A GH12 endoglucanase from A. aculeatus F-50 was expressed in P. pastoris, crystallized and diffracted to a resolution of 1.6 Å.
Keywords: cellulase, endoglucanase, Aspergillus, industrial enzyme
Abstract
Cellulose is the most abundant renewable biomass on earth, and its decomposition has proven to be very useful in a wide variety of industries. Endo-1,4-β-d-glucanase (EC 3.2.1.4; endoglucanase), which can catalyze the random hydrolysis of β-1,4-glycosidic bonds to cleave cellulose into smaller fragments, is a key cellulolytic enzyme. An endoglucanase isolated from Aspergillus aculeatus F-50 (FI-CMCase) that was classified into glycoside hydrolase family 12 has been found to be effectively expressed in the industrial strain Pichia pastoris. Here, recombinant FI-CMCase was crystallized. Crystals belonging to the orthorhombic space group C2221, with unit-cell parameters a = 74.2, b = 75.1, c = 188.4 Å, were obtained by the sitting-drop vapour-diffusion method and diffracted to 1.6 Å resolution. Initial phase determination by molecular replacement clearly shows that the crystal contains two protein molecules in the asymmetric unit. Further model building and structure refinement are in progress.
1. Introduction
Cellulose is the most abundant renewable biomass on earth; it is the major component of plant cell walls and constitutes 35–50% of plant dry mass (Sakuragi et al., 2011 ▶). Cellulose is a polysaccharide that is composed of β-1,4-glycosidic bond-linked glucose units. The hydrolysis of cellulose requires three main groups of enzymes: endo-1,4-β-d-glucanases (endoglucanases; EC 3.2.1.4), cellobiohydrolases (EC 3.2.1.91) and β-glycosidases (EC 3.2.1.21) (Lynd et al., 2002 ▶). Among these, endoglucanase, which randomly hydrolyzes β-glycosidic bonds to cleave cellulose into smaller fragments, is a key cellulolytic enzyme. Endoglucanases have been widely applied in various industries, such as the manufacture of animal feed, the food industry, the textile industry and biofuel production (Kuhad et al., 2011 ▶). Owing to their great economic potential, obtaining structural information on endoglucanases is of great interest in order to better understand of the catalytic mechanisms and properties of these enzymes. In addition, the three-dimensional protein structure is an important basis for subsequent protein engineering to improve the efficiency of enzymes. Previously, we have succeeded in applying structure-based rational design to improve the performance of several industrial enzymes (Huang et al., 2012 ▶, 2014 ▶; Wu et al., 2014 ▶), including an endoglucanase from a thermophilic bacterium (Cheng et al., 2012 ▶).
Endoglucanases have been found in various microorganisms, including fungi and bacteria (Doi, 2008 ▶). Aspergillus aculeatus F-50 is known to produce many cellulolytic enzymes, including three endoglucanases (FI-CMCase, FII-CMCase and FV-CMCase; Murao et al., 1979 ▶). Among them, FI-CMCase is the most abundant cellulase and plays an important role in cellulose hydrolysis (Murao et al., 1988 ▶). FI-CMCase belongs to glycoside family 12 (GH12) in the CAZy database according to protein sequence similarity. The enzyme was subsequently expressed and characterized in Escherichia coli (Ooi et al., 1993 ▶) and Saccharomyces cerevisiae (Minamiguchi et al., 1995 ▶; Ooi et al., 1994 ▶), with an optimal pH of 5.0 and an optimal temperature of 323 K. FI-CMCase has been crystallized, but no protein structure has been reported for reasons that are not known (Hata et al., 1994 ▶; Yoshizawa et al., 2011 ▶).
Pichia pastoris is a very useful vehicle which has been extensively utilized to express various recombinant heterologous proteins (Macauley-Patrick et al., 2005 ▶; Li et al., 2007 ▶). P. pastoris has several advantages compared with other expression systems, including well established molecular-manipulation protocols, rapid growth to high cell density with low cost and secretion of pure heterologous proteins into the culture broth (Damasceno et al., 2012 ▶). These features confer P. pastoris with high potential for industrial enzyme production. Recently, we found that the FI-CMCase can be efficiently expressed in P. pastoris and the recombinant protein displays high activity towards carboxymethyl cellulose (CMC) and filter paper. These results substantiate the production of FI-CMCase on an industrial scale using P. pastoris. In the present study, recombinant FI-CMCase expressed in P. pastoris was crystallized.
