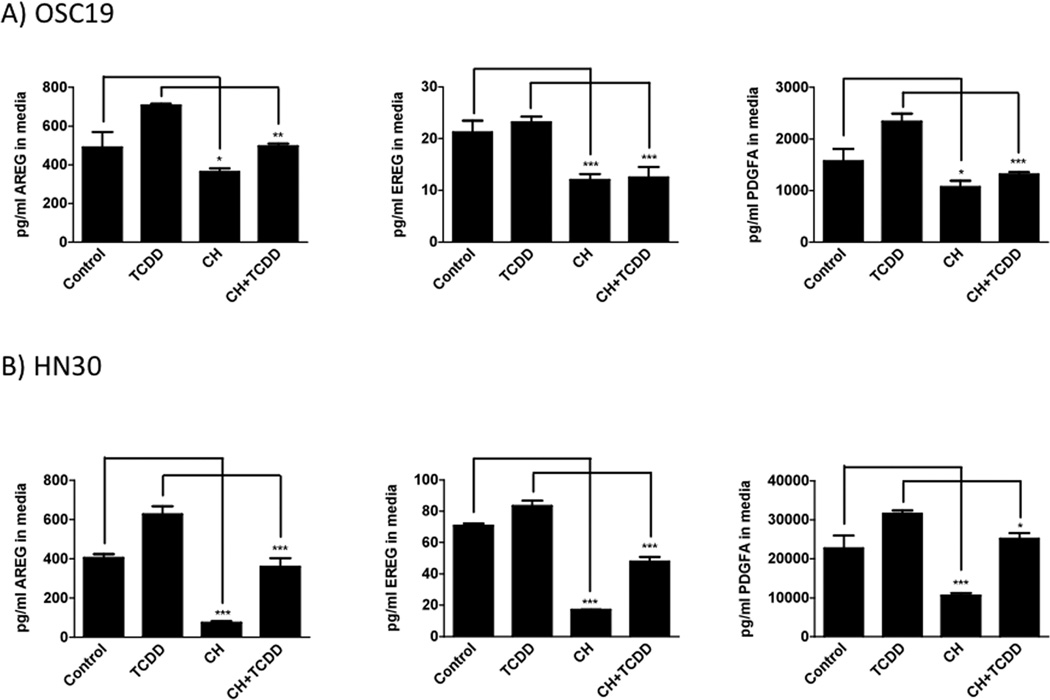
Basal protein and TCDD-induced secreted protein levels of AREG, EREG and PDGFA were significantly attenuated by CH223191 treatment in both OSC-19 and HN30 cells (Figure 2). Greater attenuation of constitutive expression of all three growth factors was observed in HN30 cells. TCDD-induced significant attenuation of FGF2 secreted protein expression in OSC-19 cell culture medium with no reduction of either basal or TCDD-induced expression observed in HN30 cell culture medium (Figure S5).
Figure 2.
Secreted protein expression levels of growth factors modulated by the AHR.
OSC-19 (A) and HN30 (B) cells were treated with either TCDD (10 nM), CH223191 (500 nM) or both for 24 h. ELISAs were used to quantitate growth factor levels.
The AHR antagonist, CH22391 is not cytotoxic
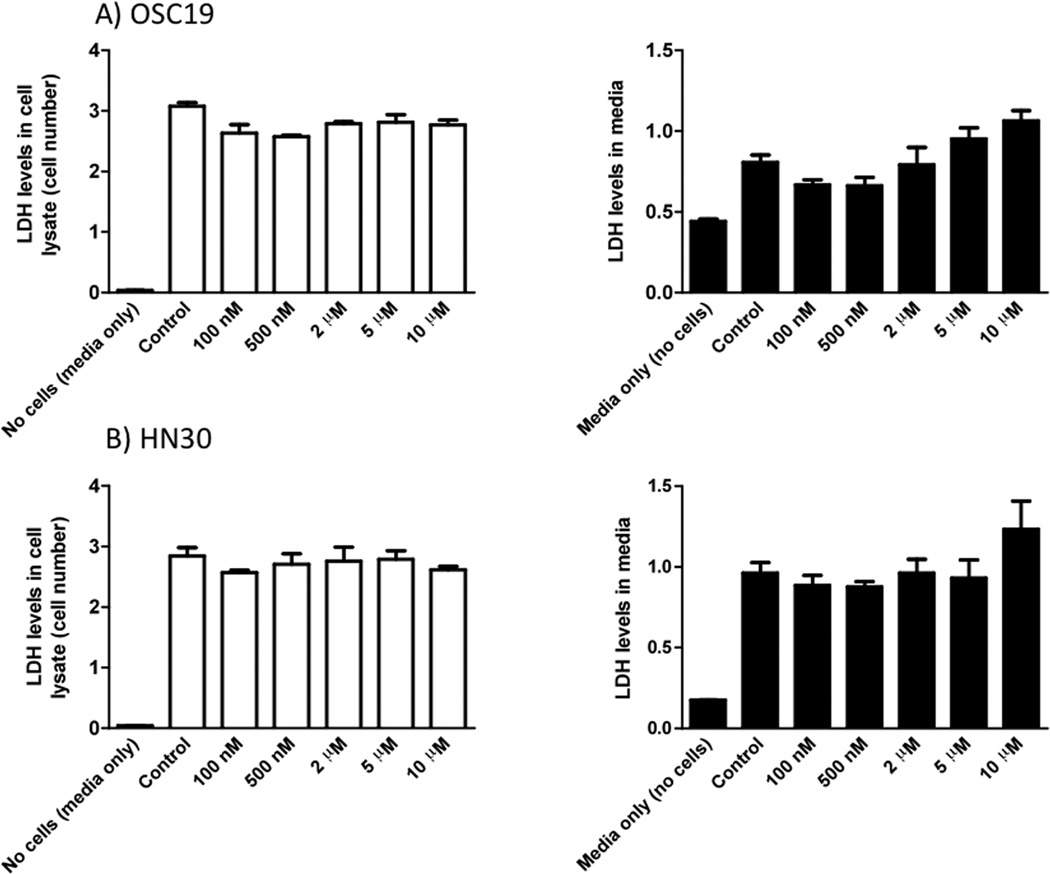
A 500 nM concentration of the AHR antagonist, CH223191, which caused attenuation of basal and/or TCDD induced growth factor protein levels did not influence LDH levels, either in cell lysates (a measure of cell number and cytotoxicity) or media (a measure of cell viability and membrane integrity) following treatment of OSC-19 or HN30 cells for 24 h (Figure 3). Although, the highest dose of CH223191 (10 µM) appeared to cause an increase in LDH levels in culture media, suggesting a slight decrease in cell viability at this concentration. However, it should be noted that this dose is 20-fold higher than the concentration of antagonist used for these studies. TCDD (10 nM final concentration) was also tested, using the LDH leakage assay, for its ability to induce toxicity and no toxicity after 24 h was observed (unpublished data).
Figure 3.
AHR antagonists are not cytotoxic. Analysis of LDH levels in whole cell lysates and cell culture media 24 h post treatment in both OSC-19 and HN30 cells.
In silico analysis identifies DRE-like elements in upstream promoters of AHR regulated growth factors
