Abstract
The canonical Wnt/β-catenin pathway is activated in triple-negative breast cancer (TNBC). The activation of this pathway leads to the expression of specific target genes depending on the cell/tissue context. Here, we analyzed the transcriptome of two different TNBC cell lines to define a comprehensive list of Wnt target genes. The treatment of cells with Wnt3a for 6h up-regulated the expression (fold change > 1.3) of 59 genes in MDA-MB-468 cells and 241 genes in HCC38 cells. Thirty genes were common to both cell lines. Beta-catenin may also be a transcriptional repressor and we found that 18 and 166 genes were down-regulated in response to Wnt3a treatment for 6h in MDA-MB-468 and HCC38 cells, respectively, of which six were common to both cell lines. Only half of the activated and the repressed transcripts have been previously described as Wnt target genes. Therefore, our study reveals 137 novel genes that may be positively regulated by Wnt3a and 104 novel genes that may be negatively regulated by Wnt3a. These genes are involved in the Wnt pathway itself, and also in TGFβ, p53 and Hedgehog pathways. Thorough characterization of these novel potential Wnt target genes may reveal new regulators of the canonical Wnt pathway. The comparison of our list of Wnt target genes with those published in other cellular contexts confirms the notion that Wnt target genes are tissue-, cell line- and treatment-specific. Genes up-regulated in Wnt3a-stimulated cell lines were more strongly expressed in TNBC than in luminal A breast cancer samples. These genes were also overexpressed, but to a much lesser extent, in HER2+ and luminal B tumors. We identified 72 Wnt target genes higher expressed in TNBCs (17 with a fold change >1.3) which may reflect the chronic activation of the canonical Wnt pathway that occurs in TNBC tumors.
Introduction
Breast cancer is one of the most common tumors in women. It is a complex, heterogeneous disease comprising several subgroups of pathologies with different patient outcomes [1–3]. Triple-negative breast cancer (TNBC), closely related to basal-like breast cancer (BLBC), is characterized by an absence of estrogen receptor (ER) and progesterone receptor (PR) expression and a lack of human epidermal growth factor receptor 2 (HER2) overexpression/amplification. TNBC itself constitutes a heterogeneous group of breast cancer [4–6], which is highly proliferative and genetically instable, and associated with a poor prognosis. Unlike other breast cancer subtypes, such as luminal (expressing ER and PR) and HER2-overexpressing (HER2+) tumors, TNBC cannot be treated with targeted therapies, such as tamoxifen or anti-HER2 antibodies. TNBC patients are therefore treated exclusively with conventional cytotoxic therapies, but about half of them present relapse and metastasis within the first three to five years after treatment [7]. Therefore, treatment of patients with TNBC remains a major challenge for oncologists and alternative treatments to conventional chemotherapies are needed to improve their survivals.
The Wnt signaling pathway mediates biological processes such as cell adhesion, migration, proliferation, differentiation and survival [8–10]. It consists of two main arms: the canonical (Wnt/β-catenin) and the non-canonical pathways, which differ in terms of their dependence on β-catenin [11,12]. The activation of the canonical Wnt pathway leads to the stabilization of β-catenin which translocates to the nucleus and induces the expression of Wnt target genes. Besides its function in normal cells/tissues, Wnt signaling can become deregulated during human disease. The best documented example is the tumorigenesis of colorectal cancer [13]. The Wnt/β-catenin pathway is also activated in human breast cancer, in particular in the TNBC/BLBC breast cancer subtype that is associated with poor prognosis [14]. Indeed, the activated form of β-catenin has been observed in breast cancer [15–18], and is frequently found in the TNBC/BLBC subtype [14,19–23]. The aberrant activation of the Wnt/β-catenin pathway in mice leads to mammary carcinogenesis [24], and transgenic mice expressing a constitutively active form of β-catenin in the mammary gland develop basal-like tumors [25], suggesting a crucial role for the canonical Wnt pathway in TNBC/BLBC tumorigenesis. Mutations of genes encoding intracellular components of the canonical pathway, including APC (encoding adenomatous polyposis coli), CTNNB1 (encoding β-catenin) and AXIN, are frequent in colorectal and hepatocellular cancers [11,12,26], but are rare in breast cancer [22–24,27–30]. Instead, deregulated expression of cell surface components such as LRP6 or FZD7 transmembrane receptors may be responsible for the activation of the Wnt pathway in TNBC/BLBC [14,19–23].
Nuclear localization of β-catenin and the expression of Wnt target genes reflect the activation of the Wnt/β-catenin pathway. Nevertheless, the detection of nuclear β-catenin is experimentally challenging and dependent on the tissues and the cell lines tested. An alternative way to evaluate the activation of the canonical Wnt pathway is to measure the expression of Wnt/β-catenin target genes. However, Wnt target genes vary substantially depending on the cellular/tissue context. The exceptions are AXIN2 and NKD1 which are considered as universal Wnt target genes [12]. The role of β-catenin in the transcriptional activation of its target genes is well documented. However, recent studies have also reported a link between β-catenin and transcriptional repression, which is an underestimated aspect of the Wnt signaling [31–33]. Several methodological approaches have been undertaken to identify Wnt target genes in different cellular or tissue contexts: stimulation of cells with Wnt3a or Wnt1 ligand (recombinant protein or plasmid) [34,35]; depletion of β-catenin (siRNA) [36] or Tcf (dominant negative construct) [37]; overexpression of active β-catenin (plasmid) [36]; evaluation of Wnt signaling activity (nuclear staining of β-catenin [38,39], mutations of the CTNNB1 gene [40]); and screening to identify binding sites for the Tcf transcription factor in DNA sequences [34]. Wnt target genes have been mostly examined in colon and also in ovarian and liver cancers [37–39], but are not frequently examined in breast cancer. Some Wnt target genes are components of the Wnt pathway itself. Such targets are mostly inhibitors (e.g., AXIN2 and NKD1) and probably prevent the uncontrolled activation of the pathway through negative feedback loops. Therefore, the characterization of β-catenin target genes in breast tissue may lead to the discovery of new regulators of the Wnt pathway and improve our understanding of TNBC tumorigenesis. We thus used microarrays to investigate the expression of 19,738 transcripts following Wnt3a stimulation in two TNBC cell lines, HCC38 and MDA-MB-468, and we report a comprehensive list of genes that are activated or repressed in breast cancer by Wnt3a. Pathway analysis revealed that the Wnt target genes were mainly associated with the Wnt, TGFβ, p53 and Hedgehog pathways. The comparison of our list of Wnt target genes with those previously identified in fibroblast and epithelial cell lines confirms that the identity of Wnt target genes is highly dependent on the cellular/tissue context. We examined the expression level of our lists of Wnt target genes in 130 human breast tumors. We found that 72 Wnt target genes (17 with a fold change > 1.3) may reflect the activation of the canonical Wnt pathway in a more chronic situation.
Materials and Methods
Triple-negative cell lines
BT20, BT549, HCC38, HCC70, HCC1187, HCC1937, MDA-MB-157, MDA-MB-231 and MDA-MB-468 cells were purchased in May 2006 and May 2008 from the American Type Culture Collection (LGC Standards, Molsheim, France). Cells were characterized by DNA and RNA microarrays [41,42] and authenticated in 2013 by short tandem repeat profiling (data not shown).
BT20 cells were cultured in MEM(Eagle) (Sigma, Saint Quentin Fallavier, France) containing 1% Glutamax (Invitrogen), 10% fetal bovine serum (FBS, Invitrogen, Cergy Pontoise, France), 1.5g/L sodium bicarbonate (Invitrogen), 0.1mM non-essential amino-acids (Invitrogen) and 1mM sodium pyruvate (Invitrogen). BT549 and MDA-MB-468 cells were maintained in RPMI-1640 containing Glutamax (Invitrogen) and supplemented with 10% FBS. HCC38, HCC70, HCC1187, HCC1937 cells were cultured in RPMI-1640 with Glutamax containing 10% FBS, 1.5g/L sodium bicarbonate, 10mM Hepes (Invitrogen) and 1mM sodium pyruvate. MDA-MB-157 cells were maintained in Leibovitz's L-15 medium containing Glutamax (Invitrogen) and supplemented with 10% FBS and 10mM Hepes. MDA-MB-231 cells were cultured in DMEM-F12 with Glutamax (Invitrogen) containing 10% FBS. Antibiotics were added to all media (100U/mL penicillin and 100μg/mL streptomycin). Cells were cultured at 37°C in a 5% CO2 humidified incubator. For all experiments, cells were used up to the 15th passage after thawing.
Transfection of plasmid DNA
The Wnt-responsive element-luciferase reporter WRE was kindly provided by the Galapagos company (Romainville, France), the mutant-responsive element-luciferase variant MRE by Ron Smits (Rotterdam, The Netherlands) [43] and the pRL-TK plasmid by the Servier company (Croissy-sur-Seine, France). The construct pRK5-SK-β-cateninΔGSK and empty vector were obtained from René Bernards (Ultrecht, The Netherlands) [44] and Maria Carla Parrini (Institut Curie Paris, France), respectively.
FuGENE HD reagent (Promega, Charbonnières-les-Bains, France) was used to transiently transfect cells with plasmids according to the manufacturer’s recommendations.
Compounds
Recombinant human Wnt3a (R&D Systems, Lille, France) was reconstituted at 10μg/mL in PBS containing 0.1% BSA, then used in experiments at a final concentration of 100ng/mL. This concentration of Wnt3a is routinely used to activate the Wnt pathway in mammary cells, in particular in TNBC cell lines [35,45–48]. We have shown that this concentration of Wnt3a leads to the activation of the Wnt signaling pathway (S1 Fig).