2. Materials and methods
2.1. Protein preparation
The gene encoding FI-CMCase (GenBank accession No. X52525.1) from A. aculeatus F-50 was chemically synthesized and then cloned into the vector pPICZαA (Invitrogen) using the EcoRI and NotI restriction sites. The recombinant plasmid was linearized by PmeI and transformed into P. pastoris strain X33 (Invitrogen) by electroporation. The transformants were selected on YPD plates (1% yeast extract, 2% peptone, 2% dextrose, 2% agar) containing 100 µg ml−1 Zeocin and incubated at 303 K for 2 d. The protein expression of the transformants was tested by small-scale expression as follows. The selected colonies were inoculated into 5 ml YPD medium (1% yeast extract, 2% peptone, 2% dextrose) and then amplified in 50 ml BMGY medium [1% yeast extract, 2% peptone, 100 mM potassium phosphate pH 6.0, 1.34% yeast nitrogen base (YNB) with ammonium sulfate without amino acids, 4 × 10−5% biotin, 1% glycerol] at 303 K for 24 h. The cells were harvested by centrifugation and resuspended in 20 ml BMMY medium (1% yeast extract, 2% peptone, 100 mM potassium phosphate pH 6.0, 1.34% YNB with ammonium sulfate without amino acids, 4 × 10−5% biotin, 0.5% methanol) to induce protein expression. Subsequently, the transformants with higher expression levels were chosen for scaled-up expression. The cell stock was grown at 303 K in 100 ml YPD medium containing 100 µg ml−1 Zeocin for 24 h. The cells were then transferred into 900 ml YPD medium. After a further 24 h, the cells were harvested and then resuspended in 1 l BMMY medium. A total of 0.5% methanol was supplemented every 24 h to induce protein expression for four consecutive days. The supernatant was collected by centrifugation and then purified on an FPLC system using a diethylaminoethyl (DEAE) Sepharose Fast Flow column (GE Healthcare, Uppsala, Sweden). The proteins were dialyzed twice against 5 l buffer consisting of 25 mM Tris–HCl pH 7.5 and loaded onto a DEAE column. Next, the purified proteins were eluted using a gradient of 0–250 mM NaCl in 25 mM Tris pH 7.5. The purified protein was finally concentrated to 10 mg ml−1 in 25 mM Tris–HCl pH 7.5, 150 mM NaCl using an Amicon Ultra-15 Centricon (Millipore) and the purity was checked by SDS–PAGE analysis (>95%).
2.2. Crystallization and data collection
Initial crystallization screening was performed manually using 768 different reservoir conditions from kits from Hampton Research (Laguna Niguel, California, USA), including Crystal Screen, Crystal Screen 2, Crystal Screen Cryo, Crystal Screen Lite, MembFac, Natrix, Index, SaltRx, SaltRx 2, PEG/Ion, PEG/Ion 2, Quick Screen and Grid Screens (Ammonium Sulfate, MPD, Sodium Chloride, Sodium Malonate, PEG 6000 and PEG/LiCl); all crystallization experiments were conducted at 298.15 K using the sitting-drop vapour-diffusion method. In general, 1 µl FI-CMCase protein solution (10 mg ml−1 in 25 mM Tris–HCl, 150 mM NaCl pH 7.5) was mixed with 1 µl reservoir solution in 24-well Cryschem plates (Hampton Research) and equilibrated against 500 µl reservoir solution. Initial crystals of FI-CMCase were obtained within 10 d using Crystal Screen condition No. 45 [0.2 M zinc acetate dihydrate, 0.1 M sodium cacodylate pH 6.5, 18%(w/v) polyethylene glycol 8000]. The optimized crystallization condition for all crystals mentioned here consisted of 0.3 M zinc acetate dihydrate, 0.1 M sodium cacodylate pH 6.5, 19%(w/v) polyethylene glycol 8000. Within 10–12 d, the crystals reached dimensions of about 0.6 × 0.2 × 0.1 mm. An X-ray diffraction data set was collected to 1.6 Å resolution on beamline BL13C1 of the National Synchrotron Radiation Research Center (NSRRC), Hsinchu, Taiwan. During data collection, the oscillation range was 0.5°, the exposure time was 5 s and the crystal-to-detector distance was 220 mm. The diffraction images were processed using HKL-2000 (Otwinowski & Minor, 1997 ▶). Data-collection statistics are given in Table 1 ▶.
Table 1. Data-collection statistics for the FI-CMCase crystal.
Values in parentheses are for the highest resolution shell.
| Beamline | BL13C1, NSRRC |
| Wavelength () | 0.97622 |
| Resolution () | 251.60 (1.661.60) |
| Space group | C2221 |
| Unit-cell parameters | |
| a () | 74.2 |
| b () | 75.1 |
| c () | 188.4 |
| No. of measured reflections | 461227 (43891) |
| No. of unique reflections | 70702 (6858) |
| Completeness (%) | 99.7 (97.3) |
| R merge † (%) | 7.5 (45.9) |
| Mean I/(I) | 39.8 (5.2) |
| Multiplicity | 6.5 (6.4) |
| Mosaicity () | 0.420.53 |
| Detector | ADSC Q315r |
| X-ray beam size (m) | 200 |
| Oscillation range (o) | 0.5 |
| Time of exposure (s) | 5 |
| Crystal-to-detector distance (mm) | 220 |
R
merge = 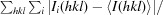
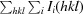 .
