The therapeutic antibody omalizumab inhibits the binding of IgE to its receptors. Two crystal structures of the omalizumab Fab fragment have been determined at 1.9 and 3.0 Å resolution.
Keywords: IgE, Fab fragment, therapeutic antibody
Abstract
Omalizumab is a humanized anti-IgE antibody that inhibits the binding of IgE to its receptors on mast cells and basophils, thus blocking the IgE-mediated release of inflammatory mediators from these cells. Omalizumab binds to the Fc domains of IgE in proximity to the binding site of the high-affinity IgE receptor Fc∊RI, but the epitope and the mechanisms and conformations governing the recognition remain unknown. In order to elucidate the molecular mechanism of its anti-IgE activity, the aim was to analyse the interaction of omalizumab with human IgE. Therefore, IgE Fc C∊2–4 was recombinantly produced in mammalian HEK-293 cells. Functionality of the IgE Fc was proven by ELISA and mediator-release assays. Omalizumab IgG was cleaved with papain and the resulting Fab was purified by ion-exchange chromatography. The complex of IgE Fc with omalizumab was prepared by size-exclusion chromatography. However, crystals containing the complex were not obtained, suggesting that the process of crystallization favoured the dissociation of the two proteins. Instead, two structures of the omalizumab Fab with maximum resolutions of 1.9 and 3.0 Å were obtained. The structures reveal the arrangement of the CDRs and the position of omalizumab residues known from prior functional studies to be involved in IgE binding. Thus, the structure of omalizumab provides the structural basis for understanding the function of omalizumab, allows optimization of the procedure for complex crystallization and poses questions about the conformational requirements for anti-IgE activity.
1. Introduction
Several immune-mediated diseases, including allergic hypersensitivity reactions (Finkelman et al., 2010 ▶), can be linked to circulating IgE antibodies. Although they are the least abundant type of antibodies, IgE antibodies exhibit a variety of peculiarities regarding structure and effector functions, with major functional consequences. They act as a key molecule in a network of proteins including the high-affinity IgE receptor Fc∊RI, the low-affinity receptor CD23 and the IgE- and Fc∊RI-binding protein galectin-3 (Gould & Sutton, 2008 ▶). On cross-linking by allergens, IgE bound to Fc∊RI on mast cells and basophils triggers degranulation, the release of pro-inflammatory mediators and immediate reactions (Gould & Sutton, 2008 ▶).
IgE is an evolutionary conserved, heavily glycosylated, heterotetrameric structure containing two light chains and two ∊ heavy chains. The ∊ heavy-chain constant region has four constant domains as found in IgY, with the C∊2 domain being essential for intramolecular contacts.
The human Fc∊RI is expressed as a tetramer on mast cells and basophils and as a trimer lacking the signal-amplifying β-subunit on antigen-presenting cells (APCs) such as dendritic cells, Langerhans cells, monocytes, eosinophils, platelets and smooth muscle cells (Iikura et al., 2001 ▶; Kinet, 1999 ▶; Kraft & Kinet, 2007 ▶). The extracellular domains of the ligand-binding α chain display an affinity for IgE in the range of 1011 M −1, providing the basis for efficient loading of IgE with long-term stability on effector cells and resulting in a half-life of approximately 10 d (Chang, 2000 ▶).
In order to therapeutically reduce increased levels of IgE and excess mast-cell degranulation, different strategies including the development of antagonistic anti-IgE antibodies (Peng, 2009 ▶) have been pursued. Omalizumab (rhuMab E25; Xolair) is a humanized κ IgG1 antibody and the only IgE-neutralizing antibody approved for clinical use in the treatment of allergic asthma. With annual sales exceeding one billion dollars in 2013 and EU approval in 2014 for the treatment of chronic spontaneous urticaria, omalizumab is an important drug. It targets an epitope within the constant regions of the ∊ heavy chain (Zheng et al., 2008 ▶; Hunt et al., 2012 ▶) that is thought to be in the proximity of the binding site and therefore prevents the interaction of serum IgE with the Fc∊RI on effector cells (Presta et al., 1993 ▶, 1994 ▶; Shields et al., 1995 ▶). Besides reducing the serum IgE concentration, anti-IgE treatment also reversed phenotypic and functional effects of IgE-enhanced Fc∊RI levels on basophils (MacGlashan et al., 1997 ▶) or mast cells (Gomez et al., 2007 ▶). Consequently, histamine release and even the chronic inflammatory process during the late phase were reduced in skin-prick tests (Holgate et al., 2005 ▶; MacGlashan et al., 1997 ▶; van Neerven et al., 2006 ▶).
Nevertheless, not all patients with allergic asthma benefit from anti-IgE treatment. Failure may be caused by the formation of long-lasting pharmacologically active IgE–omalizumab complexes (Hamilton et al., 2005 ▶) that hamper the reliable determination of the serum IgE concentration and the correct dosing of anti-IgE (Braren et al., 2011 ▶). In vitro studies demonstrated that a reduction in free IgE to less than 10 ng ml−1 (4.16 IU ml−1) was required to prevent IgE-mediated cross-linking of Fc∊RI and subsequent effector-cell activation. This sensitivity, however, differs among individuals (MacGlashan et al., 1986 ▶). Case reports also demonstrated a dramatic improvement of chronic urticaria in patients with allergic asthma under omalizumab treatment. Efficacy and safety were subsequently verified in large phase III studies (Maurer et al., 2013 ▶).
