The p38 MAP kinases are an important family of drug targets for a myriad of inflammatory, as well as other, diseases. Presented here is the crystal structure of the active form of MAPK13 (p38d) as well as a comprehensive analysis of all known apo inactive and active p38 MAPK structures, revealing a common mode of activation as well as some unique structural features.
Keywords: MAPK13, p38 MAP kinases
Abstract
The p38 MAP kinases (p38 MAPKs) represent an important family centrally involved in mediating extracellular signaling. Recent studies indicate that family members such as MAPK13 (p38δ) display a selective cellular and tissue expression and are therefore involved in specific diseases. Detailed structural studies of all p38 MAPK family members are crucial for the design of specific inhibitors. In order to facilitate such ventures, the structure of MAPK13 was determined in both the inactive (unphosphorylated; MAPK13) and active (dual phosphorylated; MAPK13/pTpY) forms. Here, the first preparation, crystallization and structure determination of MAPK13/pTpY are presented and the structure is compared with the previously reported structure of MAPK13 in order to facilitate studies for structure-based drug design. A comprehensive analysis of inactive versus active structures for the p38 MAPK family is also presented. It is found that MAPK13 undergoes a larger interlobe configurational rearrangement upon activation compared with MAPK14. Surprisingly, the analysis of activated p38 MAPK structures (MAP12/pTpY, MAPK13/pTpY and MAPK14/pTpY) reveals that, despite a high degree of sequence similarity, different side chains are used to coordinate the phosphorylated residues. There are also differences in the rearrangement of the hinge region that occur in MAPK14 compared with MAPK13 which would affect inhibitor binding. A thorough examination of all of the active (phosphorylated) and inactive (unphosphorylated) p38 MAPK family member structures was performed to reveal a common structural basis of activation for the p38 MAP kinase family and to identify structural differences that may be exploited for developing family member-specific inhibitors.
1. Introduction
The mitogen-activated protein kinases (MAPKs) are crucially involved in signal transduction in response to several extracellular stimuli. In mammalian cells, there are four well characterized MAPK families, ERK1/2, ERK5, JNK and p38 MAPK; these are serine/threonine kinases that catalyze the reversible phosphorylation of proteins. Their central involvement in crucial signaling pathways has made them important drug targets.
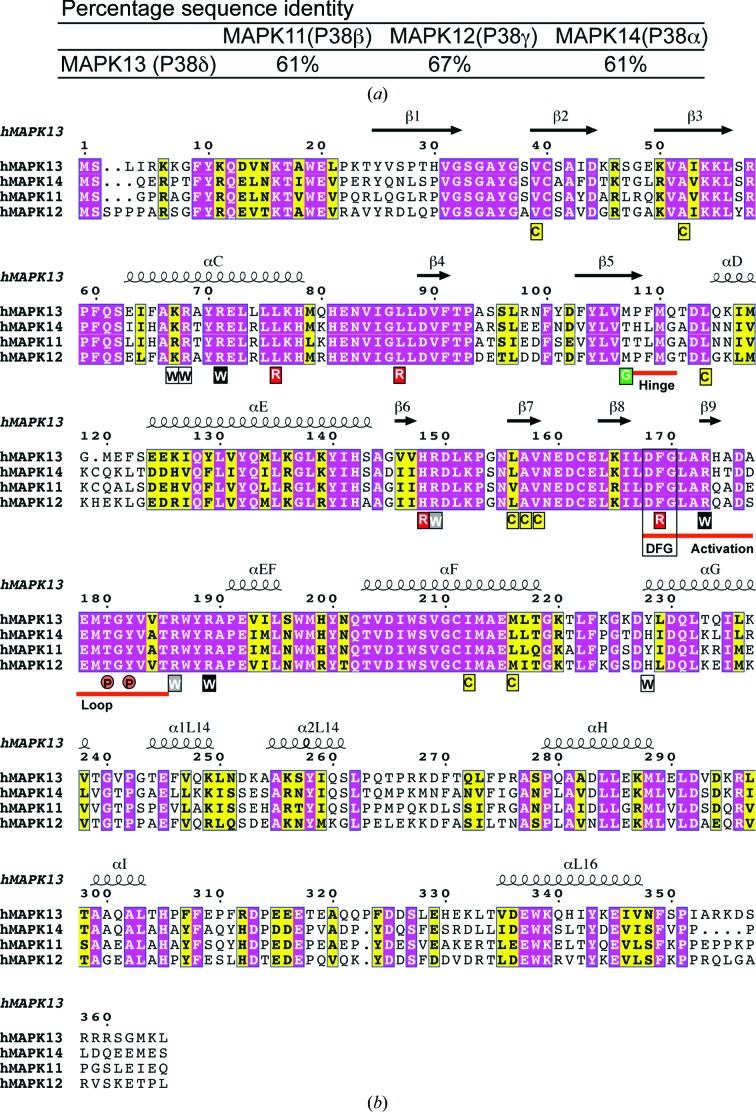
The p38 MAPKs consist of four family members: p38α (MAPK14), p38β (MAPK11), p38γ (MAPK12) and p38δ (MAPK13). These p38 MAPKs are highly similar in sequence, and all are activated by dual phosphorylation mediated by the MAPK kinases MKK3 and MKK6 (Cuenda & Rousseau, 2007 ▶). Within the family, they can be divided into two subgroups based on sequence similarity, cellular expression patterns and substrate specificity: MAPK14 (p38α) and MAPK11 (p38β) represent one subgroup, and MAPK12 (p38γ) and MAPK13 (p38δ) represent the other. MAPK14 (UniProt ID L7RSM2) and MAPK11 (Q15759) display the highest sequence identity (75%). MAPK14 was the first family member to be identified; it is fairly ubiquitously expressed and is centrally involved in pro-inflammatory signaling, thereby making it the focus of several drug-development efforts, while MAPK11 expression is more specific and its function appears to be redundant with that of MAPK14. In contrast, MAPK12 (P53778) and MAPK13 (O15264) are more divergent in sequence (62 and 61% identical to MAPK14, respectively), display more tissue-specific expression patterns and therefore contribute to more restricted functions (Risco & Cuenda, 2012 ▶). This tightly controlled expression pattern suggests that distinct biological pathways may be targeted by inhibiting specific isoforms. In this vein, MAPK13 has recently emerged as a potential disease-specific drug target. MAPK13 regulates the insulin secretion and survival of pancreatic β-cells, implying a central role in diabetes (Sumara et al., 2009 ▶). MAPK13 is also pivotal in neutrophil chemotaxis pathways, contributing to acute respiratory distress syndrome (ARDS; Ittner et al., 2012 ▶). In addition, studies in knockout mice indicate that MAPK13 contributes to pathways activated in chronic inflammatory conditions (Risco et al., 2012 ▶), is crucial for the development of arthritis in certain models (Criado et al., 2014 ▶) and is centrally involved in tumorogenesis in colitis-associated colon cancer models (Del Reino et al., 2014 ▶). Along these lines, we have observed that a signaling pathway progressing through MAPK13 mediates mucus overproduction in chronic inflammatory lung disease, and that MAPK13 inhibitors can block inflammatory mucus production in cell-culture models (Alevy et al., 2012 ▶).
