Abstract
The principles of cancer immunoediting have set the foundations for understanding the dual host-protective and tumor sculpting actions of immunity on cancer and establishing the basis for novel individualized cancer immunotherapies. During cancer immunoediting, the host immune system shapes tumor fate in three phases through the activation of innate and adaptive immune mechanisms. In the first phase, Elimination, transformed cells are destroyed by a competent immune system. Sporadic tumor cells that manage to survive immune destruction may then enter an Equilibrium phase where editing occurs. The Escape phase represents the third and final phase of the process, where immunologically sculpted tumors begin to grow progressively, become clinically apparent and establish an immunosuppressive tumor microenvironment. This review focuses on important recent developments that have enhanced our understanding of each phase of the cancer immunoediting process, summarizes the discovery of new predictive and prognostic biomarkers and discusses development of novel and objectively effective cancer immunotherapies.
Introduction
The last two decades have seen the end of the long-standing argument about whether the immune system has positive, negative or null effects on tumor development. Recent work from many labs have unequivocally documented that immunity can, in fact, facilitate cellular transformation, prevent or control tumor outgrowth and shape the immunogenicity of tumors. These three apparently paradoxical functions of the immune system are separable based on their temporal occurrence during tumor formation, the nature of the transforming event, the particular components of immunity involved in each process, and in the nature of the tumor specific antigens expressed in the transformed cell. Whereas one body of work has clearly established the capacity of chronic inflammation to initiate and promote cancer [1], a second set of studies from other laboratories, including our own, has revealed that an intact immune system can prevent/control and shape/promote cancer by a process we call ‘Cancer Immunoediting’ [2,3]. The evolution of the cancer immunoediting concept from the older and perhaps more controversial ‘cancer immunosurveillance’ hypothesis has helped interpret the predictive and prognostic significance of immune infiltrates into tumors.
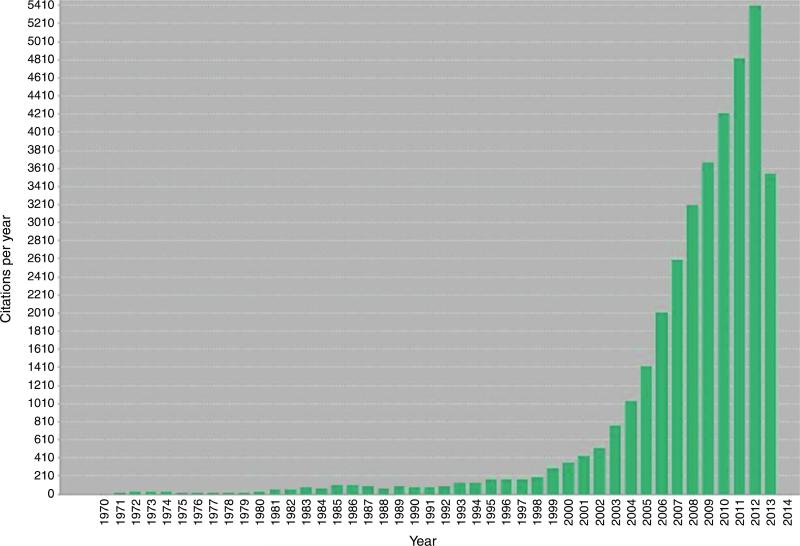
The immune surveillance theory originally proposed more than 50 years ago by Burnet and Thomas predicted that the immune system acted as a sentinel in recognizing and eliminating nascent transformed cells [4]. Extensive work over the past 15 years revealed that this surveillance function of immunity was only a part of the story and prompted us to refine and extend the concept into one we call ‘cancer immunoediting’ to more accurately describe the many facets of immune system–tumor interactions [2,3]. This dynamic process, whereby the immune system not only protects against cancer development but also shapes the character of emerging tumors, is composed of three phases — Elimination, Equilibrium and Escape — and has been extensively reviewed elsewhere [5,6]. Whereas the Elimination phase has largely been inferred from mouse tumor model studies, evidence for the Equilibrium and Escape phases have come from analyses of cancers in both mice and humans. Consequently, escape from immune control is now recognized to be one of the ‘Hallmarks of Cancer’ [7]. The emergence of cancer immunoediting as a framework to understand the extent of the immune system's interaction with cancer, has, in part, prompted a recent burgeoning of the scientific literature discussing this process as demonstrated by dramatically increased citation (Figure 1). Herein we review recent findings that have added to our understanding of cancer immunoediting and discuss the relevance of this process to cancer immunotherapy.
Figure 1.
Number of ‘cancer immunosurveillance or cancer immunoediting’ citations per year demonstrating the importance and increased interest in the field over recent years (derived from search of above terms in ISI Web of Science).
Immunoediting
The role of the immune system in shaping the immunogenicity of tumors has now been unequivocally established. Tumors arising in RAG2-deficient mice (lacking T, B and NKT cells) are, as a group, more immunogenic than those derived from immunocompetent hosts [5]. A central principle of cancer immunoediting is that T-cell recognition of tumor antigens drives the immunological elimination or sculpting of a developing cancer. However, until recently, little was known about the antigens expressed in nascent tumor cells, whether they are sufficient to induce anti-tumor immune responses, or whether their expression can be altered following interaction with the immune system.
Recently, we used a genomics approach to determine the mutational landscape of a highly immunogenic, unedited sarcoma cell line derived from methylcholanthrene (MCA)-treated Rag2−/− mice, which represent good models of nascent primary tumor cells. By combining exome sequence analysis with MHC class I prediction algorithms, a point mutation in Spectrin- β2 was identified and validated as the source of a neo-epitope in the unedited Rag2−/− derived d42m1 MCA sarcoma that functioned as a major immunodominant rejection antigen [8]. This study showed that cancer immunoediting was the consequence of a T-cell-dependent immunoselection process leading to the outgrowth of tumor cell clones lacking immunodominant rejection antigens that displayed reduced immunogenicity.
A similar conclusion was reached by DuPage et al. using a genetic mouse model of sarcomagenesis. This model employed immunocompetent and immunodeficient mice engineered to express a constitutively activated, oncogenic form of Kras and a floxed p53 tumor suppressor gene [9,10]. Intramuscular injection of a lentivirus encoding the cre-recombinase plus strong class I model epitopes (SIINFEKL and SIYRYYGL) led to the outgrowth of SIINFEKL and SIYRYYGL expressing sarcomas in immunodeficient mice. By contrast, the appearance of sarcomas in lentivirus transduced immunocompetent mice was delayed and those that grew out lacked expression of the model antigens. In this model, editing of the SIINFEKL and SIYRYYGL expression was the result of a T cell-dependent process that resulted in the epigenetic silencing of the exogenously introduced genes encoding the model antigens.
