Abstract
Developing translational biomarkers is a priority for psychiatry research. Task-independent functional brain imaging is a relatively novel technique that allows examination of the brain’s intrinsic networks, defined as functionally and (often) structurally connected populations of neurons whose properties reflect fundamental neurobiological organizational principles of the central nervous system. The ability to study the activity and organization of these networks has opened a promising new avenue for translational investigation, because they can be analogously examined across species and disease states. Interestingly, imaging studies have revealed shared spatial and functional characteristics of the intrinsic network architecture of the brain across species, including mice, rats, non-human primates, and humans. Using schizophrenia as an example, we show how intrinsic networks may show similar abnormalities in human diseases and animal models of these diseases, supporting their use as biomarkers in drug development.
Keywords: biomarker, connectivity, graph theory, neuroimaging, resting state, schizophrenia
Why ‘task-independent’ functional imaging?
A major obstacle facing psychiatry research is the lack of effective translational biomarkers, or biological indicators of disease state. These assays are not only essential for improving our understanding of the neurobiological mechanisms that underlie disease, but also for providing screening tools to increase the probability of success for investigational compounds as they enter clinical trials.
To that end, investigators have long been interested in using functional magnetic resonance imaging (fMRI) to study neuronal function across species. fMRI is a technique in which the detection of magnetic field disruptions due to the flow of deoxygenated blood is used as a surrogate measure of localized neuronal activity. Great advantages of fMRI are its safety, noninvasiveness, and high spatial resolution. Early attempts at using fMRI as a translational tool were hampered, however, by limitations in its analysis methods. Early fMRI studies in humans were almost entirely ‘task’ based (e.g., a working memory task), because the fMRI signal – the blood oxygen level-dependent (BOLD) response – could only be interpreted as a comparison between conditions (e.g., task versus no task) using a general linear model (GLM)-based approach. This limitation severely restricted the utility of fMRI in animal studies, not only due to limitations in cognitive ability, but also because many animals (e.g., rodents) were required to be restrained, sedated, or anesthetized during scanning.
Fortunately, recent advances in fMRI analysis methods have enabled researchers to quantify and understand brain function in terms of intrinsic brain networks that are present across all cognitive states, including during rest, sedation, anesthesia, and sleep [1–3]. Intrinsic networks are defined as functionally and (often) structurally connected areas whose activity is thought to reflect fundamental neurobiological organizational principles of the central nervous system. Intrinsic networks are frequently referred to as resting state networks, although they can be extracted regardless of the mental state of the subject. Intrinsic networks are identified methodologically by either seed or independent component analysis (ICA) data-driven based methods (Box 1), and consist of large populations of neurons that demonstrate low-frequency (<0.1 Hz) synchronous BOLD responses [4]. An additional advantage of these techniques is that, unlike traditional GLM-based analysis, they do not impose prior constraints on the time course of the BOLD response, which may vary between individuals [5,6]. This flexibility may help explain why, remarkably, multiple anatomically distinct networks are consistently extracted, reflecting a map of intrinsic functional brain connectivity [7]. These networks may be specialized for functions such as executive function, salience processing, and introspection [8,9]. The activity and functional connectivity of intrinsic networks are dramatically altered in neuropsychiatric diseases such as Alzheimer’s disease (AD) [10], bipolar disorder [11], autism [12], attention deficit hyperactivity disorder (ADHD) [13], obesity [14,15], and schizophrenia [11,16], supporting their potential utility as biomarkers.
Box 1. Extracting intrinsic networks from fMRI data.
Two methods are commonly used to extract intrinsic networks from fMRI data: seed-based functional connectivity and ICA. These methods are conceptually identical across species.
Seed-based functional connectivity. In this technique, the correlation coefficients between one time series of data (the ‘seed’) and many other time series (the ‘targets’) are extracted [64]. Higher correlation coefficients imply more synchronous activity and therefore higher functional connectivity between the seed and a target. For example, the time series of the average BOLD response in the hippocampus may be correlated with the time series of the other major brain areas. An averaged correlation coefficient between the seed and all other areas may also be calculated to yield a value for overall connectivity of the seed.
ICA. The goal of ICA is to identify statistically independent patterns of BOLD response within the brain [64]. These independent patterns are then classified into networks based on the anatomical localization of their components. Networks identified by ICA show synchronous fMRI BOLD response with each other, as well as asynchronous response with other networks. The level of neuronal activity within an ICA-extracted network can be estimated by the magnitude of the signal fluctuations within that component [65].