Given the importance of MAPK13 as a potential specific drug target in chronic inflammatory lung disease (and possibly in other diseases), we initiated a structure-based drug-design project targeting this kinase by determining the structure of the inactive form of the enzyme and comparing it with two inhibitor-bound complexes (Alevy et al., 2012 ▶). In order to develop a more complete understanding of the structural basis for activation of MAPK13, we developed methods to produce activated (dual phosphorylated on Thr180 and Tyr182) MAPK13 (referred to in the following as MAPK13/pTpY), determined the X-ray crystal structure of MAPK13/pTpY and compared it with our previous structure of inactive unphosphorylated MAPK13 (MAPK13). In addition, we compared these with previously determined structures of other inactive (MAPK14 and MAPK11; Wang et al., 1997 ▶; Patel et al., 2009 ▶) and active (dual phosphorylated; MAPK14 and MAPK12; Bellon et al., 1999 ▶, Zhang et al., 2011 ▶) p38 MAPKs. These studies reveal a universal structural basis for activation for the p38 MAPK family.
2. Materials and methods
2.1. Expression constructs
Full-length human MAPK13 consists of 365 amino acids and contains several basic residues at the C-terminus (six of the last 12 residues are either Arg or Lys), and this full-length protein (1–365) would not readily crystallize in initial trials. Thus, for crystallization, a slightly truncated MAPK13 construct (1–352) was designed and cloned into pET-28a as an N-terminally His6-tagged construct using the NdeI and XhoI restriction-endonuclease sites, as described previously (Alevy et al., 2012 ▶). The construct was confirmed by sequencing.
2.2. Protein expression and purification
The construct was transformed into Escherichia coli Rosetta2 (DE3) cells (Stratagene) and colonies were grown on a plate with kanamycin selection. Cultures for protein expression were grown in LB medium using chloramphenicol (40 µg ml−1) and kanamycin (50 µg ml−1) selection. Typically, 8 × 1 l cultures were grown at 37°C until the OD600 reached 0.8–1.0. Protein expression was then induced at 30°C by the addition of 0.5 mM IPTG, and each 1 l of medium was enriched with 10 ml saturated glucose solution during protein expression. Protein expression was carried out at 30°C for 4 h.
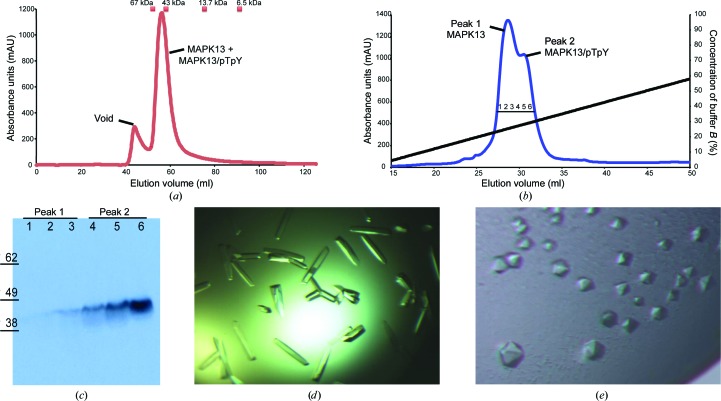
Cell pellets were harvested by centrifugation (typically yielding 5–10 g cell paste per litre of culture) and suspended in lysis buffer suitable for nickel–nitrilotriacetic acid (Ni–NTA) chromatography (50 mM K2HPO4 pH 8.0, 300 mM NaCl, 10 mM imidazole, 10% glycerol, 10 mM β-mercaptoethanol). The cells were lysed by the addition of 0.5 mg ml−1 lysozyme and DNAse I followed by sonication. The clarified lysate was passed over Ni–NTA, which was washed with lysis buffer containing 20 mM imidazole and the proteins were then eluted with 250 mM imidazole. The protein was further purified by gel-filtration chromatography on an ÄKTA FPLC. The protein was run over a Superdex 75 16/60 prep-grade column in a buffer consisting of 20 mM HEPES pH 7.5, 150 mM NaCl, 0.001% NaN3, 5 mM dithiothreitol, 10% glycerol. The protein (at this point still a mixture of MAPK13 and MAPK13/pTpY) eluted as a single peak correlating to a monomeric molecular weight (Fig. 1 ▶ a). This peak was harvested and then dialyzed against a buffer consisting of 20 mM Tris pH 8.0, 10 mM NaCl, 1 mM DTT, 10% glycerol (buffer A). The protein was then subjected to anion-exchange chromatography using a Mono Q 5/50 GL column (GE Life Sciences). The protein was bound onto the column using buffer A and then eluted off using a gradient of 0–60% buffer B (20 mM Tris pH 8.0, 1 M NaCl, 1 mM DTT, 10% glycerol) over 40 column volumes. This resulted in the separation of MAPK13 and MAPK13/pTpY (Fig. 1 ▶ b). The eluted fractions were analyzed by Western blot using anti-phospho-p38 antibody (AF869), which was raised against a dual-phosphorylated p38 activation-loop peptide (Millipore; Fig. 1 ▶ c). These preparations yielded an average of 6 mg MAPK13 and 4 mg MAPK13/pTpY per litre of culture (i.e. roughly a 3:2 ratio of MAPK13:MAPK13/pTpY).