The clinical relevance of the above findings has been supported or confirmed in both preclinical and human studies. First our study, together with a distinct, independently published study by the Sahin group [11] established the principle that tumor exome analysis provides an opportunity to rapidly identify tumor specific mutational antigens thus forming the basis for development of individualized cancer immunotherapies. Subsequent work by others has indeed extended this finding to human cancer patients [12,13]. Second, a recent report, which followed the progression of a melanoma patient whose tumor was initially positive for the antigens NY-ESO-1, MAGE-C1 and Melan-A, showed that treatment with a vaccine targeting NY-ESO-1 lead to the outgrowth of tumors that lacked NY-ESO-1 but not MHC Class I, MAGE-C1 and Melan-A. This study, together with others like it, provides evidence that immunoediting also occurs as a consequence of immunotherapy in human cancer patients [14]. Additionally, analysis of patients in a phase I clinical trial receiving an NY-ESO vaccine showed that participants who relapsed had NY-ESO or MHC class I negative tumors [15]. Together these studies point out that cancer immunoediting is a process that occurs in both mice and humans. In addition, they reveal the crucial need to target multiple tumor antigens during cancer immunotherapy in order to prevent outgrowth of tumor cell variants that lack expression of individual tumor specific antigens. Although these studies have established the importance of adaptive immunity, and particularly T cells, in the editing process, other recent studies have revealed that innate immunity can also edit tumor immunogenicity. To assess the role of the innate immune system in cancer immunoediting, the tumorigenicity and immunogenicity of MCA-induced sarcomas derived from WT, Rag2−/−, and Rag2−/− x γc−/− (lacking all lymphocytes, including NK cells) were recently compared. MCA- induced sarcoma incidence was greatest in Rag2−/− x γc−/− mice, lower in Rag2−/− mice and much lower in WT mice. Tumor cells generated in Rag2−/− x γc−/− mice showed an increased regressor frequency (rejected when transplanted into immunocompetent host) compared with cell lines derived from Rag2−/− mice. Regressor cell lines were never isolated from MCA-treated WT mice [16]. These results suggest that in the absence of adaptive immunity, innate cells present in Rag2−/− mice but absent in Rag2−/− x γc−/− mice can shape, at least to some degree, the immunogenicity of tumors. In Rag2−/− mice, NK cells (presumably activated by local elaboration of endogenous IL-12) can produce IFN-γ that in turn induces activation of CD45+CD11b+MHCIIhiCD206loLy6Clo M1 macrophages, which act as important effectors of cancer immunoediting. These results show that the degree to which a tumor undergoes immunoediting is dependent on the degree of immunocompetence of the host [5,16]. Further work is needed to define the structures that are recognized on tumor cells by activated M1 macrophages.
Elimination
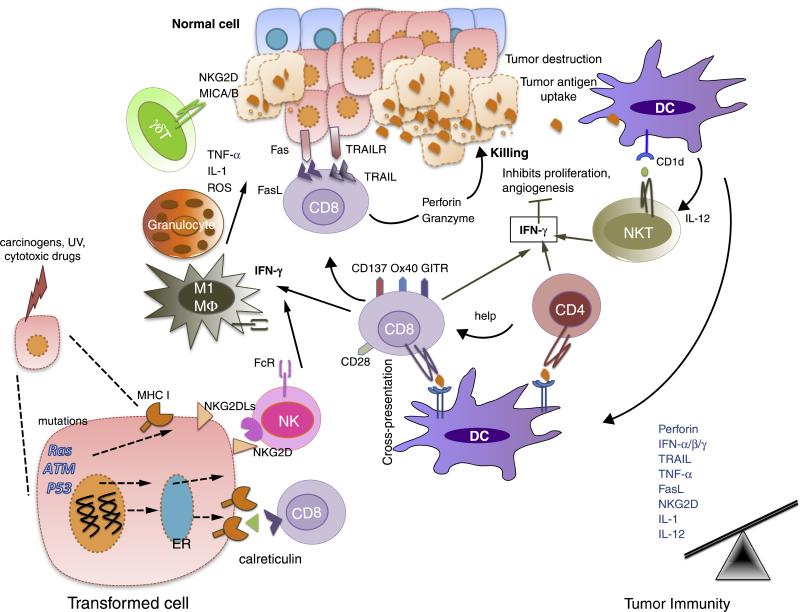
Previous reviews have extensively described the studies that support the Elimination phase of cancer immunoediting and have summarized the mechanisms underlying the host protective phase of cancer immunoediting [5,6,17]. Immunodeficient mice develop more carcinogen-induced and spontaneous cancers than wild-type mice, and tumors derived from immunodeficient mice are more immunogenic than those from immunocompetent mice. Here the role of host effector molecules, such as IFN-γ, perforin, Fas/FasL, and TRAIL; recognition molecules such as NKG2D; and an intact lymphocyte compartment in protective anti-tumor immunity, are well recognized [5,6,17,18] (Figure 2). Both type I (IFN-α/β) and type II interferons (IFN-γ) are required for development of anti-tumor immune responses but play distinct roles in the cancer immunoediting process. Whereas IFN-γ targets both tumor cells and hematopoietic cells, IFN- α/β acts primarily on host cells. Two recent studies showed that type I IFNs are required for initiation of the early anti-tumor response and act on CD8α/CD103+ DCs to enhance cross-presentation of tumor antigens to CD8+ T cells [18,19]. Type I IFN sensitivity in granulocytes, macrophages and NK cells, all of which express type I IFN receptors, was not required for tumor rejection [18].
Figure 2.