.
3. Results and discussion
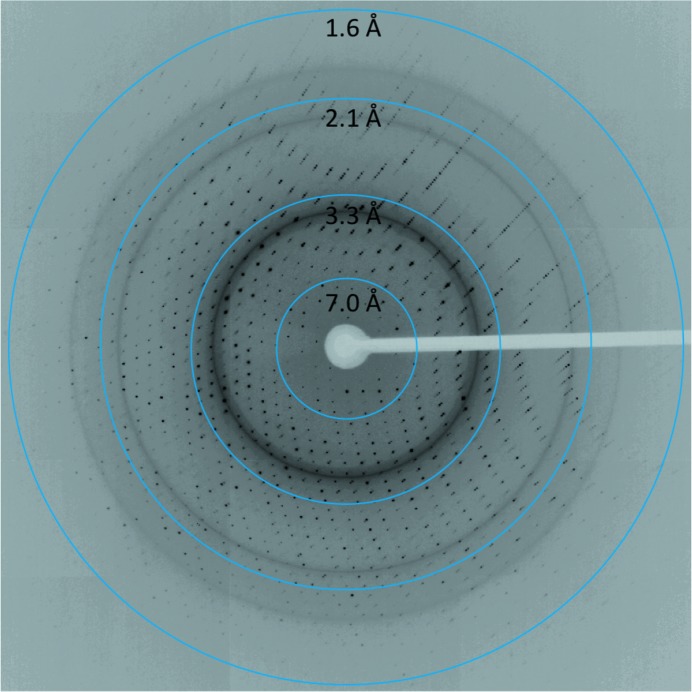
The FI-CMCase crystal (Fig. 1 ▶) was obtained in 0.3 M zinc acetate dihydrate, 0.1 M sodium cacodylate pH 6.5, 19%(w/v) polyethylene glycol 8000. Prior to data collection at 100 K, the crystal was mounted in a cryoloop and flash-cooled in liquid nitrogen with a slightly modified cryoprotectant which consisted of 0.3 M zinc acetate dihydrate, 0.1 M sodium cacodylate pH 6.5, 21%(w/v) polyethylene glycol 8000, 10% glycerol. Based on the diffraction pattern (Fig. 2 ▶), these FI-CMCase crystals belonged to the orthorhombic space group C2221, with unit-cell parameters a = 74.2, b = 75.1, c = 188.4 Å, α = β = γ = 90°. Assuming the presence of two molecules in the asymmetric unit, the Matthews coefficient V M (Matthews, 1968 ▶) is 2.64 Å3 Da−1 and the estimated solvent content is 53.42%.
Figure 1.
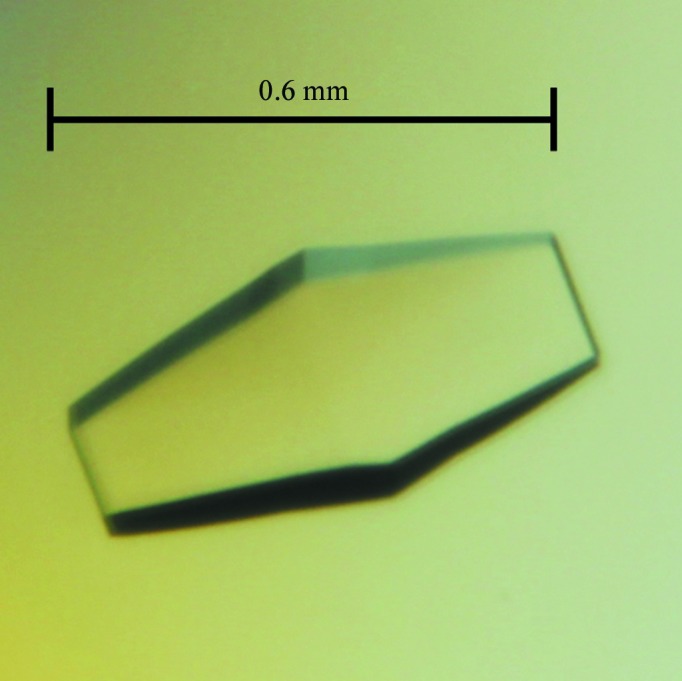
A crystal of FI-CMCase. The crystal reached approximate dimensions of 0.6 × 0.2 × 0.1 mm in 10–12 d.
Figure 2.
A diffraction pattern of the FI-CMCase crystal.