These results clearly indicate that AHR activity modulates the expression of the growth factors examined, which lead to the hypothesis that these factors are directly regulated through the presence of DRE in their promoters. Thus, starting at the transcription start site and going up to 2500 bases upstream in the regulatory region these growth factors were scanned for the presence of DRE-like elements by searching for the DRE consensus sequence, ‘G/T N T/G CGTG A/C’ in all possible orientations. A total of 20 DRE-like consensus sequences were found distributed across the four growth factors (Figure 4). The sequence logo obtained following a search with the above consensus sequence across all four growth factor genes, along with a display of the exact locations of the DRE-like elements, can be found in the supplementary data (Figure S6, Table S2).
Figure 4.
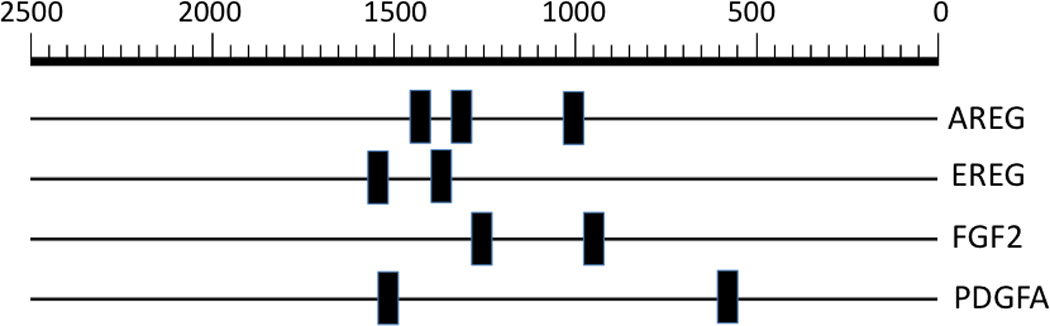
All growth factors identified possess DRE-like elements in their upstream promoters. Location of the consensus sequence ‘G/T N T/G CGTG A/C’, in all possible orientations, up to 2500 bp upstream of the transcription start site in AREG, EREG, FGF2 and PDGFA. Sequence logos for the respective consensus sequences along with a table displaying exact locations of the various consensus sequences in the promoter regions of AREG, EREG, FGF2 and PDGFA can be found in the supplementary data. The March 2006 (NCBII36/hg18) assembly of the human genome was used for this analysis.
Attenuation of AHR levels mitigates the expression of AHR regulated growth factors
OSC-19 cells transfected with AHR siRNA exhibited a 47% and 90% attenuation of AHR protein levels, respectively, relative to cells transfected with control siRNAs (Figure 5). Subsequent analysis of gene expression, following knock down of AHR expression, exhibited a repression of constitutive and/or TCDD induced gene expression of multiple growth factors, in OSC-19 (Figure 6). The expression of CYP1A1 is shown for reference of AHR activity. Treatment with AHR ligands failed to exhibit a significant level of repression of basal or induced levels of FGF2 (Figure S7). We were unable to obtain a sufficient level of AHR knockdown in HN30 cells.
Figure 5.