Antibodies
Primary antibodies used were mouse monoclonal anti-β-catenin (clone 14/beta-catenin, BD Transduction Laboratories, Le Pont de Claix, France; 1:1,000) or anti-active-β-catenin (clone 8E7, Millipore, Molsheim, France; 1:500), rabbit monoclonal anti-LRP5 (clone D5G4, Cell Signaling Technology, Ozyme, Saint Quentin en Yvelines, France; 1:1,000), anti-LRP6 (clone C5C7, Cell Signaling Technology; 1:500), and mouse monoclonal anti-actin (clone AC-15, Sigma; 1:5,000). The secondary antibodies used were horseradish peroxidase-conjugated anti-mouse or anti-rabbit IgG (Jackson ImmunoResearch, Interchim, Montluçon, France; 1:20,000).
SDS-PAGE and western blotting
Cells were lysed in Laemmli buffer containing 50mM Tris (pH 6.8), 2% sodium dodecyl sulfate (SDS), 5% glycerol, 2mM 1,4-dithio-DL-threitol, 2.5mM ethylenediaminetetraacetic acid, 2.5mM ethylene glycol tetraacetic acid, 2mM sodium orthovanadate, 10mM sodium fluoride and a cocktail of protease (Roche) and phosphatase (Pierce, Perbio, Brebières, France) inhibitors. Protein concentration in each sample was determined with the reducing agent compatible version of the BCA Protein Assay kit (Pierce). Equal amounts of total protein were fractionated under reducing conditions by SDS–PAGE and then blotted onto PVDF membranes (Bio-Rad, Marnes-la-Coquette, France). The membranes were blocked with 5% BSA or 10% skimmed milk in TBS containing 0.1% Tween 20 (TBS-T), hybridized with the primary antibody of interest overnight at 4°C. Membranes were washed in TBS-T and then hybridized with the secondary antibody for one hour at room temperature. Antibodies were diluted in TBS-T containing 5% BSA or 10% skimmed milk. After washing with TBS-T, immune complexes on membranes were detected by enhanced chemiluminescence (Amersham, GE Healthcare, Orsay, France). Actin was used as loading control.
Beta-catenin reporter assay
Cells were transiently transfected with either the reporter plasmid WRE (containing Tcf-binding sites driving the transcription of the firefly luciferase enzyme) or the mutant variant MRE (containing inactive Tcf-binding sites) as described above. Co-transfection with pRL-TK, which encodes a Renilla luciferase gene downstream from a minimal HSV-TK promoter, was systematically performed to normalize for transfection efficiency. Eight hours after transfection, cells were washed and cultured overnight in culture medium without serum, then Wnt3a was added for 3, 6, 9 or 12 hours.
For experiments in which the construct pRK5-SK-β-cateninΔGSK or pRK5-SK was used, cells were co-transfected with WRE and pRL-TK or MRE and pRL-TK. Under these conditions, cells were cultured for 6, 12 or 24 hours.
After treatment (Wnt3a stimulation or overexpression of active β-catenin), cells were lysed, and a luciferase assay was performed with the Dual-Luciferase Reporter Assay Kit (Promega), according to the manufacturer's instructions.
Triplicates for each condition were included in the experiment and the experiment was repeated at least twice.
Microarray analysis in cell lines
Cells were seeded in six-well plates, serum starved overnight, and then treated with Wnt3a for the indicated times (6, 12 or 24 hours). Triplicates for each condition were included in the experiment. Total RNA was extracted with the RNeasy Mini Kit from Qiagen (Courtaboeuf, France) following the manufacturer’s recommendations. After RNA quality and quantity controls, samples were hybridized onto Gene st 1.1 Affymetrix chips. Samples were processed as described on the website of the company. The data were analyzed with the brainarray HuGene11stv1_Hs_ENTREZG version 14 custom chipset definition file for the HuGene11stv1 affymetrix array [49]. The data were first log2 transformed and normalized with RMA [50]. A linear model was fitted with limma [51] including three factors that were treated as fixed effects: cell line (either HCC38 or MDA-MB-468), time (6, 12 or 24 hours) and treatment (Wnt3a stimulated or control) and all possible interaction terms. For each cell line at each time point, the significance of differences between Wnt3a-treated and control cells was determined and a correction for multiple testing was applied with Benjamini & Hochberg’s methodology [52]. Only genes with significantly different expression (P < 0.05), with a log2 fold change superior to 0.3785 (i.e. fold change > 1.3) or inferior to -0.3785 (i.e. fold change < 1.3), were selected. Pathway enrichment analysis was carried out with GeneTrail [53]. A hypergeometric test, corrected for multiple testing with the Benjamini-Hochberg method, was used to assess the significance of the over-representation of biological annotations among gene lists. The transcriptomic data of Wnt3a-treated cells are available in Gene Expression Omnibus (GEO) (accession number: GSE65238).
Quantitative real time reverse transcription-polymerase chain reaction (qRT-PCR)
Cells were seeded in six-well plates, serum starved overnight, and then treated with Wnt3a for 3, 6, 9 or 12 hours. For experiments in which cells were transfected with the construct pRK5-SK-β-cateninΔGSK or the empty vector pRK5-SK, cells were cultured for 12, 24 or 48 hours. Triplicates for each condition were included in the experiment and the experiment was repeated at least three times.
Total RNA was extracted with the RNeasy Mini Kit (Qiagen). For each reaction, 50 to 100ng RNA was combined with components of KAPA SYBR FAST One-Step qRT-PCR Kit (KapaBiosystems, Clinisciences, Nanterre, France) and the Quantitect primers (Qiagen) to a final volume of 20μL, according to instructions of the manufacturers. The reaction mix was subjected to qRT-PCR performed with the 7900HT apparatus and SDS2.4 software (Applied Biosystems) with the following settings: 5 min at 42°C (RT step), 5 min incubation at 95°C followed by a three-step cycling program with 40 cycles of 15 sec at 95°C, 30 sec at 60°C and 30 sec at 72°C (PCR step). A post-PCR dissociation analysis step was included according to instrument guidelines to distinguish specific from non-specific amplification products. All data were normalized to endogenous actin expression.
Tissue samples and microarray data
The human samples used in this study have been previously described [41,42]. RNA microarray (Affymetrix U133 Plus 2.0) performed on 41 TNBC, 30 HER2+, 30 luminal B (LB), 29 luminal A (LA) and 11 healthy tissue breast samples, have also been previously described [41,42] (GEO accession number: GSE65216).
Analysis of the enrichment of the up- and down-regulated Wnt target genes in human breast tumor samples
We analyzed whether the Wnt target genes we identified were enriched in human tumor samples. We compared the gene expression data obtained in TNBC cell lines stimulated with Wnt3a with that obtained in our cohort of 130 human breast cancer samples [41,42]. The two experiments were not done with the same arrays: the cell line (Gene st 1.1, Affymetrix) and the tumor (U133 Plus 2.0, Affymetrix) experiments contained 19738 and 11543 genes, respectively. We restricted our study to the 11262 genes present on both arrays. Of note, AXIN2 was not found in our tumor dataset. We applied a FDR (False Discovery Rate) cut-off of 0.05 and a log-2 fold change threshold of 0.
We used the Fisher exact test to assess the significance of the intersection between the genes that were up- or down-regulated upon the stimulation of cell lines with Wnt3a, and those that were more strongly or more poorly expressed in tumors (TNBC, HER2+, LB) than in LA samples. Several gene sets were considered. For the cell line experiment, we considered the up- or down-regulated genes, the different time points (6, 12 and 24 hours) and the two TNBC cell lines. For the breast tumors, we considered the genes that were more strongly or more poorly expressed in the three different subgroups of tumors (TNBC, HER2+ and LB) than in LA samples. Each subgroup was considered separately and all comparisons between the different breast cancer subtypes were performed.
The comparisons between tumors and cell lines were performed with data obtained in HCC38 and MDA-MB-468 cell lines. However, there was no significant overlap between the MDA-MB-468 and the tumor datasets, possibly because of the very low number of genes found to be differentially expressed in Wnt3a-stimulated MDA-MB-468 cells. Therefore, we focused only on the data obtained with HCC38 cells.
The gene lists were as follows:
MDA-MB-468 cells: 6h up-regulated (66 genes), 12h up-regulated (74 genes), 24h up-regulated (20 genes), 6h down-regulated (34 genes), 12h down-regulated (43 genes), 24h down-regulated (12 genes).
HCC38 cells: 6h up-regulated (419 genes), 12h up-regulated (907 genes), 24h up-regulated (653 genes), 6h down-regulated (256 genes), 12h down-regulated (707 genes), 24h down-regulated (530 genes).
Tumor samples (relative to LA): TNBC up-regulated (3497 genes), HER2+ up-regulated (2110 genes), LB up-regulated (1587 genes), TNBC down-regulated (2971 genes), HER2+ down-regulated (2001 genes), LB down-regulated (1184 genes).
Other lists of genes up- or down-regulated in tumors: TNBC vs HER2+ (up-regulated: 2242 genes; down-regulated: 1865 genes), TNBC vs LB (up-regulated: 2775 genes; down-regulated: 2725 genes), HER2+ vs LB (up-regulated: 581 genes; down-regulated: 581 genes).
In addition, we selected the Wnt target genes that were up-regulated at both the earliest (6h) and the latest time (24h) point after Wnt3a stimulation, to identify potential up-regulated Wnt target genes that could reflect the chronic activation of the Wnt pathway in human cancer. The analysis was performed with the 133 Wnt target genes up-regulated in HCC38 cells, which displayed a higher number of up-regulated genes than in MDA-MB-468 cells (only 13 genes were up-regulated at both 6h and 24h). Of these 133 genes that were up-regulated in HCC38 cells at both 6h and 24h after Wnt3a stimulation, 72 were more strongly expressed in TNBC than in LA tumors. We applied a fold change of > 1.3, which was defined for the cell line experiment, to select the most differently expressed Wnt target genes. Seventeen out of 72 genes were more strongly expressed in TNBC than in LA tumors, at this fold change. We generated a heatmap of the genes ordered by their P value in the t-test.