Elimination is a phase of cancer immunoediting where both the innate and adaptive immune system together detect and destroy early tumors before they become clinically visible. Normal cells (blue) are transformed into tumor cells by carcinogens and other genotoxic insults along with the failure of intrinsic tumor suppressor mechanisms (e.g. p53, ATM). These tumor cells express stress-induced molecules such as surface calreticulin, tumor antigens in context of MHC class I molecules, and/or NKG2D ligands recognized by CD8+ effector cells and NK cells, respectively. DCs can also take up and cross-present tumor antigens to T cells including NKT cells (glycolipid antigens presenting via CD1d). These activated effector cells release IFN-γ that can mediate anti-tumor effects by inhibiting tumor cell proliferation and angiogenesis. CD8+ T cells can induce tumor cell apoptosis by interacting with Fas and TRAIL receptors on tumor cells, or by secreting perforin and granzymes. Effector T cells express co-stimulatory molecules such as CD28, CD137, GITR, OX40 that enhance their proliferation and survival. γδ T cells can also recognize and kill tumors expressing NKG2D ligands (MICA/B in humans). Innate immune cells such as macrophages (M1) and granulocytes also contribute to anti-tumor immunity by secreting TNF-α, IL-1, IL-12 and ROS. In the Elimination phase, the balance is towards anti-tumor immunity due to an increase in expression of tumor antigens, MHC class I, Fas and TRAIL receptor on tumor cells and perforin, granzymes, IFN-α/β/γ, IL-1, IL-12, TNF-α in the tumor microenvironment.
Another recently published interesting study reported that aneuploidy, and particularly tetraploidization, while contributing to oncogenesis, increases tumor cell immunogenicity leading to immune elimination of these cells. Hyperploid cells have a constitutively active endoplasmic reticulum stress response, resulting in the aberrant cell surface exposure of calreticulin and increased cellular immunogenicity [20]. The immunosurveillance/elimination mechanism for control of ploidy was dependent on CD4+ and CD8+ T cells, IFN-γ and the IFN-γ receptor which is consistent with other experimental models of immunosurveillance/elimination (reviewed in [17]). These provocative results suggest that hyperploidy in carcinogen-induced and oncogene-induced cancer provides a recognition function for this mechanism of immune elimination of tumors.
Clearly one of the confounding issues in the study of Elimination is that the read-out is the absence of tumors (i.e. a negative result). Thus, there is always a question of whether tumors had originally formed and were eliminated or were never there to begin with. The absences of models that provide positive evidence for elimination have not been forthcoming until recently. Croxford et al. have now shown that early-stage B cells in Eμ-myc mice are tumourigenic and sharply regress in the periphery between 6 and 9 weeks of age [21]. In this model, sustained myc expression induces DNA damage (via the serine/threonine protein kinase ATM) and the DNA damage response can induce ligands that enhance immune recognition. Regression of early-stage transformed B cells was impaired by blockade of DNAM-1, a lymphocyte receptor for one such ligand (CD155), or deletion of T cells and NK1.1+ cells. Studies of tumor incidence in Eμ-myc mice under these conditions remain to be reported.
In addition to conventional elimination mechanisms involving adaptive immunity, new pathways of elimination have been recently reported that rely predominantly on cells of the innate immune system. In a mouse model of liver carcinoma, NK cells were shown to eliminate senescent tumor cells in a manner that was dependent on tumor cell intrinsic expression of p53 [22]. Upon expression of p53, tumor cells underwent senescence and secreted various interleukins (IL-6, IL-12 and IL-15) and chemokines such as CCL2 that recruited NK cells to senescent tumors. NK cells eliminated these lesions via mechanisms involving tumor cell expression of NKG2D ligands. NKG2D ligands can be induced on tumor cells by a variety of other stimuli, including Ras signaling and the DNA damage response [23,24]. Similarly, in a model of K-ras induced hepatocellular carcinoma, it was shown that premalignant senescent hepatocytes were cleared by liver-infiltrating K-ras-specific CD4+ T cells with the help of macrophages [25]. Impaired surveillance of senescent tissue resulted in liver cancer development [25].
Indirect evidence for an important innate role of NK cells in Elimination has also been uncovered. Dysfunctional telomeres suppress tumor progression by activating cell- intrinsic programs that lead to growth arrest. Increased levels of TRF2, a key factor in telomere protection, are observed in various human malignancies and contribute to oncogenesis. Recently, it was shown a high level of TRF2 in tumor cells decreased their ability to recruit and activate NK cells [26]. By screening for TRF2-bound genes, it was found that HS3ST4, a gene encoding for the heparan sulphate (glucosamine) 3-O-sulphotransferase 4, was regulated by TRF2 and inhibited the recruitment of NK cells in an epistatic relationship with TRF2. Overall, these results revealed a TRF2-dependent pathway that is tumor-cell intrinsic and regulates the host protective functions of NK cells against cancer development.
Another recent series of studies point to a possible, druggable role for macrophages in innate tumor elimination. Tumor cells have been shown to constitutively express CD47 that functions as a ‘don't eat me’ signal upon interacting with the SIRPα inhibitory receptor on macrophages. Monoclonal antibody blockade of CD47 led to phagocyte dependent elimination of tumor cells [27]. However, since red blood cells also express CD47, such a treatment could potentially lead to catastrophic anemia. Therefore, as an alternative approach, high affinity soluble human SIRPα variants were designed and were shown in various mouse cancer models to inhibit the interaction of macrophage SIRPα with tumor cell expressed CD47 [28]. These variants induced no toxicity and acted synergistically with tumor-specific therapeutic antibodies (acting via antibody-dependent cellular cytotoxicity) in mouse models of lymphoma and HER2+ breast carcinoma.
Equilibrium
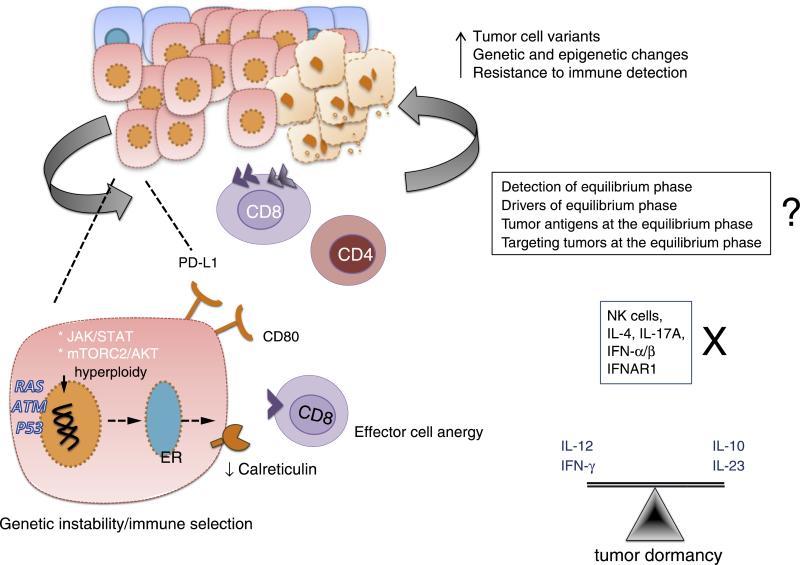
The molecular mechanisms that trigger immune-mediated tumor dormancy (Equilibrium phase) are poorly understood because this phase has been difficult to model in mice and have been described only anecdotally in humans (reviewed in [5]). A landmark study in 2007 demonstrated the role of adaptive Th1-like immunity in immune-mediated dormancy of fibrosarcoma [29]. A follow up study using the same mouse model of MCA- induced fibrosarcoma and p53 mutant tumors has shown that immune-mediated tumor dormancy may be a very prolonged process [30]. Importantly, the balance of IL-12 promoting elimination, and IL-23 (sharing the common subunit IL-12 (p40) promoting persistence, maintains tumors in equilibrium (Figure 3) [30]. Although a minor tumor-promoting role for IL-10 was also uncovered, many other pathways (e.g. IL-4, IL-17A, TNF, IFN-αβ) were shown to be dispensable for this phase.
