HutB from V. cholerae has been cloned, overexpressed, purified and crystallized in a monomeric form. Crystals belonging to space group P43212 produced diffraction data to 2.4 Å resolution.
Keywords: Vibrio cholerae, iron uptake, periplasmic haem-binding protein, ABC transporter
Abstract
The mechanism of haem transport across the inner membrane of pathogenic bacteria is currently insufficiently understood at the molecular level and no information is available for this process in Vibrio cholerae. To obtain structural insights into the periplasmic haem-binding protein HutB from V. cholerae (VcHutB), which is involved in haem transport through the HutBCD haem-transport system, at the atomic level, VcHutB was cloned, overexpressed and crystallized using 1.6 M ammonium sulfate as a precipitant at pH 7.0. X-ray diffraction data were collected to 2.4 Å resolution on the RRCAT PX-BL-21 beamline at the Indus-2 synchrotron, Indore, India. The crystals belonged to space group P43212, with unit-cell parameters a = b = 62.88, c = 135.8 Å. Matthews coefficient calculations indicated the presence of one monomer in the asymmetric unit, with an approximate solvent content of 45.02%. Molecular-replacement calculations with Phaser confirmed the presence of a monomer in the asymmetric unit.
1. Introduction
Vibrio cholerae, the causative agent of the diarrhoeal disease cholera (Faruque et al., 1998 ▶; Kaper et al., 1995 ▶), encodes a large number of nutrient-transport systems, reflecting the importance of these nutrients for its growth, survival and pathogenicity. In Gram-negative bacteria, the outer membrane forms a permeability barrier which restricts the passage of both nutrients and toxic environmental agents (Kadner, 1990 ▶; Moeck & Coulton, 1998 ▶). Therefore, most nutrients cross the outer membrane into the periplasmic space by diffusion through porins, while larger nutrients are transported into the periplasm via specific high-affinity active-transport systems. Transport of the essential nutrients through the periplasm and across the inner membrane is facilitated by a periplasmic binding protein-dependent ABC (ATP-binding cassette) transport system. In these systems, the periplasmic ligand-binding protein (PLBP) binds and delivers the ligand to the inner membrane permease (Oldham et al., 2008 ▶).
Successful pathogenic bacteria must acquire one such essential nutrient, iron, from the host. Because the concentration of free iron in tissue is as low as 10−12 M (Clarke et al., 2002 ▶), most pathogenic bacteria have developed elaborate mechanisms to obtain iron at concentrations sufficient for their growth. The main strategy for iron acquisition involves the synthesis and secretion of small iron-chelating molecules termed siderophores (Braun et al., 1998 ▶; Byers & Arceneaux, 1998 ▶; Earhart, 1996 ▶). Additionally, numerous Gram-negative bacterial pathogens have developed a sophisticated mechanism for recruiting host haem iron (Eakanunkul et al., 2005 ▶; Tong & Guo, 2007 ▶).
The first step of haem acquisition takes place through a TonB-dependent cell-surface receptor. Haem transport across the periplasmic space involves an ABC transporter system made up of a soluble PLBP, an inner membrane haem permease and an ATPase. Finally, upon crossing the inner membrane, the haem is broken down by haem oxygenase and iron is released into the cytosol. The mechanism of haem transport by haemin ABC transporters through the inner membrane is currently insufficiently understood at the molecular level. Ho et al. (2007 ▶) solved the crystal structures of two haem-binding PLBPs, ShuT from Shigella dysenteriae and PhuT from Pseudomonas aeruginosa, which share a common architecture with the haem binding in a narrow cleft between the N- and C-terminal binding domains (Ho et al., 2007 ▶). A comparison of PhuT/ShuT with the vitamin B12 binding protein BtuF and the hydroxamic-type siderophore binding protein FhuD demonstrates that PhuT/ShuT more closely resemble BtuF, which reflects the closer similarity in ligands (haem and vitamin B12) compared with the ligands of FhuD (Ho et al., 2007 ▶). The periplasmic binding protein HmuT from Yersinia pestis (YpHmuT), however, binds two stacked haems in a central binding cleft that is larger than those of the homologous periplasmic haem-binding proteins ShuT and PhuT (Mattle et al., 2010 ▶), pointing towards a variable mode of haem binding by PLBPs.