We used the Fisher exact test to assess the significance of the overlap between two gene lists. For all analyses comparing cell lines and tumors, we used an adjusted (Benjamini Hochberg) P value cut off of 5% and no fold change threshold.
Statistical analyses for in vitro experiments
Data are presented as the mean ± standard deviation (SD). Differences between groups were determined with Student's t-test. Results were considered significant at a P value lower than 0.05.
Results and Discussion
The canonical Wnt pathway is activated in triple-negative breast cancer cell lines
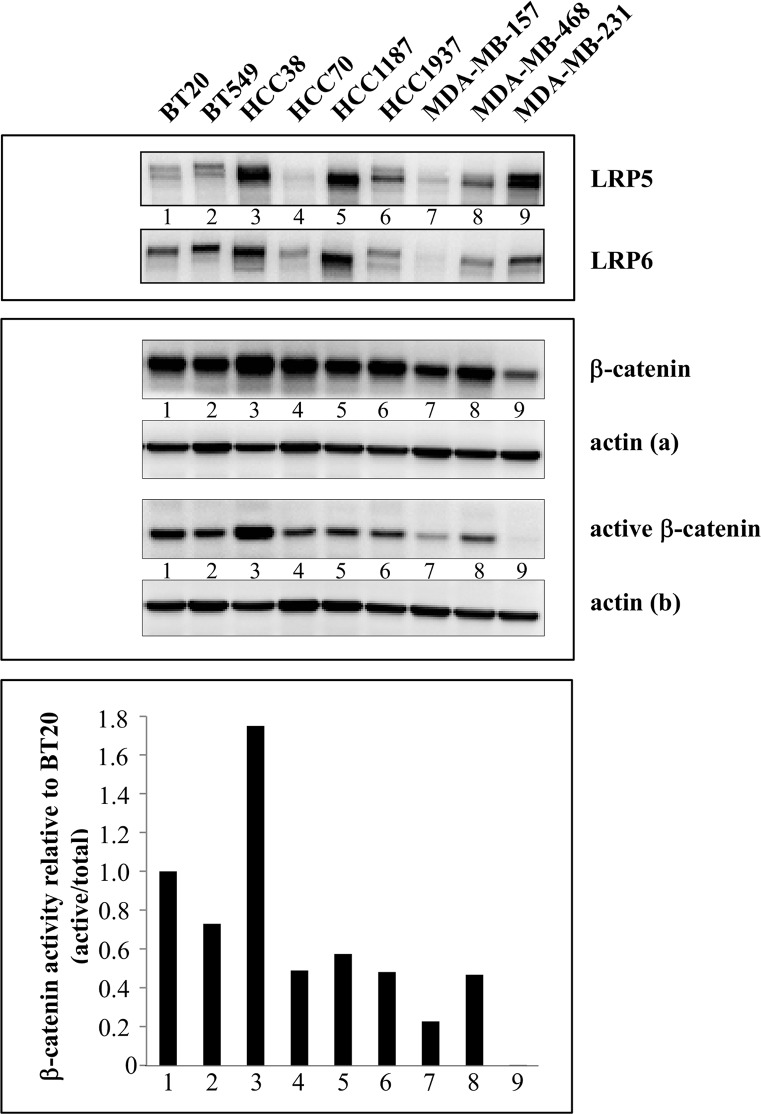
We sought to evaluate Wnt activity in TNBC cell lines; therefore, we measured the abundance of total β-catenin and its unphosphorylated, active form in TNBC cell lines under resting conditions (Fig 1). The abundance of the total and active forms of β-catenin varied in the different cell lines (Fig 1). Except for MDA-MB-231, the canonical Wnt pathway was active in all tested TNBC cell lines in unstimulated conditions, and was most active in HCC38 cells (Fig 1).
Fig 1. Expression of β-catenin, LRP5 and LRP5 in TNBC cell lines.
The abundance of LRP5, LRP6 and β-catenin (total and active forms) was evaluated in different TNBC cell lines by western blotting. Actin was used as a loading control: actin (a) for LRP5 and beta-catenin; actin (b) for LRP6 and active beta-catenin.
We examined whether Wnt3a, a Wnt ligand commonly used to stimulate the canonical Wnt pathway, could stimulate Wnt/β-catenin signaling beyond basal levels observed in HCC38 and MDA-MB-468 cell lines, which displayed differences in their basal Wnt activity (Fig 1). Tcf-mediated transcriptional activity was measured with a reporter plasmid that expresses the luciferase gene under the control of several Tcf binding sites (see Material and Methods section). For both cell lines, Wnt/β-catenin activity was higher than in control cells as early as 3h following the incubation of cells with Wnt3a (S1 Fig). This activation was highest 6h post-treatment and started to decrease at 12h (S1 Fig), indicating that the transcriptional activity of β-catenin is optimal 6 hours following the initial stimulus. Although the transfection of HCC38 cells was not optimal (not shown), luciferase activity was higher in HCC38 cells than in MDA-MB-468 cells (S1 Fig), indicating that Wnt3a activates the Wnt pathway more strongly in HCC38 cells than in MDA-MB-468 cells. This may be because the Wnt3a receptors, LRP5 and LRP6, are more strongly expressed in HCC38 cells than in MDA-MB-468 cells (Fig 1).
Altogether, these results indicate that the canonical Wnt pathway is active in TNBC cell lines, and that Wnt3a further stimulates it.
Genes activated by Wnt3a in HCC38 and MDA-MB-468 triple-negative breast cancer cell lines
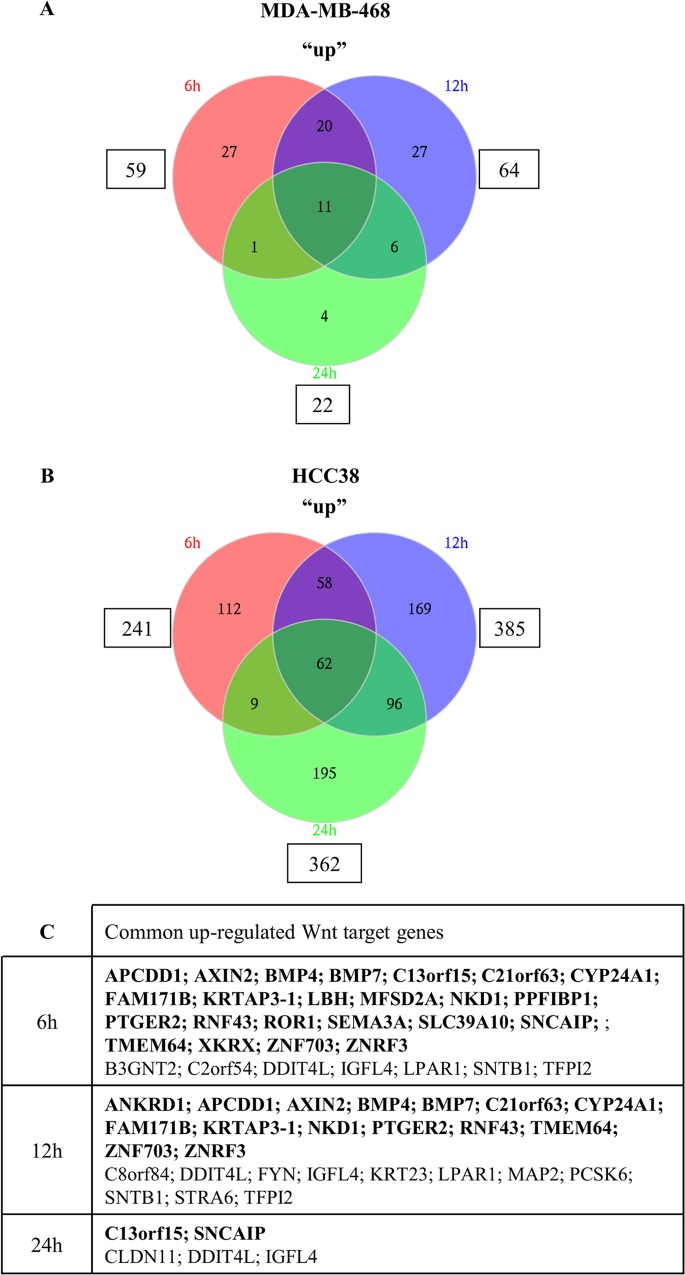
The expression of Wnt target genes appears to be cell/tissue context dependent [31,32]. We therefore performed expression profiling with Gene st 1.1 Affymetrix chips in HCC38 and MDA-MB-468 cell lines stimulated with recombinant Wnt3a to identify a comprehensive list of Wnt target genes in breast cancer cells. A total of 19,738 transcripts were included on the array. The incubation of cells with Wnt3a for 6h, 12h, or 24h up-regulated (fold change > 1.3) the expression of 59, 64, and 22 genes, respectively in MDA-MB-468 cells (Fig 2A, S1 Dataset) and of 241, 385 and 362 genes, respectively in HCC38 cells (Fig 2B, S1 Dataset).
Fig 2. Wnt3a-dependent gene activation in MDA-MB-468 and HCC38 cells.
Venn diagrams indicate the number of genes that were up-regulated in MDA-MB-468 (A) or HCC38 cells (B) following Wnt3a stimulation for 6, 12 or 24 hours. Genes found in common in both cell lines are listed in the table and known Wnt target genes are shown in bold (C).
