Abstract
Resilience to host inflammation and other perturbations is a fundamental property of gut microbial communities, yet the underlying mechanisms are not well understood. We have found that human gut microbes from all dominant phyla are resistant to high levels of inflammation-associated antimicrobial peptides (AMPs) and have identified a mechanism for lipopolysaccharide (LPS) modification in the phylum Bacteroidetes that increases AMP resistance by four orders of magnitude. Bacteroides thetaiotaomicron mutants that fail to remove a single phosphate group from their LPS were displaced from the microbiota during inflammation triggered by pathogen infection. These findings establish a mechanism that determines the stability of prominent members of a healthy microbiota during perturbation.
Human gut microbial communities reside in an open ecosystem subject to disruptions ranging from dietary change to toxin exposure and pathogen invasion. Many of these perturbations, and functional disorders such as irritable bowel syndrome, are accompanied by nonspecific immune responses that impart a disruptive influence on community structure and function (1, 2). Host inflammatory mechanisms to remove harmful organisms and restrict bacteria to the lumen commonly target conserved molecular patterns found on pathogens and commensals alike, yet healthy gut microbial communities can remain stable for years in humans (3).
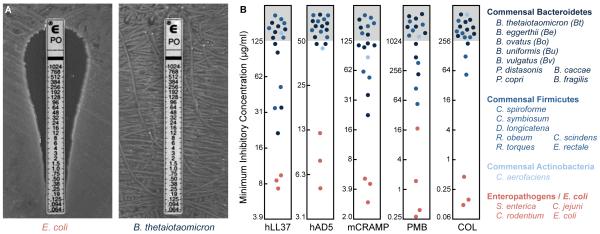
Because cationic antimicrobial peptides (AMPs) represent one of the most prominent and nonspecific components of the host response to pathogens, we determined the minimum inhibitory concentration (MIC) of polymyxin B (PMB) for a common human gut commensal, Bacteroides thetaiotaomicron. PMB is a bacterial cationic AMP with a similar mode of action to mammalian AMPs (4). Strikingly, this commensal exhibits 680- to 2400-fold increased resistance to PMB by comparison with mammalian enteropathogens or Escherichia coli (Fig. 1A, fig. S1, and table S1). A survey of 17 prominent human commensals, representing the three major bacterial phyla in the gut (5), revealed that resistance against multiple inflammation-associated human and murine AMPs is a general feature of the human gut microbiota (Fig. 1B, fig. S1, and table S1).
Fig. 1. Human gut commensals are highly resistant to cationic AMPs.
(A) MIC of PMB against Escherichia coli and Bacteroides thetaiotaomicron. (B) MICs of human and mouse inflammation-induced AMPs and bacterial surrogates against prominent human gut commensal bacteria (blue) and enteropathogens (red). MICs of PMB and colistin were determined using E-test strips; others were determined using the microtiter broth dilution method. See also fig. S1 and table S1.
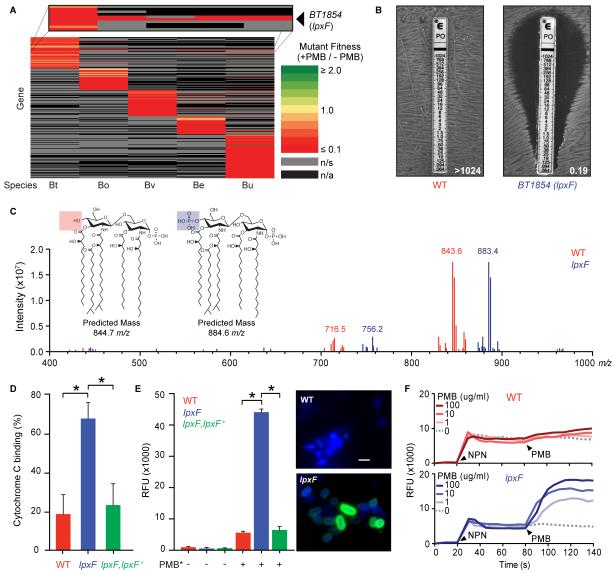
We screened transposon mutant populations of five human gut commensal species for genes required for fitness in the presence of PMB. This approach identified a single gene (encoded by BT1854 in B. thetaiotaomicron) that mediated PMB resistance in all species tested (Fig. 2A, table S2-4). Targeted deletion and complementation of BT1854 confirmed this phenotype for multiple AMPs (Fig. 2B and table S1).
