Abstract
BACKGROUND:
The potential role of environmental Mycobacterium tuberculosis in the epidemiology of TB remains unknown. We investigated the transmission of M tuberculosis from humans to the environment and the possible transmission of M tuberculosis from the environment to humans.
METHODS:
A total of 1,500 samples were collected from three counties of the Tehran, Iran metropolitan area from February 2012 to January 2014. A total of 700 water samples (47%) and 800 soil samples (53%) were collected. Spoligotyping and the mycobacterial interspersed repetitive units-variable number of tandem repeats typing method were performed on DNA extracted from single colonies. Genotypes of M tuberculosis strains isolated from the environment were compared with the genotypes obtained from 55 patients with confirmed pulmonary TB diagnosed during the study period in the same three counties.
RESULTS:
M tuberculosis was isolated from 11 of 800 soil samples (1%) and 71 of 700 water samples (10%). T family (56 of 82, 68%) followed by Delhi/CAS (11 of 82, 13.4%) were the most frequent M tuberculosis superfamilies in both water and soil samples. Overall, 27.7% of isolates in clusters were related. No related typing patterns were detected between soil, water, and clinical isolates. The most frequent superfamily of M tuberculosis in clinical isolates was Delhi/CAS (142, 30.3%) followed by NEW-1 (127, 27%). The bacilli in contaminated soil (36%) and damp water (8.4%) remained reculturable in some samples up to 9 months.
CONCLUSIONS:
Although the dominant M tuberculosis superfamilies in soil and water did not correspond to the dominant M tuberculosis family in patients, the presence of circulating genotypes of M tuberculosis in soil and water highlight the risk of transmission.
The presence of Mycobacterium tuberculosis in the environment and its potential role in the epidemiology of TB has been debated since the early 20th century.1 Musehold1 started the debate showing the presence of virulent TB bacteria in the sewage of a TB sanatorium in 1900.
TB is characterized as being transmitted by aerosols as a consequence of direct contact with a patient with pulmonary TB. However, TB transmission could occur without patient contact. Johnson et al2 demonstrated that exposure to medical waste resulted in TB infection in three patients. To date, few studies have assessed the environment as a source of M tuberculosis infection.3 Transmission of M tuberculosis from the environment is possible as TB bacilli have been isolated from sputum or carpet up to 19 days, wood over 88 days, and moist and dry soil up to 4 weeks following contamination.4,5 Furthermore, not only can M tuberculosis survive in soil, but it also remains virulent.6 As there are 13.7 active and 8.8 million newly diagnosed TB cases each year, it is quite likely that soil and water can become contaminated with M tuberculosis through expectoration.7
To conduct a systematic study of the potential role of transmission of M tuberculosis from the environment to humans, we report the isolation, identification, and genotyping of M tuberculosis isolates from soil and water samples from the Tehran, Iran metropolitan area.
Materials and Methods
This study was reviewed and approved by the institutional review board of the National Research Institute of Tuberculosis and Lung Disease of Iran (approval number of MRC-2011/023).
Sample Collection and Preparation
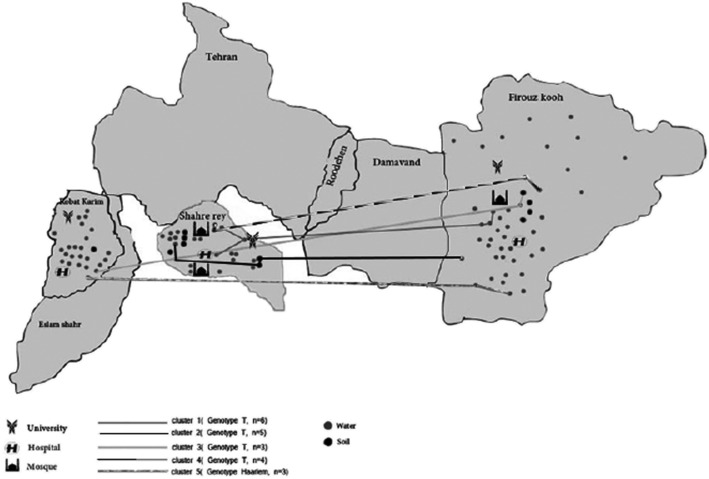
In total, 700 soil samples and 800 water samples were collected from three counties of Tehran metropolitan areas—Robat Karim, Firuzkuh, and Shahr-e-Ray—from February 2012 to January 2014 (Fig 1). We collected 5 to 7 g of soil sample from depths of 3 to 5 cm, suspended it in a 50-mL sterile tube, and processed it using a modified Engbaek method.8‐10 Fifty to 100 mL of water from different sources (200 from damp waters, 100 from tap waters, and 500 samples from running water on raceway systems) were collected. The raceway system is a cement canal (10-70 m length and 20-70 cm width) used for transporting the wastewater or sustainable rainwater to the central recycling and treatment sector. The raceway system is designed for almost all streets, lanes, and alleys in the Tehran metropolitan area.