The difference in the number of up-regulated mRNAs between the two cell lines is consistent with the data obtained with the reporter assay showing that Wnt3a activates the Wnt pathway to a greater extent in HCC38 cells than in MDA-MB-468 cells (S1 Fig). In HCC38 cells, more genes were up-regulated at 12h and 24h after stimulation with Wnt3a than at 6h (Fig 2B, S1 Dataset). These genes may be “direct” or “secondary” Wnt target genes. However, one can postulate that the gene list found at the earliest time point (6h) is likely more enriched in “direct” Wnt target genes than those at later time points (12h, 24h). Therefore, we next focused on the genes up-regulated at 6h. Half of the target genes (30 out of 59) identified in MDA-MB-468 cells incubated for 6h with Wnt3a were also up-regulated in HCC38 cells after the same duration of treatment (Fig 2C, S2 Dataset). Among the 30 common genes, 23 were previously reported as Wnt target genes (Fig 2C, Table 1).
Table 1. Literature search for the 36 Wnt target genes (30 “up” and “6 down”) identified in both TNBC cell lines stimulated with Wnt3a for 6h.
| Gene Symbol (official symbol) | ID | Wnt target gene | Role in Wnt/β-catenin signaling pathway | References |
|---|---|---|---|---|
| APCDD1 | 147495 | yes | inhibitor | [36,40,54–58] |
| AXIN2 | 8313 | yes | inhibitor | [35,37,56–63] |
| B3GNT2 | 10678 | ? | ||
| BMP4 | 652 | yes | ? | [37,38,57,64] |
| BMP7 | 655 | yes | inhibitor | [57,65] |
| C13orf15 (RGCC) | 28984 | yes | ? | [58] |
| C21orf63 (EVA1C) | 59271 | yes | ? | [58] |
| C2orf54 | 79919 | ? | ||
| CYP24A1 | 1591 | yes | ? | [38] |
| DDIT4L | 115265 | ? | ||
| FAM171B | 165215 | yes | ? | [58] |
| IGFL4 | 444882 | ? | ||
| KRTAP3-1 | 83896 | yes | ? | [57] |
| LBH | 81606 | yes | ? | [66] |
| LPAR1 | 1902 | ? | ||
| MFSD2A | 84879 | yes | ? | [58] |
| NKD1 | 85407 | yes | inhibitor | [58,59,67–70] |
| PPFIBP1 | 8496 | yes | ? | [58] |
| PTGER2 | 5732 | yes | ? | [71] |
| RNF43 | 54894 | yes | inhibitor | [37,57,58,72,73] |
| ROR1 | 4919 | yes | ? | [57] |
| SEMA3A | 10371 | yes | ? | [35,37,56,59–63] |
| SLC39A10 | 57181 | yes | ? | [37] |
| SNCAIP | 9627 | yes | ? | [58] |
| SNTB1 | 6641 | ? | ||
| TFPI2 | 7980 | ? | ||
| TMEM64 | 169200 | yes | ? | [58] |
| XKRX | 402415 | yes | ? | [57] |
| ZNF703 | 80139 | yes | ? | [37,58] |
| ZNRF3 | 84133 | yes | inhibitor | [37,57,58,73,74] |
| C10orf81 (PLEKHS1) | 79949 | ? | ||
| CLDN8 | 9073 | ? | ||
| FBXO32 | 114907 | yes | ? | [40,58] |
| PCDH8 | 5100 | ? | ||
| PPP1R3C | 5507 | ? | ||
| TLR1 | 7096 | ? |
As expected, the “universal” Wnt-induced genes AXIN2 and NKD1 [12] were up-regulated in both TNBC cell lines (Fig 2C, S2 Dataset). These two genes showed the highest fold change in HCC38 cells incubated with Wnt3a: 10.2 for NKD1 (P = 2.2x10-23) and 9.86 for AXIN2 (P = 6.4x10-19) (S2 Dataset). In addition to AXIN2 [35,37,56–63] and NKD1 [58,59,68–70], the 21 other previously reported Wnt target genes (Fig 2C, Table 1) that were up-regulated in both cell lines were: APCDD1 [36,40,54–58], BMP4 [37,38,57,64], BMP7 [57], C13orf15 [58], C21orf63 [58], CYP24A1 [38], FAM171B [58], KRTAP3-1 [57], LBH [66], MFSD2A [58], PPFIBP1 [58], PTGER2 [71], RNF43 [37,57,58,72,73], ROR1 [57], SEMA3A [35,37,56,59–63], SLC39A10 [37], SNCAIP [58], TMEM64 [58], XKRX [57], ZNF703 [37,58] and ZNRF3 [37,57,58,73]. Therefore, we report seven novel Wnt target genes among the genes that were up-regulated in both cell lines: B3GNT2, C2orf54, DDIT4L, IGFL4, LPAR1, SNTB1 and TFPI2 (Table 1, Fig 2C). In addition to the 30 genes common to both cell lines at the 6h time point, which could be considered as specific to breast cancer cells, 29 and 211 Wnt target genes were specifically up-regulated in MDA-MB-468 and in HCC38 cells, respectively (Fig 2, S1 Dataset).
Altogether, this analysis revealed 270 genes that are potentially activated by Wnt3a at 6h; 133 (49%) were previously described as Wnt-induced genes and 137 are putative novel Wnt target genes (S3 Dataset).
Genes repressed by Wnt3a in HCC38 and MDA-MB-468 triple-negative breast cancer cell lines
Although β-catenin is most commonly thought of as a transcriptional activator, several reports indicate that it may also be a transcriptional repressor [12,31,33,75]. Few studies have investigated this aspect of the canonical Wnt signaling pathway and it remains poorly understood. Genes down-regulated by Wnt signaling have been detected in human hepatoma cells displaying deregulated Wnt/β-catenin activities [39], in Wnt3a-stimulated human thymocytes, murine C3H10T1/2, NIH3T3 and PC12 cells [56,62,76], in human CTNNB1-mutated Wilms tumors [40], in intestinal tumors from Apc(Min) mice [77], and in a mouse mammary epithelial cell line stimulated with Wnt1 [34]. Some genes repressed by Wnt signaling are also mentioned in the “Wnt Home Page” website (http://www.stanford.edu/group/nusselab/cgi-bin/wnt/target_genes).
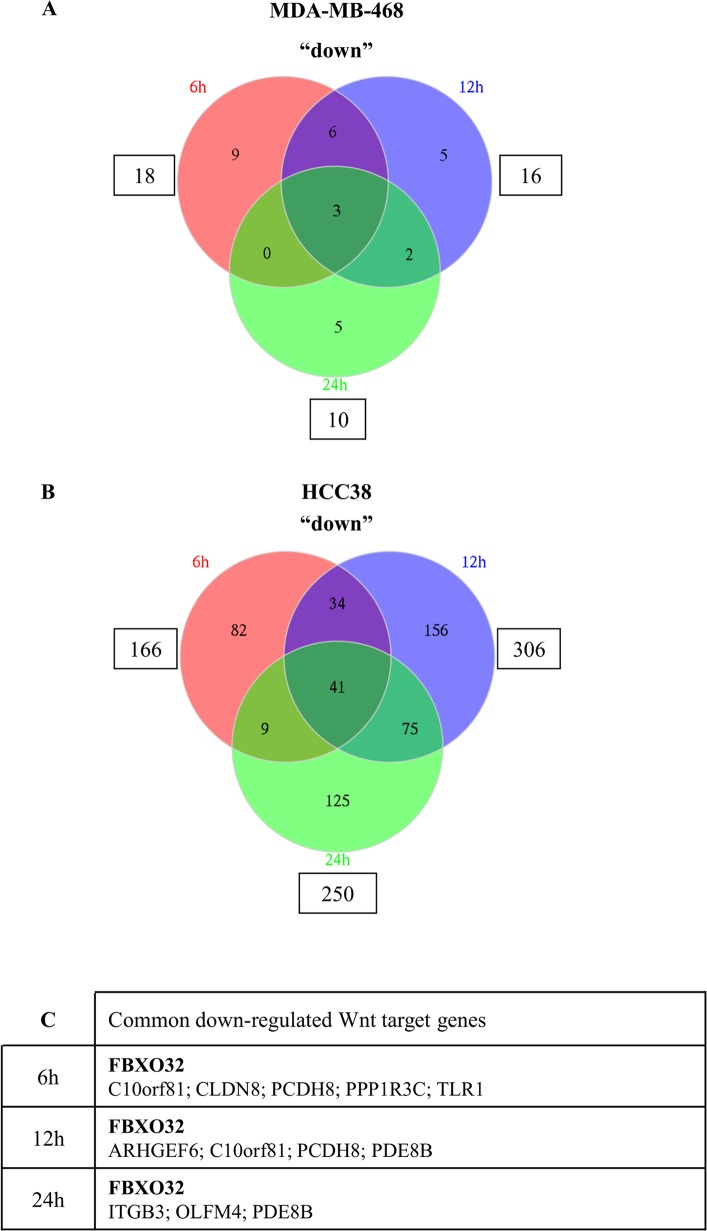
In agreement with these reports, we found that Wnt3a down-regulated the expression of several genes in TNBC cell lines. The incubation of cells with Wnt3a for 6h, 12h, or 24h down-regulated (fold change < 1.3) the expression of 18, 16, and 10 genes, respectively in MDA-MB-468 cells (Fig 3A, S1 Dataset) and 166, 306 and 250 genes, respectively in HCC38 cells (Fig 3B, S1 Dataset).
Fig 3. Wnt3a-dependent gene repression in MDA-MB-468 and HCC38 cells.
Venn diagrams indicate the number of genes that were down-regulated in MDA-MB-468 (A) or HCC38 cells (B) following Wnt3a stimulation for 6, 12 or 24 hours. Genes found in common in both cell lines are listed in the table and known Wnt target genes are shown bold (C).