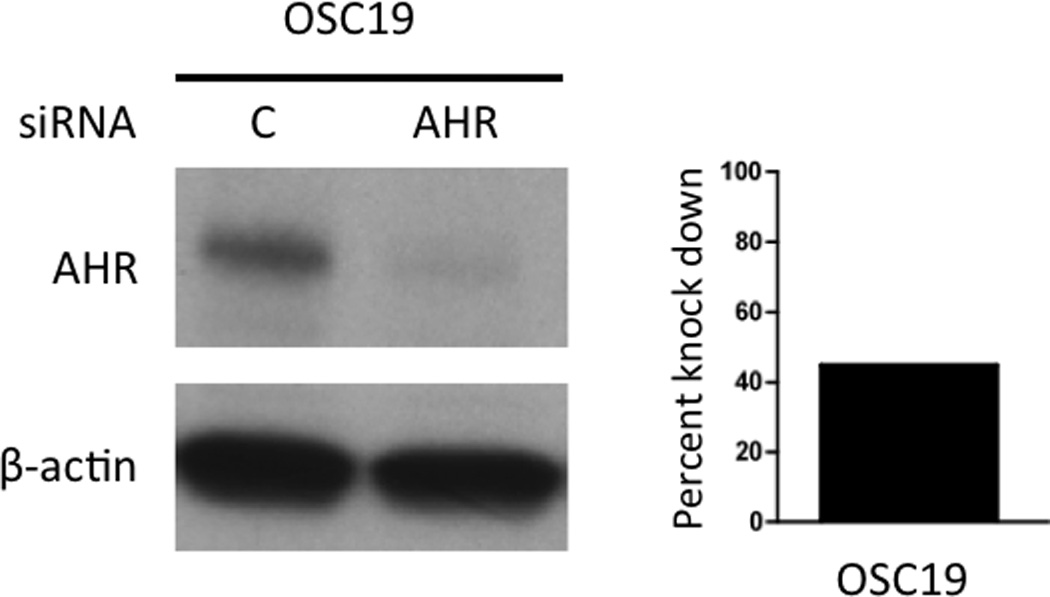
Attenuation of AHR expression in OSC-19 cells. (A) Immunoblot analysis following knock down of AHR by electroporation with siRNA. Results were normalized to the expression of β actin and the percent of AHR repression determined.
Figure 6.
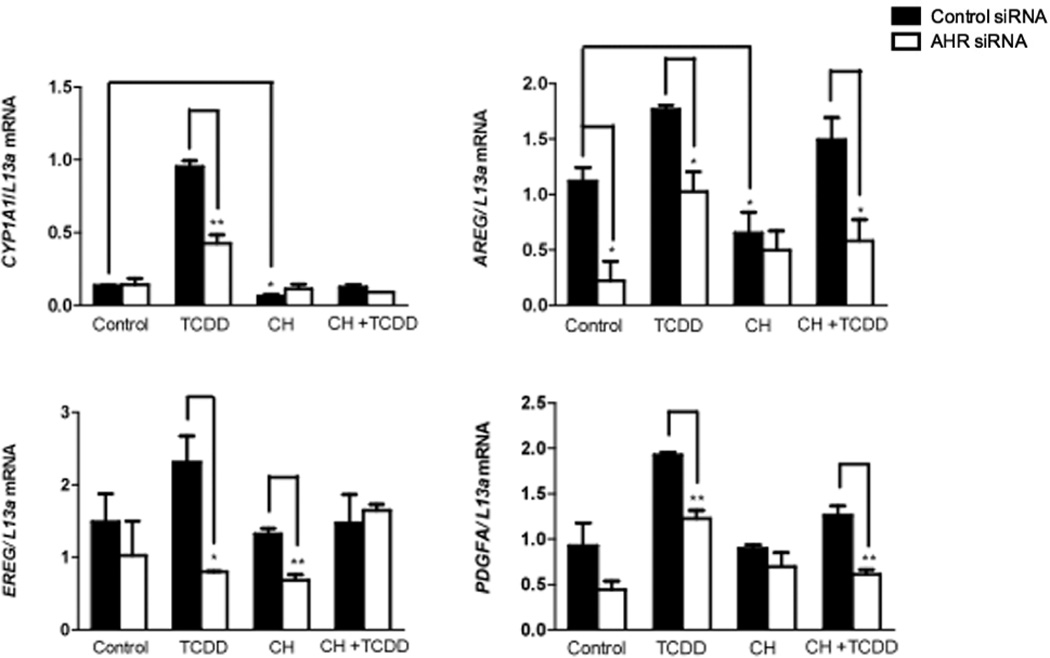
Repression of AHR levels mitigates expression of multiple growth factors. Gene expression analysis of CYP1A1 and other growth factors following electroporation of OSC-19 cells.