Figure 1.
Purification and crystallization of unphosphorylated MAPK13 and dual-phosphorylated MAPK13 (MAPK13/pTpY). (a) Representative size-exclusion chromatogram displaying a single peak for the mixture of MAPK13 and MAPK13/pTpY eluting from a Superdex 75 16/60 column. Peak center elution volumes for molecular-weight calibration standards are marked as squares. (b) Representative ion-exchange chromatogram displaying peaks for the MAPK13 and MAPK13/pTpY crystallization constructs. (c) Western blot of the collected ion-exchange fractions from (a) using anti-phospho-p38 antibody. (d) Crystals of unphosphorylated MAPK13. The largest crystals are 0.05 × 0.2 µm in size. (e) Crystals of MAPK13/pTpY. The largest crystals are 0.15 × 0.15 µm in size.
2.3. Crystallization of MAPK13/pTpY and MAPK13
MAPK13/pTpY was exchanged into buffer consisting of 20 mM HEPES pH 7.5, 150 mM NaCl, 0.001% NaN3, 1 mM dithiothreitol, 10% glycerol and concentrated using an Amicon spin concentrator (Millipore). MAPK13/pTpY would not crystallize under similar conditions to MAPK13. Therefore, we initiated crystallization trials using broad commercial screens including The JCSG Core I–IV Suites (Qiagen), The PEGs I and II Suites (Qiagen), Crystal Screen (Hampton Research) and Index (Hampton Research), followed by optimization. Crystals were grown at 17°C using the hanging-drop vapour-diffusion method. Hexagonal crystals of MAPK13/pTpY were grown by mixing protein solution (at 10 mg ml−1) with reservoir solution (100 mM bis-tris pH 6.2–6.6, 21% PEG 3350, 200 mM NaCl) in a 1:1 ratio (Fig. 1 ▶ e). The total drop volume was 2 µl. Orthorhombic crystals of nonphosphorylated MAPK13 (Fig. 1 ▶ d) have been described previously (Alevy et al., 2012 ▶).
2.4. Data collection, processing and structure determination
X-ray diffraction data were collected on beamline 4.2.2 at the Advanced Light Source (ALS) using a NOIR-1 CCD detector and an 85 × 120 µm beam size. Data were indexed and processed with HKL-2000 (Otwinowski & Minor, 1997 ▶). A molecular-replacement solution was obtained using BALBES (Long et al., 2008 ▶). The best solution utilized phosphorylated MAPK12 (p38γ/pTpY; PDB entry 1cm8; Bellon et al., 1999 ▶) as a probe. Solutions were not obtained using unphosphorylated full-length MAPK13 (PDB entry 4exu; Alevy et al., 2012 ▶) and could only be obtained by searching separately with the N- and C-lobe domains. This is owing to the large degree of relative motion between the N- and C-lobes that occurs upon phosphorylation. The solution had a Q-factor of 0.829 and a final R work and R free of 0.267 and 0.328, respectively. The model was completed in Coot (Emsley et al., 2010 ▶) using σA-weighted 2F obs − F calc maps, and refinement was carried out in PHENIX (Adams et al., 2010 ▶). Because of the moderate resolution, torsion-angle NCS restraints were used throughout. Data-collection and refinement statistics are summarized in Table 1 ▶. Following all refinements, model bias in electron-density maps was reduced using the kicked-map feature in PHENIX. The final model for both chains included residues 15–351 (residues 1–14 and 352 could not be located in electron-density maps and were not built). The average error in the coordinates from the Luzzati plot was calculated using SFCHECK (Vaguine et al., 1999 ▶) in the CCP4i interface (Potterton et al., 2003 ▶; Winn et al., 2011 ▶). LSQ was used within Coot to perform and calculate r.m.s.d.s of Cα superpositions. Motion between domains upon phosphorylation was analyzed using DynDom using the domain-select mode (Hayward & Berendsen, 1998 ▶). All molecular-graphics figures were produced using PyMOL (Schrödinger). All crystallographic software was provided from the latest distributions of the SBGrid (Morin et al., 2013 ▶).
Table 1. Data-collection and refinement statistics for MAPK13/pTpY.
Values in parentheses are for the highest resolution shell.
| Data-collection statistics | |
| Space group | P65 |
| Unit-cell parameters () | a = 107.23, b = 107.23, c = 125.53 |
| X-ray source | ALS 4.2.2 |
| Wavelength () | 1.0070 |
| Resolution () | 50.02.60 (2.692.60) |
| R merge (%) | 0.088 (0.590) |
| Completeness (%) | 97.9 (91.5) |
| Multiplicity | 8.9 (5.5) |
| I/(I) | 19.8 (2.0) |
| Wilson B factor (2) | 48.61 |
| Refinement statistics | |
| R work | 0.205 (0.265) |
| R free | 0.255 (0.318) |
| No. of amino-acid residues | 674 |
| No. of waters | 186 |
| R.m.s.d., bond lengths () | 0.005 |
| R.m.s.d., angles () | 1.015 |
| Average B factor (2) | |
| Protein (chain A/chain B) | 57.2 (51.2/63.2) |
| Water | 47.8 |
| Ramachandran plot | |
| Favored (%) | 93.0 |
| Allowed (%) | 6.1 |
| Outliers (%) | 0.9 |
| Poor rotamers (No./%) | 5/0.84 |
| Clashscore (score/percentile) | 7.79/99th |
| MolProbity score (score/percentile) | 1.87/98th |
| Luzzati error | 0.3067 |
| PDB code | 4myg |
3. Results and discussion
3.1. Production, purification and crystallization of active MAPK13/pTpY
Large amounts of pure, active (phosphorylated) forms of MAPKs are required for structural, biophysical and enzymatic studies. For other p38 MAPKs, this has previously been accomplished either by using eukaryotic expression systems capable of post-translational modifications such as insect cells (Pargellis et al., 2002 ▶) or by overexpression and purification from bacteria followed by directed phosphorylation by treatment with a constitutively active MKK6 mutant (Bellon et al., 1999 ▶). Here, we demonstrate that MAPK13/pTpY is produced by autophosphorylation during expression in E. coli and that the unphosphorylated and phosphorylated MAPK13 can be separated using ion-exchange chromatography (Figs. 1 ▶ b and 1 ▶ c), producing protein of sufficient purity for the crystallization of both forms (Figs. 1 ▶ d and 1 ▶ e). We have reported previously that λ-phosphatase can be used to dephosphorylate MAPK13 (Alevy et al., 2012 ▶); however, this treatment is not very efficient (only increasing the yield of MAPK13 by about 10%) and is unnecessary to produce either MAPK13 or MAPK13/pTpY suitable for crystallization. The expression and purification scheme outlined here is sufficient for these purposes. Previous studies have indicated that activated MAPKs may dimerize and that this dimerization contributes to kinase activity. We observe that MAPK13/pTpY co-elutes with MAPK13 in the context of a gel-filtration chromatography experiment, eluting at a roughly monomeric weight (Fig. 1 ▶ a). Thus, within the context of a gel-filtration chromatography experiment, MAPK13 appears to be monomeric in both the active and the inactive forms.