Wnt3a down-regulated the expression of more genes in HCC38 cells than in MDA-MB-468 cells, consistent with our previous observation that more genes are affected by Wnt3a in HCC38 cells than in MDA-MB-468 cells (Fig 2). One third (six out of 18) of the transcripts down-regulated in MDA-MB-468 cells incubated with Wnt3a for 6h were also down-regulated in HCC38 cells after the same treatment: C10orf81, CLDN8, FBXO32, PCDH8, PPP1R3C and TLR1 (Fig 3C, S2 Dataset). Among them, C10orf81, FBXO32 and PCDH8 were also down-regulated in both cell lines after 12h of Wnt3a treatment (Fig 3C, S2 Dataset). The expression of FBXO32 in stimulated cells after 24h of treatment was still lower than in resting control cells (Fig 3C, S2 Dataset). Except for FBXO32 [40,58], none of the genes down-regulated in both TNBC cell lines have been previously identified as Wnt target genes (Table 1). In addition to these genes, 12 and 160 Wnt target genes were specifically down-regulated at 6h in MDA-MB-468 and in HCC38 cells, respectively (Fig 3, S1 Dataset).
Altogether, 178 potential targets were retrieved from our analysis; only 74 of them (42%) including FBXO32 have been previously described as Wnt target genes (S3 Dataset). Therefore, our study revealed 104 putative novel target genes that are down-regulated in TNBC cells stimulated with Wnt3a (S3 Dataset).
Validation of Wnt target genes in HCC38 and MDA-MB-468 triple-negative breast cancer cell lines
We sought to confirm that the Wnt targets identified by microarray analysis were deregulated in TNBC cells stimulated with Wnt3a; therefore, we examined the expression level of several genes by qRT-PCR. We selected early Wnt target genes (6h) found in both cell lines (“up”: APCDD1, AXIN2, NKD1 and DDIT4L; “down”: PCDH8 and PPP1R3C) (Fig 2C, Fig 3C, S2 Dataset). The magnitude of change in gene expression following Wnt3a treatment differed between qRT-PCR and microarray analysis, but the overall pattern was similar (Table 2 and Table 3, S2 and S3 Figs).
Table 2. qRT-PCR analysis of gene expression in MDA-MB-468 cells treated with Wnt3a.
| Wnt3a treatment | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| 3h | 6h | 9h | 12h | ||||||
| GENE SYMBOL | Fold change | +/- SD | Fold change | +/- SD | Fold change | +/- SD | Fold change | +/- SD | |
| P value | P value | P value | P value | ||||||
| up | APCDD1 | 2.9 * | 0.7 | 3.7 * | 0.9 | 2.7 * | 0.6 | 2.4 * | 0.3 |
| <0.001 | <0.001 | <0.001 | <0.001 | ||||||
| AXIN2 | 6.6 * | 1.5 | 3.5 * | 1.0 | 2.7 * | 0.6 | 2.4 * | 0.6 | |
| <0.001 | <0.001 | <0.001 | <0.001 | ||||||
| DDIT4L | 14.0 * | 5.7 | 9.0 * | 1.9 | 7.0 * | 1.3 | 6.3 * | 1.8 | |
| <0.001 | <0.001 | <0.001 | <0.001 | ||||||
| NKD1 | 1.5 * | 0.6 | 3.0 * | 1.1 | 1.9 * | 0.5 | 1.4 * | 0.2 | |
| 0.024 | <0.001 | <0.001 | 0.002 | ||||||
| down | PCDH8 | 1.2 | 0.3 | -1.9 * | 0.4 | -1.3 | 0.4 | -1.8 * | 0.4 |
| 0.241 | <0.001 | 0.152 | <0.001 | ||||||
| PPP1R3C | -1.8 * | 0.5 | -1.8 * | 0.3 | -1.8 * | 0.5 | -1.4 * | 0.3 | |
| <0.001 | <0.001 | <0.001 | <0.001 | ||||||
Fold change is relative to expression level in control cells (treated with PBS containing BSA). Results are expressed as the mean of three different experiments performed in triplicate. The significant P values are represented by a star (Student’s t test):
* P<0.05 (i.e. higher or lower expression versus control condition).
Table 3. qRT-PCR analysis of gene expression in HCC38 cells treated with Wnt3a.
| Wnt3a treatment | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| 3h | 6h | 9h | 12h | ||||||
| GENE SYMBOL | Fold change | +/- SD | Fold change | +/- SD | Fold change | +/- SD | Fold change | +/- SD | |
| P value | P value | P value | P value | ||||||
| up | APCDD1 | 2.2 * | 0.9 | 5.1 * | 2.1 | 4.4 * | 1.7 | 3.3 * | 0.7 |
| 0.002 | <0.001 | <0.001 | <0.001 | ||||||
| AXIN2 | 17.3 * | 4.4 | 27.0 * | 11.9 | 16.7 * | 4.6 | 11.7 * | 1.5 | |
| <0.001 | <0.001 | <0.001 | <0.001 | ||||||
| DDIT4L | 7.5 * | 2.3 | 10.3 * | 4.8 | 7.8 * | 4.4 | 4.2 * | 0.6 | |
| <0.001 | <0.001 | <0.001 | <0.001 | ||||||
| NKD1 | 4.6 * | 1.4 | 21.8 * | 12.4 | 20.6 * | 12.6 | 10.9 * | 5.4 | |
| <0.001 | <0.001 | <0.001 | <0.001 | ||||||
| down | PCDH8 | -2.3 * | 0.6 | -10.1 * | 4.3 | -5.7 * | 2.7 | -6.5 * | 3.6 |
| <0.001 | <0.001 | <0.001 | <0.001 | ||||||
| PPP1R3C | -4.1 * | 0.6 | -4.8 * | 1.5 | -4.3 * | 1.5 | -3.4 * | 0.9 | |
| <0.001 | <0.001 | <0.001 | <0.001 | ||||||
Fold change is relative to expression level in control cells (treated with PBS containing BSA). Results are expressed as the mean of three different experiments performed in triplicate. The significant P values are represented by a star (Student’s t test):
* P<0.05 (i.e. higher or lower expression versus control condition).
The expression of APCDD1, AXIN2, DDIT4L and NKD1 was up-regulated in MDA-MB-468 cells within the first 3-6h of Wnt3a treatment, and was subsequently down-regulated later on (Table 2). The expression of PPP1R3C was down-regulated at 3h of Wnt3a treatment and that of PCDH8 was down-regulated at 6h of Wnt3a treatment (Table 2). We also carried out similar time-course experiments with Wnt3a in HCC38 cells (Table 3). The expression of APCDD1, AXIN2, DDIT4L and NKD1 was up-regulated at 3h and reached a maximum level at 6h (Table 3). By contrast, the levels of PPP1R3C and PCDH8 transcripts were lower in cells treated with Wnt3a for 3h than in resting cells, and reached their lowest levels at 6h (Table 3). The expression of PPP1R3C and PCDH8 was more strongly down-regulated in HCC38 cells than in MDA-MB-468 cells, consistent with the microarray data (S2 Dataset, S2 and S3 Figs). Overall, qRT-PCR experiments on a selection of genes validated the microarray data and both analyses revealed a time-course of target gene expression similar to that reported by others [32,78]. The chronological order and duration of Wnt target gene activation/repression may be crucial for the controlled course of events downstream from the activated Wnt/β-catenin pathway in breast cancer. However, further investigation is required to explore this issue.
We sought to confirm that the regulation of the expression of the Wnt target genes was β-catenin-dependent; therefore, we transfected MDA-MB-468 cells with a construct encoding a transcriptionally active form of β-catenin, which carries a mutation in the GSK3β phosphorylation sites, rendering it refractory to the destruction complex. The experiment was not performed in HCC38 cells because these cells were harder to transfect. Luciferase assays showed that the expression of active β-catenin in MDA-MB-468 cells resulted in a very high level of Wnt activation in cells co-transfected with the WRE reporter plasmid (S4 Fig). The transcriptional activity of β-catenin was higher at 6h after transfection than in control cells, reached a peak at 12h and stayed constant until 24h.
Next, we measured the abundance β-catenin mRNA (to evaluate the efficiency of transfection, data not shown) and that of selected Wnt3a target genes by qRT-PCR in cells expressing the active, mutant form of β-catenin. The transfection of active β-catenin in MDA-MB-468 cells resulted in the up-regulation of APCDD1, AXIN2, DDIT4L and NKD1 and the down-regulation of PPP1R3C expression (Table 4).
Table 4. qRT-PCR analysis of gene expression in MDA-MB-468 expressing the active, mutant form of β-catenin.
| expression of constitutively active β-catenin | |||||||
|---|---|---|---|---|---|---|---|
| 12h | 24h | 48h | |||||
| GENE SYMBOL | Fold change | +/- SD | Fold change | +/- SD | Fold change | +/- SD | |
| P value | P value | P value | |||||
| up | APCDD1 | 1.5 * | 0.4 | 2.3 * | 0.7 | 3.5 * | 0.6 |
| 0.003 | <0.001 | <0.001 | |||||
| AXIN2 | 2.2 * | 0.6 | 2.8 * | 0.7 | 4.4 * | 1.1 | |
| <0.001 | <0.001 | <0.001 | |||||
| DDIT4L | 2.3 * | 0.7 | 8.8 * | 4.2 | 11.4 * | 4.8 | |
| <0.001 | <0.001 | <0.001 | |||||
| NKD1 | 1.1 | 0.3 | 1.6 * | 0.4 | 2.2 * | 0.6 | |
| 0.557 | <0.001 | <0.001 | |||||
| down | PCDH8 | -1.2 | 0.3 | 1.7 * | 0.6 | -1.8 | 1.1 |
| 0.201 | 0.010 | 0.076 | |||||
| PPP1R3C | -1.1 | 0.3 | -1.6 * | 0.8 | -1.6 * | 0.3 | |
| 0.309 | 0.048 | <0.001 | |||||
Fold change is relative to expression level in control cells (transfected with empty vector). Results are expressed as the mean of three different experiments performed in triplicate. The significant P values are represented by a star (Student’s t test):
* P<0.05 (i.e. higher or lower expression versus control condition).