Figure 1 –
Water and soil sampling locations in Tehran metropolitan areas.
Water samples were decontaminated with cetylpyridinium chloride (final concentration of 0.05%) for 30 min and digested using a standard protocol.11,12 The final water and soil sediments were acid-fast stained (Ziehl-Neelsen) and cultured by inoculating three Löwenstein-Jensen medium bottles with sediments (200 μL of sediment/tube). Bottles were sealed and incubated at 37°C, 25°C, and 42°C for 12 weeks. The inoculated cultures were checked for growth every 3 to 4 days. Acid-fast colonies were identified by standard phenotypic tests (including niacin and nitrate tests). A molecular analysis was performed on single colonies derived from subcultures of original isolates as previously described.13
Patients
We randomly selected 458 patients with culture-positive pulmonary TB who were residents of the sampled counties and were diagnosed during the study period. Of 458 patients, 25 were from Robat Karim, 10 were from Firuzkuh, 20 were from Shahr-e-Ray, and 413 were from Tehran city. Demographic information including nationality of patients was collected. M tuberculosis isolates from patients were genotyped and compared with M tuberculosis isolates from water and soil.
DNA Extraction and Species Identification From Acid-Fast Bacilli-Positive Cultures
The DNA was extracted from heat-inactivated colony suspensions using a QIAGEN DNA Extraction kit. Species identification was performed by using 16S-23S RNA gene spacer polymerase chain reaction (PCR)-restriction fragment length polymorphism as discussed elsewhere.14 The amplified products were digested with HaeIII and CfoI restriction enzymes and electrophoresed on 8% polyacrylamide gel. Mutations resulting in resistance to rifampin and isoniazid were identified by multiplex PCR as previously published.14
Genotyping of the Isolates by Spoligotyping and Mycobacterial Interspersed Repetitive Units-Variable Number of Tandem Repeats
The clinical and environmental isolates (468 and 82, respectively) were characterized using spoligotyping and mycobacterial interspersed repetitive units-variable number of tandem repeats (MIRU-VNTR) analysis.15,16 MIRU-VNTR genotyping was performed by PCR amplification of a panel of 15 M tuberculosis MIRU loci using primers described in the MIRU-VNTR standard protocol.15,17 The PCR products were run on 1.5% agarose gel, and the number of MIRU-VNTR copies for each isolate was determined. MIRU-VNTR analysis was performed using a MIRU-VNTR plus database.18 The discrimination power was evaluated by the Hunter and Gaston index (HGI).19
To avoid any laboratory errors, the positive and negative control groups were included in each experimental setup. The positive control samples consisted of standard M tuberculosis H37RV, Mycobacterium fortuitum type 1, Mycobacterium kansasii, and Mycobacterium avium.
Results
M tuberculosis Isolation From Soil and Water
Acid-fast bacteria were isolated from 568 samples (37.8%), 608 were acid-fast bacilli smear, and culture negative (40.0%) and the remaining were contaminated (324 samples, 21.6%). From 568 mycobacterial-positive samples, 219 isolates (38.5%) were rapidly growing, and 349 strains (61.4%) were slowly growing mycobacteria. The most frequently isolated species among the slowly growing mycobacteria were Mycobacterium farcinogenes (87, 24.9%) and M tuberculosis (82, 23.4%). The highest frequency of M tuberculosis isolates was in Firuzkuh city (36 of 186, 19.3%), followed by Robat Karim (22 of 149, 14.7%) and Shahr-e-Ray (24 of 233, 10.3%) (Fig 1). M tuberculosis was more frequently recovered from water (71, 86.5%) than soil (11, 13.4%) (P < .05). The majority of M tuberculosis isolates were recovered from raceway systems (56 of 500, 11.2%) or dump water (15 of 200, 7.5%). Samples from tap or drinking water (n = 100) lacked any mycobacteria. Three multidrug-resistant M tuberculosis (MDR-TB) (3.6%), four monodrug-resistant strains (three isoniazid and one rifampin, 4.8%), and 58 pan-susceptible strains (70%) were detected among the water and soil isolates. We had no reproducible results on drug-resistant pattern for 17 samples. Samples of water and soil recultured 3 months from the original collection showed that 45% of soil and 15% of water that had originally been positive remained so. Samples were reculturable for M tuberculosis 6 months after the initial sampling, but had a lower level of positivity rate (soil, 36%; water, 8.4%) (P < .05).
