To the Editor
Recent analyses of several databases of allergens provide evidence for structural similarities of allergenic proteins.1 The 707 allergens with known sequences belong to only 184 (~2%) of the 9318 protein families (Pfams).2 Furthermore, of the rare Pfams that contain an allergen, 81 contain multiple allergens and 10 Pfams with the most allergens contain 300 (42%) allergens. The congruence of pollen allergens with structural families is even more apparent. Of the 157 pollen allergens with known sequences, 93 (59%) reside in just 5 Pfams,3 suggesting that pollen allergens share a very limited number of relatively unique structures. Identifying the common structural features of allergens may help to elucidate their unique structural elements and potentially the mechanism(s) for their allergenicity.
The goal of our research was to use the highly allergenic mountain cedar (Juniperus ashei, Cupressaceae) pollen as a model for characterizing the allergenicity of individual proteins and to identify the structural elements that are required for allergic sensitization or reactions.4 In the current study, we first quantified patient IgE antibodies to crude extract of cedar pollen and to purified Jun a 1,5 a major mountain cedar allergen, using ImmunoCap technology (Phadia, Uppsala, Sweden). The vast majority (median, 93%) of IgE antibodies to cedar pollen in the serum from 35 allergic subjects (34 subjects; see Table E2 in this article’s Online Repository at www.jacionline.org) reacted with Jun a 1 (see Fig E1 in this article’s Online Repository at www.jacionline.org).
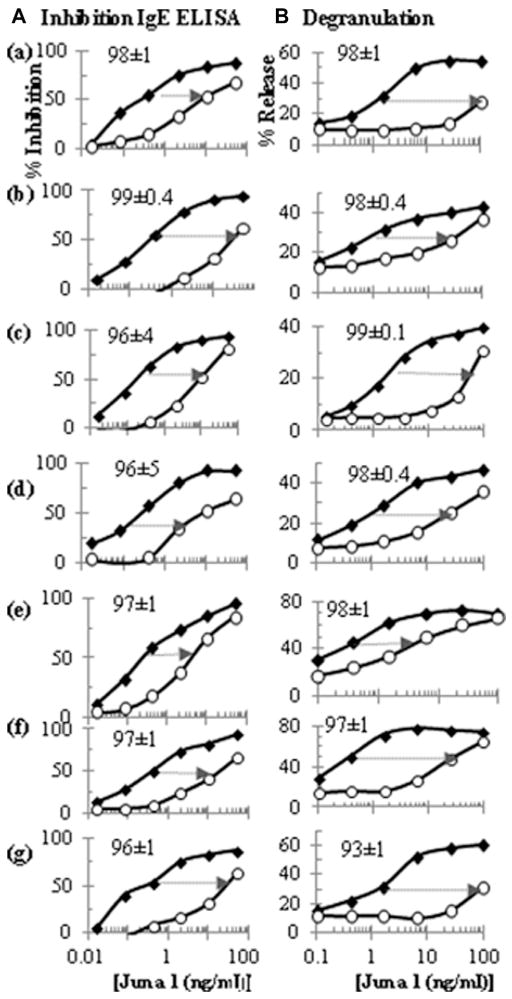
To define the fine specificity and complexity of these antibodies, we chose 7 sera with high concentrations of IgE anti–Jun a 1 antibodies and adequate serum volume to assess the relative amount of IgE binding to linear versus discontinuous (conformational) epitopes. We first used ELISA inhibition assays, in which binding of IgE to wells coated with native Jun a 1 was inhibited by prior incubation of the sera with either native or denatured Jun a 1. Our previous study showed that exposing Jun a 1 to 6 mol/L guanidine-HCl caused irreversible denaturation of Jun a 1.6 More than 96% of the IgE reactivity to native Jun a 1 was lost after denaturation of the allergen (Fig 1, A). To assess the biological activity of IgE antibodies to conformational and linear epitopes on Jun a 1, we next tested the ability of the same sera to sensitize and induce degranulation of cultured mast cells that express human FcεR1 (RBL SX-38)7 after stimulating with native or denatured Jun a 1. Fig 1, B, shows a dominance of IgE antibodies to conformational epitopes in mast cell degranulation, similar to that seen in ELISAs, indicating that IgE antibodies to conformational epitopes of Jun a 1 should be particularly effective in mediating clinical allergic reactions to cedar pollen.
FIG 1.