Figure 3.
In the Equilibrium phase of cancer immunoediting, the immune system holds the tumor in a state of functional dormancy. Some tumor cells undergo genetic and epigenetic changes and due to constant immune pressure, tumor cell variants evolve that resist immune recognition (antigen loss or defects in antigen-presentation) and induce immunosuppression (PDL1). The Equilibrium phase is a balance between anti-tumor (IL-12, IFN-γ) and tumor promoting cytokines (IL-10, IL-23). The adaptive immune system is required to maintain tumor in a functionally dormant state while NK cells and cytokines such as IL-4, IL-17A and IFN-α/β are dispensable.
Another recent report compared the cellular environment of tumors in equilibrium versus those that escape and found high proportions of CD8+ T cells, NK cells, γδT cells and low proportions of NKT cells, Foxp3+ Treg cells, and MDSCs were associated with maintaining occult cancer in an immune-mediated equilibrium state [31]. This study further supports the concept that the relative balance of immunosuppressive cells and immune cells capable of manifesting antitumor effector functions in the tumor microenvironment is associated with maintaining tumor cells in a state of immune-mediated dormancy. It remains to be determined what shifts the balance in favor of immune escape or elimination following equilibrium but since editing occurs in this phase, it is likely that evolution of tumor cell variants that either lack key tumor specific antigens or the capacity to present them is an important factor that contributes to the decision process. Additional support for the occurrence of the Equilibrium phase comes from the finding that tumor antigen-specific T cells can arrest the growth of experimentally induced pancreatic tumors (Tag-induced multistage carcinogenesis) in mice by a coordinated interaction between IFN-γ and TNF [32]. In the absence of either TNFR or IFN-γ, the same T cells promoted angiogenesis and multistage carcinogenesis. It was further shown that, the combination of IFN-γ and TNF drive Tag-expressing cancers into senescence by inducing permanent growth arrest in G1/G0, activation of p16INK4a, and downstream Rb hypophosphorylation at Ser795 [33]. This cytokine-induced senescence requires STAT1 and TNFR1 signaling in addition to p16INK4a. Since IFN-γ and TNF induce senescence in numerous murine and human cancers, this may be a general mechanism for arresting cancer progression.
Identification of occult cancer in equilibrium in mice and humans remains a significant technical challenge, but advances in imaging technology with suitable antigen markers may allow circulating tumor cells and niches to be explored further.
Escape
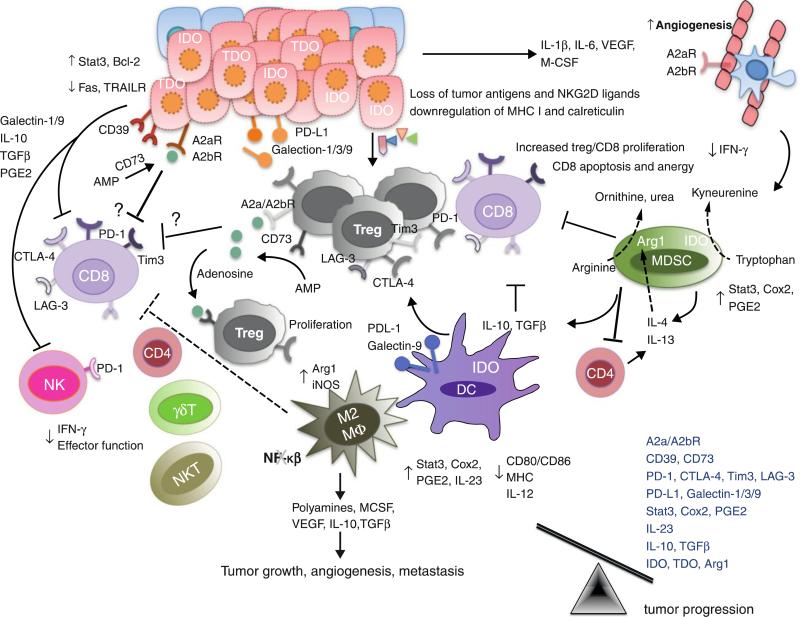
Tumor cell escape can occur through many different mechanisms including: reduced immune recognition (such as the absence of strong tumor antigens, or loss of MHC class I, class I-like, or co-stimulatory molecules), increased resistance or survival (such as increased expression of STAT-3 or anti-apoptotic molecule Bcl2), or development of an immunosuppressive tumor microenvironment (cytokines such as VEGF, TGF-β; immunoregulatory molecules such as IDO, PD-1/PD-L1, Tim-3/ galectin-9, LAG-3). These mechanisms have been extensively discussed elsewhere [5,6,17,34–36] and are summarized in Figure 4. In the past few years, we have witnessed a growing list of new moieties that contribute to tumor-induced immunosuppression, such as CD73 [37], adenosine receptors [38], and new B7 family checkpoint molecules including VISTA [39,40] and BTLA [41,42].
Figure 4.