Perhaps the greatest advantage of task-independent fMRI, however, is its translational utility. Because it does not require animals to perform a task, they can be either sedated or restrained during scanning, providing suitable conditions for analysis of intrinsic networks using methods analogous to those used for human data. Furthermore, traditional implanted-electrode recordings of brain activity are labor-intensive, invasive, spatially restricted, and only permit the simultaneous study of one or two isolated regions. By contrast, fMRI provides noninvasive whole-brain coverage of neuronal response, allowing the researcher to understand the brain as a dynamic, integrated system of connections within and between networks of many regions. Using fMRI, researchers have analyzed intrinsic brain network activity from a variety of organisms, including mice, rats, and non-human primates. Perhaps the most significant aspect of these findings is that core features of intrinsic networks have been conserved across species, suggesting that their fundamental organization may have been evolutionarily selected for over time. These similarities present the intriguing possibility that disruptions in the networks observed in disease states may be replicated in animal models, highlighting the translational utility of the approach. Ultimately, intrinsic networks may become invaluable biomarkers by which to measure the neurobiological effects of investigational and other compounds of interest.
Accordingly, this review focuses on two major topics. First, it examines recent findings characterizing core intrinsic networks across species to illustrate the degree to which these networks have been conserved. We do not argue that these networks are topographically identical – indeed, large differences in brain size, neo-cortex/paleocortex ratio, and cognitive function between species preclude any notion of sameness – but rather illustrate that analogous methods can be used across species to identify brain networks that share common features. Second, by using schizophrenia – a devastating disorder with well-known intrinsic network abnormalities – as an example, this review illustrates how task-independent fMRI might be used as a translational tool for drug discovery.
Intrinsic brain networks: from mice to men
The default mode network (DMN)
The DMN is the most widely studied and well-characterized intrinsic network. The DMN was discovered when researchers observed that activity in several brain areas was synchronously reduced during cognitive tasks and consequently increased at rest. Connectivity analyses later confirmed that these regions constituted an intrinsic functional network [17,18]. Due to its tendency to be down-modulated during many tasks, and therefore be active as a default, the network was coined the DMN. The human DMN consists of anterior (medial prefrontal cortex/orbito-frontal cortex/anterior cingulate) and posterior (inferior parietal/posterior cingulate/precuneus) brain areas [17] (Figure 1). The hippocampus/medial temporal lobe is considered an accessory hub of the network. The DMN is readily and reproducibly detectable regardless of the analytic technique used, and irrespective of the cognitive state of the individual, be it during an effortful task, rest, or even during sleep [1]. The functions of the DMN are not completely understood. The network is particularly active during actions that are self-referential: for example, reflecting on the past, planning for the future, or monitoring internal state [17]. Because the network also shows activity while under anesthesia [19,20] and during the early stages of sleep [2], however, its activity does not necessarily imply awake, self-referential thinking.
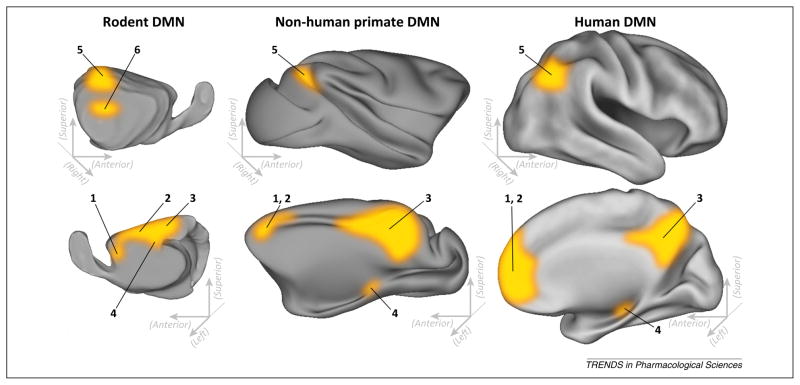
Figure 1.
Comparison of putative default networks of the rat, non-human primate, and human brain. DMN regions are displayed on surface renderings of the right hemisphere, in lateral (top) and medial (bottom) views; for rats [21,22] (left), macaques and marmoset monkey [24,25] (center), and human brains [8,17–19] (right). Numbered labels are based on regional homologs between species. 1: orbitofrontal cortex; 2: cingulate gyrus (in rats) or anterior cingulate cortex (monkey and human); 3: retrosplenial cortex (rat) or retrosplenial/posterior cingulate cortex (monkey and human); 4: hippocampus; 5: posterior parietal cortex; and 6: auditory/temporal association cortex (rat only). Adapted from [19,22]. Abbreviation: DMN, default mode network.