Fig. 2. Lipid A dephosphorylation mediates AMP resistance in prominent human gut bacteria.
(A) Heat map of the relative fitness of transposon mutant strains in the presence and absence of PMB. Columns indicate human gut bacterial species tested (abbreviations from Fig. 1B); rows indicate gene orthologs with insertions displaying a significantly altered fitness (q<0.05) in the presence of PMB in at least one species. n/s, not significantly altered; n/a, no ortholog. (B) BT1854 (LpxF) is necessary for PMB resistance in B. thetaiotaomicron. Representative MIC assessed using the E-test method is shown. (C) LpxF is required for dephosphorylation of lipid A. FT-ICR MS analysis of lipid A isolated from wildtype (red) and lpxF deletion mutant (blue) B. thetaiotaomicron revealed a shift of the predominant peak by ~40 m/z, consistent with the gain of a single phosphate group. Inset shows predicted structures and m/z values of the doubly deprotonated ([M-2H]2−) ions for monophosphorylated (red) and bis-phosphorylated (blue) penta-acylated lipid A. Minor peaks are consistent with tetra-acylated lipid A structures (fig. S2C-D). (D) Deletion of lpxF increases cationic cytochrome C binding to B. thetaiotaomicron, indicating altered surface charge. (E) Increased PMB-Oregon Green (PMB*) binding to lpxF deletion mutant cells measured by fluorescence quantification (left panel) and microscopy (right panels). Error bars represent standard deviation and asterisks indicate significance (p<0.01). Representative fluorescence microscopy images of bacterial cells incubated with PMB* (green) are shown as an overlay with 4′, 6′ diamino-2-phenylindole (DAPI) (blue). Scale bar indicates 2 μm. (F) LpxF protects the outer membrane from PMB perturbation. 1-N-phenylnaphthylamine (NPN) uptake profiles of select strains were measured followed by challenge with the indicated concentration of PMB. Arrowheads indicate the addition of NPN (20s) and PMB (80s). Readings were taken in 5s intervals. Each experiment was performed in triplicate with representative results shown. See figure S3 for complementation.
BT1854 has limited homology to the phospholipid biosynthetic enzyme PgpB, a phosphatidylglycerol phosphatase (6) that is not implicated in AMP resistance. However, an unusual PgpB homolog, LpxF, catalyzes the removal of the negatively charged 4′-phosphate group from the lipopolysaccharide (LPS) lipid A anchor in a small, phylogenetically heterogeneous group of AMP-resistant pathogens (7-9). Notably, B. thetaiotaomicron produces an under-phosphorylated lipid A structure when compared to most gram-negative organisms (10) (fig. S2A-B).
Mass spectrometry (MS) analysis of lipid A extracted from B. thetaiotaomicron revealed a predominant peak at m/z 843.6, consistent with the published (10) doubly-deprotonated ([M-2H]2−) ion structure of penta-acylated 4′-dephosphorylated lipid A (predicted exact mass: 844.7m/z) (Fig. 2C). In contrast, MS analysis of lipid A extracted from the B. thetaiotaomicron BT1854 deletion mutant revealed a peak consistent with the (M-2H)2− ion of penta-acylated, bis-phosphorylated lipid A (predicted exact mass: 884.6m/z) (Fig. 2C). An additional minor peak consistent with a tetra-acylated structure is observed for all strains (Fig. 2C and fig. S2C-D). Complementation restores the wildtype lipid A profile (fig. S2E). Based on these results, we designated this protein as LpxF.