Despite the clinical relevance of targeting IgE, the structural basis of the interaction of omalizumab with IgE still remains to be defined. The crystal structures of complexes between the IgE Fc and Fc∊RI (Holdom et al., 2011 ▶) and CD23 (Dhaliwal et al., 2012 ▶) revealed that the Fc moiety of IgE adopts a bent structure that undergoes conformational changes upon binding to Fc∊RI and CD23. We attempted to determine the structure of the complex between the Fab of omalizumab and the Fc moiety of IgE. Although the complex appeared to be very stable prior to crystallization, it dissociated during crystallization. Here, we report two crystal structures of the omalizumab Fab in distinct space groups and describe the architecture of the complementarity-determining regions (CDRs) of this therapeutic antibody. Common crystal-packing interactions involving the CDRs apparently favour the dissociation of the complex under rather different conditions of crystallization.
2. Materials and methods
2.1. Macromolecule production
2.1.1. Omalizumab Fab
The Fab fragment was obtained by incubating a therapeutic preparation of omalizumab (Novartis) and papain (Sigma–Aldrich) at 1:500(w:w) for 16 h at 37°C. Papain was inactivated by the addition of iodoacetamide to a final concentration of 30 mM. The digest was then dialysed against 50 mM sodium acetate pH 5 (buffer A) prior to loading onto a 1 ml Mono S column (GE Healthcare) equilibrated with buffer A. The column was eluted with a linear gradient from 0 to 280 mM NaCl in buffer A. The fractions containing the omalizumab Fab as identified by SDS–PAGE were pooled and dialyzed against 10 mM HEPES pH 7.2, 100 mM NaCl. The sequence of the omalizumab Fab is presented in the Supporting Information.
2.1.2. IgE Fc cloning and expression
The ∊ heavy-chain constant regions C∊2–4 were amplified from an IgE expression vector originally constructed from a human cDNA library using the primers TGATCATTTAAATGTGTCCAGTGTGCCAGGGACTTCAC and TCCCGGTAAACATCACCACCATTGAGTTTAAACGATC. The cDNA was introduced into an expression vector providing a human immunoglobulin signal sequence via SmiI and MssI restriction enzymes (Braren et al., 2007 ▶). The sequence of the IgE Fc construct is presented in the Supporting Information. Human embryonic kidney (HEK-293) cells were cultivated in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10%(v/v) heat-inactivated foetal calf serum (FCS), 100 IU ml−1 penicillin and 100 µg ml−1 streptomycin. For expression, HEK-293 cells were transfected with 3 µg plasmid DNA using Nanofectin (GE Healthcare) according to the recommendations of the manufacturer. Stably transfected cells were selected by the addition of Zeocin and showed an expression of 10–15 mg l−1. Expression was pursued by cultivating cells in roller flasks (Greiner), from which the supernatant was collected and subjected to purification. A C-terminal histidine tag enabled the purification of the IgE Fc via nickel-based affinity chromatography. The cellular supernatant was loaded onto a 1 ml HisTrap Excel column (GE Healthcare) equilibrated with PBS (500 mM NaCl, 40 mM Na2HPO4, 10 mM NaH2PO4 pH 8.0). After washing with 10 ml PBS and 20 ml 5% PBS–imidazole (100 mM NaCl, 40 mM Na2HPO4, 10 mM NaH2PO4, 300 mM imidazole pH 7.4), the protein was eluted using a 5–100% gradient of PBS–imidazole. The IgE Fc was further purified using a Superdex 200 10/300 GL size-exclusion column (GE Healthcare) previously equilibrated with running buffer consisting of 10 mM HEPES pH 7.2, 100 mM NaCl.
The binding of the IgE Fc to Fc receptor and antibodies was assessed by ELISA. For Fc∊RI and omalizumab, purified IgE Fc (50 µg ml−1) was coated on microtitre plates (Greiner) at 4°C and blocked with 40 mg ml−1 milk powder in PBS. Thereafter, solubilized Fc∊RI produced as an IgY Fc fusion protein (Braren et al., 2011 ▶), omalizumab and the omalizumab Fab at a concentration of 10 µg ml−1 were incubated for 2 h at room temperature. After washing, binding to the IgE Fc was detected using alkaline phosphatase-conjugated anti-IgY antibody, anti-human κ antibody and anti-human IgG antibody diluted 1:30 000 and 30 µl substrate solution.