3.2. Structure of MAPK13/pTpY and comparison with MAPK13
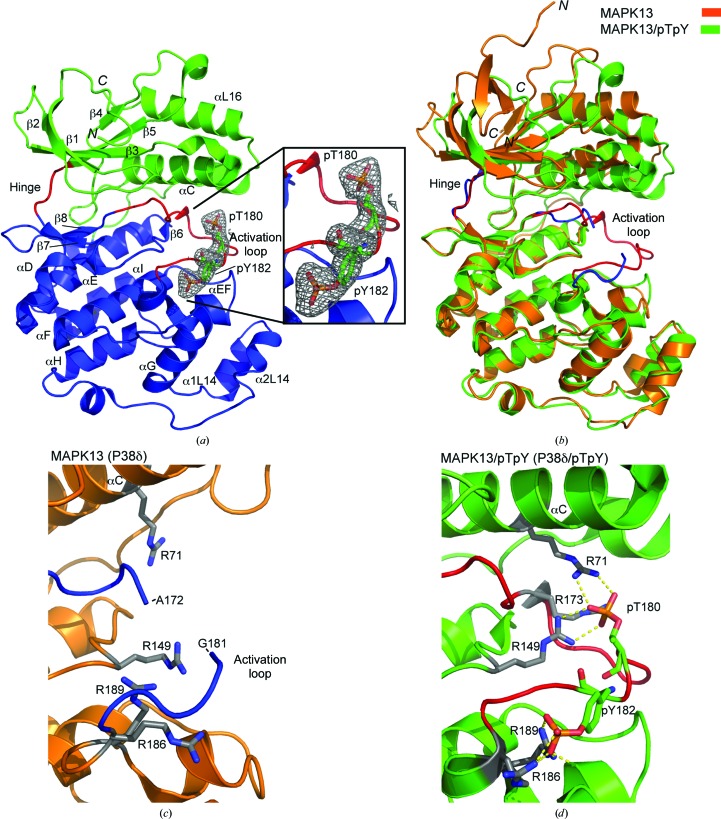
To gain an understanding of the structural basis for the activation of MAPK13, and a more complete understanding of p38 MAPK family activation in general, we determined the structure of MAPK13/pTpY and compared it with our previously determined structure of inactive MAPK13 (Alevy et al., 2012 ▶). In our previously determined MAPK13 structure (PDB entry 4exu) there is one molecule in the asymmetric unit. There are two molecules in the asymmetric unit of the MAPK13/pTpY structure that are nearly identical (the same residues are built; r.m.s.d. on Cα atoms of 0.94 Å); thus, only the A chain will be discussed and used in structural comparisons throughout this manuscript. While the crystals of MAPK13 diffracted to high resolution (1.70 Å), the crystals of MAPK13/pTpY diffracted to moderate resolution (2.60 Å; see Table 1 ▶); however, the phosphorylation sites and covalently bound phosphates, as well as the entire activation loop, were well resolved in the electron-density maps (Fig. 2 ▶ a). The structures display the standard MAPK topology consisting of N- and C-lobes encompassing a catalytic groove between them. Nearly the entire protein could be placed in electron-density maps for both MAPK13 (2–172 and 181–351) and MAPK13/pTpY (15–351). The main difference between the two structures occurs in the relative orientation of the N- and C-lobes and also in the activation loop (Fig. 2 ▶ b). The two phosphorylated residues in the activation loop are coordinated by basic side chains from both lobes of the kinase, and this results in a more compact catalytic site and overall structure. The largest difference between the two structures occurs in the activation loop; much of this region is notably absent from electron-density maps and is therefore likely to be disordered in the MAPK13 structure, but it is very well ordered in the MAPK13/pTpY structure (Figs. 2 ▶ c and 2 ▶ d). This is in contrast to MAPK14, where the activation loop is clearly observed in both the inactive MAPK14 and activated (MAPK14/pTpY) structures (see, for example, Fig. 5b). We have previously demonstrated that these crystals of inactive MAPK13 are useful for the determination of MAPK13–inhibitor complexes via soaking for the purposes of structure-based drug-design studies (Alevy et al., 2012 ▶), since the activation loop is disordered and is not involved in crucial crystal contacts, and therefore is amenable to the conformational adjustments required for inhibitor binding.
Figure 2.