Immunoglobulin binding to conformational and linear epitopes of Jun a 1. The % of binding due to discontinuous epitopes is indicated for each serum. These were computed from the relative amount of native (◆) and guanidine-denatured Jun a 1 (○) required to inhibit by 50% the binding of patient IgE to native Jun a 1 on ELISA plates (A) and to induce mast cell degranulation of RBL SX-38 cells sensitized with patient sera (B). RBL, Rat basophilic leukemia.
To identify and enumerate the conformational epitopes of Jun a 1 that are recognized by the patients’ IgE, we produced a panel of mAbs and selected those that (1) reacted with native but not denatured Jun a 1 and (2) were inhibited from binding to Jun a 1 by human sera, indicating that these mouse IgG antibodies recognize conformational structures that are also recognized by IgE antibodies in the sera from our highly sensitized patients (see Fig E2 in this article’s Online Repository at www.jacionline.org). Within this panel of antibodies, we identified 4 distinct (non-competing) groups (G1–G4) on the basis of their cross-inhibition in ELISAs (see Table E1 in this article’s Online Repository at www.jacionline.org). Additional characteristics of these antibodies and the types of epitopes they recognize are shown in Fig E2. Of particular interest was the finding that the epitope for G1 was dependent on a structure maintained by a disulfide bond (Fig E2, B). We have previously reported that guanidine treatment of Jun a 1 results in irreversible loss of α-helical structures.6 Together these observations suggest that epitope G1 and potentially other (G2–G4) conformational epitopes are located in one or both of the α-helical regions, near the N- and C-terminal regions, and contain disulfide bonds (Cys7-Cys21 and Cys285-Cys291).8 However, further studies will be required to confirm the importance of these regions and map each of the mAbs to specific conformational elements.
To test whether our mAbs G1 to G4 bind to the linear epitopes we previously described, we performed inhibition assays with mAb KW-S91,6 which we have previously mapped to a region of Jun a 1 that contains linear epitopes we termed 2 and 3.9 This antibody caused partial inhibition of E2 and E3 binding (17% and 27% vs 83% for autologous binding of KW-S91). This suggests that the epitope for these mAbs may reside in the extensive parallel helical region of Jun a 1, near the linear epitopes 2 and 3.
We next used the mAbs G1 to G4 in competition experiments to determine the frequency with which the IgE antibodies from the 7 subjects recognized G1 to G4 epitopes (Fig 2). Interestingly, the IgE antibody responses of 5 of the 7 sera were strongest against epitopes G2 or G3, suggesting that these may represent dominant epitopes. This pattern is also reflected in the mean intensity of the IgE responses to the 4 epitopes, shown in Fig 2, B.
FIG 2.
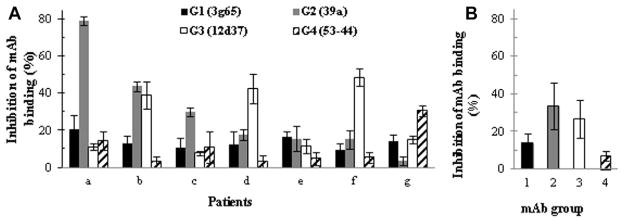
Relative binding of patients’ antibodies to discontinuous epitopes defined by mAbs G1 to G4. Binding of serum IgE from 7 patients with cedar pollinosis (a–g) and (A) mean of 7 patients to 4 distinct discontinuous epitopes of Jun a 1 (B). The results indicate the frequency and relative intensity of the patients’ responses to each of the 4 epitopes defined by mAbs G1 to G4.
Our findings indicate that the IgE response to mountain cedar pollen is strongly focused on a single protein Jun a 1. The major targets of the patients’ IgE were at least 4 discontinuous regions, brought together by protein folding of this highly dominant allergen. We have also found that purified Jun a 1 is predominantly monomeric (zonal ultracentrifugation indicated >95% monomers; see Fig E3 in this article’s Online Repository at www.jacionline.org). Thus, IgE antibodies from each of our patients’ serum must bind to at least 2 different epitopes10 to degranulate mast cells or basophils. This is consistent with the results of our mAb inhibition studies, which indicate that Jun a 1 displays at least 4 discrete IgE epitopes and that each of our patients’ sera contained antibodies to 2 or more of these discontinuous epitopes.