V. cholerae has multiple iron-acquisition systems, including the utilization of haem and haemoglobin, the synthesis and transport of the catechol-type siderophore vibriobactin and the transport of several exogenous siderophores such as enterobactin. Analysis of the hut genes indicated that hutBCD, which are predicted to encode a periplasmic binding protein (HutB), a cytoplasmic membrane permease (HutC) and an ATPase (HutD), are required to reconstitute the V. cholerae haem-transport system in Escherichia coli (Occhino et al., 1998 ▶). However, mechanistic details of the transport of nutrients in V. cholerae are restricted to the structure of the vibriobactin binding protein ViuP in apo and holo forms (Li et al., 2012 ▶), and the molecular mechanism of haem transport in V. cholerae is as yet unknown. Here, we report the cloning, overexpression, crystallization and preliminary X-ray crystallographic analysis of the periplasmic binding protein HutB (VcHutB) from V. cholerae O395 strain in the apo form.
2. Materials and methods
2.1. Cloning, overexpression and purification of VcHutB
The hutB gene was cloned into the kanamycin-resistant pET-28a(+) vector (Novagen) using specific primers (forward primer 5′-GGAATTCGCATATGCGGATCGTCAGTGCAGGAAGTGCAG-3′ and reverse primer 5′-CATTCGGGATCCTCAGGGGTACAGCAGAGTTTGAATAC-3′). The primers were synthesized (NeuProCell) with NdeI and BamHI restriction-enzyme sites. Chromosomal DNA of V. cholerae strain O395, isolated using the protocol described in the Molecular Biology Laboratory Manual of UMBC (http://userpages.umbc.edu/~jwolf/method1.htm), was used as the template to amplify the region encoding hutB. The 762 bp hutB PCR amplicon and the pET-28a(+) vector were double-digested with the restriction enzymes NdeI and BamHI and the digested products were purified from 1.0% agarose gel using a gel-extraction kit (Invitrogen). These two double-digested DNA fragments were then ligated using T4 DNA ligase. The clones were selected appropriately using E. coli XL1-Blue cells with kanamycin resistance. The construct was verified by restriction-digestion analysis and DNA sequencing. VcHutB (residues 24–277 excluding the signal peptide; 254 amino acids) was then overexpressed in E. coli BL21(DE3) cells in the presence of the antibiotic kanamycin as a fusion protein with a 6×His tag at the N-terminus followed by a thrombin cleavage site (Fig. 1 ▶ a).
Figure 1.
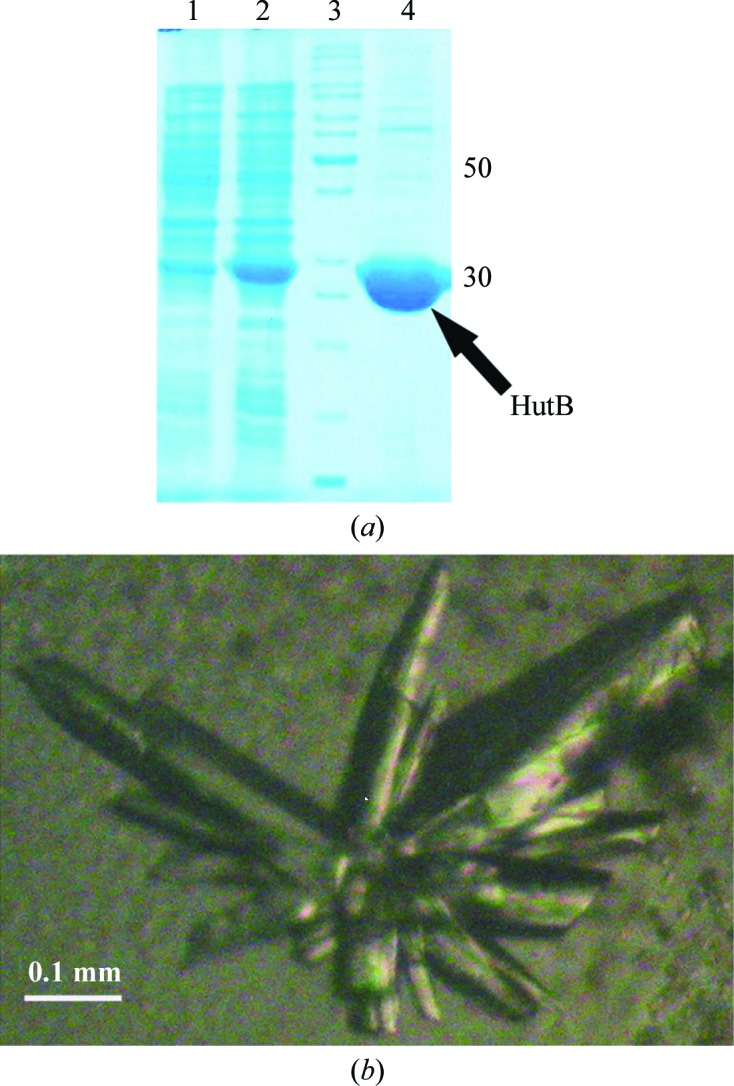
(a) 12% SDS–PAGE: lanes 1 and 2, uninduced and induced expression of 6×His-tagged VcHutB, respectively; lane 3, molecular-weight markers (labelled in kDa); lane 4, purified 6×His-tagged VcHutB. (b) Crystals of VcHutB grown in the presence of precipitant solution consisting of 1.6 M ammonium sulfate, 0.1 M HEPES pH 7.0.