The affinity of the omalizumab Fab was determined by surface plasmon resonance (SPR) measurements using a T200 Biacore system. The IgE Fc was covalently coupled to a total of 1040 resonance units (RU) on a CM5 sensor chip (GE Healthcare) using standard NHS–EDC coupling procedures. Measurements were performed at 25°C in buffer consisting of 10 mM monosodium phosphate, 40 mM disodium phosphate, 100 mM NaCl pH 7.4 supplemented with 0.01% Tween 20. For kinetic analyses, increasing concentrations of the omalizumab Fab were injected at a flow rate of 25 µl min−1. The association phase was monitored for 600 s and the dissociation phase was monitored for 600 s. Sensor surfaces were regenerated by subsequent injection of 1 M Tris buffer pH 10. The dissociation constant at equilibrium K d was calculated using the Biacore T200 evaluation software.
2.1.3. Cellular mediator-release assays
In vitro degranulation was analyzed as described previously (Hecker et al., 2011 ▶). After sensitization of RBL-SX38 cells with rIgE, Fc and washing with incomplete Tyrode’s buffer, cross-linking was achieved by addition of goat anti-human IgE and incubation for 60 min at 37°C. β-Hexosaminidase release of viable versus lysed cells was assessed using p-nitrophenyl N-acetylglucosaminide (Sigma–Aldrich) as a substrate for 60 min at 37°C. The reaction was quenched by 0.1 M carbonate buffer pH 10 and the absorbance was analyzed at 405 nm.
2.2. Crystallization
For complex formation, the omalizumab Fab and the IgE Fc were mixed in a 2.1:1 molar ratio. The complex was then purified by size exclusion on a 24 ml Superdex 200 10/300 GL column equilibrated in 10 mM HEPES pH 7.2, 100 mM NaCl. The purified and concentrated complex (6 mg ml−1) was used for crystallization screening with the commercial screens Index, PEGRx, PEGRx 2, SaltRx, Natrix, Natrix 2 (Hampton Research), Structure Screen I + II, MIDAS and JCSG-Plus (Molecular Dimensions). Using a Mosquito crystallization robot, 200 nl complex solution was mixed with 200 nl reservoir solution and the resulting drop was equilibrated against 60 µl reservoir solution. Crystals grew at room temperature under several conditions. The initial conditions were optimized with tailored screens prepared using the Mimer software and the associated robot (Brodersen et al., 2013 ▶).
2.3. Data collection and processing
Crystals in 0.1 M HEPES pH 7.5, 70% MPD as well as crystals in 0.1 M HEPES pH 7, 30%(w/v) polyvinyl pyrrolidone K15, 0.1 M Li2SO4 were briefly soaked in the respective reservoir buffer supplemented with 5%(v/v) glycerol and were subsequently cooled in liquid nitrogen. Diffraction data were collected on beamline P14 at PETRA III and were reduced with XDS (Kabsch, 2010 ▶). Data-collection statistics are summarized in Table 1 ▶.
Table 1. Data-collection statistics.
Values in parentheses are for the outer shell.
| PDB code | 4x7t | 4x7s |
|---|---|---|
| Wavelength () | 0.9755 | 0.9763 |
| Space group | P21 | P212121 |
| a, b, c () | 85.47, 73.49, 87.04 | 64.86, 73.41, 140.18 |
| , , () | 90, 116.46, 90 | 90, 90, 90 |
| Resolution range | 45.413.00 (3.1073.000) | 48.611.90 (1.9681.900) |
| Total No. of reflections | 65442 (6150) | 342486 (26150) |
| No. of unique reflections | 19417 (1912) | 53406 (5169) |
| Completeness (%) | 99.37 (99.32) | 99.77 (98.52) |
| Multiplicity | 3.4 (3.2) | 6.4 (5.1) |
| I/(I) | 5.80 (1.11) | 12.73 (0.71) |
| R r.i.m. | 0.202 | 0.074 |
| Overall B factor from Wilson plot (2) | 57.16 | 41.49 |
2.4. Structure solution and refinement
The unpublished structure of the omalizumab Fab (PDB entry 2xa8; C. H. Huang, F. H. A. Hung, C. Lim, T. W. Chang & C. Ma, unpublished work) was used as a search model in molecular replacement using Phaser (McCoy et al., 2007 ▶). Manual model building was carried out in Coot (Emsley & Cowtan, 2004 ▶). Refinement of positions and individual B factors was performed using phenix.refine (Afonine et al., 2012 ▶). For crystal form 1 containing two Fab molecules in the asymmetric unit related by noncrystallographic symmetry (NCS), we implemented torsion-angle restraints between the two NCS molecules. MolProbity (Chen et al., 2010 ▶) was used for structure validation following each refinement cycle. Figures were generated using PyMOL (v.1.3r1; Schrödinger) and the elbow angle was calculated with phenix.fab_elbow_angle. Refinement statistics are summarized in Table 2 ▶.
Table 2. Structure-refinement statistics.