Several AHR ligands induce the expression of putative AHR mediated growth factors
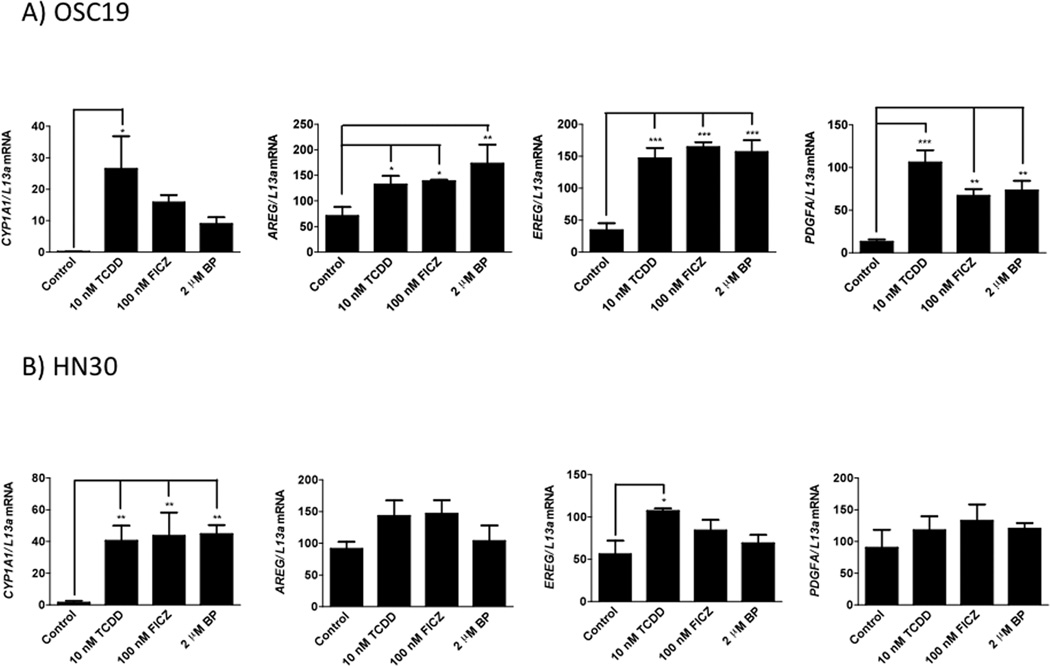
In addition to TCDD, two other AHR agonists, FICZ and benzo(a)pyrene, also significantly induced the expression of AREG, EREG and PDGFA in OSC-19 cells. These genes were also induced to varying extents in HN30 cells, though none of these were significant compared to vehicle control (Figure 7). The relatively high AHR levels ion HN30 may mediate a high basal transcriptional activity and thus a mutated induction upon addition of AHR agonists.
Figure 7.
Putative AHR regulated growth factors exhibit induction with other prototypic AHR ligands. FICZ (100 nM, 24 h) and benzo(a)pyrene (2 µM, 24h), two AHR ligands also cause varying levels of induction to the four growth factor genes in both OSC-19 (A) and HN30 (B) cells. CYP1A1 expression is shown for comparison.
The expression of FGF2 was enhanced by TCDD and FICZ exposure, but was not altered by benzo(a)pyrene (Figure S8).
CH223191 attenuates TCDD+IL1B-induced expression of multiple growth factor genes
Given previous reports of regulation of inflammatory gene signaling by AHR, along with the possible role of AHR in mediating growth factor expression observed here, the role of AHR in regulating the expression of various growth genes, following treatment with TCDD+IL1B was examined with or without the addition of CH223191. Treatment with TCDD+IL1B in combination caused a significant increase in the expression of all growth factors examined, relative to vehicle control. CH223191 attenuated TCDD+IL1B-induced expression of multiple growth factor genes in both OSC-19 and HN30 cells (Figure 8). However, the expression of FGF2 though induced significantly by treatment with TCDD+IL1B, was not mitigated by CH223191 (Figure S9).
Figure 8.
CH223191 attenuated TCDD+IL1B induced gene expression of multiple growth factor genes in both OSC-19 (A) and HN30 (B) cells. Treatment with TCDD alone caused a significant increase in the expression of most growth factor genes relative to vehicle control. However, treatment with IL1B alone did not produce a similar effect. Treatment with TCDD and IL1B in combination caused a significant increase in the expression of all growth factors examined, relative to vehicle control. CYP1A1 expression is shown for comparison.
DISCUSSION
Head and neck cancers are currently the 8th most common type of cancer in the U.S., accounting for ~ 3–5% of all cancers [15,25,26]. In 2012 it is estimated that nearly 52,610 people will develop head and neck cancer and about 11,500 will die from it [26,27]. Most head and neck cancers are squamous cell carcinomas, originating in the epithelium of the upper aerodigestive tract. These comprise nearly 90% of all head and neck cancers. Tobacco use and alcohol consumption have long known to be risk factors [25]. However, more recent increases in HNSCCs have been linked to exposure to human papilloma virus type 16 and 18 [14,25]. Treatment options for HNSCCs is complicated and often calls for a multidisciplinary approach [25]. Even those who survive face a lifetime risk of succumbing to cardiac or respiratory problems as well as second primary tumors that arise at a rate of 3–5% a year, thereby putting the overall survival rate of patients with recurrent or metastatic HNSCCs to less than a year [14,15]. Apart from surgery and radiation therapy, commonly used for the treatment of early stage cancer, various classes of chemotherapeutic agents such as platinum compounds, antimetabolites or taxanes are used when patients have unresectable tumors or when organ preservation is critical [14,16]. Novel therapies under trial also include, mammalian target of rapamycin (mTOR), and epidermal growth factor inhibitors. However, despite aggressive treatment protocols, the prognosis of most patients, especially those with recurrent HNSCCs remains poor, calling for novel approaches and targets that could possibly be used in combination with current protocols for a better prognosis [15].