Genotypic Analysis of M tuberculosis Isolates From Soil and Water
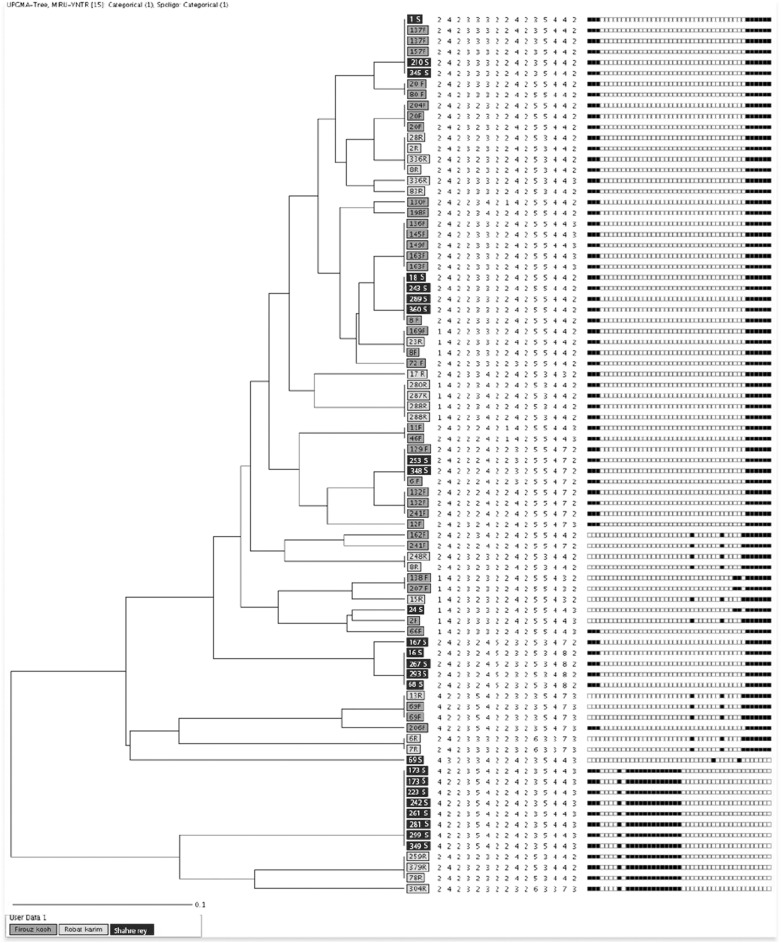
For molecular typing, the colonies from each culture positive slant were first subcultured into Löwenstein-Jensen culture media and then the single colonies were used for DNA extraction. By spoligotyping, 96% of strains were classified into four clusters. The largest genotypes were T superfamily with 56 isolates (68.29%), followed by Delhi/CAS (11, 13.41%), Beijing (six, 7.32%), and Haarlem (six, 7.32%) lineages. The other identified classes were Canetti (one, 1.2%), NEW-1 (one, 1.2%), and Uganda family (one, 1.2%). A combination of both MIRU-VNTR with spoligotyping increased the level of discrimination between isolates. For example, a T superfamily with 56 isolates by spoligotypes subdivided into 11 clusters (n = 40 strains) and 16 single isolates. In total, 65 strains (79.2%) classified into 18 clusters by using both PCR-typing methods. Minimum and maximum number of isolates per cluster ranged from two to eight strains. From an epidemiologic point of view, the majority of strains in clusters (13 of 18, 72%) were isolated from within one local area in the same county (Fig 2). Twenty-eight percent of those clustered were formed by isolates that were collected from different counties. Four of these clusters belong to the T superfamily with minimum (n = 2) and maximum (n = 5) differences in locus Mtub04, MIRU10, MIRU40, MIRU26, MIRU16, and ETR-A, respectively (Fig 3). As shown in Figures 1 and 3, the majority of interconnected clusters (n = 3 with 15 isolates) were collected from soil and water samples from two counties (Firuzkuh and Shahr-e-Ray). Overall, the allelic diversities of 15 MIRU-VNTR loci for soil and water isolates showed the locus MIRU 10 with highly discriminative power (> 0.6). Whereas loci Mtub04, MIRU40, MIRU16, ETR-A, MIRU26, MIRU31, QUB26, and QUB4156 were moderately discriminative (≥ 0.3, < 0.6), loci ETR-C, MIRU04, Mtub21, QUB11b, Mtub30, and Mtub39 were poorly discriminative loci (< 0.3) (Table 1).