Structure of MAPK13/pTpY and comparison with MAPK13. (a) Ribbon diagram of MAPK13/pTpY color-coded as follows: N-lobe, green; C-lobe, blue; hinge, red; activation loop, red. Simulated-annealing OMIT F obs − F calc electron density contoured at 3.0σ for pThr180 and pTyr182 is shown in the inset. (b) Superposition of MAPK13pTpY (green with red activation loop and hinge) and MAPK13 (orange with blue activation loop and hinge). MAPK13/pTpY was superposed using the C-lobes. (c, d) Coordination of pThr180 and pTyr182 and comparison of structure of the activation loop between the active and inactive forms. Hydrogen bonds are shown as yellow dashed lines. ‘Warhead’ arginines involved in direct interactions with pThr180 and pTyr182 are shown with grey C atoms. Hydrogen bonds are shown as yellow dashed lines. Note that a major portion of the MAPK13 activation loop is disordered, displays no visible electron density and therefore could not be placed in the inactive structure.
3.3. Comparison of activation phosphate coordination in p38 MAPK/pTpY structures
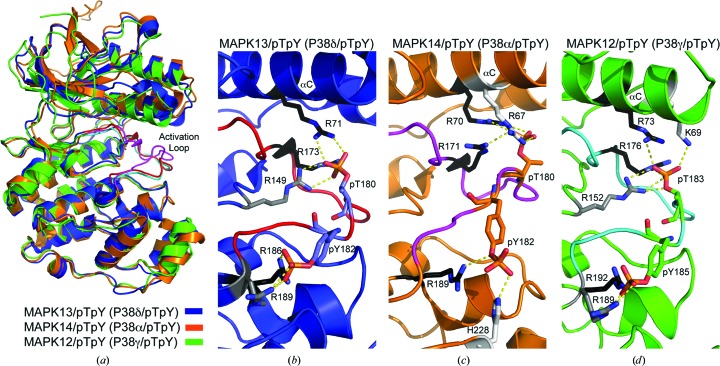
The activation loop is the nexus of conformational differences between unphosphorylated and phosphorylated structures. Phosphorylation of Thr180 and Tyr182 induces conformational changes in the activation loop in order to stabilize the large negative charges introduced by the phosphate groups. Packing analysis suggests that this conformational change is not greatly influenced by crystal packing. This coordination involves basic residues from both the N- and C-lobes, which in turn drives reorientation of catalytic site residues and, ultimately, closure between the two lobes, producing a conformation that is compatible with substrate binding and/or catalysis. Given the crucial importance of phosphate coordination in producing the catalytically active form, we examined the known p38 MAPK/pTpY structures in order to compare their activation-loop conformations and coordination of phosphate groups. Using superpositions of the C-lobes, MAPK13/pTpY and MAPK12/pTpY displayed nearly identical activation-loop conformations (r.m.s.d. on Cα of 0.23 Å for activation-loop residues), while MAPK14/pTpY displayed a very different activation-loop conformation (r.m.s.d. on Cα of 2.24 Å for activation-loop residues) (Fig. 3 ▶). Surprisingly, the basic residues utilized to coordinate the phosphates were not invariant in the structures of the p38 MAPK family, even though nearly all of the residues used are sequence-invariant throughout the family and in proximity (Figs. 3 ▶ and 4 ▶ b). For coordination of the pThr moiety, two invariant arginines (Arg71 and Arg173 in MAPK13) are used in all three structures, while a third (Arg149 in MAPK13) was utilized in two of the structures (Figs. 3 ▶ b, 3 ▶ c, 3 ▶ d and 4 ▶ b). Additionally, this moiety is coordinated by two unique residues in both MAPK14/pTpY (Arg67; Fig. 3 ▶ c) and MAPK12/pTpY (Lys69; Fig. 3 ▶ d). For coordination of the pTyr moiety, one invariant arginine was used by all three of the kinases (Arg186 in MAPK13) as well as an additional invariant arginine in MAPK13/pTpY and MAPK12/pTpY only (Arg189 in MAPK13). A unique histidine (His228), which is not conserved in MAPK13 or MAPK12, was also utilized to coordinate the pTyr phosphate in MAPK14/pTpY (Fig. 3 ▶ c). Taken altogether, these observations are surprising in that the p38 MAPK family display a high degree of sequence identity (e.g. 61% identity between MAPK13 and MAPK14, the most divergent pair; Fig. 4 ▶ a) yet achieve stabilization of the active form of the kinase using different structural elements. This is owing to a redundancy of basic residues available in this region and is also likely to be contributed to by the different configurations of N-lobes in the structures. These observations suggest that despite the high degree of sequence identity, structural differences can exist between the active forms of the kinases, and this could contribute to selective activation and possibly be exploited to design specific inhibitor strategies.
Figure 3.
Comparison of activation-loop phosphate-group coordination in p38 MAPK/pTpY crystal structures. (a) Overlay of MAP13/pTpY (PDB entry 4myg; blue with red activation loop), MAP14/pTpY (PDB entry 3py3; orange with magenta activation loop) and MAP12/pTpY (PDB entry 1cm8; green with cyan activation loop) superposed using the C-lobes. Note the similarity in activation-loop conformations in MAPK13/pTpY and MAPK12/pTpY. (b, c, d) Coordination of pThr and pTyr phosphate groups. ‘Warhead’ basic residues involved in direct interactions with phosphate groups are colored according to their conserved use in the three structures (black, used in all three; grey, used in two; white, used in one). Hydrogen bonds are shown as yellow dashed lines.
Figure 4.
Sequence comparison of p38 MAP kinases. (a) Table of percentage sequence identity of MAPK13 compared with other p38 MAPK family members. (b) Sequence alignment of human p38 MAPKs. Invariant and conserved residues are highlighted with magenta and yellow backgrounds, respectively. The secondary structure of phosphorylated human MAPK13 is shown above the alignment. The hinge region, DFG motif, gatekeeper (G), dual phosphorylation site (P), activation loop, R-spine residues (R) and C-spine (C) are shown. Warhead basic residues (W) are colored according to their conserved use to coordinate activation-loop phosphates in the p38 MAPK/pTpY structures compared here (black, used in all three; grey, used in two; white, used in one). This figure was generated using ESPript3.0 (Gouet et al., 2003 ▶).