Based on its hypothesized functions (for example, self-reflection), one might speculate that the DMN is a uniquely human network, without analogs in other species. Surprisingly, however, striking similarities in DMN architecture exist between humans, rats [21,22] and non-human primates [19,23] (Figure 1). An early task-independent fMRI study in macaque monkeys found that temporoparietal and medial prefrontal areas demonstrated correlated response with a posterior cingulate seed [19]. Additional evidence that this network may be functionally analogous to the human DMN was provided by a recent meta-analysis that observed reduced activity of these brain regions across 15 sensory processing and cognitive tasks [24], as well a study that observed down-modulation of the posterior cingulate during an attention task [25]. In rats, Upadhyay and colleagues found correlated response between an anterior cingulate seed and the retrosplenial cortex/posterior cingulate, bilateral parietal cortex, temporal association cortex, and hippocampus [21]. A second study by Lu and coworkers that used ICA found similar results [22], although these researchers found more extensive correlations with the medial ridge of the cingulate cortex. Expansive cingulate involvement in the rat DMN is anatomically distinctive from the non-human primate and human DMN, suggesting that the DMN areas recruited across these species are not identical. Indeed, the primary distinctive feature of the human DMN is increased involvement of anterior regions, possibly indicative of an evolutionary adaptation that facilitates complex spontaneous (stimulus-independent) cognition [17].
The unique features of the DMN in different species, however, do not preclude the translational applications of examining the network. Indeed, the anatomical similarities of the DMN, in combination with the reduced activity of the network during tasks in both humans and non-human primates, suggest that the network may serve similar functions across species. Future meta-analyses in rats demonstrating down-modulation of the DMN during sensory stimulation would support the hypothesis that the DMN serves comparable task-negative functions.
Other intrinsic networks
In addition to the DMN, several other intrinsic networks have been identified in the human brain based on their functional localization. These include the sensorimotor network, the executive control network, the visual networks, the auditory networks, the temporo-parietal network, the cerebellar network, and the frontoparietal ventral and dorsal attention networks [7]. Of these networks, the most conserved across species are the cerebellar, somatosensory, motor, auditory, and visual networks [23,26–29]. The monkey ventral attention network is also considerably homologous to the human version [23,26]. These findings suggest that along with the DMN, these networks may be the most ancient, evolutionarily conserved intrinsic networks [30]. Other higher-order networks such as the dorsal attention network, the salience network, and the executive network are substantially reduced, have transplanted components, or are absent in the non-human primate and rodent brain [26–30]. The relative contribution of subcortical regions, such as the basal ganglia, to these networks may be greater in lower-order species such as rats, possibly due to more poorly developed neocortical structures in these animals [27]. A small number of networks are also more readily observable in rodents, such as a putative olfactory network [28] and an autonomic network that includes the hypothalamus [27].
Notably, task-independent fMRI has been combined with pharmacological interventions to identify networks associated with specific neurotransmitter systems. In one rodent fMRI study, injection of the selective serotonin reuptake inhibitor fluoxetine identified a serotonergic network encompassing the raphe nucleus, striatal, and medial temporal regions [31]. Furthermore, injection of D-amphetamine, a potentiator of dopaminergic transmission, identified a dopaminergic network encompassing regions associated with the mesolimbic and mesocortical pathways [31]. As expected, functional connectivity of this network was disrupted by the D3 receptor antagonist SB277011A [32]. These studies demonstrate how neurotransmitter systems spanning the entire brain can be identified, examined, and modulated using task-independent fMRI in combination with pharmacologic manipulation.
Complex network architecture
In addition to individual intrinsic networks, task-independent fMRI can be used to study the interaction of multiple networks or brain regions. Indeed, over the past decade, neuroscientists have become increasingly interested in understanding the functional organization of the brain as a dynamic system of interconnected regions. The emergent properties of this organization fundamentally define the complex network architecture of the brain. These features are well conserved across species and are altered in disease states, supporting their potential utility in translational drug discovery. To understand complex network architecture, researchers primarily use techniques adapted from graph theory, or the mathematical study of a system based on nodes and the edges between them [33]. In a neurobiological framework, a node is defined as an anatomical region (e.g., brain structure) and an edge is defined as a connection between two nodes that demonstrate correlated activity. Analysis of complex network architecture complements other methods of connectivity analysis by revealing the functional overarching organizational principles of the brain, such as: (i) efficient, low-cost information transfer; and (ii) a modular organization that maximizes local processing ability while facilitating global, long distance processing [34].
One striking feature of functional brain architecture that previous studies have demonstrated to be remarkably conserved across species – humans, non-human primates, rats, and even nematodes – is its ‘small worldness’ [19,34–37]. A small world network has two fundamental characteristics: (i) high clustering, with dense local connections but sparse long-distance connections; and (ii) high efficiency, that is, a short distance between any two nodes chosen at random. These features enable small world networks to minimize the time and distance required to travel between any two random locations in the network while simultaneously maximizing local processing and minimizing ‘wiring cost’ (in the brain, this term is best conceptualized as axonal length). For an analogy, consider a global corporation with headquarters in major cities. Most communication occurs between members of local branches of the corporation. When a corporation-wide decision must be made, however, a single member of a local branch will communicate with one member of each other branch in order to pass the message efficiently across the entire company. Less adaptive networks include lattice networks, which have high clustering but low efficiency, and random networks, which have high efficiency but low clustering (Figure 2).