Figure 2 –
Dendrogram of soil and water Mycobacterium tuberculosis strains, which shows clustered and single strains. MIRU-VNTR = mycobacterial interspersed repetitive units-variable number of tandem repeats; UPGMA = unweighted pair group method with arithmetic mean.
Figure 3 –
Radial tree of M tuberculosis for clinical isolates and soil and water isolates. See Figure 2 legend for expansion of abbreviations.
TABLE 1 ] .
Allelic Diversity Index of Mycobacterium tuberculosis MIRU Loci for Clinical Samples and Soil and Water Isolates
| Locus Name | HGI for Clinical Samples | DIa | HGI for Water Samples | DIa | HGI for Soil Samples | DIa |
| Mtub04 | 0.68 | High | 0.45 | Moderate | 0.55 | Moderate |
| ETR-C | 0.6 | High | 0.23 | Poor | 0.34 | Moderate |
| MIRU04 | 0.08 | Poor | 0.0 | Poor | 0.0 | Poor |
| MIRU40 | 0.54 | Moderate | 0.44 | Moderate | 0.41 | Moderate |
| MIRU10 | 0.78 | High | 0.6 | High | 0.61 | High |
| MIRU16 | 0.73 | High | 0.49 | Moderate | 0.41 | Moderate |
| Mtub21 | 0.73 | High | 0.12 | Poor | 0.0 | Poor |
| QUB11b | 0.58 | Moderate | 0.04 | Poor | 0.08 | Poor |
| ETR-A | 0.65 | High | 0.36 | Moderate | 0.0 | Poor |
| Mtub30 | 0.5 | Moderate | 0.0 | Poor | 0.0 | Poor |
| MIRU26 | 0.79 | High | 0.39 | Moderate | 0.41 | Moderate |
| MIRU31 | 0.64 | High | 0.43 | Moderate | 0.08 | Poor |
| Mtub39 | 0.58 | Moderate | 0.07 | Poor | 0.0 | Poor |
| QUB26 | 0.81 | High | 0.48 | Moderate | 0.23 | Moderate |
| QUB4156 | 0.76 | High | 0.48 | Moderate | 0.41 | Moderate |
DI = discriminatory index; HGI = Hunter and Gaston index; MIRU = mycobacterial interspersed repetitive unit.
DI, high (> 0.6), moderate (≥ 0.3, < 0.6), poor (< 0.3).
Genotypic Analysis of M tuberculosis Isolates From Patients
Based on molecular typing, major identified lineages belonged to Delhi/CAS (142, 30.34%), NEW-1 (127, 26.92%), Beijing (75, 16.03%), and Haarlem (5%) (Table 2). Using MIRU-VNTR and spoligotyping, 202 strains (43.1%) classified into 77 clusters and another 266 strains were single (56.9%). A direct epidemiologic link could be established for 30 patients in nine clusters (11.6%). The allelic diversities of 15 MIRU-VNTR loci for patient isolates showed that loci MIRU16, ETR-A, MIRU31, ETR-C, MIRU10, MIRU26, Mtub04, QUB26, QUB4156, and MTUB21 were highly discriminative (> 0.6), loci MTUB30, MIRU40, Mtub39, and QUB 11b were moderately discriminative (≥ 0.3, < 0.6), and locus MIRU04 was poorly discriminative (< 0.3). The most discriminative locus was QUB26 (HGI = 0.81).
TABLE 2 ] .
Distribution of Identified M tuberculosis Lineages in Clinical Samples and Soil and Water Isolates Using the MIRU-VNTR Plus Database
| M tuberculosis Family | Patients’ Samples, No. (%) | Soil Samples, No. (%) | Water Samples, No. (%) |
| Delhi/CAS | 142 (30) | 3 (27) | 8 (11) |
| NEW-1 | 127 (27) | … | 1 (1) |
| Beijing | 75 (16) | 1 (9) | 5 (7) |
| T | 26 (6) | 7 (64) | 49 (69) |
| Haarlem | 26 (5) | … | 6 (8) |
| Ghana | 21(4) | … | … |
| URAL | 17 (4) | … | … |
| LAM | 11 (2) | … | … |
| Uganda II | 6 (1) | … | … |
| Uganda I | 5 (1) | … | 1 (1) |
| S | 5 (1) | … | … |
| TUR | 4 (0.8) | … | … |
| Bovis | 2 (0.4) | … | … |
| West African 1 | 1 (0.2) | … | … |
| Canetti | … | … | 1 (1) |
MIRU-VNTR = mycobacterial interspersed repetitive units-variable number of tandem repeats. See Table 1 legend for expansion of other abbreviation.