Characterization of wildtype and lpxF mutant strains showed that LpxF increased resistance to inflammation-associated AMPs by neutralizing the negative charge of the cell (Fig. 2D), decreasing AMP binding at the bacterial surface (Fig. 2E and fig. S3A), and reducing AMP-dependent membrane disruption (Fig. 2F and fig. S3B).
If resistance to inflammation-associated AMPs determines Bacteroidetes fitness in the gut, then a strain with reduced AMP resistance should be outcompeted by an isogenic wildtype strain in a mammalian host during inflammation. We established a gnotobiotic model of gut inflammation by colonizing germfree mice with a mixture of wildtype, lpxF deletion mutant, and complemented (lpxF,lpxF+) B. thetaiotaomicron strains 7d prior to infection with Citrobacter rodentium, a murine enteropathogen that mimics human gastrointestinal infection (11), or an avirulent tir mutant incapable of inducing colitis (fig. S4A-C) (12).
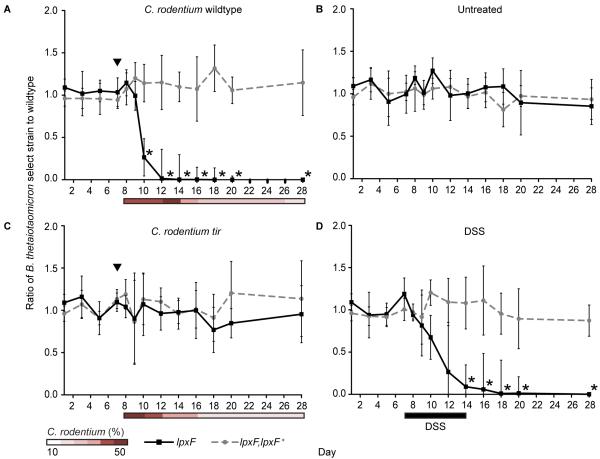
We next monitored the relative abundance of each strain over time in these animals. The B. thetaiotaomicron lpxF deletion mutant was rapidly displaced by wildtype and lpxF,lpxF+ complemented strains in mice infected with C. rodentium (Fig. 3A and table S5). Displacement occurred at the onset of inflammation (fig. S5A and S5B) and concurrent with increased AMP secretion measured from colonic tissue explants (fig. S5C). By contrast, the lpxF deletion mutant was able to persist in mice that had not been infected with C. rodentium (Fig. 3B) and in mice infected with the non-inflammatory C. rodentium tir mutant, which colonizes the gut to similar levels as the wildtype pathogen (Fig. 3C). The lpxF mutant was also outcompeted in gnotobiotic mice exposed to dextran sulfate sodium (DSS), a chemical inducer of intestinal inflammation, at the onset of inflammation (Fig. 3D, fig. S4A-C, and fig. S5). Exposure in vitro to numerous AMPs, but not C. rodentium or DSS, recapitulates the in vivo results (fig. S4D-E). LpxF thus appears to be a general determinant of resilience during inflammation and not a specific requirement for response to C. rodentium.
Fig. 3. AMP resistance determines the resilience of a prominent human gut symbiont in gnotobiotic mice with colitis.
Germfree mice (n=5/group) were colonized with wildtype, the lpxF deletion mutant (solid lines), and complemented (lpxF,lpxF+; dashed lines) B. thetaiotaomicron strains 7 days prior to initiation of inflammation by C. rodentium infection (A), no further treatment (B), infection with a non-inflammatory C. rodentium tir mutant (C), or exposure to 3% DSS ad libitum for 7 days (black bar) (D). Infection with C. rodentium strains is indicated by black arrowheads and relative C. rodentium abundances are reported as percent of total fecal DNA (shaded bars); error bars represent standard deviation and asterisks indicate significant (p<0.01) differences.