Figure 2.
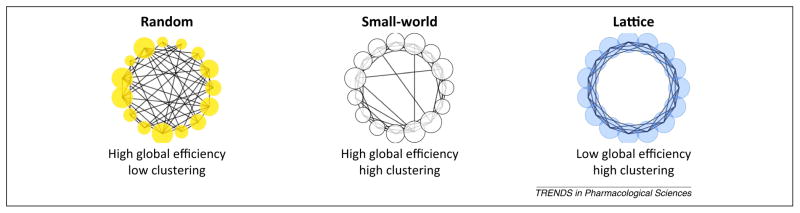
Illustrative examples of lattice, random, and small-world networks. A lattice network is characterized by high clustering (each node is highly connected to its neighbors) but low efficiency (long path length). A random network is characterized by high efficiency (short path length) but low clustering. A small-world network maximizes both efficiency and clustering while minimizing wiring cost (the number of edges). The rodent, non-human primate, and human brain all have small-world properties that may be disrupted in human as well as animal models of neurologic and psychiatric diseases. Adapted from [48].
Other concepts used to describe network architecture are modularity, betweenness centrality, and hub. Modularity describes the degree to which local nodes are connected to each other. The modules identified in this analysis are conceptually similar to components from an ICA. Betweenness centrality measures the number of shortest path lengths that pass through a node. Nodes with high betweenness centrality are considered hubs; these are the most crucial for efficient information transfer between local clusters (groups of interconnected nearest neighbors). Across species, the brain shows high modularity, with clusters connected by a limited number of hubs primarily localized to the medial prefrontal and retrospenial/posterior cingulate cortices [22,24,38]. Interestingly, these regions are primary components of the DMN, suggesting that the DMN is a major contributor to the overall functional architecture of the brain. In addition, the evolutionary conservation of these metrics suggests that they may be key guiding principles in brain organization, possibly due to limitations in brain size, metabolic cost, and the time required to transfer information [34].
Task-independent fMRI as a translational tool: example in schizophrenia
In this final section, to illustrate how task-independent animal fMRI may be used as a translational tool for therapeutic development, we discuss its potential applications in schizophrenia research.
Development of functional neuroimaging biomarkers –that is, stable, reproducible, clinically relevant measures of brain function that predict treatment response – is inherently difficult in psychiatric disorders such as schizophrenia due to their genotypic and phenotypic heterogeneity. Nonetheless, one of the most widely studied and consistently reported neurobiological abnormalities in schizophrenia patients is hyperactivity and hyperconnectivity of the DMN, both within and between its components [1,39,40]. Less-severe but otherwise similar DMN changes have been reported in first-degree relatives [41] and people at risk for schizophrenia [42], suggesting that: (i) these changes are not an ancillary effect of antipsychotic medication; and (ii) a threshold for DMN dysfunction may exist that is crossed in schizophrenia. Related to this last point, DMN hyperactivity and hyperconnectivity may predict the severity of positive, negative, and cognitive symptoms in schizophrenia patients [40,43,44].
Despite the potential utility of DMN dysfunction as a biomarker for schizophrenia (reviewed in [40]), to the best of our knowledge only one study to date has examined the effect of an investigational compound on DMN function in the illness. Using a double-blind, crossover, placebo-controlled design, Tregellas et al. observed reduced posterior DMN activity in schizophrenia patients after 4 weeks treatment with the nicotinic α7 receptor partial agonist 3-(2,4-dimethoxybenzylidine) anabaseine (DMXB-A) [45]. This effect was associated with improved symptoms after drug administration.
The recent findings with DMXB-A suggest that abnormalities in DMN function may predict treatment response in schizophrenia, supporting its utility as a biomarker. We suggest that examining DMN activity may be an effective method of screening compounds in early stages of therapeutic development, increasing the probability that these compounds will demonstrate efficacy in more expensive, late-stage trials. This screening process may eventually include animal models of schizophrenia as well human patients. As emphasized in this review, a great advantage of studying the DMN is that it can be readily examined in different species. Furthermore, numerous animal models exist for schizophrenia. These include pharmacological models (using dopaminergic agonists and glutamatergic antagonists), developmental models (ventral hippocampal lesion, perinatal behavioral and neurotoxic stress, disrupted in schizophrenia-1 mutant mice, neuregulin-1 mutant mice) and combinations of the above (e.g., a ‘two-hit’ model that includes genetic mutation combined with an early-life stressor) (reviewed in [46]). An important area for future investigation is to use fMRI to examine DMN activity in these models relative to healthy animals. Based on findings in schizophrenia, it is possible to speculate that several of these models will demonstrate DMN hyperactivity and connectivity in their medial components (e.g., posterior cingulate). Interestingly, a recent study found that ketamine, a glutamate receptor antagonist that is used to model schizophrenia in rodents (e.g., cause hyperlocomotor activity, social deficits, and working memory deficits), increases connectivity between the posterior cingulate/retrosplenial cortex and hippocampus in rats in a dose-dependent manner [47]. This study suggests that a pharmacological rodent model of schizophrenia may mimic DMN features observed in human patients.