Genotypic Comparison of M tuberculosis Isolates From the Environment and Patients
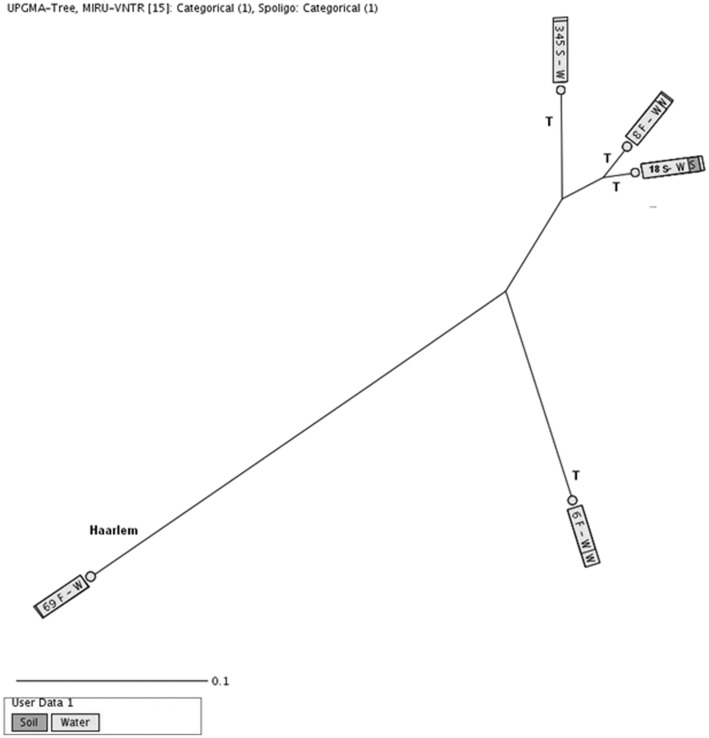
Cluster analysis based on spoligotyping and MIRU-VNTR revealed no similar genotypes between environmental and clinical isolates of M tuberculosis. The differences in the allelic diversities of 15 MIRU-VNTR ranged from a minimum of two to a maximum of 10 that we found in a Haarlem with a lower number of differences, that is, two loci (MIRU16 and Mtub21) and T superfamilies with a higher number of differences (Mtub04, ETRC, MIRU40, MIRU10, MIRU16, QUB11b, ETRA, MIRU31, Mtub39, and QUB26).
Discussion
In this study, mycobacteria were isolated from 568 of 1,500 soil and water samples (37.8%) from the Tehran metropolitan area. The majority of samples isolated were slow-growing mycobacteria. M tuberculosis was isolated from 82 soil and water samples (5%). Three isolated M tuberculosis strains (3.6%) were MDR-TB. Some of the soil and water samples remained culturable for M tuberculosis at least for 6 months after sampling. No genotyping similarity between M tuberculosis isolates from the environment and isolates from patients was found.
Isolation of M tuberculosis from soil and water raises the possible risk of infection from the environment. This study further confirms the ability of M tuberculosis to remain viable in water and soil for an extended period time. Such sources of infection have been proven for the nonobligate pathogens of the Mycobacterium genus, particularly for M avium.20
The discovery that 11% of water samples from raceway systems yielded M tuberculosis suggests these widely used systems can serve as a source of M tuberculosis infection. The mechanism of M tuberculosis survival in the race water remained unclear. Given that water tends to flow slowly in these channels, such a water-collecting system by itself may encourage the growth of microorganisms and possibly M tuberculosis. We have not studied the cohabitation of M tuberculosis and other organisms in the water (such as amoebae) and its potential role in survival of M tuberculosis. A number of mycobacterial species have been shown to grow in amoebae, including Mycobacterium ulcerans21 and M avium.22 Interestingly, it was shown that M tuberculosis is an amoeba-resistant pathogen and can survive into amoeba cytoplasmic vacuoles.23
The discovery that MDR-TB isolates were present in 3.6% of samples is an alarming sign for communities with a high burden of MDR-TB, which needs prompt global attention. Given the fact MDR-TB is a growing problem in the world, the epidemiologic role of the environment should be evaluated.
Based on molecular studies, the most frequent M tuberculosis superfamilies in both water and soil samples belong to the T family (56 of 82, 68%). Fifty-six isolates with T lineages had a minimum of one and a maximum of five differences in allelic diversities of 15 MIRU-VNTR loci, respectively. Another frequent superfamily was Delhi/CAS by 11 isolates (13.4%). Overall, the molecular typing of isolated strains had a 79% clustering rate. This finding supports the presence of a common source of M tuberculosis strains for water and soil. We speculate that the practice of watering soil on the raceway system is the most probable reason for soil contamination. This finding has important epidemiologic implications which need further exploration.