For overexpression, 10 ml LB broth was inoculated with a single colony and was grown overnight at 310 K. 1 l LB broth was inoculated with 10 ml of the overnight culture and the culture was grown at 310 K until the OD600 reached 0.6. The cells were then induced with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). After induction at 310 K for 3 h, the cells were harvested at 4500g for 20 min and the pellet was resuspended in 7 ml ice-cold lysis buffer consisting of 50 mM Tris–HCl pH 7.0, 300 mM NaCl. Phenylmethylsulfonyl fluoride (final concentration of 1 mM) and 1 mg ml−1 lysozyme were added to the resuspended solution and the cells were lysed by sonication on ice. The cell lysate was then centrifuged (12 000g for 50 min) at 277 K and the supernatant was collected. Ni2+–NTA (Qiagen) resin was equilibrated with the lysis buffer and the supernatant containing 6×His-tagged VcHutB was immobilized on the resin for Ni2+–NTA-based immobilized metal-ion affinity chromatography. The resin was washed with wash buffers consisting of the components of the lysis buffer with increasing concentrations of imidazole (5 and 10 mM) and the 6×His-tagged VcHutB was eluted with elution buffer with an imidazole concentration of 150 mM. The purity of the eluted fractions was checked using 12% SDS–PAGE and the fractions were found to be nearly homogeneous. The eluted protein was dialyzed against Tris buffer consisting of 50 mM Tris–HCl pH 7.0, 300 mM NaCl and concentrated using an Amicon Ultra centrifugation unit (molecular-weight cutoff 10 kDa). The homogeneity of the purified protein was checked using 12% SDS–PAGE (Fig. 1 ▶ a).
2.2. Crystallization
For crystallization, VcHutB was concentrated to 25 mg ml−1 using an Amicon Ultra centrifugation unit (molecular-weight cutoff 10 kDa) in buffer consisting of 50 mM Tris–HCl pH 7.0, 300 mM NaCl. The concentration of the protein was measured spectrophotometrically using a calculated extinction coefficient of 4470 M −1 cm−1. Initial crystallization trials were performed by the hanging-drop vapour-diffusion method in 24-well crystallization trays (Hampton Research, Laguna Niguel, California, USA) using Grid Screen PEG 6000, Grid Screen Ammonium Sulfate, Crystal Screen (Jancarik & Kim, 1991 ▶) and Crystal Screen 2 from Hampton Research. Typically, 2 µl of protein solution was mixed with an equal volume of the screening solution and equilibrated against 600 µl of the latter. Initially, tiny crystals were observed in the presence of 20% PEG 6000 at pH 6.0 (a condition from Grid Screen PEG 6000) and 1.6 M ammonium sulfate pH 7.0 incubated at 293 K for two to three weeks. These crystallization conditions were further optimized. Finally, equilibration of a mixture of 3 µl protein solution with 2 µl precipitant solution consisting of 0.8 M ammonium sulfate, 0.1 M HEPES pH 7.0 against a reservoir solution consisting of 500 µl 1.6 M ammonium sulfate, 0.1 M HEPES pH 7.0 at 293 K for 21 d produced diffraction-quality crystals with a longest dimension of 0.4 mm (Fig. 1 ▶ b).
2.3. Data collection and processing
Crystals of VcHutB were looped out using 0.4 mm LithoLoops (Molecular Dimensions), soaked in cyroprotectant consisting of 25%(v/v) glycerol, 1 M ammonium sulfate, 0.1 M HEPES pH 7.0, 150 mM NaCl and immediately flash-cooled in liquid nitrogen. The quality of the crystals was judged and the cooling conditions were optimized using a MAR Research image-plate detector (345 mm) and Cu Kα radiation generated by a Bruker–Nonius FR591 rotating-anode generator equipped with Osmic MaxFlux confocal optics. Diffraction images were then collected from a cryocooled crystal (100 K) using a MAR225 CCD (Rayonix) detector on the RRCAT PX-BL-21 beamline at the Indus-2 synchrotron, Indore, India with an oscillation range of 1°. A total of 360 images were collected. The data were indexed and integrated using XDS (Kabsch, 2010 ▶) and subsequently scaled using AIMLESS from CCP4 (Winn et al., 2011 ▶). Data-collection and processing parameters are given in Table 1 ▶.