The AHR has recently been linked to several cancers and has been found to differentially impact a number of endpoints such as apoptosis, proliferation, cell growth and differentiation [4,5]. The AHR has also been implicated in tumor initiation, promotion and progression. Both tumor promotion and progression is thought to be driven in part by a dysregulation of cell-cell contact, which in turn is a critical regulator of proliferation, differentiation and cellular motility, leading to enhanced epithelial to mesenchymal transition (EMT) [5]. Additionally, AHR has been found to be activated by several exogenous and endogenous ligands with many of the exogenous compounds being ubiquitous environmental pollutants, such as benzo(a)pyrene, which is generated from the combustion of organic matter (e.g. cigarette smoke) [3]. Such activation could lead to high constitutive levels of AHR transcriptional activity, in turn triggering several downstream effects mediated by the AHR. We have recently determined that HN30 cells have 7-fold more AHR than human epidermal keratinocytes, further enhancing constitutive AHR transcriptional potential [13]. For example, activation of the AHR pathway has been linked to enhanced cancer cell invasiveness and poor prognosis as reported in cancers, such as those of the stomach and upper urinary tract [28,29]. Furthermore, another study reported enhanced AHR expression, caused by siRNA-mediated knock down of the aryl hydrocarbon receptor repressor, resulting in enhanced anchorage independent growth of multiple cancer cell lines [30]. However, the role of the AHR in cancer has been controversial and appears to be cell-type or context-dependent. For example, in breast tumors a high basal level of AHR expression has been linked with aggressive growth and metastatic phenotype [31]. Conversely, another study reported that further activation of the AHR by exogenous AHR ligands actually inhibits invasive and metastatic phenotype in a panel of breast cancer cell lines [32]. In cancers of the liver and pituitary gland the AHR has been reported to function as a tumor suppressor [33,34]. Since all these studies point to a role of the AHR in cancer progression, this would suggest that the AHR is a target to modulate various aspects of tumor cell survival and outgrowth, including HNSCCs.
Previous studies from this laboratory have established a role for the AHR in mediating both constitutive and cytokine inducible expression of the pro-inflammatory cytokine, IL6, in multiple HNSCC cell lines, thereby suggesting a role for AHR in contributing to the aggressive phenotype of these cells [11]. The role of AHR in mediating cytokine inducible IL6 was also shown in MCF-7 breast cancer cells and the endocervical cancer cell line, ECC-1 [12]. In this regard, prior reports from the literature have linked high IL6 expression to the promotion of enhanced cellular proliferation and increased EMT in head and neck cancers [35]. Recently our laboratory found that antagonism of the AHR, either through use of AHR antagonists or siRNA-mediated abrogation of AHR, to significantly mitigate pro-survival IL6 expression and also greatly attenuate the migratory and invasive behavior of these HNSCC cells [13]. Agarose spot migration assays and Boyden chamber invasion assays revealed nearly a 92–95% decrease in migratory potential and a 60–70% reduction in invasive capacity. In addition, AHR antagonism has also been found to attenuate the expression of drug efflux pumps such as ABCG2, thereby possibly helping to increase retention times of cancer chemotherapeutic drugs.
Therefore, given previous evidence regarding the role of AHR in HNSCC, this study sought to use a whole genome expression profiling based approach to identify other novel targets of AHR associated with the aggressive phenotype of head and neck cancers. Of the 58 probesets altered in common across both the HNSCC cell lines, OSC-19 and HN30, only about 24 appeared to be up-regulated by TCDD and down-regulated by CH223191, relative to the basal expression levels. A number of these altered targets have been documented to be associated with the progression of various cancers. Some of these include MMP1 and HAS3, both found associated with non-small cell lung cancer [36,37] and cervical cancer [38], FOSL1, which increases cell growth of MCF-7 cells [39], RUNX2 found to be important in prostate cancer [40], and ANGPTL4 found to promote colorectal cancer progression [41]. Clearly additional studies’ examining the significance of these findings is needed.
In this report we chose to focus on the growth factor targets of AHR activity. Growth factors constitute polypeptide factors that stimulate cellular proliferation, mediate anti-apoptotic activity and are required for normal cell growth and development [42]. Dysregulation of signaling pathways that mediate normal functions of growth factors is often found in cancer, thereby making them an attractive target for cancer therapy [43]. Paracrine activity of growth factors and cytokines, in combination or individually, has been reported to play a prominent role in the accumulation of genetic insults that lead to malignancy [18]. Of the three growth factor targets identified in this study: epiregulin (EREG), fibroblast growth factor 2 (FGF2), and platelet derived growth factor A (PDGFA), EREG has been reported to be over-expressed in oral squamous cell carcinomas and is a predictor of a poor prognosis [44]. Another study reported higher expression of FGF2 and FGFR in HNSCC cell lines [45]. PDGFA, however, has not been reported to be associated with head and neck cancers. AREG has been reported to be over-expression in certain head and neck cancers [44]. Except for one report examining mouse EREG, none of the growth factors reported here have been previously shown to be regulated by AHR activity [46].