During the Escape phase of cancer immunoediting, the immune system fails to restrict tumor outgrowth and tumor cells emerge causing clinically apparent disease. In this phase, tumor cells evade immune recognition (loss of tumor antigens, MHC class I or co-stimulatory molecules), express molecules of increased resistance (STAT-3), survival (anti-apoptotic molecule bcl2) and immunosuppression (IDO, TDO, PD-L1, galectin-1/3/9, CD39, CD73, adenosine receptors) and secrete cytokines VEGF, TGF-β, IL-6, M-CSF that enhance angiogenesis. Furthermore, MDSCs, M2 macrophages and DCs may also express immunoregulatory molecules such as arginase, iNOS and IDO and secrete immunosuppressive cytokines IL-10 and TGF-β that can inhibit CD8+ proliferation or induce apoptosis. MDSCs and IDO expressing DCs also induces the generation of regulatory T cells. IDO, arginase, CD39 and CD73 are immunoregulatory enzymes whereas IDO catabolize tryptophan to kyneurenine, arginase catabolize L-arginine to ornithine and urea, CD39 metabolise ATP to AMP which can further be metabolised to adenosine by CD73. Adenosine can bind to adenosine receptors — A2aR and A2bR expressed on tumor cells, endothelial cells and immune cells. T cells including Tregs may express inhibitory receptors such as PD-1, CTLA-4, Tim-3 and LAG-3 that suppresses anti-tumor immune response and favor tumor outgrowth. In the Escape phase, the balance is skewed towards tumor progression due to the presence of immunosuppressive cytokines and molecules such as IL-10, TGF-β, VEGF, IDO, PD-L1.
Targeted immunotherapies based on immune escape mechanisms.
Immunotherapy has recently emerged as a viable and potentially transformative approach to cancer treatment. However, therapeutic intervention often fails because of the plasticity of cells within the tumor microenvironment. Thus, approaches that involve combinations that target multiple pathways may prove synergistic and often are capable of generating a systemically effective memory response [43]. Many preclinical approaches have been extensively reviewed and a number of promising approaches are soon to enter the clinic [34,44,45]. In particular, the recent clinical combination of Ipilimumab (anti-CTLA-4) [46] and Nivolumab (anti-PD-1) [47,48] may have revolutionized thinking about the treatment strategy for melanoma patients [49]. This combination has shown responses in almost half of the metastatic melanoma patients, for which conventional therapies have failed [49]. The use of combination therapies provides increased opportunities for more effective beneficial clinical effects but also carry an increased risk of immunopathology. Therefore, it will be important in the future to seek a balance between tumor immunity and inflammatory pathology [50]. Tumor immunity and inflammatory pathology are closely related, but nonetheless separable and pre-clinical models that can tease out the mechanisms underlying this double edged sword of combination therapies will be invaluable for determining which combinations provide an increased therapeutic index.
Immune cells in tumors: predictive and prognostic significance
One significant advantage of measurable anti-tumor effects of immunotherapies in patients is our emerging capability to undertake genomic studies of tumors and the host to define key parameters that differentiate between responders from non-responders, and develop new approaches to stratify patients and their tumors. We have witnessed a tremendous explosion in the identification of immune signatures for various types of human cancer [51–54]. Similar signatures are observed within cancers with a better prognostic outcome, and cancers with an increased likelihood to respond to therapy or undergo complete regression. The parameters establishing the immune contexture are comprised of the density of CD3+, CD8+, and CD45RO+ T cells and their location at the tumor center and invasive margin combined with the quality of the tertiary lymphoid islets in the affected organ (the ‘Immunoscore’). These features are associated with an activated Th1 signature, including IFN-γ, STAT1, IL-12, IRF1, T-bet, perforin, granzymes, CXCR3 and CCR5 ligand chemokines, CXCL9, CXCL10 and CCL5, and adhesion molecules (MAD- CAM1, ICAM1 and VCAM1) [52]. There are many examples where recent work in the field has suggested that tumor infiltrates can be successfully used as a prognostic biomarker to predict the outcome of treatment [55,56]. Furthermore, in some cancers, these features have been found to be a more powerful prognostic indicator for tumor staging than previous pathological criteria. Other signatures have emerged from patients receiving IL-2 [57], MAGE-A3 vaccination [58], and Ipilimumab [59], and many targeted and conventional therapies also display similar lymphocyte signatures [60–62].
Role of microbiota in tumor growth and response to therapy
Another recent, clinically relevant finding concerning the interaction of immunity with cancer is the emerging role of the host microbiota during tumor formation and progression. Infiltrating Th1 cells and CD8+ cytotoxic T cells constitute a positive prognostic sign in colorectal cancer. By contrast, myeloid cells and Th17 cells promote tumourigenesis, and a Th17 expression signature in stage I/II colorectal cancer is associated with a drastic decrease in disease-free survival [63]. Many epithelial cancers develop proximally to microbial communities, which are only physically separated from immune cells by an epithelial barrier. Barrier deterioration induced by colorectal-cancer-initiating genetic lesions results in adenoma invasion by microbial products that trigger tumor-elicited inflammation, which in turn drives tumor growth. IL-23 mainly produced by tumor-associated myeloid cells that are likely to be activated by microbial products, is key in this process [64]. More recently, additional studies have demonstrated the importance of the host microbiome to carcinogenesis and tumor response to therapy [65].
Conclusion — the influences of cancer immunoediting on cancer immunotherapy
Recognizing cancer immunoediting and understanding the mechanisms that underpin it have provided the justification for many new immune-based cancer treatments. Some of these treatments are demonstrating remarkable responses in cancer patients, alone or in combination. In cancer, the immune system is not ignorant of the presence disease, but rather is actively suppressed by it. The challenge now lies in determining which patients are most suitable to receive these immunotherapies and how we can use information about their tumors and tumor microenvironments to inform us about the most effective treatments on a personalized basis. In spite of the recent success of immunotherapy in various human cancers, central questions remain unanswered. Even though we now have a better understanding of the mechanisms of tumor escape and equilibrium, questions still remain as to why some tumors escape immune control while others do not. Is the strength of the TCR response to antigen central in determining whether nascent cells are eliminated and why do some tumor clones further progress into equilibrium and escape? What has occurred in a large proportion of patients who have no obvious or apparent immune reaction with their cancer? Did they ever develop one? If not, can one be engineered? In those patients with an immune reaction, what is the simplest combination of therapies to achieve disease free survival? Will we eventually be able to develop personalized cancer immunotherapies designed specifically for an individual cancer patient and their individual tumors? Surely, with the recent explosion in our understanding of cancer immunogenomics and our rapidly expanding recognition of and ability to manipulate immune components that positively and negatively affect tumor immunity, we can expect significant improvements in the next few years in cancer treatment in general and cancer immunotherapy in particular.