In addition to the DMN, other features of intrinsic network dynamics are beginning to be understood in schizophrenia as well as animal models of the illness. For example, other intrinsic networks such as the salience network [48–50], auditory network [51,52], and cortical–subcortical networks [53] are disrupted in schizophrenia and related to symptom severity [16]. Analysis of complex network architecture has observed reduced local and global efficiency in schizophrenia patients, as demonstrated by reduced modularity, longer path lengths, lower clustering coefficients, and disruptions in small world brain architecture that is characteristic of the healthy brain [54–58]. These network abnormalities may predict the severity of positive and negative symptoms in the illness [57] and have demonstrated an inverse correlation with antipsychotic dose [55]. With regards to animal models of schizophrenia, Dawson and coworkers recently observed that subchronic (5 days) injection of the N-methyl-D-aspartate (NMDA) receptor antagonist phencyclidine (PCP) reduced small world properties of the rat brain, as shown by increased path length and reduced clustering [59]. Furthermore, PCP reduced betweenness centrality as demonstrated by a reduced number of network hubs after treatment relative to saline injection [59]. Another study found that a second putative feature of schizophrenia – low modularity – was increased in rats after injection of D-amphetamine, fluoxetine, or nicotine [60]. All three of these drugs have been investigated for the treatment of cognitive and/or negative symptoms in schizophrenia, and compounds with related pharmacology are in various stages of clinical development. These results suggest that the effects of these compounds on network architecture may have clinical relevance, supporting their use as pre-clinical screening tools for drug discovery.
Importantly, task-independent fMRI does not supersede the use of other imaging techniques in drug development. Indeed, for investigational compounds, an important early step is target validation in animals, using techniques such as positron emission tomography (PET). Like task-independent fMRI, PET data can be examined across species using similar methodology and subject conditions (e.g., at rest). After target engagement is confirmed, task-independent animal fMRI can be used as an additional screening tool to examine potentially clinically relevant downstream neurobiological effects. These studies may be performed concurrently with behavioral assays, and only compounds that pass all three screening measures may be allowed to progress further along the drug development pipeline. Consequently, in humans, examination of intrinsic networks by task-independent fMRI as well as proof of target engagement by PET may be added (in conjunction with clinical measures) as primary outcomes to early stage clinical trials. In summary, using this approach, a drug must demonstrate efficacy across a range of fully translational and clinical assays before it enters Phase III, increasing its likelihood of success.
Concluding remarks
A major crisis looms for drug development in psychiatric disorders and the patients for whom new treatments are intended to help. On the one hand, effective interventions for the symptoms that most affect quality of life, such as cognitive symptoms of schizophrenia [61], are almost entirely lacking. On the other hand, rising costs of research combined with a low probability of success for compounds entering clinical trials (as of 2009, <4% for self-originated central nervous system drugs) [62] has resulted in many companies either downsizing or dropping developmental platforms for these disorders altogether [63]. Clearly, new strategies must be developed to more efficiently screen investigational compounds in preclinical and early stage clinical trials, reducing costs and providing incentive for institutions to expand, rather than contract, psychiatric drug development programs.
In this review, we have highlighted a relatively novel approach to functional neuroimaging – analysis of task-independent brain function – and demonstrated how analogous methods can be used in translational fashion from rodents to man. It must be emphasized, however, that its suggested utility as an early screening device must at this point remain speculative, because few studies have examined how intrinsic networks such as the DMN are altered in animal models of psychiatric disease. Clearly, additional research needs to be performed in these areas, particularly using developmental animal models that may closely mirror the developmental and clinical time course of psychiatric illnesses. If these models are further established as reliable indicators of disease state, they hold great promise as invaluable tools for therapeutic development.