CH223191, a potent antagonist of AHR [47], mitigated both gene expression and protein levels of AREG, EREG and PDGFA, while TCDD, an AHR agonist, mediated their enhanced expression, thereby suggesting that these genes are modulated by AHR activity. In contrast, the induction and mitigation of gene expression or protein levels of FGF2 was not as significant as the other 3 growth factors studied. Interestingly, FGF2 has been reported to enhance fibroblast-independent tumor growth in head and neck cancers [48] and was found here to possess multiple DRE-like elements in its promoter region. There are other reports supporting this concept of repressing AHR activity as a means of modulating cancer cell signaling, for example, the recent reports of Src-mediated cross-talk between AHR and epidermal growth factor receptors in stimulating colon cancer cell proliferation [49] and c-Jun mediated cross-talk between AHR and matrix metalloproteinases in gastric cancers [28]. Thus, the ability of constitutive AHR to participate in growth factor expression would suggest that the AHR plays an important role in the facilitating several signaling pathways that play a direct role in tumor cell malignancy and aggressive behavior.
The use of siRNA to repress AHR expression provided additional evidence of the importance of the AHR in mediating AREG, EREG and PDGFA expression. We did not pursue the significance of each individual target gene to the phenotype observed in HNSCC cells considering the large number of genes that are influenced by AHR antagonist treatment. The tumor microenvironment typically involves both aberrant growth factor signaling and pro-inflammatory gene expression. In light of this information and our previous reports on the role of AHR in regulating inflammatory gene signaling [10,11], the expression of various growth factor genes was examined in the presence of an inflammatory mediator, IL1B, and an AHR agonist. A significant increase in expression of various growth factor genes in the presence of TCDD and IL1B in combination along with an attenuation of their expression upon co-treatment with CH223191, further point to the utility of regulating multiple pro-survival and pro-metastatic ‘cancer pathways’ through the repression of AHR activity. HNSCC is a complex and difficult to treat disease that requires multiple treatment strategies. Thus, considering that AHR antagonism would be a relatively non-toxic treatment, this approach offers considerable potential as ‘adjuvant therapy’ to existing treatment regimens to greatly improve disease outcomes of recurrent, highly metastatic HNSCCs.
Supplementary Material
ACKNOWLEDGEMENT
This work was supported in part by funds from the DuPont Haskell Global Centers for Health and Environmental Sciences, Newark, DE and National Institutes of Health ES004869 and ES019964. Also we thank the expertise of the Penn State Genomics Core Facility staff for their assistance with processing of the microarrays.
Abbreviations
- AHR
aryl hydrocarbon receptor
- AREG
amphiregulin
- ARNT
aryl hydrocarbon nuclear translocator
- BP
benzo(a)pyrene
- DRE
dioxin response element
- EREG
epiregulin
- FICZ
6-formylindolo[3,2-b]carbazole
- HNSCC
head and neck squamous cell carcinoma
- TCDD
2,3,7,8 tetrachlorodibenzo-p-dioxin
References
- 1.Beischlag TV, Luis Morales J, Hollingshead BD, Perdew GH. The aryl hydrocarbon receptor complex and the control of gene expression. Crit Rev Eukaryot Gene Expr. 2008;18:207–250. doi: 10.1615/critreveukargeneexpr.v18.i3.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Denison MS, Soshilov AA, He G, DeGroot DE, Zhao B. Exactly the same but different: promiscuity and diversity in the molecular mechanisms of action of the aryl hydrocarbon (dioxin) receptor. Toxicol Sci. 2011;124:1–22. doi: 10.1093/toxsci/kfr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stejskalova L, Dvorak Z, Pavek P. Endogenous and exogenous ligands of aryl hydrocarbon receptor: current state of art. Curr Drug Metab. 2011;12:198–212. doi: 10.2174/138920011795016818. [DOI] [PubMed] [Google Scholar]
- 4.Abel J, Haarmann-Stemmann T. An introduction to the molecular basics of aryl hydrocarbon receptor biology. Biol Chem. 2010;391:1235–1248. doi: 10.1515/BC.2010.128. [DOI] [PubMed] [Google Scholar]
- 5.Dietrich C, Kaina B. The aryl hydrocarbon receptor (AhR) in the regulation of cell-cell contact and tumor growth. Carcinogenesis. 2010;31:1319–1328. doi: 10.1093/carcin/bgq028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramadoss P, Marcus C, Perdew GH. Role of the aryl hydrocarbon receptor in drug metabolism. Expert Opin Drug Metab Toxicol. 2005;1:9–21. doi: 10.1517/17425255.1.1.9. [DOI] [PubMed] [Google Scholar]