Acknowledgements
We apologize to all the investigators whose research could not be appropriately cited due to space limitations. R. D. Schreiber is supported by NIH grants from the National Cancer Institute, the Cancer Research Institute, and the Ludwig Institute for Cancer Research. M. J. Smyth is supported by a NHMRC Australia Fellowship and Program Grant. D. Mittal is supported by a Susan G. Komen Breast Cancer Foundation Program Grant. M.M. Gubin is supported by an NIH training grant from the National Cancer Institute.
References and recommended reading
- 1.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shankaran V, Ikeda H, Bruce AT, White JM, Swanson PE, Old LJ, Schreiber RD. IFNgamma and lymphocytes prevent primary tumor development and shape tumor immunogenicity. Nature. 2001;410:1107–1111. doi: 10.1038/35074122. [DOI] [PubMed] [Google Scholar]
- 3.Dunn GP, Koebel CM, Schreiber RD. Interferons, immunity and cancer immunoediting. Nat Rev Immunol. 2006;6:836–848. doi: 10.1038/nri1961. [DOI] [PubMed] [Google Scholar]
- 4.Burnet M. Cancer; a biological approach. I. The processes of control. Br Med J. 1957;1:779–786. doi: 10.1136/bmj.1.5022.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science. 2011;331:1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 6.Dunn GP, Old LJ, Schreiber RD. The immunobiology of cancer immunosurveillance and immunoediting. Immunity. 2004;21:137–148. doi: 10.1016/j.immuni.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 7.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 8.Matsushita H, Vesely MD, Koboldt DC, Rickert CG, Uppaluri R, Magrini VJ, Arthur CD, White JM, Chen YS, Shea LK, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482:400–404. doi: 10.1038/nature10755. [This study demonstrates that highly antigenic mutant proteins of an unedited tumor can be mapped with exome sequencing and that T-cell-dependent immunoselection represents one mechanism of cancer immunoediting.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DuPage M, Mazumdar C, Schmidt LM, Cheung AF, Jacks T. Expression of tumor-specific antigens underlies cancer immunoediting. Nature. 2012;482:405–409. doi: 10.1038/nature10803. [This study illustrates that primary sarcomas were edited to become less immunogenic through the selective outgrowth of cells that were able to escape T lymphocyte attack.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DuPage M, Cheung AF, Mazumdar C, Winslow MM, Bronson R, Schmidt LM, Crowley D, Chen J, Jacks T. Endogenous T cell responses to antigens expressed in lung adenocarcinomas delay malignant tumor progression. Cancer Cell. 2011;19:72–85. doi: 10.1016/j.ccr.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castle JC, Kreiter S, Diekmann J, Lower M, van de Roemer N, de Graaf J, Selmi A, Diken M, Boegel S, Paret C, et al. Exploiting the mutanome for tumor vaccination. Cancer Res. 2012;72:1081–1091. doi: 10.1158/0008-5472.CAN-11-3722. [DOI] [PubMed] [Google Scholar]
- 12.Robbins PF, Lu YC, El-Gamil M, Li YF, Gross C, Gartner J, Lin JC, Teer JK, Cliften P, Tycksen E, et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med. 2013;19:747–752. doi: 10.1038/nm.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Rooij N, van Buuren MM, Philips D, Velds A, Toebes M, Heemskerk B, van Dijk LJ, Behjati S, Hilkmann H, El Atmioui D, et al. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J Clin Oncol. 2013;10:e439–e442. doi: 10.1200/JCO.2012.47.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.von Boehmer L, Mattle M, Bode P, Landshammer A, Schafer C, Nuber N, Ritter G, Old L, Moch H, Schafer N, et al. NY-ESO-1-specific immunological pressure and escape in a patient with metastatic melanoma. Cancer Immunol. 2013;13:12. [PMC free article] [PubMed] [Google Scholar]
- 15.Nicholaou T, Chen W, Davis ID, Jackson HM, Dimopoulos N, Barrow C, Browning J, Macgregor D, Williams D, Hopkins W, et al. Immunoediting and persistence of antigen-specific immunity in patients who have previously been vaccinated with NY-ESO-1 protein formulated in ISCOMATRIX. Cancer Immunol Immunotherapy. 2011;60:1625–1637. doi: 10.1007/s00262-011-1041-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Sullivan T, Saddawi-Konefka R, Vermi W, Koebel CM, Arthur C, White JM, Uppaluri R, Andrews DM, Ngiow SF, Teng MW, et al. Cancer immunoediting by the innate immune system in theabsence of adaptive immunity. J Exp Med. 2012;209:1869–1882. doi: 10.1084/jem.20112738. [This study demonstrates the role of innate immunity in cancer immunoediting.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vesely MD, Kershaw MH, Schreiber RD, Smyth MJ. Natural innate and adaptive immunity to cancer. Annu Rev Immunol. 2011;29:235–271. doi: 10.1146/annurev-immunol-031210-101324. [DOI] [PubMed] [Google Scholar]
- 18.Diamond MS, Kinder M, Matsushita H, Mashayekhi M, Dunn GP, Archambault JM, Lee H, Arthur CD, White JM, Kalinke U, et al. Type I interferon is selectively required by dendritic cells for immune rejection of tumors. J Exp Med. 2011;208:1989–2003. doi: 10.1084/jem.20101158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuertes MB, Kacha AK, Kline J, Woo SR, Kranz DM, Murphy KM, Gajewski TF. Host type I IFN signals are required for antitumor CD8+ T cell responses through CD8{alpha}+ dendritic cells. J Exp Med. 2011;208:2005–2016. doi: 10.1084/jem.20101159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Senovilla L, Vitale I, Martins I, Tailler M, Pailleret C, Michaud M, Galluzzi L, Adjemian S, Kepp O, Niso-Santano M, et al. An immunosurveillance mechanism controls cancer cell ploidy. Science. 2012;337:1678–1684. doi: 10.1126/science.1224922. [DOI] [PubMed] [Google Scholar]