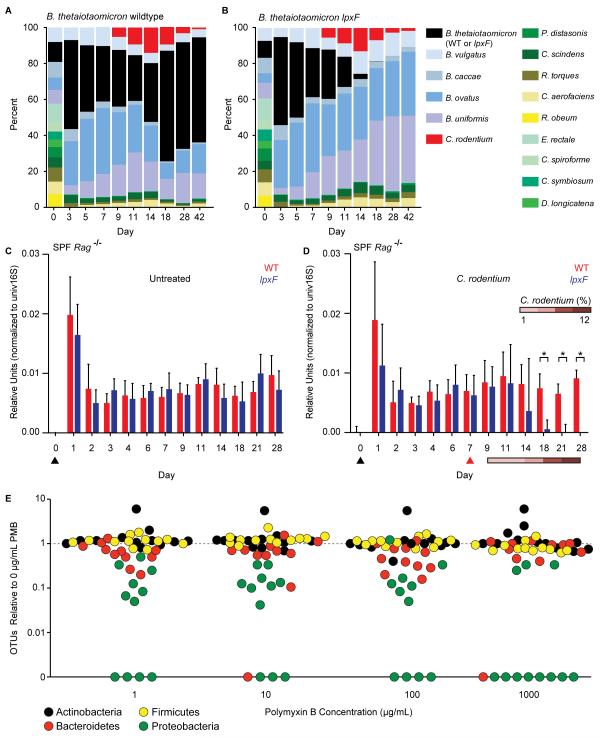
If resistance to AMPs is also important for resilience in the context of a multi-phylum human gut microbiota residing in the mammalian gut environment, then a mutant sensitive to AMPs should exhibit a loss of resilience in this community. We colonized germfree mice with 14 bacterial species, including wildtype B. thetaiotaomicron, that represent the three dominant phyla observed in the human gut (tables S6-S8) (13). Strikingly, this community remained largely stable through the course of C. rodentium infection (Fig. 4A) and associated inflammatory events. To determine whether LpxF is required for this stability during perturbation, we replaced B. thetaiotaomicron with the lpxF deletion mutant. Unlike wildtype B. thetaiotaomicron, the lpxF mutant was rapidly and specifically displaced from the defined human gut microbiome upon C. rodentium infection and consequent host inflammation (Fig. 4B and fig. S6A). By contrast, in the absence of C. rodentium infection both the B. thetaiotaomicron wildtype and lpxF mutant-containing communities remained stable for the duration of the experiment (fig. S6B).
Fig. 4. AMP resistance determines commensal resilience in the context of a human or murine gut microbiota.
(A, B) Germfree mice (n=5/group) were colonized with 14 prominent human gut microbes, including B. thetaiotaomicron wildtype or lpxF deletion mutant, 7 days prior to infection with C. rodentium; community composition was monitored by species-specific quantitative polymerase chain reaction (qPCR) and reported as median percent of total. Uninfected controls are shown in fig. S6B. (C, D) Specific pathogen free Rag−/− mice (n=5/group) were gavaged (black arrowhead) with either B. thetaiotaomicron wildtype or lpxF deletion mutant; in (D), mice were infected with C. rodentium 7 days later (red arrowhead). Colonization levels were assessed by qPCR from fecal DNA. C. rodentium levels are reported percentage of total fecal DNA. Error bars indicate standard deviation and asterisks indicate significance (p<0.05). (E) AMP resistance is a general feature of human gut Bacteroidetes, Firmicutes, and Actinobacteria. Fecal samples from 12 unrelated, healthy human donors were cultured on varying PMB concentrations (x-axis) and the number of species-level phylotypes (OTUs) belonging to each phylum observed from each donor (points, colored by phylum) was normalized to the number observed in culture in the absence of PMB. A weighted (abundance) analysis provides similar results (fig. S8B).