References
- 1.Whitfield-Gabrieli S, Ford JM. Default mode network activity and connectivity in psychopathology. Annu Rev Clin Psychol. 2012;8:49–76. doi: 10.1146/annurev-clinpsy-032511-143049. [DOI] [PubMed] [Google Scholar]
- 2.Fukunaga M, et al. Large-amplitude, spatially correlated fluctuations in BOLD fMRI signals during extended rest and early sleep stages. Magn Reson Imaging. 2006;24:979–992. doi: 10.1016/j.mri.2006.04.018. [DOI] [PubMed] [Google Scholar]
- 3.Jordan D, et al. Simultaneous electroencephalographic and functional magnetic resonance imaging indicate impaired cortical top-down processing in association with anesthetic-induced unconsciousness. Anesthesiology. 2013;119:1031–1042. doi: 10.1097/ALN.0b013e3182a7ca92. [DOI] [PubMed] [Google Scholar]
- 4.Fox MD, et al. The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proc Natl Acad Sci USA. 2005;102:9673–9678. doi: 10.1073/pnas.0504136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aguirre GK, et al. The variability of human, BOLD hemodynamic responses. Neuroimage. 1998;8:360–369. doi: 10.1006/nimg.1998.0369. [DOI] [PubMed] [Google Scholar]
- 6.Handwerker DA, et al. Variation of BOLD hemodynamic responses across subjects and brain regions and their effects on statistical analyses. Neuroimage. 2004;21:1639–1651. doi: 10.1016/j.neuroimage.2003.11.029. [DOI] [PubMed] [Google Scholar]
- 7.Smith SM, et al. Correspondence of the brain’s functional architecture during activation and rest. Proc Natl Acad Sci USA. 2009;106:13040–13045. doi: 10.1073/pnas.0905267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raichle ME, Snyder AZ. A default mode of brain function: a brief history of an evolving idea. Neuroimage. 2007;37:1083–1090. doi: 10.1016/j.neuroimage.2007.02.041. [DOI] [PubMed] [Google Scholar]
- 9.Seeley WW, et al. Dissociable intrinsic connectivity networks for salience processing and executive control. J Neurosci. 2007;27:2349–2356. doi: 10.1523/JNEUROSCI.5587-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y, et al. Regional homogeneity, functional connectivity and imaging markers of Alzheimer’s disease: a review of resting-state fMRI studies. Neuropsychologia. 2008;46:1648–1656. doi: 10.1016/j.neuropsychologia.2008.01.027. [DOI] [PubMed] [Google Scholar]
- 11.Whalley HC, et al. Review of functional magnetic resonance imaging studies comparing bipolar disorder and schizophrenia. Bipolar Disord. 2012;14:411–431. doi: 10.1111/j.1399-5618.2012.01016.x. [DOI] [PubMed] [Google Scholar]
- 12.Minshew NJ, Keller TA. The nature of brain dysfunction in autism: functional brain imaging studies. Curr Opin Neurol. 2010;23:124–130. doi: 10.1097/WCO.0b013e32833782d4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castellanos FX, Proal E. Large-scale brain systems in ADHD: beyond the prefrontal-striatal model. Trends Cogn Sci. 2012;16:17–26. doi: 10.1016/j.tics.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tregellas JR, et al. Altered default network activity in obesity. Obesity (Silver Spring) 2011;19:2316–2321. doi: 10.1038/oby.2011.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Garcia I, et al. Alterations of the salience network in obesity: a resting-state fMRI study. Hum Brain Mapp. 2013;34:2786–2797. doi: 10.1002/hbm.22104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karbasforoushan H, Woodward ND. Resting-state networks in schizophrenia. Curr Top Med Chem. 2012;12:2404–2414. doi: 10.2174/156802612805289863. [DOI] [PubMed] [Google Scholar]
- 17.Buckner RL, et al. The brain’s default network: anatomy, function, and relevance to disease. Ann N Y Acad Sci. 2008;1124:1–38. doi: 10.1196/annals.1440.011. [DOI] [PubMed] [Google Scholar]
- 18.Greicius MD, et al. Functional connectivity in the resting brain: a network analysis of the default mode hypothesis. Proc Natl Acad Sci USA. 2003;100:253–258. doi: 10.1073/pnas.0135058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vincent JL, et al. Intrinsic functional architecture in the anaesthetized monkey brain. Nature. 2007;447:83–86. doi: 10.1038/nature05758. [DOI] [PubMed] [Google Scholar]