- 21.Croxford JL, Tang ML, Pan MF, Huang CW, Kamran N, Phua CM, Chng WJ, Ng SB, Raulet DH, Gasser S. ATM-dependent spontaneous regression of early Emu-myc-induced murine B-cell leukemia depends on natural killer and T cells. Blood. 2013;121:2512–2521. doi: 10.1182/blood-2012-08-449025. [The authors describe a setting where early immune control of B cell lymphoma can be monitored.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iannello A, Thompson TW, Ardolino M, Lowe SW, Raulet DH. p53-dependent chemokine production by senescent tumor cells supports NKG2D-dependent tumor elimination by natural killer cells. J Exp Med. 2013;210:2057–2069. doi: 10.1084/jem.20130783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gasser S, Orsulic S, Brown EJ, Raulet DH. The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature. 2005;436:1186–1190. doi: 10.1038/nature03884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu XV, Ho SS, Tan JJ, Kamran N, Gasser S. Ras activation induces expression of Raet1 family NK receptor ligands. J Immunol. 2012;189:1826–1834. doi: 10.4049/jimmunol.1200965. [DOI] [PubMed] [Google Scholar]
- 25.Kang TW, Yevsa T, Woller N, Hoenicke L, Wuestefeld T, Dauch D, Hohmeyer A, Gereke M, Rudalska R, Potapova A, et al. Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature. 2011;479:547–551. doi: 10.1038/nature10599. [DOI] [PubMed] [Google Scholar]
- 26.Biroccio A, Cherfils-Vicini J, Augereau A, Pinte S, Bauwens S, Ye J, Simonet T, Horard B, Jamet K, Cervera L, et al. TRF2 inhibits a cell-extrinsic pathway through which natural killer cells eliminate cancer cells. Nat Cell Biol. 2013;15:818–828. doi: 10.1038/ncb2774. [DOI] [PubMed] [Google Scholar]
- 27.Willingham SB, Volkmer JP, Gentles AJ, Sahoo D, Dalerba P, Mitra SS, Wang J, Contreras-Trujillo H, Martin R, Cohen JD, et al. The CD47-signal regulatory protein alpha (SIRPa) interaction is a therapeutic target for human solid tumors. Proc Natl Acad Sci U S A. 2012;109:6662–6667. doi: 10.1073/pnas.1121623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weiskopf K, Ring AM, Ho CC, Volkmer JP, Levin AM, Volkmer AK, Ozkan E, Fernhoff NB, van de Rijn M, Weissman IL, et al. Engineered SIRPalpha variants as immunotherapeutic adjuvants to anticancer antibodies. Science. 2013;341:88–91. doi: 10.1126/science.1238856. [This study demonstrates the ability of a new therapuetic adjuvant target- ing SIRPa to enhance the anti-tumor activity of antibodies operating via their Fc..] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koebel CM, Vermi W, Swann JB, Zerafa N, Rodig SJ, Old LJ, Smyth MJ, Schreiber RD. Adaptive immunity maintains occult cancer in an equilibrium state. Nature. 2007;450:903–907. doi: 10.1038/nature06309. [DOI] [PubMed] [Google Scholar]
- 30.Teng MW, Vesely MD, Duret H, McLaughlin N, Towne JE, Schreiber RD, Smyth MJ. Opposing roles for IL-23 and IL-12 in maintaining occult cancer in an equilibrium state. Cancer Res. 2012;72:3987–3996. doi: 10.1158/0008-5472.CAN-12-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu X, Peng M, Huang B, Zhang H, Wang H, Xue Z, Zhang L, Da Y, Yang D, Yao Z, et al. Immune microenvironment profiles of tumor immune equilibrium and immune escape states of mouse sarcoma. Cancer Lett. 2013;340:124–133. doi: 10.1016/j.canlet.2013.07.038. [DOI] [PubMed] [Google Scholar]
- 32.Muller-Hermelink N, Braumuller H, Pichler B, Wieder T, Mailhammer R, Schaak K, Ghoreschi K, Yazdi A, Haubner R, Sander CA, et al. TNFR1 signaling and IFN-gamma signaling determine whether T cells induce tumor dormancy or promote multistage carcinogenesis. Cancer Cell. 2008;13:507–518. doi: 10.1016/j.ccr.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 33.Braumuller H, Wieder T, Brenner E, Assmann S, Hahn M, Alkhaled M, Schilbach K, Essmann F, Kneilling M, Griessinger C, et al. T-helper-1-cell cytokines drive cancer into senescence. Nature. 2013;494:361–365. doi: 10.1038/nature11824. [DOI] [PubMed] [Google Scholar]
- 34.Mellman I, Coukos G, Dranoff G. Cancer immunotherapy comes of age. Nature. 2011;480:480–489. doi: 10.1038/nature10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gajewski TF, Fuertes M, Spaapen R, Zheng Y, Kline J. Molecular profiling to identify relevant immune resistance mechanisms in the tumor microenvironment. Curr Opin Immunol. 2011;23:286–292. doi: 10.1016/j.coi.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mantovani A, Sica A. Macrophages, innate immunity and cancer: balance, tolerance, and diversity. Curr Opin Immunol. 2010;22:231–237. doi: 10.1016/j.coi.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 37.Beavis PA, Stagg J, Darcy PK, Smyth MJ. CD73: a potent suppressor of antitumor immune responses. Trends Immunol. 2012;33:231–237. doi: 10.1016/j.it.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 38.Beavis PA, Divisekera U, Paget C, Chow MT, John LB, Devaud C, Dwyer K, Stagg J, Smyth MJ, Darcy PK. Blockade of A2A receptors potently suppresses the metastasis of CD73+ tumors. Proc Natl Acad Sci U S A. 2013;110:14711–14716. doi: 10.1073/pnas.1308209110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang L, Rubinstein R, Lines JL, Wasiuk A, Ahonen C, Guo Y, Lu LF, Gondek D, Wang Y, Fava RA, et al. VISTA, a novel mouse Ig superfamily ligand that negatively regulates T cell responses. J Exp Med. 2011;208:577–592. doi: 10.1084/jem.20100619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ceeraz S, Nowak EC, Noelle RJ. B7 family checkpoint regulators in immune regulation and disease. Trends Immunol. 2013;34:556–563. doi: 10.1016/j.it.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hobo W, Norde WJ, Schaap N, Fredrix H, Maas F, Schellens K, Falkenburg JH, Korman AJ, Olive D, van der Voort R, et al. B and T lymphocyte attenuator mediates inhibition of tumor-reactive CD8+ T cells in patients after allogeneic stem cell transplantation. J Immunol. 2012;189:39–49. doi: 10.4049/jimmunol.1102807. [DOI] [PubMed] [Google Scholar]