Values in parentheses are for the outer shell.
| PDB code | 4x7t | 4x7s |
|---|---|---|
| Final R cryst | 0.193 (0.338) | 0.200 (0.419) |
| Final R free | 0.236 (0.380) | 0.233 (0.406) |
| Cruickshank DPI () | 0.436 | 0.113 |
| No. of non-H atoms | ||
| Protein | 6698 | 3349 |
| Ions | 10 | 0 |
| Water | 0 | 182 |
| Total | 6708 | 3531 |
| R.m.s. deviations | ||
| Bonds () | 0.006 | 0.004 |
| Angles () | 1.11 | 0.89 |
| Average B factor (2) | ||
| Protein | 64.70 | 53.90 |
| Ions | 61.60 | |
| Ramachandran plot (%) | ||
| Favoured regions | 96.8 | 96.8 |
| Additionally allowed | 3.0 | 3.2 |
| Outliers | 0.2 | 0 |
3. Results and discussion
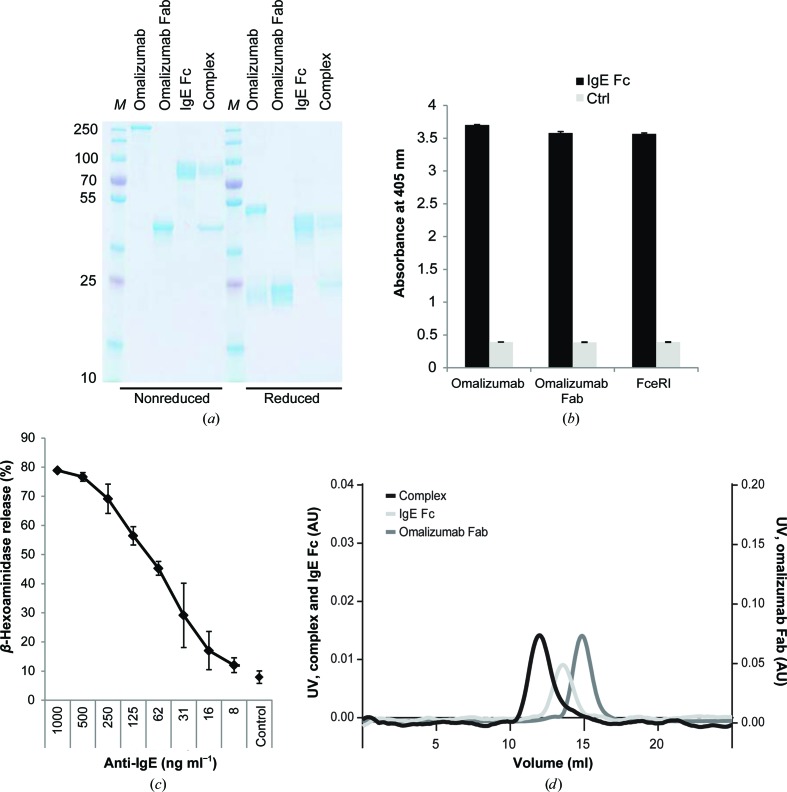
The omalizumab Fab was obtained by cleavage of the parental 160 kDa IgG using papain and showed the expected molecular mass of approximately 45 kDa on SDS–PAGE after purification (Fig. 1 ▶ a). Recombinant expression of IgE Fc C∊2–4 was performed by stable transfection of HEK-293 cells. SDS–PAGE analyses of the IgE Fc purified from the supernatant by Ni2+-affinity chromatography indicated proper assembly of the homodimer (Fig. 1 ▶ a). All molecules also showed the expected bands under reducing conditions. Although the glycosylation sites within the IgE Fc have been subjected to mutagenesis in recent studies to generate more homogenous protein, we retained all sites in our construct. The broad band observed on SDS–PAGE suggested that IgE was successfully glycosylated. The functionality of the IgE Fc was demonstrated by ELISA using the omalizumab IgG and Fab as well as solubilized Fc∊RI α chain extracellular domains fused to IgY Fc (Fig. 1 ▶ b). SPR studies of the omalizumab Fab and the IgE Fc used for crystallization revealed a K d of 1 × 10−8 M −1 (data not shown). This is fully in line with published data on the omalizumab Fab affinity (K d of 1.5 × 10−8 M −1) for interaction with full-length IgE (Putnam et al., 2008 ▶). Biological activity and binding to cellular Fc∊RI was assessed by applying the IgE Fc to humanized RBL-2H3 cells transfected with Fc∊RI. IgE Fc-mediated receptor cross-linking using anti-IgE antibodies resulted in pronounced activation and release of β-hexosaminidase in a concentration range similar to that of IgE from mammalian cells (Fig. 1 ▶ c). Overall, these data suggest that the IgE Fc and the omalizumab Fab are properly folded and fully functional. Analytical size-exclusion chromatography and SDS–PAGE of the mixed protein clearly indicated the formation of a stable complex in solution (Figs. 1 ▶ a and 1 ▶ d).
Figure 1.