Regarding PCDH8, the results were less clear as we noticed that the abundance of PCDH8 mRNA in cells expressing activated β-catenin was higher at 24h, and tended to be lower 48h post-transfection (Table 4). This may be due to the cell line used for the experiment because PCDH8 was found to be highly down-regulated in HCC38 (max fold change = -10.1) and not to such an extent in MDA-MB-468 (max fold change = -1.9) upon Wnt3a stimulation (Table 2, Table 3). These results indicate that the expression of APCDD1, AXIN2, DDIT4L, NKD1, PPP1R3C and potentially PCDH8 is regulated by Wnt3a through the activation of β-catenin.
These data confirmed that the analyzed genes are indeed Wnt/β-catenin target genes (some confirmation is required for PCDH8 in another cell line) and that the genes identified in our microarray analysis at the earliest time point (6h) may therefore also be Wnt/β-catenin target genes. However, additional systematic experiments are required to confirm this hypothesis.
Analysis of pathways deregulated in HCC38 and MDA-MB-468 triple-negative breast cancer cell lines upon Wnt3a treatment
The Wnt/β-catenin pathway activates but also represses the transcription of genes encoding products that may positively or negatively regulate the canonical Wnt pathway. Indeed, 34 out of 270 genes that were up-regulated in cells stimulated with Wnt3a for 6h, and 12 out of 178 genes that were down-regulated under the same conditions, encode known activators or inhibitors of this signaling pathway (S3 Dataset). Expression changes of these Wnt regulators are necessary to maintain Wnt signaling and to avoid the uncontrolled activation of the canonical pathway.
Although most Wnt target genes identified in both TNBC cell lines (24 out of 36, Table 1) have been previously described in other studies, their role in the Wnt/β-catenin pathway needs to be established. The exceptions to this are APCDD1, AXIN2, BMP7, NKD1, RNF43 and ZNRF3, which are already known to regulate negatively the canonical pathway (S3 Dataset). Among the 12 potential novel Wnt target genes identified in HCC38 and MDA-MB-468 cells (Table 1, Fig 2C and Fig 3C), a link with the Wnt/β-catenin pathway was previously reported only for DDIT4L, LPAR1 and TLR1 [79–83].
REDD2 (product of the gene DDIT4L) activates the TSC (tuberous sclerosis complex) complex and negatively regulates mTOR signaling [79,84]. However, Wnt has also been shown to promote mTOR activity by preventing the activation of the TSC complex through two mechanisms: the inhibition of GSK3β, which is a known activator of the TSC complex, and the activation of the small GTPase Rac1 [80]. Additional experiments would be required to examine whether REDD2 induced by Wnt3a in TNBC cells attenuates mTOR activity that is initially stimulated by Wnt signaling (negative feedback loop).
In colon cancer cells, the binding of lysophosphatidic acid (LPA) to its receptors LPA2 and LPA3 but not to LPA1 (product of the gene LPAR1) activates the Wnt/β-catenin pathway [81]. Thus, it is possible that LPA1 plays a role in the aberrant activation of β-catenin signaling in TNBC cells. Further analyses are needed to confirm this hypothesis.
Only one paper mentions a link between Wnt and TLR1, reporting that Wnt3a suppresses pro-inflammatory responses to TLR1/2 ligands in dendritic cells [82].
We used KEGG analysis to examine enriched pathways among the genes positively or negatively regulated by Wnt3a in each cell line to explore the effect of Wnt signaling on cellular functions (S4 Dataset).
We focused mainly on HCC38 cells because the number of deregulated genes was very different between the two TNBC cell lines and higher in HCC38 cells (Fig 2 and Fig 3). Wnt target genes that were up-regulated in HCC38 cells stimulated for 6h with Wnt3a were associated with the TGFβ, Wnt, p53 and Hedgehog signaling pathways, whereas the down-regulated Wnt target genes were associated with calcium and mTOR signaling pathways (S4 Dataset). Some of these pathways (e.g., TGFβ and p53 pathways) were still deregulated even after 12h or 24h of treatment. Many Wnt target genes play a role in Wnt regulation (S3 Dataset); therefore, we expected to find the Wnt signaling pathway among the affected pathways. Interestingly, the pathways affected in both HCC38 cells and MDA-MB-468 cells (S4 Dataset) control proliferation (e.g., Hedgehog, Jak-STAT, p53 and Wnt signaling), apoptosis/autophagy (e.g., p53 and mTOR signaling) and inflammation/immune responses (e.g., TGFβ and toll-like receptor signaling, and cytokine-cytokine receptor interactions). These results are not surprising given the well documented crosstalk between Wnt signaling and these pathways [80,85–92]. Moreover, Hedgehog, TGFβ, Jak-STAT and toll-like receptor signaling pathways are deregulated in breast cancer and play a role in its development [93–96]. Altogether, these data may provide insight into the effect of aberrant Wnt/β-catenin signaling on biological processes involved in the development of TNBC/BLBC.
Comparison of our list of Wnt target genes identified in HCC38 and MDA-MB-468 triple-negative breast cancer cell lines with previously published lists
We compared our list of Wnt target genes with previously published lists (activated and repressed genes were compared separately) (S5 Dataset) [34,35,37–40,56–63,97,98] to determine whether the Wnt target genes that we identified were specific to breast cancer cells.
First, we compared our lists of Wnt target genes to those reported in NIH3T3 fibroblasts [56,97] and in C3H10T1/2 mesenchymal cells [35,37,56,59–63] stimulated with Wnt3a. Of the Wnt target genes identified in C3H10T1/2 cells, four out of the 21 up-regulated genes (AHR, AXIN2, SEMA3A and TGFB3), but none of the 19 repressed genes responded to Wnt3a in TNBC cells [35,37,56,59–63]. Eleven genes that were up-regulated in response to Wnt3a in fibroblasts (AHR, APCDD1, ARL4C, AXIN2, FAS, FZD7, GADD45G, IRS1, KLF5, TGFB3 and WNT11) and three genes that were down-regulated (CITED2, PDGFRA and PTGFR) were also found in our study [56,97].
Second, the comparison was performed with Wnt target genes identified in cancer cells other than breast tumors. Nine of 76 Wnt target genes up-regulated in ovarian endometrioid adenocarcinoma (BMP4, CCND1, CYP24A1, FGF9, IRS1, MSX2, PCSK6, SEMA3C and SFN) were also identified in our study [38]. Thirteen out of 208 Wnt target genes overexpressed in colorectal tumors [37] were also up-regulated by Wnt signaling in TNBC cells: AXIN2, CDC25A, KIAA1199, LEF1, MET, MYB, PHLDA1, SLC19A2, SLC25A19, SLC39A10, SOX4, ZNF703 and ZNRF3. Several other studies have also investigated the Wnt signature in the intestine. Herbst et al. identified deregulated genes in DLD1 and SW480 cells [58] and compared the potential target genes with data published for LS174T cells [57]. Among the 193 genes commonly regulated by β-catenin in the three colorectal tumor cell lines (61 “up” and 132 “down”) [58], 12 were also overexpressed (APCDD1, AXIN2, C10orf2, CDC25A, DEPDC7, EDAR, FGF9, KIAA1199, NAV3, RNF43, VSNL1 and ZNRF3) and one was repressed (ELF3) in response to Wnt3a in our TNBC cells. FBXO32, which was identified in both TNBC cell lines, was down-regulated in DLD1, not in SW480 cells [58]. Among the 122 target genes identified as overexpressed in Wilms tumors, only APCDD1 was common to our study [40]. FUT8 was the only Wnt target gene among 54 up-regulated genes in hepatoma cells that was also up-regulated in response to Wnt3a in TNBC cells [39]. AXIN2 and IRS1 were the only Wnt target genes in breast cancer cells that were also identified in hepatocellular carcinoma cells [63].
Finally, we compared our Wnt target genes to those found in normal and cancerous mammary cells. One study examined gene expression in the mouse mammary epithelial cell line C57MG in response to Wnt3a [35]; 16% of Wnt3a-induced genes (AXIN2, AHR, CCND1, EFNB2, IRS1 and KLF5) and 8% of the repressed genes (ANGPT1) found in this study were also identified in our TNBC cells. Another study examined gene expression in C57MG cells stimulated by Wnt1, another canonical ligand [34]. Interestingly, IRF7 was the only common gene that responded to both Wnt3a and Wnt1 in these cells (see list of “down” genes) [34,35]. We found that EGR1, FAS and SLC7A2 were up-regulated and DIO2, ETS1 and VEGFA, were down-regulated in TNBC in response to Wnt3a, consistent with reports in Wnt1-treated C57MG cells [34,35]. Among the 248 target genes identified in normal murine mammary gland epithelial cells treated with Wnt3a, seven were common to our study (“up”: AHR, AXIN2, CCND1, GADD45G and HS3ST1; “down”: MMP13 and TNFAIP2) [98].
Overall, these observations show that very few Wnt target genes identified in breast cancer cells were common to other tissues; thus, only a handful can be considered as ‘universal’ target genes e.g. APCDD1, AXIN2 and IRS1 (S5 Dataset). Unexpectedly, NKD1, another well-known Wnt target gene, was reported in only one [58] out of the 13 selected studies (S5 Dataset). Although they are derived from the same tissue, the TNBC cell lines HCC38 and MDA-MB-468 display strong differences regarding the number and the identity of Wnt target genes. Similar observations were made in the colorectal cancer lines DLD1 and SW480 depleted in β-catenin [58] (S5 Dataset). In addition, C57MG mammary epithelial cells treated with Wnt1 [34] or Wnt3a [35], two canonical ligands from two different classes, display different gene expression profiles (S5 Dataset), suggesting that a particular Wnt ligand affects the expression of a specific set of Wnt target genes.