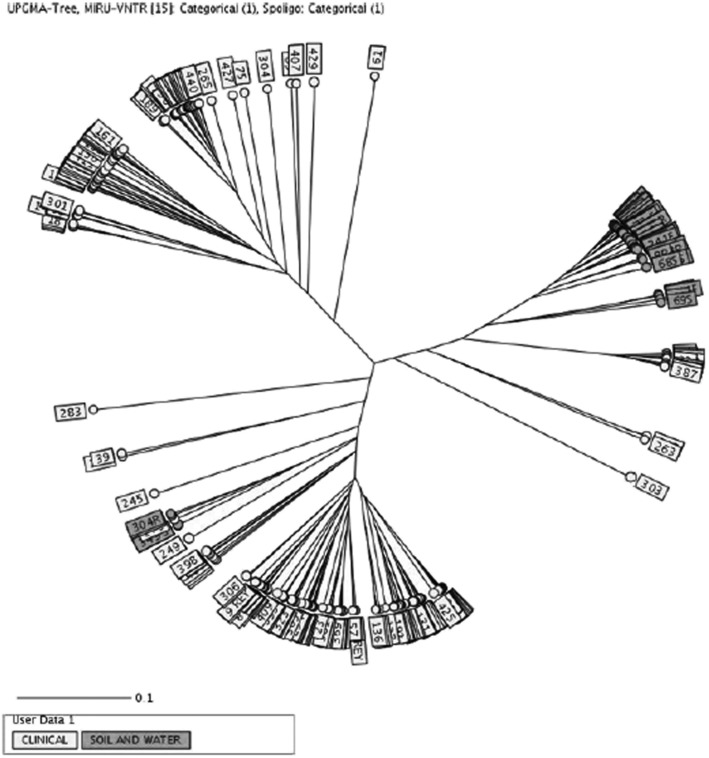
The isolation of M tuberculosis from soil and water samples raises an important question about the risk of transmission in people who living in nearby areas. Our analysis of the clinical M tuberculosis isolates showed the majority of them were belong to Delhi/CAS family (142, 30.3%). As shown in Figure 4, no genotypic similarity was found between isolated strains from soil and water and isolates from patients. Several explanations for this are possible. The first explanation is that transmission from the environment to nearby humans is simply not occurring. Another explanation may lie in the small sample size of this study, which failed to capture patient isolates also found in soil and water. The short study period may also explain our failure to connect environmental and clinical isolates, which probably would require an extension of the study period of at least 3 to 5 years. Generally, a genomic molecular marker(s) with a low rate of mutations is needed for a true judgment of transmission in tuberculosis epidemiologic studies.24 Wirth et al25 looked at the genetic diversity and mutation rates in different lineages of M tuberculosis by using the MIRU-VNTR genetic maker. They showed that the probability of demonstrating a repeat change over periods of up to 7 years was 1% for the five most variable loci. This corresponds to a single locus mutation rate of 1.4 × 10−3 per year. Overall, they estimated the posterior mean for the mutation rate to be 10−3.91.25
Figure 4 –
Genotypic similarity between isolated strains from soil and water and isolates from patients. See Figure 2 legend for expansion of abbreviations.
In the present study, the most common M tuberculosis isolated from water and soil samples belonged to the T superfamily (68.29%). However, the T family strains were isolated only from 5% of patients with pulmonary TB. The T superfamily is characterized by the absence of spacers 33, 36, and Duchêne et al26 suggested the African origin of these alleles. We previously demonstrated that the T family was a circulating strain in Iran, Turkey, and Iraq.16,27 The isolation of T, CAS, and Haarlem superfamilies of M tuberculosis from clinical and environmental samples draws our attention to the potential risk of nature contamination by humans.
The potential transmission of M tuberculosis from water and soil to domestic animals should also been evaluated. The transmission of human M tuberculosis to animals could happen and has previously been reported from different countries in Asia, Europe, and Africa.28‐30 Our study highlights the potential role of soil and water as a source of infection of domestic and wild animals with human M tuberculosis strains in the TB endemic countries.
The primary limitations of this study are small sample size and short study period. As previously discussed, the small sample size may have caused patients with isolates also found in the environment to have been missed. Also, the short study period may have likewise undermined the detection of this phenomenon. To detect M tuberculosis cross-transmission between the environment and humans, a longitudinal study with a longer period of time is necessary. Another problem in this study was the high contamination rate of environmental samples. Generally, the contamination rate for clinical samples is about 1% to 3% in our laboratory. The high contamination rate in the early phase of the study was improved with additional training for our laboratory personnel.