Table 1. Data-collection and data-processing parameters.
Values in parentheses are for the outermost resolution shell.
| Beamline | PX-BL-21, Indus-2 |
| Wavelength () | 0.98 |
| Oscillation () | 1 |
| No. of frames | 132 |
| Space group | P43212 |
| Unit-cell parameters () | a = b = 62.88, c = 135.8 |
| Resolution () | 37.02.4 (2.492.40) |
| No. of molecules in asymmetric unit | 1 |
| Mathews coefficient V M (3Da1) | 2.24 |
| Solvent content (%) | 45.02 |
| No. of observations | 108572 (10718) |
| No. of unique reflections | 11192 (1134) |
| Multiplicity | 9.7 (9.5) |
| Mosaicity () | 0.40 |
| Completeness (%) | 99.2 (97.5) |
| R merge † (%) | 6.1 (71.7) |
| Average I/(I) | 29.9 (3.3) |
R
merge = 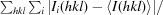
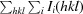 , where Ii(hkl) is the observed intensity of the ith measurement of reflection hkl and I(hkl is the mean intensity of reflection hkl calculated after scaling.
, where Ii(hkl) is the observed intensity of the ith measurement of reflection hkl and I(hkl is the mean intensity of reflection hkl calculated after scaling.
3. Results
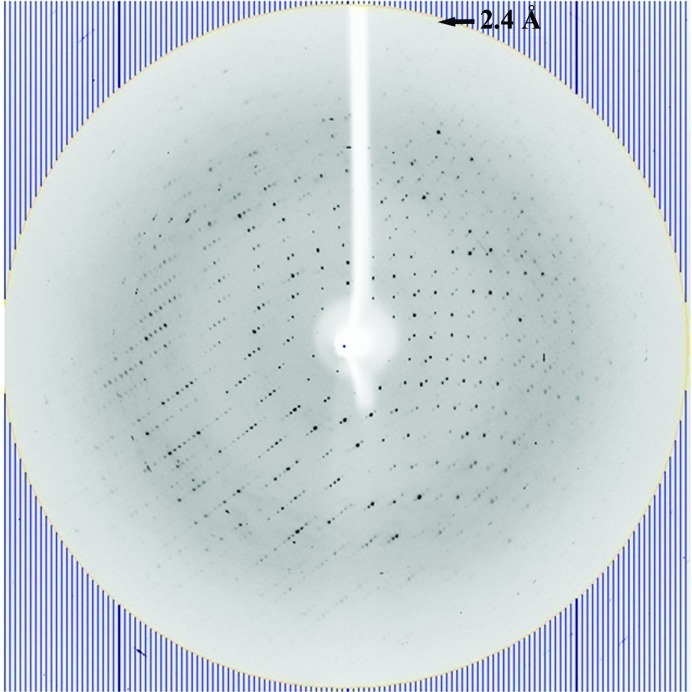
The crystals of VcHutB were of good quality and diffraction data were collected to 2.4 Å resolution. Although the R merge in the highest resolution bin was 71.7%, the 〈I/σ(I)〉 and the completeness in the highest resolution bin were 3.3 and 97.5%, respectively; therefore, the data were processed to 2.4 Å resolution (Table 1 ▶). A typical diffraction image is shown in Fig. 2 ▶.
Figure 2.
X-ray diffraction image of the VcHutB. The crystal diffracted to 2.4 Å resolution.
The VcHutB crystals belonged to the orthorhombic crystal system, with unit-cell parameters a = b = 62.88, c = 135.8 Å (Table 1 ▶). Calculation of the Matthews coefficient indicated the most probable presence of only one molecule (considering the molecular weight of 30 kDa) in the asymmetric unit, with a V M (Matthews, 1968 ▶) of 2.24 Å Da−1 and a solvent content of 45.02%.