- 20.Greicius MD, et al. Persistent default-mode network connectivity during light sedation. Hum Brain Mapp. 2008;29:839–847. doi: 10.1002/hbm.20537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Upadhyay J, et al. Default-mode-like network activation in awake rodents. PLoS ONE. 2011;6:e27839. doi: 10.1371/journal.pone.0027839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu H, et al. Rat brains also have a default mode network. Proc Natl Acad Sci USA. 2012;109:3979–3984. doi: 10.1073/pnas.1200506109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Belcher AM, et al. Large-scale brain networks in the awake, truly resting marmoset monkey. J Neurosci. 2013;33:16796–16804. doi: 10.1523/JNEUROSCI.3146-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mantini D, et al. Default mode of brain function in monkeys. J Neurosci. 2011;31:12954–12962. doi: 10.1523/JNEUROSCI.2318-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayden BY, et al. Electrophysiological correlates of default-mode processing in macaque posterior cingulate cortex. Proc Natl Acad Sci USA. 2009;106:5948–5953. doi: 10.1073/pnas.0812035106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hutchison RM, et al. Resting-state networks in the macaque at 7 T. Neuroimage. 2011;56:1546–1555. doi: 10.1016/j.neuroimage.2011.02.063. [DOI] [PubMed] [Google Scholar]
- 27.Becerra L, et al. Robust reproducible resting state networks in the awake rodent brain. PLoS ONE. 2011;6:e25701. doi: 10.1371/journal.pone.0025701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jonckers E, et al. Functional connectivity fMRI of the rodent brain: comparison of functional connectivity networks in rat and mouse. PLoS ONE. 2011;6:e18876. doi: 10.1371/journal.pone.0018876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sforazzini F, et al. Distributed BOLD and CBV-weighted resting-state networks in the mouse brain. Neuroimage. 2014;87:403–415. doi: 10.1016/j.neuroimage.2013.09.050. [DOI] [PubMed] [Google Scholar]
- 30.Mantini D, et al. Evolutionarily novel functional networks in the human brain? J Neurosci. 2013;33:3259–3275. doi: 10.1523/JNEUROSCI.4392-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwarz AJ, et al. In vivo mapping of functional connectivity in neurotransmitter systems using pharmacological MRI. Neuroimage. 2007;34:1627–1636. doi: 10.1016/j.neuroimage.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 32.Schwarz AJ, et al. Pharmacological modulation of functional connectivity: the correlation structure underlying the phMRI response to d-amphetamine modified by selective dopamine D3 receptor antagonist SB277011A. Magn Reson Imaging. 2007;25:811–820. doi: 10.1016/j.mri.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 33.Sporns O, et al. Organization, development and function of complex brain networks. Trends Cogn Sci. 2004;8:418–425. doi: 10.1016/j.tics.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 34.Bullmore E, Sporns O. The economy of brain network organization. Nat Rev Neurosci. 2012;13:336–349. doi: 10.1038/nrn3214. [DOI] [PubMed] [Google Scholar]
- 35.Watts DJ, Strogatz SH. Collective dynamics of ‘small-world’ networks. Nature. 1998;393:440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- 36.Schwarz AJ, et al. Community structure and modularity in networks of correlated brain activity. Magn Reson Imaging. 2008;26:914–920. doi: 10.1016/j.mri.2008.01.048. [DOI] [PubMed] [Google Scholar]
- 37.Liang Z, et al. Uncovering intrinsic connectional architecture of functional networks in awake rat brain. J Neurosci. 2011;31:3776–3783. doi: 10.1523/JNEUROSCI.4557-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van den Heuvel MP, Sporns O. Network hubs in the human brain. Trends Cogn Sci. 2013;17:683–696. doi: 10.1016/j.tics.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 39.Tregellas JR, et al. Intrinsic hippocampal activity as a biomarker for cognition and symptoms in schizophrenia. Am J Psychiatry. 2014;171:549–556. doi: 10.1176/appi.ajp.2013.13070981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tregellas JR. Neuroimaging biomarkers for early drug development in schizophrenia. Biol Psychiatry. 2013 doi: 10.1016/j.biopsych.2013.08.025. http://dx.doi.org/10.1016/j.biopsych.2013.08.025. [DOI] [PMC free article] [PubMed]
- 41.Liu H, et al. Schizophrenic patients and their unaffected siblings share increased resting-state connectivity in the task-negative network but not its anticorrelated task-positive network. Schizophr Bull. 2012;38:285–294. doi: 10.1093/schbul/sbq074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shim G, et al. Altered resting-state connectivity in subjects at ultra-high risk for psychosis: an fMRI study. Behav Brain Funct. 2010;6:58. doi: 10.1186/1744-9081-6-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garrity AG, et al. Aberrant “default mode” functional connectivity in schizophrenia. Am J Psychiatry. 2007;164:450–457. doi: 10.1176/ajp.2007.164.3.450. [DOI] [PubMed] [Google Scholar]