In conclusion, patients infected with TB can contaminate an ecosystem. M tuberculosis remains alive for extended periods of time, particularly in soil. Our findings suggest that water and soil exposure might be a potential environmental risk factor. Epidemiologic genomic analysis is usually performed on clinical isolates, but our findings suggest investigating soil and water samples for M tuberculosis in the TB endemic countries. This may provide a complete picture of circulating genotypes within specific regions.
Acknowledgments
Author contributions: P. F. takes responsibility for the content of the manuscript, including the data and analysis. P. F. and M. Mirsaeidi contributed to the conception, hypotheses delineation, and design of the study; A. A. V., P. F., M. Mozafari, D. M., A. M. F., S. S., and S. R. contributed to data analysis and interpretation; and P. F. and M. Mirsaeidi contributed to the writing of the article or were substantially involved in its revision before submission.
Financial/nonfinancial disclosures: The authors have reported to CHEST that no potential conflicts of interest exist with any companies/organizations whose products or services may be discussed in this article.
Role of sponsors: The sponsors had no role in the design of the study, the collection and analysis of the data, or the preparation of the manuscript.
Other contributions: We thank Joseph O. Falkinham III, PhD, and Mary Beth Allen, PhD, for their great comments.
ABBREVIATIONS
- HGI
Hunter and Gaston index
- MDR-TB
multidrug-resistant Mycobacterium tuberculosis
- MIRU-VNTR
mycobacterial interspersed repetitive units-variable number of tandem repeats
- PCR
polymerase chain reaction
Footnotes
FUNDING/SUPPORT: This study was supported by the National Research Institute of Tuberculosis and Lung Disease of Tehran, Iran. Dr Mirsaeidi is supported by the National Institutes of Health [Grant 5 T32 HL 82547-7].
Reproduction of this article is prohibited without written permission from the American College of Chest Physicians. See online for more details.
References
- 1.Musehold P. Uber die Widerstand-fahigkeit der mit dem Lungenauswur-fherausbeforderten Tuberkelbazillen in Abwassern im Flusswasser und in Kultiverten Boden. Arb Kaiserl Gesundh. 1900;17:56. [Google Scholar]
- 2.Johnson KR, Braden CR, Cairns KL, et al. Transmission of Mycobacterium tuberculosis from medical waste. JAMA. 2000;284(13):1683-1688. [DOI] [PubMed] [Google Scholar]
- 3.Norby B, Fosgate GT, Manning EJ, Collins MT, Roussel AJ. Environmental mycobacteria in soil and water on beef ranches: association between presence of cultivable mycobacteria and soil and water physicochemical characteristics. Vet Microbiol. 2007;124(1-2):153-159. [DOI] [PubMed] [Google Scholar]
- 4.Kramer A, Schwebke I, Kampf G. How long do nosocomial pathogens persist on inanimate surfaces? A systematic review. BMC Infect Dis. 2006;6:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rickards BR, Slack FH, Arms BL. Longevity of B. tuberculosis in sputum. Am J Public Hygiene. 1909;19(3):586-594. [PMC free article] [PubMed] [Google Scholar]
- 6.Ghodbane R, Mba Medie F, Lepidi H, Nappez C, Drancourt M. Long-term survival of tuberculosis complex mycobacteria in soil. Microbiology. 2014;160(pt 3):496-501. [DOI] [PubMed] [Google Scholar]
- 7.World Health Organization. Global tuberculosis report 2013. WHO website. http://apps.who.int/iris/bitstream/10665/91355/1/9789241564656_eng.pdf?ua=1. Accessed April 2014.