Protein characterization. (a) SDS–PAGE analysis of omalizumab, the omalizumab Fab, the recombinant IgE Fc and the Fab–Fc complex isolated by size-exclusion chromatography under nonreducing (left) and reducing (right) conditions. Lane M contains molecular-weight marker (labelled in kDa). (b) Immunoreactivity and functionality of the recombinant IgE Fc was assessed by ELISA using omalizumab, the Fab generated from omalizumab and a solubilized Fc∊RI fusion protein as the primary antibody and anti-human IgG, anti-human κ chain and anti-chicken IgY conjugated to alkaline phosphatase, respectively, for detection. (c) Biological activity of the recombinant IgE Fc in mediator-release assays. RBL-SX38 cells providing the human Fc∊RI were sensitized with IgE Fc. Degranulation was induced by the addition of anti-IgE and was monitored by the β-hexosaminidase activity released into the culture supernatants. Data are the mean ± standard deviation of triplicate measurements. (d) Analytical size-exclusion chromatography of the isolated omalizumab Fab IgE Fc proteins and their complex. For the latter, Fab and IgE Fc were mixed in a 2.1:1 molar ratio.
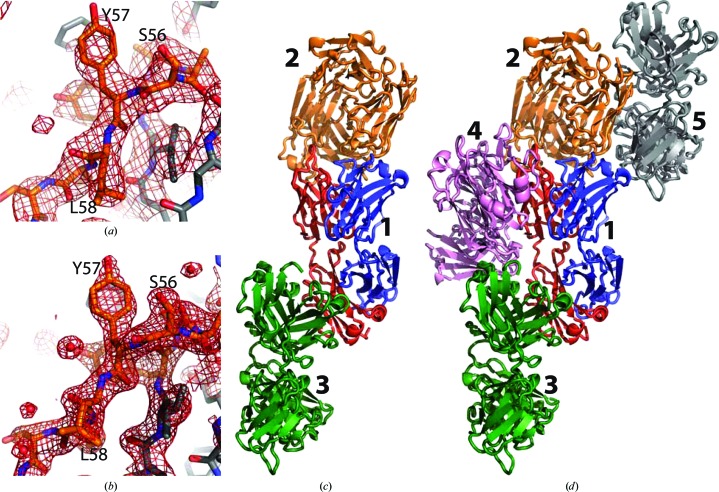
In our crystallization experiments with the omalizumab–IgE Fc complex we obtained crystals under a variety of conditions. All of these turned out to belong to two different crystal forms. Crystal form 1 possesses P21 symmetry, with diffraction data extending to 3 Å resolution, while the second crystal form displayed P212121 symmetry and diffracted to a maximum resolution of 1.9 Å (Table 1 ▶ and Figs. 2 ▶ a and 2 ▶ b). The unit-cell parameters of form 2 turned out to be virtually identical to those of PDB entry 2xa8 (a = 64.6, b = 73.9, c = 141.1 Å in space group P212121), which is based on data extending to 2.4 Å resolution. Indeed, the structure we determined of the omalizumab Fab based on form 2 is almost identical to that in PDB entry 2xa8, although more water molecules were identified in our structure. The crystal packing in crystal form 2 and in PDB entry 2xa8 is also virtually the same. In form 1 we found two NCS-related Fab molecules, but again no IgE Fc was present. In both structures we could trace 219 residues in the heavy chain and 218 residues in the light chain. The overall conformations of the three molecules in both crystal forms are fairly similar, with elbow angles between the variable and constant domains in the range 128–136°. Likewise, the antigen-recognizing VH–VL domain tandem superimpose with a root-mean-square deviation (r.m.s.d.) of 0.28–0.50 Å for the three molecules. In crystal form 2 each Fab interacts with two others (Fig. 2 ▶ c). This crystal-packing trimer is almost completely conserved in crystal form 1 for one of the molecules, but additional contacts are formed to the NCS-related Fab and a fourth Fab (Fig. 2 ▶ d).
Figure 2.
Structure determination. (a) Example of an OMIT 2mF o − DF c electron-density map from crystal form 1 after simulated annealing in which the shown residues were omitted. The map is contoured at 1σ. (b) As in (a) but from crystal form 2. (c) Crystal-packing trimer extracted from crystal form 2 with P212121 symmetry. In the central molecule marked ‘1’ the heavy and light chains are coloured blue and red, respectively. (d) Crystal packing in crystal form 1 with P21 symmetry; the trimer from crystal form 2 is shown in the same colours and numbered as in (c). Notice how molecule 1 now forms additional contacts with the two Fab molecules 4 and 5.