- 44.Whitfield-Gabrieli S, et al. Hyperactivity and hyperconnectivity of the default network in schizophrenia and in first-degree relatives of persons with schizophrenia. Proc Natl Acad Sci USA. 2009;106:1279–1284. doi: 10.1073/pnas.0809141106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tregellas JR, et al. Effects of an alpha 7-nicotinic agonist on default network activity in schizophrenia. Biol Psychiatry. 2011;69:7–11. doi: 10.1016/j.biopsych.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mouri A, et al. Animal models of schizophrenia for molecular and pharmacological intervention and potential candidate molecules. Neurobiol Dis. 2013;53:61–74. doi: 10.1016/j.nbd.2012.10.025. [DOI] [PubMed] [Google Scholar]
- 47.Gass N, et al. Sub-anesthetic ketamine modulates intrinsic BOLD connectivity within the hippocampal-prefrontal circuit in the rat. Neuropsychopharmacology. 2014;39:895–906. doi: 10.1038/npp.2013.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smucny J, Tregellas J. Nicotinic modulation of intrinsic brain networks in schizophrenia. Biochem Pharmacol. 2013;86:1163–1172. doi: 10.1016/j.bcp.2013.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moran LV, et al. Disruption of anterior insula modulation of large-scale brain networks in schizophrenia. Biol Psychiatry. 2013;74:467–474. doi: 10.1016/j.biopsych.2013.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Manoliu A, et al. Aberrant dependence of default mode/central executive network interactions on anterior insular salience network activity in schizophrenia. Schizophr Bull. 2013 doi: 10.1093/schbul/sbt037. http://dx.doi.org/10.1093/schbul/sbt037. [DOI] [PMC free article] [PubMed]
- 51.Liemburg EJ, et al. Abnormal connectivity between attentional, language and auditory networks in schizophrenia. Schizophr Res. 2012;135:15–22. doi: 10.1016/j.schres.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 52.Rotarska-Jagiela A, et al. Resting-state functional network correlates of psychotic symptoms in schizophrenia. Schizophr Res. 2010;117:21–30. doi: 10.1016/j.schres.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 53.Klingner CM, et al. Thalamocortical connectivity during resting state in schizophrenia. Eur Arch Psychiatry Clin Neurosci. 2013;264:111–119. doi: 10.1007/s00406-013-0417-0. [DOI] [PubMed] [Google Scholar]
- 54.Liu Y, et al. Disrupted small-world networks in schizophrenia. Brain. 2008;131:945–961. doi: 10.1093/brain/awn018. [DOI] [PubMed] [Google Scholar]
- 55.Rubinov M, et al. Small-world properties of nonlinear brain activity in schizophrenia. Hum Brain Mapp. 2009;30:403–416. doi: 10.1002/hbm.20517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anderson A, Cohen MS. Decreased small-world functional network connectivity and clustering across resting state networks in schizophrenia: an fMRI classification tutorial. Front Hum Neurosci. 2013;7:520. doi: 10.3389/fnhum.2013.00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu Q, et al. Altered topological properties of functional network connectivity in schizophrenia during resting state: a small-world brain network study. PLoS ONE. 2011;6:e25423. doi: 10.1371/journal.pone.0025423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alexander-Bloch AF, et al. Disrupted modularity and local connectivity of brain functional networks in childhood-onset schizophrenia. Front Syst Neurosci. 2010;4:147. doi: 10.3389/fnsys.2010.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dawson N, et al. Sustained NMDA receptor hypofunction induces compromised neural systems integration and schizophrenia-like alterations in functional brain networks. Cereb Cortex. 2014;24:452–464. doi: 10.1093/cercor/bhs322. [DOI] [PubMed] [Google Scholar]
- 60.Schwarz AJ, et al. Community structure in networks of functional connectivity: resolving functional organization in the rat brain with pharmacological MRI. Neuroimage. 2009;47:302–311. doi: 10.1016/j.neuroimage.2009.03.064. [DOI] [PubMed] [Google Scholar]
- 61.Green MF. What are the functional consequences of neurocognitive deficits in schizophrenia? Am J Psychiatry. 1996;153:321–330. doi: 10.1176/ajp.153.3.321. [DOI] [PubMed] [Google Scholar]
- 62.DiMasi JA, et al. Trends in risks associated with new drug development: success rates for investigational drugs. Clin Pharmacol Ther. 2010;87:272–277. doi: 10.1038/clpt.2009.295. [DOI] [PubMed] [Google Scholar]
- 63.Abbott A. Schizophrenia: the drug deadlock. Nature. 2010;468:158–159. doi: 10.1038/468158a. [DOI] [PubMed] [Google Scholar]
- 64.Cole DM, et al. Advances and pitfalls in the analysis and interpretation of resting-state FMRI data. Front Syst Neurosci. 2010;4:8. doi: 10.3389/fnsys.2010.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McKeown MJ, Sejnowski TJ. Independent component analysis of fMRI data: examining the assumptions. Hum Brain Mapp. 1998;6:368–372. doi: 10.1002/(SICI)1097-0193(1998)6:5/6<368::AID-HBM7>3.0.CO;2-E. [DOI] [PMC free article] [PubMed] [Google Scholar]