3.4. Comparison of structural transition upon activation in MAPK13 versus MAPK14
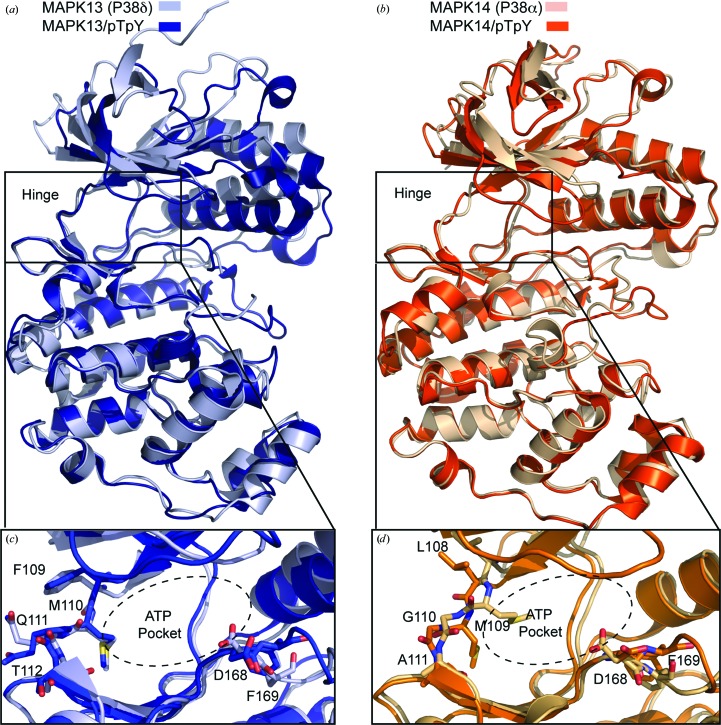
The only other p38 MAPK family member for which crystal structures of both the inactive unphosphorylated and activate pTpY forms have been determined is MAPK14, which is also the least homologous to MAPK13. Thus, we performed a comparison of the structural changes that occur between the two forms for these family members. Overall, the structural differences between the inactive and active forms of the kinases are not major; however, slightly larger changes occur upon activation of MAPK14. The r.m.s.d. for superposition of MAPK13 compared with MAPK13/pTpY was 2.38 Å (for 329 Cα atoms in common excluding the activation loop) versus 2.55 Å on comparing MAPK14 and MAPK14/pTpY (with the same 329 Cα atoms in common excluding the activation loop). The largest difference between the inactive and activated structures is the conformational rearrangement of the activation loop. This loop is disordered in MAPK13 (residues 172–180 are missing) and thus not built in the MAPK13 structure (see Fig. 2 ▶ c) and only becomes ordered in the active MAPK13/pTpY structure; however, this loop is ordered in both the MAPK14 and MAPK14/pTpY structures. The coordination of the activation-loop phosphates is carried out by basic residues from both lobes and leads to a more closed and compact structure in the activated state. We compared the relative movement of the two domains in the MAPK13 versus MAPK13/pTpY and MAPK14 versus MAPK14/pTpY structures using DynDom (Hayward & Berendsen, 1998 ▶). The difference observed in MAPK13 versus MAPK13/pTpY is most dramatic (25° relative rotation between the N- and C-lobes) compared with the MAPK14 and MAPK14/pTpY pair (only 8° relative rotation) (Figs. 5 ▶ a and 5 ▶ b).
Figure 5.
Comparison of MAPK13 and MAPK14 conformational change upon phosphorylation. (a) MAPK13/pTpY, blue; MAPK13, light blue. (b) MAPK14/pTpY, orange (PDB entry 3py3; Zhang et al., 2011 ▶); MAPK14, light orange (PDB entry 1p38; Wang et al., 1997 ▶). Note that the activation loop of MAPK14 is ordered in both the inactive and active forms, whereas the MAPK13 activation loop is disordered in the inactive form. (c, d) Comparison of the hinge and ATP-binding pocket region in (c) MAP13/pTpY and (d) MAPK14/pTpY.
The activation-loop reconfiguration also induces a large rotation of about 100° in the side chain of Asp168 in the conserved DFG motif in both MAPK13 and MAPK14, which locks the crucial Phe169 side chain into a hydrophobic pocket (Figs. 5 ▶ c and 5 ▶ d). Interestingly, there are differences in the ATP-binding site that occur in MAPK14 activation that are not present in MAPK13. In MAPK14, Met109 in the hinge region blocks the ATP-binding site; however, in MAPK14/pTpY Met109 occupies a new pocket and this conformational rearrangement allows a flip of the peptide bond owing to the presence of the adjacent Gly110 (Fig. 5 ▶ d). The hinge region of MAPK13 does not contain a glycine residue and correspondingly does not undergo large backbone changes in going from the inactive to the active form (Fig. 5 ▶ c). The arrangement of having a glycine at this position in the hinge region is not frequent, occurring in only about 9% of the human kinome. These observations suggest that targeting this structural transition could be an effective strategy to specifically target MAPK14, as has recently been demonstrated with skepinone-L (Koeberle et al., 2012 ▶).