- 8.Portaels F, De Muynck A, Sylla MP. Selective isolation of mycobacteria from soil: a statistical analysis approach. J Gen Microbiol. 1988;134(3):849-855. [DOI] [PubMed] [Google Scholar]
- 9.Tsukamura M. Differentiation of Mycobacterium tuberculosis from other mycobacteria by sodium salicylate susceptiblity. Am Rev Respir Dis. 1962;86:81-83. [DOI] [PubMed] [Google Scholar]
- 10.Engbaek HC. The sensitivity of atypical mycobacteria to sodium hydroxide and a surface active agent (“Teepol”). Acta Tuberc Pneumol Scand. 1962;42:1-9. [PubMed] [Google Scholar]
- 11.Brooks RW, George KL, Parker BC, Falkinham JO, III, Gruff H. Recovery and survival of nontuberculous mycobacteria under various growth and decontamination conditions. Can J Microbiol. 1984;30(9):1112-1117. [DOI] [PubMed] [Google Scholar]
- 12.Moghim S, Sarikhani E, Nasr Esfahani B, Faghri J. Identification of nontuberculous Mycobacteria species isolated from water samples using phenotypic and molecular methods and determination of their antibiotic resistance patterns by E-test method, in Isfahan, Iran. Iran J Basic Med Sci. 2012;15(5):1076-1082. [PMC free article] [PubMed] [Google Scholar]
- 13.Velayati AA, Farnia P, Mozafari M, et al. High prevalence of rifampin-monoresistant tuberculosis: a retrospective analysis among Iranian pulmonary tuberculosis patients. Am J Trop Med Hyg. 2014;90(1):99-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kent PT, Kubica GP. Public Health Mycobacteriology. A Guide for the Level III Laboratory. Atlanta, GA: Centers for Disease Control; 1985. [Google Scholar]
- 15.Frothingham R, Meeker-O’Connell WA. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology. 1998;144(pt 5):1189-1196. [DOI] [PubMed] [Google Scholar]
- 16.Supply P, Mazars E, Lesjean S, Vincent V, Gicquel B, Locht C. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol Microbiol. 2000;36(3):762-771. [DOI] [PubMed] [Google Scholar]
- 17.Alonso-Rodríguez N, Martínez-Lirola M, Herránz M, et al. ; INDAL-TB group. Evaluation of the new advanced 15-loci MIRU-VNTR genotyping tool in Mycobacterium tuberculosis molecular epidemiology studies. BMC Microbiol. 2008;8:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weniger T, Krawczyk J, Supply P, Niemann S, Harmsen D. MIRU-VNTRplus: a web tool for polyphasic genotyping of Mycobacterium tuberculosis complex bacteria. Nucleic Acids Res. 2010;38(Web Server issue):W326-W331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J Clin Microbiol. 1988;26(11):2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reed C, von Reyn CF, Chamblee S, et al. Environmental risk factors for infection with Mycobacterium avium complex. Am J Epidemiol. 2006;164(1):32-40. [DOI] [PubMed] [Google Scholar]
- 21.Gryseels S, Amissah D, Durnez L, et al. Amoebae as potential environmental hosts for Mycobacterium ulcerans and other mycobacteria, but doubtful actors in Buruli ulcer epidemiology. PLoS Negl Trop Dis. 2012;6:e1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ovrutsky AR, Chan ED, Kartalija M, et al. Cooccurrence of free-living amoebae and nontuberculous Mycobacteria in hospital water networks, and preferential growth of Mycobacterium avium in Acanthamoeba lenticulata. Appl Environ Microbiol. 2013;79(10):3185-3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mba Medie F, Ben Salah I, Henrissat B, Raoult D, Drancourt M. Mycobacterium tuberculosis complex mycobacteria as amoeba-resistant organisms. PLoS ONE. 2011;6(6):e20499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alland D, Whittam TS, Murray MB, et al. Modeling bacterial evolution with comparative-genome-based marker systems: application to Mycobacterium tuberculosis evolution and pathogenesis. J Bacteriol. 2003;185(11):3392-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wirth T, Hildebrand F, Allix-Béguec C, et al. Origin, spread and demography of the Mycobacterium tuberculosis complex. PLoS Pathog. 2008;4(9):e1000160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duchêne V, Ferdinand S, Filliol I, Guégan JF, Rastogi N, Sola C. Phylogenetic reconstruction of Mycobacterium tuberculosis within four settings of the Caribbean region: tree comparative analyse and first appraisal on their phylogeography. Infect Genet Evol. 2004;4(1):5-14. [DOI] [PubMed] [Google Scholar]
- 27.Merza MA, Farnia P, Salih AM, Masjedi MR, Velayati AA. First insight into the drug resistance pattern of Mycobacterium tuberculosis in Dohuk, Iraq: using spoligotyping and MIRU-VNTR to characterize multidrug resistant strains. J Infect Public Health. 2011;4(1):41-47. [DOI] [PubMed] [Google Scholar]
- 28.Chen Y, Chao Y, Deng Q, et al. Potential challenges to the Stop TB Plan for humans in China; cattle maintain M. bovis and M. tuberculosis. Tuberculosis (Edinb). 2009;89(1):95-100. [DOI] [PubMed] [Google Scholar]
- 29.Romero B, Rodríguez S, Bezos J, et al. Humans as source of Mycobacterium tuberculosis infection in cattle, Spain. Emerg Infect Dis. 2011;17(12):2393-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fetene T, Kebede N, Alem G. Tuberculosis infection in animal and human populations in three districts of Western Gojam, Ethiopia. Zoonoses Public Health. 2011;58(1):47-53. [DOI] [PubMed] [Google Scholar]