The initial phases were obtained by molecular replacement (MR) using Phaser (McCoy et al., 2007 ▶). A BLAST search with the sequence of VcHutB against the PDB showed 32, 34 and 37% sequence identity to the haem-binding PLBPs ShuT from S. dysenteriae, PhuT from P. aeruginosa and HmuT from Y. pestis, respectively. Despite this, the coordinates of chain A of ShuT from S. dysenteriae in the apo form (PDB entry 2rg7; Ho et al., 2007 ▶) produced an acceptable solution in MR. Before proceeding with the MR calculations, waters were deleted from the coordinates of the search model and the mismatched residues were mutated to alanine. One molecule of this modified search model in the asymmetric unit produced a rotation-function Z-score (RFZ) of 4.2, a translation-function Z-score (TFZ) of 10.0 and a convincing log-likelihood gain (LLG) of 88.0 in space group P43212 using data between 37.0 and 2.4 Å resolution. Interestingly, a search model consisting of the coordinates of the apo PLBP HmuT from Y. pestis did not produce any convincing solution. Initial structure refinement was carried out using PHENIX (Adams et al., 2010 ▶), which resulted in a model with an R work of 40.0% and an R free of 46.9%. The 2F o − F c map calculated with this refined model was of good quality and overlaid properly with the model. Further model building and structure refinement are in progress. Detailed structural analysis of VcHutB will provide further insights into the mechanism of haem transport in pathogenic bacteria.
Acknowledgments
We are extremely grateful to the PX-BL-21 beamline scientists Dr R. Makde, Dr B. Ghosh and Mr A. Kumar of RRCAT, Indore, India for helping us to collect the diffraction data. We are also very grateful to Professor Udayaditya Sen of SINP, Kolkata for his help and suggestions. SA is grateful to Sanjay Dey of JD’s laboratory at St Xavier’s College, Kolkata for his help and suggestions during the experiments. We are grateful to Dr J. Felix Raj SJ, Principal, St Xavier’s College, Kolkata for his encouragement and constant support. This work was supported by DAE (BRNS) grant No. 2013/37B/26/BRNS from the Government of India.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Braun, V., Hantke, K. & Köster, W. (1998). Met. Ions Biol. Syst. 35, 67–145. [PubMed]
- Byers, B. R. & Arceneaux, J. E. (1998). Met. Ions Biol. Syst. 35, 37–66. [PubMed]
- Clarke, T. E., Braun, V., Winkelmann, G., Tari, L. W. & Vogel, H. J. (2002). J. Biol. Chem. 277, 13966–13972. [DOI] [PubMed]
- Eakanunkul, S., Lukat-Rodgers, G. S., Sumithran, S., Ghosh, A., Rodgers, K. R., Dawson, J. H. & Wilks, A. (2005). Biochemistry, 44, 13179–13191. [DOI] [PubMed]
- Earhart, C. F. (1996). Escherichia Coli and Salmonella: Cellular and Molecular Biology, 2nd ed., Vol. 1. edited by F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter & H. E. Umbarger, pp. 1075–1090. Washington DC: ASM Press.
- Faruque, S. M., Albert, M. J. & Mekalanos, J. J. (1998). Microbiol. Mol. Biol. Rev. 62, 1301–1314. [DOI] [PMC free article] [PubMed]
- Ho, W. W., Li, H., Eakanunkul, S., Tong, Y., Wilks, A., Guo, M. & Poulos, T. L. (2007). J. Biol. Chem. 282, 35796–35802. [DOI] [PubMed]
- Jancarik, J. & Kim, S.-H. (1991). J. Appl. Cryst. 24, 409–411.
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kadner, R. J. (1990). Mol. Microbiol. 4, 2027–2033. [DOI] [PubMed]
- Kaper, J. B., Morris, J. G. & Levine, M. M. (1995). Clin. Microbiol. Rev. 8, 48–86. [DOI] [PMC free article] [PubMed]
- Li, N., Zhang, C., Li, B., Liu, X., Huang, Y., Xu, S. & Gu, L. (2012). J. Biol. Chem. 287, 8912–8919. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Mattle, D., Zeltina, A., Woo, J. S., Goetz, B. A. & Locher, K. P. (2010). J. Mol. Biol. 404, 220–231. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Moeck, G. S. & Coulton, J. W. (1998). Mol. Microbiol. 28, 675–681. [DOI] [PubMed]
- Occhino, D. A., Wyckoff, E. E., Henderson, D. P., Wrona, T. J. & Payne, S. M. (1998). Mol. Microbiol. 29, 1493–1507. [DOI] [PubMed]
- Oldham, M. L., Davidson, A. L. & Chen, J. (2008). Curr. Opin. Struct. Biol. 18, 726–733. [DOI] [PMC free article] [PubMed]
- Tong, Y. & Guo, M. (2007). J. Biol. Inorg. Chem. 12, 735–750. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.