Germfree mice have an immature innate immune system (14) that is largely rescued by colonization with mouse, but not human, microbiota (15). We tested whether B. thetaiotaomicron requires LpxF in animals with a complete mouse microbiota and a mature innate immune system. Because human gut Bacteroides spp. are rapidly outcompeted in specific pathogen-free (SPF) wildtype mice (16), we screened multiple strains of mice and found that B. thetaiotaomicron, like B. fragilis (16), stably colonized SPF Rag−/− animals (Fig. 4C). In these mice, the B. thetaiotaomicron lpxF deletion mutant colonized to equivalent levels as the wild type (Fig. 4C). B. thetaiotaomicron resilience during C. rodentium infection of these SPF mice remained dependent on LpxF, indicating that the inflammation caused by pathogen infection, rather than other members of the commensal mouse microbiota, mediated the requirement for AMP resistance (Fig. 4D). Many innate immune defects observed in germfree mice are also abrogated by segmented filamentous bacteria (SFB), a mouse commensal that interacts directly with the gut epithelium (15). The lpxF deletion mutant remained stable over time in gnotobiotic mice monoassociated with SFB, indicating that even a pure population of immunostimulatory commensal members of the host’s native microbiota do not induce a requirement for AMP resistance (fig. S6C).
LpxF orthologs are readily identified in all sequenced human-associated Bacteroidetes and all characterized LPS structures in this phylum reveal an under-phosphorylated lipid A structure (fig. S7), suggesting that all human gut Bacteroidetes use this mechanism to resist inflammatory perturbations. We conducted a human study to determine whether our observations from sequenced type strains extend to gut commensals captured directly from humans. To this end, we cultured 733 species-level phylotypes from 12 unrelated, healthy donors (table S9). These cultured strains represent ~98% of the donors’ original, uncultured communities at the phylum level, ~95% at the class and order levels, ~80% at the family level, and ~50% at the genus level (fig. S8A). Quantification of PMB resistance across these culture collections established that the AMP resistance profiles of fecal microbial communities isolated directly from humans mirror the phylum-level patterns observed in sequenced type strains (Fig. 4E, fig. S8B). Understanding mechanisms of microbiota stability is important for efforts to manipulate these communities for therapeutic purposes. We found that the ability of prominent human gut commensal bacteria to resist killing by AMPs also mediated stability during infection. This resilience hinges on a protein that removes a single phosphate group from the bacterial LPS to determine whether the host maintains or removes commensal bacteria in response to inflammation. In this way, commensal-encoded mechanisms for persistence in the host during inflammation complement host-encoded mechanisms for immune tolerance of the microbiota (17). As observed in certain pathogens, lipid A modification may provide additional benefits to commensal microbes beyond AMP resistance, including reduced activation of the host Toll-like receptor 4-myeloid differentiation factor 2 (TLR4-MD2) complex that recognizes common forms of bacterial LPS (7, 8, 10, 18-23). Our studies indicate that LpxF is dispensable in the absence of inflammation, suggesting that the broad conservation of this enzyme across commensal Bacteroidetes reflects selective pressures imposed by periodic inflammatory events. A delicate balance between microbial resilience and host tolerance thus allows for commensal persistence throughout a diverse range of perturbations while preventing commensal overgrowth or depletion, either of which could have deleterious effects on the host.
Supplementary Material
Acknowledgements
We thank E. Groisman, J. Galan, and the Goodman lab for helpful suggestions, C. Jacobs-Wagner for microscopy assistance, J. Leong for C. rodentium strains, and I. Ivanov for SFB. This work was supported by NIH grants DK089121, GM103574, GM105456, the Global Probiotics Council, and the Crohn’s and Colitis Foundation of America to A.L.G; NIH grants AI064184 and AI76322 to M.S.T., and Army Research Office grant W911NF-12-1-0390 to M.S.T. All human studies were conducted with approval from the Yale University Human Investigation Committee and the sequences reported in this paper have been deposited with the European Bioinformatics Institute under accession number PRJEB7697.