The crystal structure of FI-CMCase was solved by the molecular-replacement (MR) method with Phaser (McCoy et al., 2007 ▶) from the CCP4 suite (Winn et al., 2011 ▶) using the structure of the A. niger endoglucanase (65% sequence identity to FI-CMCase; PDB entry 1ks4; Khademi et al., 2002 ▶) as a search model. Initial structure refinement using REFMAC5 (Murshudov et al., 2011 ▶) resulted in a model with an R work of 33% and an R free of 35%. The initial electron-density map clearly showed that there were two monomers in the asymmetric unit. Further model building and structure refinement are in progress.
Acknowledgments
The synchrotron data collection was conducted on beamline BL13C1 of NSRRC (National Synchrotron Radiation Research Center, Taiwan) supported by the National Science Council (NSC). This work was supported by the National High Technology Research and Development Program of China (2012AA022200) and and Chinese Academy of Sciences (KSZD-EW-Z-015-2).
References
- Cheng, Y.-S., Ko, T.-P., Huang, J.-W., Wu, T.-H., Lin, C.-Y., Luo, W., Li, Q., Ma, Y., Huang, C.-H., Wang, A. H.-J., Liu, J.-R. & Guo, R.-T. (2012). Appl. Microbiol. Biotechnol. 95, 661–669. [DOI] [PubMed]
- Damasceno, L. M., Huang, C. J. & Batt, C. A. (2012). Appl. Microbiol. Biotechnol. 93, 31–39. [DOI] [PubMed]
- Doi, R. H. (2008). Ann. N. Y. Acad. Sci. 1125, 267–279.
- Hata, Y., Natori, K., Katsube, Y., Ooi, T., Arai, M. & Okada, H. (1994). J. Mol. Biol. 241, 278–280. [DOI] [PubMed]
- Huang, J.-W., Chen, C.-C., Huang, C.-H., Huang, T.-Y., Wu, T.-H., Cheng, Y.-S., Ko, T.-P., Lin, C.-Y., Liu, J.-R. & Guo, R.-T. (2014). Biochim. Biophys. Acta, 1844, 663–669. [DOI] [PubMed]
- Huang, J.-W., Cheng, Y.-S., Ko, T.-P., Lin, C.-Y., Lai, H.-L., Chen, C.-C., Ma, Y., Zheng, Y., Huang, C.-H., Zou, P., Liu, J.-R. & Guo, R.-T. (2012). Appl. Microbiol. Biotechnol. 94, 111–121. [DOI] [PubMed]
- Khademi, S., Zhang, D., Swanson, S. M., Wartenberg, A., Witte, K. & Meyer, E. F. (2002). Acta Cryst. D58, 660–667. [DOI] [PubMed]
- Kuhad, R. C., Gupta, R. & Singh, A. (2011). Enzym. Res. 2011, 280696. [DOI] [PMC free article] [PubMed]
- Li, P., Anumanthan, A., Gao, X.-G., Ilangovan, K., Suzara, V. V., Düzgüneş, N. & Renugopalakrishnan, V. (2007). Appl. Biochem. Biotechnol. 142, 105–124. [DOI] [PubMed]
- Lynd, L. R., Weimer, P. J., van Zyl, W. H. & Pretorius, I. S. (2002). Microbiol. Mol. Biol. Rev. 66, 506–577. [DOI] [PMC free article] [PubMed]
- Macauley-Patrick, S., Fazenda, M. L., McNeil, B. & Harvey, L. M. (2005). Yeast, 22, 249–270. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Minamiguchi, K., Ooi, T., Kawaguchi, T., Okada, H., Murao, S. & Arai, M. (1995). J. Ferment. Bioeng. 79, 363–366.
- Murao, S., Kanamoto, J., Sakamoto, R. & Arai, M. (1979). J. Ferment. Technol. 57, 157–162.
- Murao, S., Sakamoto, R. & Arai, M. (1988). Methods Enzymol. 160, 274–299.
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Ooi, T., Minamiguchi, K., Kawaguchi, T., Okada, H., Murao, S. & Arai, M. (1993). Biosci. Biotechnol. Biochem. 57, 1960–1961. [DOI] [PubMed]
- Ooi, T., Minamiguchi, K., Kawaguchi, T., Okada, H., Murao, S. & Arai, M. (1994). Biosci. Biotechnol. Biochem. 58, 954–956. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Sakuragi, H., Kuroda, K. & Ueda, M. (2011). J. Biomed. Biotechnol. 2011, 416931. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Wu, T.-H., Chen, C.-C., Cheng, Y.-S., Ko, T.-P., Lin, C.-Y., Lai, H.-L., Huang, T.-Y., Liu, J.-R. & Guo, R.-T. (2014). J. Biotechnol. 175, 1–6. [DOI] [PubMed]
- Yoshizawa, T., Shimizu, T., Hirano, H., Sato, M. & Hashimoto, H. (2011). Acta Cryst. F67, 830–832. [DOI] [PMC free article] [PubMed]