3.5. Comparison of conserved ‘spine’ structures in p38 MAPKs
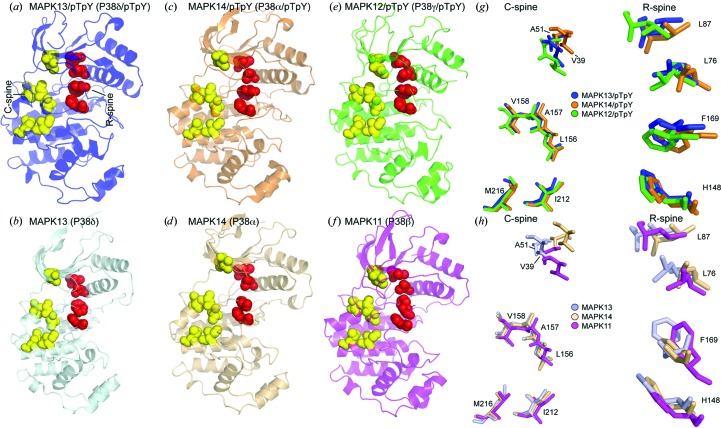
Kornev and Taylor recently performed an analysis of the conservation of spatial motifs found in activated but not in inactivated protein kinase structures and uncovered the presence of two crucial hydrophobic elements in activated kinases that they termed ‘spines’ (Taylor & Kornev, 2011 ▶). They concluded that all protein kinases contain two contiguous hydrophobic structures that link the N- and C-lobes of the protein in the activated form, that breaking these spines results in an inactive kinase structure, that many kinases display discontiguous spine structures in their inactive form, and that activation drives the assembly of contiguous spines. These two spines were termed the regulatory spine (R-spine), which consisted of four residues (two from the N-lobe, one from the C-lobe and one from the activation loop), and the catalytic spine (C-spine) consisting of seven residues (two from the N-lobe and five from the C-lobe). The R-spine is usually dynamically assembled upon reconfiguration of the activation loop after phosphorylation, whereas assembly of the R-spine is only completed upon binding of ATP, with the adenine ring docking into and completing the spine structure. Given that structures have now been determined for three p38 MAPK family members in both the activated (MAPK12/pTpY, MAPK13/pTpY and MAPK14/pTpY) and inactivated (MAPK11, MAPK13 and MAPK14) forms, we undertook the first comprehensive comparison of spine structures within this family. All of the R-spine residues are invariant within the p38 MAPK family, as are all of the C-spine residues, with the exception of conservative substitutions at two positions (Fig. 4 ▶ b). All display a discontiguous R-spine in the inactive forms (Figs. 6 ▶ b, 6 ▶ d and 6 ▶ f), which becomes contiguous after configuration of the activation loop (Figs. 6 ▶ a, 6 ▶ c and 6 ▶ e). This largely comes from the repositioning of Phe169, which takes on varied conformations in the inactive forms (Fig. 6 ▶ h) but locks into a hydrophobic pocket in the active forms (Fig. 6 ▶ g). Dislodging of this Phe side chain from this pocket, thereby breaking the C-spine, is the mechanism exploited by so-called type II and III kinase inhibitors (Karcher & Laufer, 2009 ▶). Most of the structural transitioning of the C-spine residues comes from pure translation as the kinases take on a more compact structure in the activated form. Thus, the p38 MAPKs display a dynamic assembly of spines in going from the inactive to the active state (similar to that observed in other kinases) and the largest structural change within these spines comes from reconfiguration of the Phe side chain from the DFG motif which completes the R-spine.
Figure 6.
Comparison of the assembly of ‘spine’ residues in inactive versus active p38 MAPK structures. Catalytic spine residues (C-spine) are shown as yellow CPK spheres and regulatory spine residues (R-spine) are shown as red CPK spheres. (a, b) Comparison of MAPK13/pTpY and MAPK13. (c, d) Comparison of MAPK14/pTpY and MAPK14. (e, f) Comparison of MAPK12/pTpY (PDB entry 1cm8; Bellon et al., 1999 ▶) and MAPK11 (PDB entry 3gc9; Patel et al., 2009 ▶). (g) Multiple alignment of residues comprising the two spines in p38 MAPK/pTpY structures. Alignment is based on superposition of the C-lobes. (h) Multiple alignment of residues comprising the two spines in p38 MAPK structures. Residue labeling is shown for MAPK13 in both (g) and (h).
4. Conclusion
Here, we report the first production and purification of MAPK13/pTpY suitable for structural and biophysical studies. This allowed us to determine the first structure of the activated form MAPK13/pTpY, to compare it with our previously determined structure of inactive MAPK13 and to perform the first comprehensive analysis of inactive versus active forms of p38 MAPK family members. Including the structure reported in this manuscript, there are currently structures of three p38 MAPK family members in both the inactive state, MAPK11 (Patel et al., 2009 ▶), MAPK13 and MAPK14 (Wang et al., 1997 ▶), and in the active (pTpY) state, MAPK12 (Bellon et al., 1999 ▶), MAPK13 and MAPK14 (Zhang et al., 2011 ▶). This makes this subfamily of MAP kinases the most comprehensively studied structurally, as the only other structure of an activated family member is that of ERK2 (Canagarajah et al., 1997 ▶). The major structural changes that occur in comparing the inactive with the active form of MAPK13 occur to stabilize the negative charge introduced by the two phosphates at pThr180 and pTyr182. This coordination is achieved using residues from both the N- and C-lobes of the kinase, which give the active form a more compact structure. This is consistent with what is observed on comparing MAPK14 and MAPK14/pTpY, although the interlobe rotation is more dramatic for MAPK13 (25° for MAPK13 versus 8° for MAPK14) and is closer to that observed for ERK2 (Canagarajah et al., 1997 ▶). Another major difference is in the hinge region, in which major backbone rearrangements occur in MAPK14 that are required to allow it to engage the catalytically required ATP. This rearrangement does not occur in MAPK13. This structural transition represents a possible unique feature to target for the development of specific inhibitors.
Stabilization of the major negative charges introduced by dual phosphorylation is achieved using basic residues from both lobes within the p38 MAPK family. A surprising observation from our analysis is that, despite a high degree of sequence similarity in the p38 MAPK family, different residues are utilized to carry out the coordination of the phosphate groups in the different p38 MAPKs. This suggests that the inhibition of specific p38 MAPKs may be accomplished by targeting the nonconserved residues that are critical for stabilizing the active form of the kinase.
Among the p38 MAPKs, MAPK13 appears to have a more restricted expression pattern and correspondingly has been linked to specific disease pathways. For example, we have observed that MAPK13 activity is crucial in a model of IL-13-induced mucus overproduction which upregulates the self-cleaving metalloprotease CLCA1 (Alevy et al., 2012 ▶), where self-cleavage is required to activate the CLCA1 protein (Yurtsever et al., 2012 ▶). These studies detail the first comprehensive analysis of p38 MAPK structures in the inactive and active (pTpY) forms, thus providing a framework for understanding the structural basis for activation of p38 MAPKs that could be useful in the design of specific inhibitors.