- 7.Tanos R, Patel RD, Murray IA, Smith PB, Patterson AD, Perdew GH. Aryl hydrocarbon receptor regulates the cholesterol biosynthetic pathway in a dioxin response element-independent manner. Hepatology. 2012;55:1994–2004. doi: 10.1002/hep.25571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Veldhoen M, Duarte JH. The aryl hydrocarbon receptor: fine-tuning the immune-response. Curr Opin Immunol. 2010;22:747–752. doi: 10.1016/j.coi.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 9.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 10.DiNatale BC, Schroeder JC, Francey LJ, Kusnadi A, Perdew GH. Mechanistic insights into the events that lead to synergistic induction of interleukin 6 transcription upon activation of the aryl hydrocarbon receptor and inflammatory signaling. J Biol Chem. 2010;285:24388–24397. doi: 10.1074/jbc.M110.118570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DiNatale BC, Schroeder JC, Perdew GH. Ah receptor antagonism inhibits constitutive and cytokine inducible IL6 production in head and neck tumor cell lines. Mol Carcinog. 2010;50:173–183. doi: 10.1002/mc.20702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hollingshead BD, Beischlag TV, DiNatale BC, Ramadoss P, Perdew GH. Inflammatory signaling and aryl hydrocarbon receptor mediate synergistic induction of interleukin 6 in MCF-7 cells. Cancer Res. 2008;68:3609–3617. doi: 10.1158/0008-5472.CAN-07-6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dinatale BC, Smith K, John K, Krishnegowda G, Amin SG, Perdew GH. Ah receptor antagonism represses head and neck tumor cell aggressive phenotype. Mol Cancer Res. 2012;10:1369–1379. doi: 10.1158/1541-7786.MCR-12-0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Argiris A, Karamouzis MV, Raben D, Ferris RL. Head and neck cancer. Lancet. 2008;371:1695–1709. doi: 10.1016/S0140-6736(08)60728-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Price KA, Cohen EE. Current treatment options for metastatic head and neck cancer. Curr Treat Options Oncol. 2012;13:35–46. doi: 10.1007/s11864-011-0176-y. [DOI] [PubMed] [Google Scholar]
- 16.Kurzweg T, Mockelmann N, Laban S, Knecht R. Current treatment options for recurrent/metastatic head and neck cancer: a post-ASCO 2011 update and review of last year's literature. Eur Arch Otorhinolaryngol. 2012;269:2157–2167. doi: 10.1007/s00405-012-1998-3. [DOI] [PubMed] [Google Scholar]
- 17.Kim SH, Henry EC, Kim DK, et al. Novel compound 2-methyl-2H-pyrazole-3-carboxylic acid (2-methyl-4-o-tolylazo-phenyl)-amide (CH-223191) prevents 2,3,7,8-TCDD-induced toxicity by antagonizing the aryl hydrocarbon receptor. Mol Pharmacol. 2006;69:1871–1878. doi: 10.1124/mol.105.021832. [DOI] [PubMed] [Google Scholar]
- 18.Aaronson SA. Growth factors and cancer. Science. 1991;254:1146–1153. doi: 10.1126/science.1659742. [DOI] [PubMed] [Google Scholar]
- 19.Favoni RE, de Cupis A. The role of polypeptide growth factors in human carcinomas: new targets for a novel pharmacological approach. Pharmacol Rev. 2000;52:179–206. [PubMed] [Google Scholar]
- 20.Applied Biosystems: Guide to Performing Relative Quantitation of Gene Expression Using Real-Time Quantitative PCR. 2004 [Google Scholar]
- 21.Carlson JM, Chakravarty A, DeZiel CE, Gross RH. SCOPE: a web server for practical de novo motif discovery. Nucleic Acids Res. 2007;35:W259–W264. doi: 10.1093/nar/gkm310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawashiri S, Kumagai S, Kojima K, Harada H, Yamamoto E. Development of a new invasion and metastasis model of human oral squamous cell carcinomas. Eur J Cancer B Oral Oncol. 1995;31B:216–221. doi: 10.1016/0964-1955(95)00027-f. [DOI] [PubMed] [Google Scholar]
- 23.Yoo GH, Subramanian G, Boinpally RR, et al. An in vivo evaluation of docetaxel delivered intratumorally in head and neck squamous cell carcinoma. Arch Otolaryngol Head Neck Surg. 2005;131:418–429. doi: 10.1001/archotol.131.5.418. [DOI] [PubMed] [Google Scholar]
- 24.Yoo GH, Subramanian G, Piechocki MP, et al. Effect of docetaxel on the surgical tumor microenvironment of head and neck cancer in murine models. Arch Otolaryngol Head Neck Surg. 2008;134:735–742. doi: 10.1001/archotol.134.7.735. [DOI] [PubMed] [Google Scholar]
- 25.Marur S, Forastiere AA. Head and neck cancer: changing epidemiology, diagnosis, and treatment. Mayo Clin Proc. 2008;83:489–501. doi: 10.4065/83.4.489. [DOI] [PubMed] [Google Scholar]
- 26. http://www.cancer.net/patient/Cancer+Types/Head+and+Neck+Cancer?sectionTitle=Statistics. [Google Scholar]
- 27. http://www.cancer.org/acs/groups/content/@epidemiologysurveilance/documents/document/acspc-031941.pdf. [Google Scholar]
- 28.Peng TL, Chen J, Mao W, Song X, Chen MH. Aryl hydrocarbon receptor pathway activation enhances gastric cancer cell invasiveness likely through a c-Jun-dependent induction of matrix metalloproteinase-9. BMC Cell Biol. 2009;10:27. doi: 10.1186/1471-2121-10-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishida M, Mikami S, Kikuchi E, et al. Activation of the aryl hydrocarbon receptor pathway enhances cancer cell invasion by upregulating the MMP expression and is associated with poor prognosis in upper urinary tract urothelial cancer. Carcinogenesis. 2010;31:287–295. doi: 10.1093/carcin/bgp222. [DOI] [PubMed] [Google Scholar]