Altogether, these data confirm that Wnt target genes are diverse, tissue-, cell line- and treatment-specific. Therefore, we believe it is necessary to perform similar analyses in many different breast cancer cell lines (triple negative/basal-like and luminal) after stimulation with various Wnt ligands to obtain a more complete picture of Wnt target genes in this tissue.
Wnt target genes are enriched in human breast tumor samples
We next sought to examine, in samples of human TNBC, the expression level of the Wnt target genes identified in HCC38 stimulated with Wnt3a for 6, 12 or 24h, to investigate whether the expression of these genes correlates with potential Wnt activation in these tumors. To achieve sufficient statistical power, the analyses were performed with the data obtained with HCC38 cells, and not with those obtained with MDA-MB-468 cells, because few genes were found deregulated in MDA-MB-468 cells (see Materials and Methods section for details). The comparative analyses between cell line and tumors were performed with transcriptomic data previously obtained with our cohort composed of 130 human breast cancer tumors [41,42].
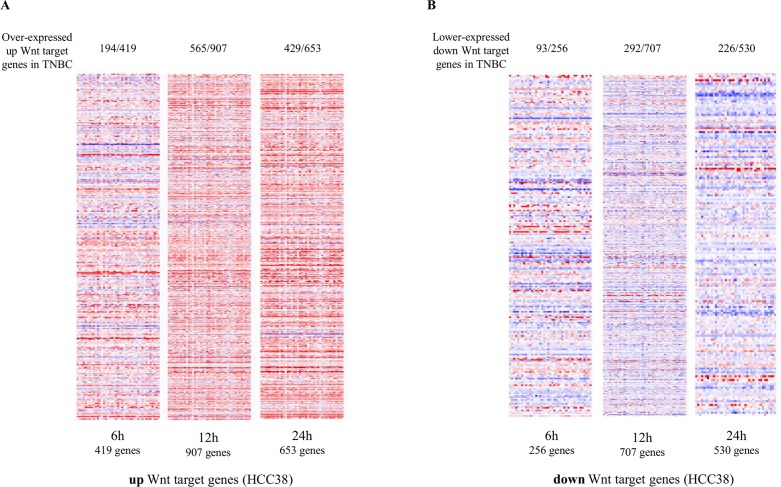
Many of the Wnt target genes that were up-regulated in Wnt3a-stimulated HCC38 cells were more strongly expressed in TNBC than in Luminal A (LA) samples (Fig 4A, Table 5). We refer to this overexpression as “enrichment” in the following analysis (down-regulated genes can similarly be defined as enriched). Genes identified in HCC38 cells stimulated with Wnt3a for 6h, 12h or 24h were enriched in TNBC samples, with the most significant enrichment corresponding to genes identified at 12h and 24h (Fig 4A, Table 5). Similarly, Wnt target genes were enriched among the genes more strongly expressed in TNBC than in HER2+ or Luminal B (LB) samples (Table 5). In addition, a significant proportion of the Wnt target genes down-regulated in HCC38 cells were more poorly expressed (and hence enriched) in TNBC than in LA samples (Fig 4B, S1 Table), or than in HER2+ or LB samples (S1 Table).
Fig 4. Wnt target genes identified in Wnt3a-stimulated HCC38 cells and their expression in TNBC samples.
We identified Wnt target genes that were differentially expressed in HCC38 cells stimulated with Wnt3a (6h, 12h or 24h), and we examined the expression level of up-regulated (A) and down-regulated (B) Wnt target genes in TNBC samples. We restricted the study to the 11262 genes present on both cell line and tumor arrays. We applied a FDR (False Discovery Rate) cut-off of 0.05 and a log-2 fold change threshold of 0. The numbers of up- regulated (A) and down- regulated (B) genes taken into account are indicated at the bottom for each time point (6h, 12h and 24h). The numbers of the Wnt target genes that were more strongly (A) or more poorly (B) expressed in TNBC than in LA samples are indicated on top of the figure. Rows: genes; columns: tumor samples. Red: more strongly expressed genes; blue: more poorly expressed genes. Genes are ordered by gene Entrez ID. The names of the genes are indicated in S8 Fig.
Table 5. Wnt target genes up-regulated in Wnt3a-stimulated HCC38 cells and their enrichment in human breast cancer samples.
| genes up-regulated in HCC38 cells | |||||
|---|---|---|---|---|---|
| Wnt3a | 6h | 12h | 24h | 6h & 24h | |
| gene nb | 419 | 907 | 653 | 133 | |
| TNBC vs LA | 3497 | 194 3.5x10 -11 | 565 3.8x10 -91 | 429 1.1x10 -78 | 72 4.0x10 -8 |
| HER2+ vs LA | 2110 | 110 1.2x10 -4 | 358 1.2x10 -52 | 288 1.8x10 -53 | 35 0.033 |
| LB vs LA | 1587 | 65 0.39 | 250 9.4x10 -29 | 230 8.7x10 -45 | 25 0.15 |
| TNBC vs HER2+ | 2242 | 142 8.9x10 -12 | 429 7.8x10 -85 | 334 5.8x10 -76 | 55 1.7x10 -8 |
| TNBC vs LB | 2775 | 171 1.1x10 -13 | 506 4.6x10 -98 | 383 1.6x10 -81 | 66 4.0x10 -10 |
| HER2+ vs LB | 581 | 36 0.0031 | 93 6.7x10 -11 | 74 1.0x10 -10 | 13 0.027 |
We assessed the significance of the overlap between the genes that were up-regulated in Wnt3a-stimulated HCC38 (6h, 12h, 24h, and those up-regulated at both 6h and 24h) and those that were strongly expressed in tumors (TNBC, HER2+, LB and LA) (gene nb). We restricted our study to the 11262 genes present on both arrays (HCC38 dataset, tumor dataset). The number of the Wnt target genes that were more strongly expressed is indicated for each tumor comparison. The associated P value is also shown underneath (in Italics). The significance of the overlap between two lists was assessed with the Fisher exact test. As an example, 194 of the 419 Wnt target genes up-regulated in Wnt3a-stimulated HCC38 cells (6h) were more strongly expressed in TNBC than in LA samples (194 of the 3497 genes more strongly expressed genes in TNBC than in LA samples) (P value = 3.5x10-11).
We also obtained similar results for the comparison of HER2+ and LB with LA samples, although the findings were less significant (Table 5, S1 Table). Wnt target genes that were up-regulated in cells stimulated with Wnt3a for 6h were more strongly expressed and enriched in HER2+ than in LA samples, but were expressed to a similar extent in LB and LA samples (Table 5). Those identified at 12h and 24h of Wnt3a stimulation were more strongly expressed and enriched in both HER2+ and LB samples than in LA samples (Table 5). Wnt target genes down-regulated in Wnt3a-stimulated cells were also more poorly expressed and enriched in HER2+ and LB samples than in LA samples (S1 Table).
In conclusion, this analysis revealed that the Wnt target genes that were up- or down-regulated in Wnt3a-stimulated HCC38 cells were highly enriched in TNBC and, to a lesser extent, in HER2+ and LB tumors. This may indicate that the Wnt pathway is activated in these three breast cancer subtypes, with the highest activation in TNBC. However, it is well established that the activation of the Wnt/β-catenin pathway induces cell proliferation, in agreement with our KEGG pathway analysis (S4 Dataset). Therefore, long exposure (24h) of cells to Wnt3a will also lead to the activation or repression of genes linked to proliferation. Breast cancer subtypes proliferate at different rates: TNBC are the most proliferative tumors, followed by HER2+, LB and LA tumors (for an example, see the expression profile of several markers of proliferation in tumors, S5 Fig). Thus, it is possible that in breast cancer subtypes, the enrichment of Wnt target genes identified in cell lines stimulated with Wnt3a for 12h or 24h reflects proliferation, which may, or may not, be related to Wnt activation. However, it is impossible to distinguish Wnt activation from proliferation, because both are closely linked.
Finally, we hypothesize that the Wnt target genes found in TNBC cell lines as soon as 6h after Wnt3a stimulation, that continue to be differentially expressed at late time points (24h) may be deregulated during the chronic activation of the Wnt pathway, such as that occurring in tumors. We focused on Wnt target genes that were up-regulated in Wnt3a-stimulated HCC38 cells because only a small number of genes were common to both time points in MDA-MB-468 cells: 133 and 13 genes were shared between the 6h and 24h time points in HCC38 and MDA-MB-468 cells, respectively (selected from a gene list restricted to those present on both the cell line and the tumor arrays; see Material and Methods for details). Of these 133 genes, 54% (72 genes) were more strongly expressed and enriched (P = 4.0x10-8) in TNBC than in LA tumors (Table 5, S6 Fig). These Wnt target genes were also enriched in TNBC vs. HER2+ (P = 1.7x10-8) or LB tumors (P = 4.0x10-10) (Table 5). Interestingly, these genes were also significantly enriched in HER2+ tumors (P = 0.033), albeit to a lesser extent than in TNBC, but were not enriched in LB tumors (P = 0.15) (Table 5). The high expression of these 72 genes may reflect an activation of the Wnt pathway in TNBC samples. When we restricted our analysis to the most strongly deregulated genes (with a fold change > 1.3), 17 genes out of 72 genes were retained (Fig 5), including 11 genes previously identified as Wnt target genes: SLC25A19, CDC6, CDC25A, C1orf109, C1orf135, PLEKHO1, PAX6, PSTPIP2, MFSD2A, FZD7, ARL4C (S3 Dataset). Among those genes, we identified the Frizzled 7 receptor (FZD7) (Fig 5). Interestingly, high levels of FZD7 reported in TNBCs have been previously proposed to drive Wnt activation in these tumors [23]. Of note, the expression profiles of most of these 17 genes appear to not be related to proliferation (S7 Fig). In conclusion, the strong expression of these 17 Wnt target genes may reflect the activation of the canonical Wnt pathway in a more chronic situation, in particular in TNBCs.