Footnotes
Supplementary Materials www.sciencemag.org Materials and Methods
References and Notes
- 1.Langhorst J, et al. Elevated human beta-defensin-2 levels indicate an activation of the innate immune system in patients with irritable bowel syndrome. Am. J. Gastroenterol. 2009;104:404–410. doi: 10.1038/ajg.2008.86. [DOI] [PubMed] [Google Scholar]
- 2.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Faith JJ, et al. The long-term stability of the human gut microbiota. Science. 2013;341:44–52. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vaara M. Agents that increase the permeability of the outer membrane. Microbiol. Rev. 1992;56:395–411. doi: 10.1128/mr.56.3.395-411.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Icho T, Raetz CR. Multiple genes for membrane-bound phosphatases in Escherichia coli and their action on phospholipid precursors. J. Bacteriol. 1983;153:722–730. doi: 10.1128/jb.153.2.722-730.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cullen TW, et al. Helicobacter pylori versus the host: remodeling of the bacterial outer membrane is required for survival in the gastric mucosa. PLoS Pathog. 2011;7:e1002454. doi: 10.1371/journal.ppat.1002454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coats SR, To TT, Jain S, Braham PH, Darveau RP. Porphyromonas gingivalis resistance to polymyxin B is determined by the lipid A 4′-phosphatase, PGN_0524. Int. J. Oral. Sci. 2009;1:126–135. doi: 10.4248/IJOS.09062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X, McGrath SC, Cotter RJ, Raetz CR. Expression cloning and periplasmic orientation of the Francisella novicida lipid A 4′-phosphatase LpxF. J. Biol. Chem. 2006;281:9321–9330. doi: 10.1074/jbc.M600435200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coats SR, et al. The lipid A phosphate position determines differential host Toll-like receptor 4 responses to phylogenetically related symbiotic and pathogenic bacteria. Infect. Immun. 2011;79:203–210. doi: 10.1128/IAI.00937-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mundy R, MacDonald TT, Dougan G, Frankel G, Wiles S. Citrobacter rodentium of mice and man. Cell. Microbiol. 2005;7:1697–1706. doi: 10.1111/j.1462-5822.2005.00625.x. [DOI] [PubMed] [Google Scholar]
- 12.Mallick EM, et al. Allele- and tir-independent functions of intimin in diverse animal infection models. Front. Microbiol. 2012;3:11. doi: 10.3389/fmicb.2012.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodman AL, et al. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe. 2009;6:279–289. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chung H, et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149:1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee SM, et al. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature. 2013;501:426–429. doi: 10.1038/nature12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vaishnava S, et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rietschel ET, et al. Bacterial endotoxin: molecular relationships of structure to activity and function. FASEB J. 1994;8:217–225. doi: 10.1096/fasebj.8.2.8119492. [DOI] [PubMed] [Google Scholar]
- 19.Cullen TW, Trent MS. A link between the assembly of flagella and lipooligosaccharide of the gram-negative bacterium Campylobacter jejuni. Proc. Natl. Acad. Sci. U. S. A. 2010;107:5160–5165. doi: 10.1073/pnas.0913451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Que-Gewirth NL, et al. A methylated phosphate group and four amide-linked acyl chains in leptospira interrogans lipid A. The membrane anchor of an unusual lipopolysaccharide that activates TLR2. J. Biol. Chem. 2004;279:25420–25429. doi: 10.1074/jbc.M400598200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanistanon D, et al. A Francisella mutant in lipid A carbohydrate modification elicits protective immunity. PLoS Pathog. 2008;4:e24. doi: 10.1371/journal.ppat.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phillips NJ, Schilling B, McLendon MK, Apicella MA, Gibson BW. Novel modification of lipid A of Francisella tularensis. Infect. Immun. 2004;72:5340–5348. doi: 10.1128/IAI.72.9.5340-5348.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Munford RS. Sensing gram-negative bacterial lipopolysaccharides: a human disease determinant? Infect. Immun. 2008;76:454–465. doi: 10.1128/IAI.00939-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.