- 42.Fourcade J, Sun Z, Pagliano O, Guillaume P, Luescher IF, Sander C, Kirkwood JM, Olive D, Kuchroo V, Zarour HM. CD8(+) T cells specific for tumor antigens can be rendered dysfunctional by the tumor microenvironment through upregulation of the inhibitory receptors BTLA and PD-1. Cancer Res. 2012;72:887–896. doi: 10.1158/0008-5472.CAN-11-2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marabelle A, Kohrt H, Sagiv-Barfi I, Ajami B, Axtell RC, Zhou G, Rajapaksa R, Green MR, Torchia J, Brody J, et al. Depleting tumor-specific Tregs at a single site eradicates disseminated tumors. J Clin Investig. 2013;123:2447–2463. doi: 10.1172/JCI64859. [This study demonstrates that antibodies can be used to target tumor infiltrative immune cells locally and thereby eliciting a systemic immune response.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vanneman M, Dranoff G. Combining immunotherapy and targeted therapies in cancer treatment. Nat Rev Cancer. 2012;12:237–251. doi: 10.1038/nrc3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel JC, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, Drake CG, Camacho LH, Kauh J, Odunsi K, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366:2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolchok JD, Kluger H, Callahan MK, Postow MA, Rizvi NA, Lesokhin AM, Segal NH, Ariyan CE, Gordon RA, Reed K, et al. Nivolumab plus ipilimumab in advanced melanoma. N Engl J Med. 2013;369:122–133. doi: 10.1056/NEJMoa1302369. [Remarkable demonstration of the therapeutic potential of combining cancer immunotherapeutics.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dranoff G. Immunotherapy at large: balancing tumor immunity and inflammatory pathology. Nat Med. 2013;19:1100–1101. doi: 10.1038/nm.3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Angell H, Galon J. From the immune contexture to the Immunoscore: the role of prognostic and predictive immune markers in cancer. Curr Opin Immunol. 2013;25:261–267. doi: 10.1016/j.coi.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 52.Galon J, Angell HK, Bedognetti D, Marincola FM. The continuum of cancer immunosurveillance: prognostic, predictive, and mechanistic signatures. Immunity. 2013;39:11–26. doi: 10.1016/j.immuni.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 53.Mlecnik B, Tosolini M, Charoentong P, Kirilovsky A, Bindea G, Berger A, Camus M, Gillard M, Bruneval P, Fridman WH, et al. Biomolecular network reconstruction identifies T-cell homing factors associated with survival in colorectal cancer. Gastroenterology. 2010;138:1429–1440. doi: 10.1053/j.gastro.2009.10.057. [DOI] [PubMed] [Google Scholar]
- 54.Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. 2013;14:1014–1022. doi: 10.1038/ni.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Balachandran VP, Cavnar MJ, Zeng S, Bamboat ZM, Ocuin LM, Obaid H, Sorenson EC, Popow R, Ariyan C, Rossi F, et al. Imatinib potentiates antitumor T cell responses in gastrointestinal stromal tumor through the inhibition of Ido. Nat Med. 2011;17:1094–1100. doi: 10.1038/nm.2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Delahaye NF, Rusakiewicz S, Martins I, Menard C, Roux S, Lyonnet L, Paul P, Sarabi M, Chaput N, Semeraro M, et al. Alternatively spliced NKp30 isoforms affect the prognosis of gastrointestinal stromal tumors. Nat Med. 2011;17:700–707. doi: 10.1038/nm.2366. [DOI] [PubMed] [Google Scholar]
- 57.Weiss GR, Grosh WW, Chianese-Bullock KA, Zhao Y, Liu H, Slingluff CL, Jr, Marincola FM, Wang E. Molecular insights on the peripheral and intratumoral effects of systemic high-dose rIL- 2 (aldesleukin) administration for the treatment of metastatic melanoma. Clin Cancer Res. 2011;17:7440–7450. doi: 10.1158/1078-0432.CCR-11-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ulloa-Montoya F, Louahed J, Dizier B, Gruselle O, Spiessens B, Lehmann FF, Suciu S, Kruit WH, Eggermont AM, Vansteenkiste J, et al. Predictive gene signature in MAGE-A3 antigen-specific cancer immunotherapy. J Clinl Oncol. 2013;31:2388–2395. doi: 10.1200/JCO.2012.44.3762. [DOI] [PubMed] [Google Scholar]
- 59.Ji RR, Chasalow SD, Wang L, Hamid O, Schmidt H, Cogswell J, Alaparthy S, Berman D, Jure-Kunkel M, Siemers NO, et al. An immune-active tumor microenvironment favors clinical response to ipilimumab. Cancer Immunol Immunotherapy. 2012;61:1019–1031. doi: 10.1007/s00262-011-1172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Halama N, Michel S, Kloor M, Zoernig I, Benner A, Spille A, Pommerencke T, von Knebel DM, Folprecht G, Luber B, et al. Localization and density of immune cells in the invasive margin of human colorectal cancer liver metastases are prognostic for response to chemotherapy. Cancer Res. 2011;71:5670–5677. doi: 10.1158/0008-5472.CAN-11-0268. [DOI] [PubMed] [Google Scholar]
- 61.Ignatiadis M, Singhal SK, Desmedt C, Haibe-Kains B, Criscitiello C, Andre F, Loi S, Piccart M, Michiels S, Sotiriou C. Gene modules and response to neoadjuvant chemotherapy in breast cancer subtypes: a pooled analysis. J Clin Oncol. 2012;30:1996–2004. doi: 10.1200/JCO.2011.39.5624. [DOI] [PubMed] [Google Scholar]
- 62.Denkert C, Loibl S, Noske A, Roller M, Muller BM, Komor M, Budczies J, Darb-Esfahani S, Kronenwett R, Hanusch C, et al. Tumor-associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J Clin Oncol. 2010;28:105–113. doi: 10.1200/JCO.2009.23.7370. [DOI] [PubMed] [Google Scholar]
- 63.Tosolini M, Kirilovsky A, Mlecnik B, Fredriksen T, Mauger S, Bindea G, Berger A, Bruneval P, Fridman WH, Pages F, et al. Clinical impact of different classes of infiltrating T cytotoxic and helper cells (Th1, th2, treg, th17) in patients with colorectal cancer. Cancer Res. 2011;71:1263–1271. doi: 10.1158/0008-5472.CAN-10-2907. [DOI] [PubMed] [Google Scholar]
- 64.Grivennikov SI, Wang K, Mucida D, Stewart CA, Schnabl B, Jauch D, Taniguchi K, Yu GY, Osterreicher CH, Hung KE, et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumor growth. Nature. 2012;491:254–258. doi: 10.1038/nature11465. [This study shows that adenoma invasion by microbial products triggers tumor-elicited inflammation, which in turn drives tumor growth.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer. 2013;13:800–812. doi: 10.1038/nrc3610. [DOI] [PMC free article] [PubMed] [Google Scholar]