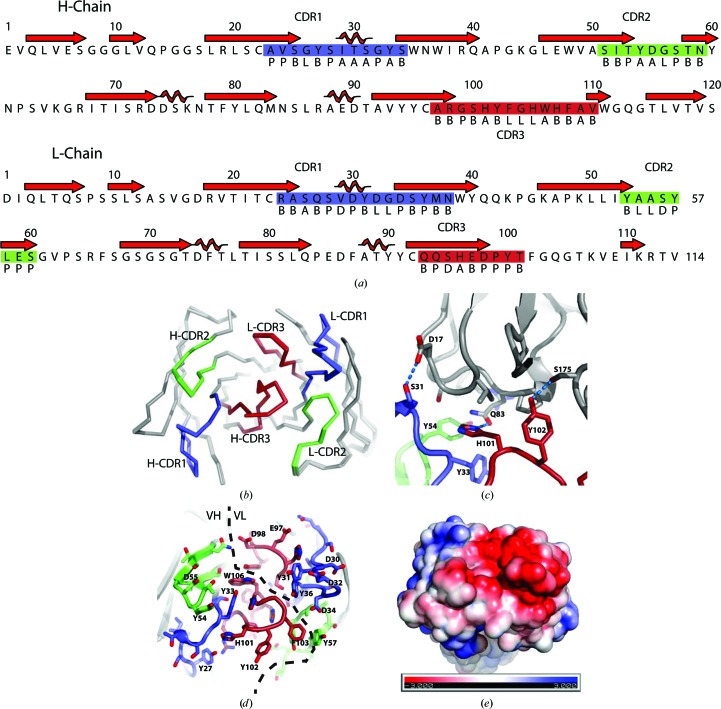
To analyze the conformations of the CDRs, we used the structure-based definition recently proposed by North et al. (2011 ▶). In the heavy chain, the CDR1 region is comprised of Ala23–Ser34 and has an H1-12-1 like conformation (Fig. 3 ▶ a). CDR2 is situated within Ser51–Asn59 and adopts the H2-9-1 conformation, while CDR3 is formed by Ala97–Val110 with the anchor region not clearly belonging to any of the classes defined by North et al. (2011 ▶). In the light chain residues Arg24–Asn38 form the CDR1 region and almost perfectly adopt the L1-15-1 conformation (Fig. 3 ▶ a). CDR2 contains residues Tyr53–Ser60 and belongs to the L2-8-1 cluster, while CDR3 Gln93–Thr101 adopts a conformation similar to the L3-9-2 cluster. Between the three Fab molecules the six CDRs superimpose with an r.m.s.d. of 0.17–0.24 Å over 67 Cα positions (Fig. 3 ▶ b). No significant differences in their CDR conformations are observed, suggesting that the CDR conformations are rather stable, although the CDRs are heavily involved in crystal-packing interactions in the two crystal forms (Figs. 2 ▶ c, 2 ▶ d and 3 ▶ c). In crystal form 2, heavy-chain CDR3 residues His101-Tyr102 and CDR1 residue Ser31 form hydrogen bonds to the light chain of a neighbouring Fab (Fig. 3 ▶ c). These interactions are partially conserved in both Fab molecules in crystal form 1. However, here the CDR1 in both molecules and in both the heavy and the light chain are also involved in crystal packing.
Figure 3.
The CDR regions of omalizumab. (a) The sequence of the two chains with the secondary structure indicated above. The CDR regions (boxed) and the Ramachandran region (below) for each residue as defined by North et al. (2011 ▶) are shown. This figure was prepared with ALINE (Bond & Schüttelkopf, 2009 ▶). (b) The three Fab molecules extracted from the two structures and superimposed through their CDR regions, which are coloured as in (a). (c) The crystal contact involving the heavy-chain CDRs and a neighbouring molecule in crystal form 2. Hydrogen bonds are indicated by dashed lines. (d) The exposed side chains of the CDR regions with a preponderance of acidic and aromatic residues. (e) Electrostatic potential mapped onto the surface calculated with the APBS PyMOL plugin (Lerner & Carlson, 2006 ▶); the Fab is shown in the same orientation as in (d).
Although the affinity between omalizumab and the IgE Fc is very high (Hayashi et al., 2007 ▶; Meno-Tetang & Lowe, 2005 ▶) and we could isolate the complex (Figs. 1 ▶ a and 1 ▶ d), we did not succeed in crystallizing the complex, suggesting that it dissociates in our crystallization drops. Based on our observation of recurring crystal-packing interactions involving the CDR regions in both crystal forms, it is likely that Fab–Fc dissociation is promoted by favourable Fab–Fab interactions competing with Fc recognition. This might be prevented by the mutation of non-CDR residues involved in Fab–Fab crystal contacts. Another plausible explanation is the inherent conformational flexibility of the Fc C∊2–C∊4 fragment that we used. This fragment has previously been crystallized alone and in complex with Fc∊RI, revealing a nonsymmetric bent structure in which the intermolecular twofold axis is degenerated (Holdom et al., 2011 ▶). In both structures C∊2 and C∊4 dimerization is observed; the C∊3 domain acts as a hinge domain and asymmetric binding of Fc∊RI to both C∊3 domains induces further bending of the Fc fragment. Recently, a structure of the Fc C∊2–C∊4 fragment in complex with an anti-∊Fab bound revealed an extended conformation with twofold symmetry (Drinkwater et al., 2014 ▶), in which each anti-∊Fab interacts primarily with one C∊3 domain but also with the connected C∊2 domain, the C∊2–C∊3 linker and the C∊3 domain of the other Fc chain. Fluorescence spectroscopy indicated that both the bent nonsymmetric and the extended symmetric conformation exist in solution (Drinkwater et al., 2014 ▶), and omalizumab binding induces unbending of the Fc (Hunt et al., 2012 ▶), but whether this leads to a fully extended and symmetric conformation of the Fc is unknown. Hence, our crystallization conditions might have promoted the bent Fc conformation, thereby disfavouring complex formation with the omalizumab Fab.