In conclusion, this study indicates that several conformational epitopes on Jun a 1 are major targets for the IgE response to mountain cedar pollen. This finding suggests that these antibodies are produced in the upper airway after exposure to inhaled, native Jun a 1. This makes Jun a 1 an excellent candidate for the development of new approaches for preventing allergic reactions. These might entail the development of agents that selectively alter the display of dominant epitopes, which may impede epitope spreading. On a more basic level, our findings are consistent with the concept that allergens, as a group, share a relatively small number of structures, which, along with other factors, such as their abundance and stability in the human environment, make them unique among proteins. The characteristics of the allergic response to mountain cedar pollen also make Jun a1 an excellent prototype for identifying the structural basis of allergenicity.
Supplementary Material
Acknowledgments
We thank Christopher C. Q. Chin, PhD, and J. Ching Lee, PhD, for the ultracentrifugation analysis of Jun a 1.
This work was supported by the National Institute of Allergy and Infectious Diseases (grant no. R01AI052428 to R.M.G. and grant no. K08AI055792 to T.M.-H.), the American Lung Association (to T.M.-H.), and John Sealy Memorial Endowment Fund from the University of Texas Medical Branch.
Footnotes
Disclosure of potential conflict of interest: R. M. Goldblum has received grants from the National Institutes of Health and has received consulting fees, travel support, and payment for lectures from Merck. B. Ning has a patent for antibody-mediated modulation of allergy. T. Midoro-Horiuti has received a grant from the National Institutes of Health (K08AI055792); has received consulting fees, travel support, and payment for lectures from Merck; and has a patent for antibody-mediated modulation of allergy. The rest of the authors declare that they have no relevant conflicts of interest.
References
- 1.Ivanciuc O, Midoro-Horiuti T, Schein CH, Xie L, Hillman GR, Goldblum RM, et al. The property distance index PD predicts peptides that cross-react with IgE antibodies. Mol Immunol. 2009;46:873–83. doi: 10.1016/j.molimm.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ivanciuc O, Garcia T, Torres M, Schein CH, Braun W. Characteristic motifs for families of allergenic proteins. Mol Immunol. 2009;46:559–68. doi: 10.1016/j.molimm.2008.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Radauer C, Breiteneder H. Pollen allergens are restricted to few protein families and show distinct patterns of species distribution. J Allergy Clin Immunol. 2006;117:141–7. doi: 10.1016/j.jaci.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 4.Liu Z, Bhattacharyya S, Ning B, Midoro-Horiuti T, Czerwinski EW, Goldblum RM, et al. Plant-expressed recombinant mountain cedar allergen Jun a 1 is allergenic and has limited pectate lyase activity. Int Arch Allergy Immunol. 2010;153:347–58. doi: 10.1159/000316345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Midoro-Horiuti T, Goldblum RM, Kurosky A, Goetz DW, Brooks EG. Isolation and characterization of the mountain cedar (Juniperus ashei) pollen major allergen, Jun a 1. J Allergy Clin Immunol. 1999;104:608–12. doi: 10.1016/s0091-6749(99)70331-3. [DOI] [PubMed] [Google Scholar]
- 6.Varshney S, Goldblum RM, Kearney C, Watanabe M, Midoro-Horiuti T. Major mountain cedar allergen, Jun a 1, contains conformational as well as linear IgE epitopes. Mol Immunol. 2007;44:2781–5. doi: 10.1016/j.molimm.2005.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiegand TW, Williams PB, Dreskin SC, Jouvin MH, Kinet JP, Tasset D. High-affinity oligonucleotide ligands to human IgE inhibit binding to Fc epsilon receptor I. J Immunol. 1996;157:221–30. [PubMed] [Google Scholar]
- 8.Czerwinski EW, Midoro-Horiuti T, White MA, Brooks EG, Goldblum RM. Crystal structure of Jun a 1, the major cedar pollen allergen from Juniperus ashei, reveals a parallel beta-helical core. J Biol Chem. 2005;280:3740–6. doi: 10.1074/jbc.M409655200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Midoro-Horiuti T, Mathura V, Schein CH, Braun W, Chin CCQ, Yu S, et al. Major linear IgE epitopes of mountain cedar pollen allergen Jun a 1 map to the pectate lyase catalytic site. Mol Immunol. 2003;40:555–62. doi: 10.1016/s0161-5890(03)00168-8. [DOI] [PubMed] [Google Scholar]
- 10.Scholl I, Kalkura N, Shedziankova Y, Bergmann A, Verdino P, Knittelfelder R, et al. Dimerization of the major birch pollen allergen Bet v 1 is important for its in vivo IgE-cross-linking potential in mice. J Immunol. 2005;175:6645–50. doi: 10.4049/jimmunol.175.10.6645. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.