- 30.Zudaire E, Cuesta N, Murty V, et al. The aryl hydrocarbon receptor repressor is a putative tumor suppressor gene in multiple human cancers. J Clin Invest. 2008;118:640–650. doi: 10.1172/JCI30024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlezinger JJ, Liu D, Farago M, et al. A role for the aryl hydrocarbon receptor in mammary gland tumorigenesis. Biol Chem. 2006;387:1175–1187. doi: 10.1515/BC.2006.145. [DOI] [PubMed] [Google Scholar]
- 32.Hall JM, Barhoover MA, Kazmin D, McDonnell DP, Greenlee WF, Thomas RS. Activation of the aryl-hydrocarbon receptor inhibits invasive and metastatic features of human breast cancer cells and promotes breast cancer cell differentiation. Mol Endocrinol. 2010;24:359–369. doi: 10.1210/me.2009-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fan Y, Boivin GP, Knudsen ES, Nebert DW, Xia Y, Puga A. The aryl hydrocarbon receptor functions as a tumor suppressor of liver carcinogenesis. Cancer Res. 2010;70:212–220. doi: 10.1158/0008-5472.CAN-09-3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jaffrain-Rea ML, Angelini M, Gargano D, et al. Expression of aryl hydrocarbon receptor (AHR) and AHR-interacting protein in pituitary adenomas: pathological and clinical implications. Endocr Relat Cancer. 2009;16:1029–1043. doi: 10.1677/ERC-09-0094. [DOI] [PubMed] [Google Scholar]
- 35.Yadav A, Kumar B, Datta J, Teknos TN, Kumar P. IL-6 promotes head and neck tumor metastasis by inducing epithelial-mesenchymal transition via the JAK-STAT3-SNAIL signaling pathway. Mol Cancer Res. 2011;9:1658–1667. doi: 10.1158/1541-7786.MCR-11-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chow G, Tauler J, Mulshine JL. Cytokines and growth factors stimulate hyaluronan production: role of hyaluronan in epithelial to mesenchymal-like transition in non-small cell lung cancer. J Biomed Biotechnol. 2010;2010:485468. doi: 10.1155/2010/485468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hart K, Landvik NE, Lind H, Skaug V, Haugen A, Zienolddiny S. A combination of functional polymorphisms in the CASP8, MMP1, IL10 and SEPS1 genes affects risk of non-small cell lung cancer. Lung Cancer. 2011;71:123–129. doi: 10.1016/j.lungcan.2010.04.016. [DOI] [PubMed] [Google Scholar]
- 38.Rajkumar T, Sabitha K, Vijayalakshmi N, et al. Identification and validation of genes involved in cervical tumourigenesis. BMC Cancer. 11:80. doi: 10.1186/1471-2407-11-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pennanen PT, Sarvilinna NS, Toimela T, Ylikomi TJ. Inhibition of FOSL1 overexpression in antiestrogen-resistant MCF-7 cells decreases cell growth and increases vacuolization and cell death. Steroids. 76:1063–1068. doi: 10.1016/j.steroids.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 40.Zhang H, Pan Y, Zheng L, et al. FOXO1 inhibits Runx2 transcriptional activity and prostate cancer cell migration and invasion. Cancer Res. 71:3257–3267. doi: 10.1158/0008-5472.CAN-10-2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim SH, Park YY, Kim SW, Lee JS, Wang D, DuBois RN. ANGPTL4 induction by prostaglandin E2 under hypoxic conditions promotes colorectal cancer progression. Cancer Res. 2011;71:7010–7020. doi: 10.1158/0008-5472.CAN-11-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goustin AS, Leof EB, Shipley GD, Moses HL. Growth factors and cancer. Cancer Res. 1986;46:1015–1029. [PubMed] [Google Scholar]
- 43.Witsch E, Sela M, Yarden Y. Roles for growth factors in cancer progression. Physiol Behav. 2010;25:85–101. doi: 10.1152/physiol.00045.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shigeishi H, Higashikawa K, Hiraoka M, et al. Expression of epiregulin, a novel epidermal growth factor ligand associated with prognosis in human oral squamous cell carcinomas. Oncol Rep. 2008;19:1557–1564. [PubMed] [Google Scholar]
- 45.Marshall ME, Hinz TK, Kono A, et al. Fibroblast growth factor receptors are components of autocrine signaling networks in head and neck squamous cell carcinoma cells. Clin Cancer Res. 2011;17:5016–5025. doi: 10.1158/1078-0432.CCR-11-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patel RD, Kim DJ, Peters JM, Perdew GH. The aryl hydrocarbon receptor directly regulates expression of the potent mitogen epiregulin. Toxicol Sci. 2006;89:75–82. doi: 10.1093/toxsci/kfi344. [DOI] [PubMed] [Google Scholar]
- 47.Zhao B, Degroot DE, Hayashi A, He G, Denison MS. CH223191 is a ligand-selective antagonist of the Ah (Dioxin) receptor. Toxicol Sci. 2010;117:393–403. doi: 10.1093/toxsci/kfq217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Z, Hartman YE, Warram JM, et al. Fibroblast growth factor receptor mediates fibroblast-dependent growth in EMMPRIN-depleted head and neck cancer tumor cells. Mol Cancer Res. 2011;9:1008–1017. doi: 10.1158/1541-7786.MCR-11-0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xie G, Peng Z, Raufman JP. Src-mediated aryl hydrocarbon and epidermal growth factor receptor cross talk stimulates colon cancer cell proliferation. Am J Physiol Gastrointest Liver Physiol. 2012;302:G1006–G1015. doi: 10.1152/ajpgi.00427.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.