Our crystal structure reveals a striking feature of omalizumab: the CDRs present a number of aromatic and acidic side chains providing the surface with a strong overall negative net charge, especially on the light-chain CDRs (Fig. 3 ▶ e). The importance of the negative charge is underscored by data indicating that mutation of Asp34 in L-CDR1 strongly decreased the binding to IgE, whereas simultaneous mutation of Glu97/Asp98 in L-CDR3 had a significant but less detrimental effect (Presta et al., 1993 ▶). Interestingly, mutation of the surface-exposed heavy-chain His101, which we observe to be engaged in a crystal contact, also decreased IgE binding (Presta et al., 1993 ▶). The protruding tyrosines are also excellent candidates for good interacting residues as they are simultaneously capable of stacking with particular aromatic groups and forming hydrogen bonds to polar groups in the IgE Fc. These features correlate quite well with the omalizumab epitope comprising IgE Fc residues 424-HLP-426 (Zheng et al., 2008 ▶), as these residues in the C∊3 domain are in the near-vicinity of the Fc∊RI binding site and with several positive arginine and lysine side chains in the IgE–Fc∊RI complex. The epitope is also close to the linker connecting the C∊2 and C∊3 domains, in agreement with the ability of omalizumab to unbend the C∊2–C∊4 fragment (Hunt et al., 2012 ▶).
In conclusion, the present study provides the structural basis for understanding the function of omalizumab and rationalizes several known properties of the antibody. Importantly, it also underscores the difficulties in crystallizing a complex in which one or more of subunits in the complex has a high propensity for crystallization. In this case, mutation of the omalizumab Fab to remove crystal contacts involving CDR regions might increase the chance of crystallizing the complex with the IgE Fc in order to finally gain a detailed structure-based understanding of how the antibody prevents IgE binding to Fc∊RI.
Supplementary Material
PDB reference: omalizumab Fab, 4x7s
PDB reference: 4x7t
Sequences of the omalizumab Fab and the recombinant Ig Fc.. DOI: 10.1107/S2053230X15004100/rl5093sup1.pdf
Acknowledgments
We would like to thank the staff of beamline P14 at PETRA III for support during data collection. This project was supported by the Danish Research Council for Technology and Production, the Novo–Nordisk Foundation through a Hallas–Møller Fellowship to GRA.
References
- Afonine, P. V., Grosse-Kunstleve, R. W., Echols, N., Headd, J. J., Moriarty, N. W., Mustyakimov, M., Terwilliger, T. C., Urzhumtsev, A., Zwart, P. H. & Adams, P. D. (2012). Acta Cryst. D68, 352–367. [DOI] [PMC free article] [PubMed]
- Bond, C. S. & Schüttelkopf, A. W. (2009). Acta Cryst. D65, 510–512. [DOI] [PubMed]
- Braren, I., Blank, S., Seismann, H., Deckers, S., Ollert, M., Grunwald, T. & Spillner, E. (2007). Clin. Chem. 53, 837–844. [DOI] [PubMed]
- Braren, I., Greunke, K., Pilette, C., Mempel, M., Grunwald, T., Bredehorst, R., Ring, J., Spillner, E. & Ollert, M. (2011). Anal. Biochem. 412, 134–140. [DOI] [PubMed]
- Brodersen, D. E., Andersen, G. R. & Andersen, C. B. F. (2013). Acta Cryst. F69, 815–820. [DOI] [PMC free article] [PubMed]
- Chang, T. W. (2000). Nature Biotechnol. 18, 157–162. [DOI] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Dhaliwal, B., Yuan, D., Pang, M. O. Y., Henry, A. J., Cain, K., Oxbrow, A., Fabiane, S. M., Beavil, A. J., McDonnell, J. M., Gould, H. J. & Sutton, B. J. (2012). Proc. Natl Acad. Sci. USA, 109, 12686–12691. [DOI] [PMC free article] [PubMed]
- Drinkwater, N., Cossins, B. P., Keeble, A. H., Wright, M., Cain, K., Hailu, H., Oxbrow, A., Delgado, J., Shuttleworth, L. K., Kao, M. W. P., McDonnell, J. M., Beavil, A. J., Henry, A. J. & Sutton, B. J. (2014). Nature Struct. Mol. Biol. 21, 397–404. [DOI] [PMC free article] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Finkelman, F. D., Boyce, J. A., Vercelli, D. & Rothenberg, M. E. (2010). J. Allergy Clin. Immunol. 125, 312–318. [DOI] [PubMed]