Supplementary Material
PDB reference: MAPK13/pTpY, 4myg
Acknowledgments
This work was supported in part by funding from NIH R01-HL119813 (to TJB), CADET P50-HL107183, CADET II UH2-HL123429 (to MJH) and an American Heart Association Predoctoral Fellowship PRE19970008 (to ZY). The results were derived from work performed at the Advanced Light Source (ALS), Berkeley, California, USA. The ALS is supported by the Office of Basic Energy Sciences of the US DOE (DE-AC02-05CH11231).
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Alevy, Y. G., Patel, A. C., Romero, A. G., Patel, D. A., Tucker, J., Roswit, W. T., Miller, C. A., Heier, R. F., Byers, D. E., Brett, T. J. & Holtzman, M. J. (2012). J. Clin. Invest. 122, 4555–4568. [DOI] [PMC free article] [PubMed]
- Bellon, S., Fitzgibbon, M. J., Fox, T., Hsiao, H.-M. & Wilson, K. P. (1999). Structure, 7, 1057–1065. [DOI] [PubMed]
- Canagarajah, B. J., Khokhlatchev, A., Cobb, M. H. & Goldsmith, E. J. (1997). Cell, 90, 859–869. [DOI] [PubMed]
- Criado, G., Risco, A., Alsina-Beauchamp, D., Pérez-Lorenzo, M. J., Escós, A. & Cuenda, A. (2014). Arthritis Rheumatol. 66, 1208–1217. [DOI] [PubMed]
- Cuenda, A. & Rousseau, S. (2007). Biochim. Biophys. Acta, 1773, 1358–1375. [DOI] [PubMed]
- Del Reino, P., Alsina-Beauchamp, D., Escos, A., Cerezo-Guisado, M. I., Risco, A., Aparicio, N., Zur, R., Fernandez-Estevez, M., Collantes, E., Montans, J. & Cuenda, A. (2014). Cancer Res. 74, 6150–6160. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Gouet, P., Robert, X. & Courcelle, E. (2003). Nucleic Acids Res. 31, 3320–3323. [DOI] [PMC free article] [PubMed]
- Hayward, S. & Berendsen, H. J. (1998). Proteins, 30, 144–154. [PubMed]
- Ittner, A., Block, H., Reichel, C. A., Varjosalo, M., Gehart, H., Sumara, G., Gstaiger, M., Krombach, F., Zarbock, A. & Ricci, R. (2012). J. Exp. Med. 209, 2229–2246. [DOI] [PMC free article] [PubMed]
- Karcher, S. C. & Laufer, S. A. (2009). Curr. Top. Med. Chem. 9, 655–676. [DOI] [PubMed]
- Koeberle, S. C., Romir, J., Fischer, S., Koeberle, A., Schattel, V., Albrecht, W., Grutter, C., Werz, O., Rauh, D., Stehle, T. & Laufer, S. A. (2012). Nature Chem. Biol. 8, 141–143. [DOI] [PubMed]
- Long, F., Vagin, A. A., Young, P. & Murshudov, G. N. (2008). Acta Cryst. D64, 125–132. [DOI] [PMC free article] [PubMed]
- Morin, A., Eisenbraun, B., Key, J., Sanschagrin, P. C., Timony, M. A., Ottaviano, M. & Sliz, P. (2013). Elife, 2, e01456. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Pargellis, C., Tong, L., Churchill, L., Cirillo, P. F., Gilmore, T., Graham, A. G., Grob, P. M., Hickey, E. R., Moss, N., Pav, S. & Regan, J. (2002). Nature Struct. Biol. 9, 268–272. [DOI] [PubMed]
- Patel, S. B., Cameron, P. M., O’Keefe, S. J., Frantz-Wattley, B., Thompson, J., O’Neill, E. A., Tennis, T., Liu, L., Becker, J. W. & Scapin, G. (2009). Acta Cryst. D65, 777–785. [DOI] [PubMed]
- Potterton, E., Briggs, P., Turkenburg, M. & Dodson, E. (2003). Acta Cryst. D59, 1131–1137. [DOI] [PubMed]
- Risco, A. & Cuenda, A. (2012). J. Signal. Transduct. 2012, 520289. [DOI] [PMC free article] [PubMed]
- Risco, A., del Fresno, C., Mambol, A., Alsina-Beauchamp, D., MacKenzie, K. F., Yang, H. T., Barber, D. F., Morcelle, C., Arthur, J. S., Ley, S. C., Ardavin, C. & Cuenda, A. (2012). Proc. Natl Acad. Sci. USA, 109, 11200–11205. [DOI] [PMC free article] [PubMed]
- Sumara, G., Formentini, I., Collins, S., Sumara, I., Windak, R., Bodenmiller, B., Ramracheya, R., Caille, D., Jiang, H., Platt, K. A., Meda, P., Aebersold, R., Rorsman, P. & Ricci, R. (2009). Cell, 136, 235–248. [DOI] [PMC free article] [PubMed]
- Taylor, S. S. & Kornev, A. P. (2011). Trends Biochem. Sci. 36, 65–77. [DOI] [PMC free article] [PubMed]
- Vaguine, A. A., Richelle, J. & Wodak, S. J. (1999). Acta Cryst. D55, 191–205. [DOI] [PubMed]
- Wang, Z., Harkins, P. C., Ulevitch, R. J., Han, J., Cobb, M. H. & Goldsmith, E. J. (1997). Proc. Natl Acad. Sci. USA, 94, 2327–2332. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Yurtsever, Z., Sala-Rabanal, M., Randolph, D. T., Scheaffer, S. M., Roswit, W. T., Alevy, Y. G., Patel, A. C., Heier, R. F., Romero, A. G., Nichols, C. G., Holtzman, M. J. & Brett, T. J. (2012). J. Biol. Chem. 287, 42138–42149. [DOI] [PMC free article] [PubMed]
- Zhang, Y.-Y., Wu, J.-W. & Wang, Z.-X. (2011). J. Biol. Chem. 286, 16150–16162. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: MAPK13/pTpY, 4myg