- Gomez, G., Jogie-Brahim, S., Shima, M. & Schwartz, L. B. (2007). J. Immunol. 179, 1353–1361. [DOI] [PMC free article] [PubMed]
- Gould, H. J. & Sutton, B. J. (2008). Nature Rev. Immunol. 8, 205–217. [DOI] [PubMed]
- Hamilton, R. G., Marcotte, G. V. & Saini, S. S. (2005). J. Immunol. Methods, 303, 81–91. [DOI] [PubMed]
- Hayashi, N., Tsukamoto, Y., Sallas, W. M. & Lowe, P. J. (2007). Br. J. Clin. Pharmacol. 63, 548–561. [DOI] [PMC free article] [PubMed]
- Hecker, J., Diethers, A., Etzold, S., Seismann, H., Michel, Y., Plum, M., Bredehorst, R., Blank, S., Braren, I. & Spillner, E. (2011). Mol. Immunol. 48, 1236–1244. [DOI] [PubMed]
- Holdom, M. D., Davies, A. M., Nettleship, J. E., Bagby, S. C., Dhaliwal, B., Girardi, E., Hunt, J., Gould, H. J., Beavil, A. J., McDonnell, J. M., Owens, R. J. & Sutton, B. J. (2011). Nature Struct. Mol. Biol. 18, 571–576. [DOI] [PMC free article] [PubMed]
- Holgate, S. T., Djukanović, R., Casale, T. & Bousquet, J. (2005). Clin. Exp. Allergy, 35, 408–416. [DOI] [PubMed]
- Hunt, J., Keeble, A. H., Dale, R. E., Corbett, M. K., Beavil, R. L., Levitt, J., Swann, M. J., Suhling, K., Ameer-Beg, S., Sutton, B. J. & Beavil, A. J. (2012). J. Biol. Chem. 287, 17459–17470. [DOI] [PMC free article] [PubMed]
- Iikura, M., Yamaguchi, M., Hirai, K., Miyamasu, M., Yamada, H., Nakajima, T., Fujisawa, T., Ra, C., Morita, Y. & Yamamoto, K. (2001). Int. Arch. Allergy Immunol. 124, 470–477. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kinet, J.-P. (1999). Annu. Rev. Immunol. 17, 931–972. [DOI] [PubMed]
- Kraft, S. & Kinet, J.-P. (2007). Nature Rev. Immunol. 7, 365–378. [DOI] [PubMed]
- Lerner, M. G. & Carlson, H. A. (2006). APBS Plugin for PyMOL. Ann Arbor: University of Michigan.
- MacGlashan, D. W. Jr, Bochner, B. S., Adelman, D. C., Jardieu, P. M., Togias, A., McKenzie-White, J., Sterbinsky, S. A., Hamilton, R. G. & Lichtenstein, L. M. (1997). J. Immunol. 158, 1438–1445. [PubMed]
- MacGlashan, D. W. Jr, Peters, S. P., Warner, J. & Lichtenstein, L. M. (1986). J. Immunol. 136, 2231–2239. [PubMed]
- Maurer, M., Rosén, K., Hsieh, H.-J., Saini, S., Grattan, C., Gimenéz-Arnau, A., Agarwal, S., Doyle, R., Canvin, J., Kaplan, A. & Casale, T. (2013). N. Engl. J. Med. 368, 924–935. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Meno-Tetang, G. M. & Lowe, P. J. (2005). Basic Clin. Pharmacol. Toxicol. 96, 182–192. [DOI] [PubMed]
- Neerven, R. J. J. van, Knol, E. F., Ejrnaes, A. & Würtzen, P. A. (2006). Int. Arch. Allergy Immunol. 141, 119–129. [DOI] [PubMed]
- North, B., Lehmann, A. & Dunbrack, R. L. Jr (2011). J. Mol. Biol. 406, 228–256. [DOI] [PMC free article] [PubMed]
- Peng, Z. (2009). Hum. Vaccin. 5, 302–309. [DOI] [PubMed]
- Presta, L. G., Lahr, S. J., Shields, R. L., Porter, J. P., Gorman, C. M., Fendly, B. M. & Jardieu, P. M. (1993). J. Immunol. 151, 2623–2632. [PubMed]
- Presta, L., Shields, R., O’Connell, L., Lahr, S., Porter, J., Gorman, C. & Jardieu, P. (1994). J. Biol. Chem. 269, 26368–26373. [PubMed]
- Putnam, W. S., Li, J., Haggstrom, J., Ng, C., Kadkhodayan-Fischer, S., Cheu, M., Deniz, Y., Lowman, H., Fielder, P., Visich, J., Joshi, A. & Jumbe, N. (2008). AAPS J. 10, 425–430. [DOI] [PMC free article] [PubMed]
- Shields, R. L., Whether, W. R., Zioncheck, K., O’Connell, L., Fendly, B., Presta, L. G., Thomas, D., Saban, R. & Jardieu, P. (1995). Int. Arch. Allergy Immunol. 107, 308–312. [DOI] [PubMed]
- Zheng, L., Li, B., Qian, W., Zhao, L., Cao, Z., Shi, S., Gao, J., Zhang, D., Hou, S., Dai, J., Wang, H. & Guo, Y. (2008). Biochem. Biophys. Res. Commun. 375, 619–622. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: omalizumab Fab, 4x7s
PDB reference: 4x7t
Sequences of the omalizumab Fab and the recombinant Ig Fc.. DOI: 10.1107/S2053230X15004100/rl5093sup1.pdf