Fig 5. Seventeen Wnt target genes up-regulated in HCC38 cells are strongly expressed in TNBC tumors and may reflect chronic activation of the Wnt signaling pathway.
To identify potentially up-regulated Wnt target genes that could reflect the chronic activation of the Wnt pathway in human cancer, we selected the Wnt target genes that were up-regulated at both the earliest (6h) and the latest (24h) time points after the stimulation of HCC38 cells with Wnt3a. Of 133 genes up-regulated in Wnt3a-stimulated HCC38 cells at both time points, 72 genes were more strongly expressed in TNBC than in LA tumors (see S6 Fig). When we restricted our analysis to the most strongly deregulated genes (with a fold change > 1.3), 17 genes out of 72 genes were retained. The genes are ordered by their P value in the t-test for the TNBC subgroup. Rows: genes; columns: tumor samples. Red: more strongly expressed genes; blue: more poorly expressed genes.
Supporting Information
(XLS)
(XLS)
(XLSX)
(XLS)
(XLSX)
MDA-MB-468 (A) or HCC38 cells (B) were transiently transfected with WRE or MRE and pRL-TK plasmids. The transcriptional activity of β-catenin/Tcf was evaluated after Wnt3a or vehicle stimulation (3, 6, 9 or 12 hours). The error bars show the standard deviation of the mean and asterisks indicate a significant P value in Student’s t test (* P<0.05, i.e. higher luciferase activity versus control condition).
(TIF)
Gene expression was evaluated in TNBC cells in the presence (red dots) or the absence (blue dots) of Wnt3a ligand and results are expressed as log2 values. The fold change between treated and control cells is indicated when significant (P < 0.05). ns: not significant.
(TIF)
Gene expression was evaluated in TNBC cells in the presence (red dots) or the absence (blue dots) of Wnt3a ligand and results are expressed as log2 values. The fold change between treated and control cells is indicated when significant (P < 0.05). ns: not significant.
(TIF)
Cells were co-transfected with pRK5-SK-β-cateninΔGSK or pRK5-SK and WRE or MRE and pRL-TK plasmids. The transcriptional activity of β-catenin/Tcf was evaluated 6, 12, and 24 hours after transfection. The error bars show the standard deviation of the mean and asterisks indicate a significant P value in Student’s t test (* P<0.05, i.e. higher luciferase activity versus control condition).
(TIF)
mRNA expression of 4 markers of proliferation are shown in TNBC, HER2+, luminal B (LB), luminal A (LA) samples as well as in normal breast tissues (norm). RNA quantifications were logarithmic (log2) transformed and illustrated by boxplots.
(TIF)
To identify potentially up-regulated Wnt target genes that could reflect the chronic activation of the Wnt pathway in human cancer, we selected the Wnt target genes that were up-regulated at both the earliest time point (6h) and the latest time point (24h) after the stimulation of HCC38 cells with Wnt3a. Of the 133 genes up-regulated in HCC38 cells at both time points, 72 were more strongly expressed in TNBC than in LA tumors. The genes are ordered by their P value in the t-test for the TNBC subgroup. Rows: genes; columns: tumor samples. Red: more strongly expressed genes; blue: more poorly expressed genes.
(TIF)
The abundance of mRNA of the 17 Wnt target genes is shown for TNBC, HER2+, luminal B (LB), luminal A (LA) samples as well as normal breast tissues (norm). The values were log2 transformed and are illustrated by boxplots.
(TIF)
Identical data that are in Fig 4 but with the names of the genes indicated.
(PDF)
(DOCX)
Acknowledgments
We are indebted to Caroline Hego and Emilie Henry for generating the microarray data. We also thank Dr Céline Baldeyron, Amélie Brisson, Dr François Lallemand and Bérengère Marty-Prouvost (Institut Curie) for helpful discussions. We acknowledge Amélie Brisson, Bérengère Marty-Prouvost and Dr David Silvestre for their critical reading of the manuscript.
Data Availability
All the transcriptomic raw and normalized data are available from the Gene Expression Omnibus database. Experiment performed in Wnt3a stimulated cell line (GEO accession number: GSE65238). Breast tumors of our cohort (GEO accession number: GSE65216).
Funding Statement
This work was supported by Institut de Recherches Servier and Institut Curie. Co-authors Francisco Cruzalegui and Gordon C. Tucker are employed by Institut de Recherche Servier. Institut de Recherche Servier provided support in the form of salaries for authors FC and GCT, but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486: 346–352. 10.1038/nature10983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98: 10869–10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490: 61–70. 10.1038/nature11412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121: 2750–2767. 10.1172/JCI45014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486: 395–399. 10.1038/nature10933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Metzger-Filho O, Tutt A, de Azambuja E, Saini KS, Viale G, Loi S, et al. Dissecting the heterogeneity of triple-negative breast cancer. J Clin Oncol. 2012;30: 1879–1887. 10.1200/JCO.2011.38.2010 [DOI] [PubMed] [Google Scholar]
- 7. Liedtke C, Mazouni C, Hess KR, Andre F, Tordai A, Mejia JA, et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol. 2008;26: 1275–1281. 10.1200/JCO.2007.14.4147 [DOI] [PubMed] [Google Scholar]
- 8. Akiyama T, Kawasaki Y. Wnt signalling and the actin cytoskeleton. Oncogene. 2006;25: 7538–7544. [DOI] [PubMed] [Google Scholar]
- 9. Lai SL, Chien AJ, Moon RT. Wnt/Fz signaling and the cytoskeleton: potential roles in tumorigenesis. Cell Res. 2009;19: 532–545. 10.1038/cr.2009.41 [DOI] [PubMed] [Google Scholar]
- 10. Teo JL, Kahn M. The Wnt signaling pathway in cellular proliferation and differentiation: A tale of two coactivators. Adv Drug Deliv Rev. 2010;62: 1149–1155. 10.1016/j.addr.2010.09.012 [DOI] [PubMed] [Google Scholar]
- 11. Polakis P. The many ways of Wnt in cancer. Curr Opin Genet Dev. 2007;17: 45–51. [DOI] [PubMed] [Google Scholar]
- 12. MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17: 9–26. 10.1016/j.devcel.2009.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149: 1192–1205. 10.1016/j.cell.2012.05.012 [DOI] [PubMed] [Google Scholar]
- 14. King TD, Suto MJ, Li Y. The Wnt/beta-catenin signaling pathway: a potential therapeutic target in the treatment of triple negative breast cancer. J Cell Biochem. 2012;113: 13–18. 10.1002/jcb.23350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lin SY, Xia W, Wang JC, Kwong KY, Spohn B, Wen Y, et al. Beta-catenin, a novel prognostic marker for breast cancer: its roles in cyclin D1 expression and cancer progression. Proc Natl Acad Sci U S A. 2000;97: 4262–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bafico A, Liu G, Goldin L, Harris V, Aaronson SA. An autocrine mechanism for constitutive Wnt pathway activation in human cancer cells. Cancer Cell. 2004;6: 497–506. [DOI] [PubMed] [Google Scholar]
- 17. Prasad CP, Gupta SD, Rath G, Ralhan R. Wnt signaling pathway in invasive ductal carcinoma of the breast: relationship between beta-catenin, dishevelled and cyclin D1 expression. Oncology. 2007;73: 112–117. 10.1159/000120999 [DOI] [PubMed] [Google Scholar]
- 18. Nakopoulou L, Mylona E, Papadaki I, Kavantzas N, Giannopoulou I, Markaki S, et al. Study of phospho-beta-catenin subcellular distribution in invasive breast carcinomas in relation to their phenotype and the clinical outcome. Mod Pathol. 2006;19: 556–563. [DOI] [PubMed] [Google Scholar]
- 19. Geyer FC, Lacroix-Triki M, Savage K, Arnedos M, Lambros MB, MacKay A, et al. beta-Catenin pathway activation in breast cancer is associated with triple-negative phenotype but not with CTNNB1 mutation. Mod Pathol. 2011;24: 209–231. 10.1038/modpathol.2010.205 [DOI] [PubMed] [Google Scholar]
- 20. Khramtsov AI, Khramtsova GF, Tretiakova M, Huo D, Olopade OI, Goss KH. Wnt/beta-catenin pathway activation is enriched in basal-like breast cancers and predicts poor outcome. Am J Pathol. 2010;176: 2911–2920. 10.2353/ajpath.2010.091125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lindvall C, Zylstra CR, Evans N, West RA, Dykema K, Furge KA, et al. The Wnt co-receptor Lrp6 is required for normal mouse mammary gland development. PLoS One. 2009;4: e5813 10.1371/journal.pone.0005813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu CC, Prior J, Piwnica-Worms D, Bu G. LRP6 overexpression defines a class of breast cancer subtype and is a target for therapy. Proc Natl Acad Sci U S A. 2010;107: 5136–5141. 10.1073/pnas.0911220107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yang L, Wu X, Wang Y, Zhang K, Wu J, Yuan YC, et al. FZD7 has a critical role in cell proliferation in triple negative breast cancer. Oncogene. 2011;30: 4437–4446. 10.1038/onc.2011.145 [DOI] [PubMed] [Google Scholar]
- 24. Brennan KR, Brown AM. Wnt proteins in mammary development and cancer. J Mammary Gland Biol Neoplasia. 2004;9: 119–131. [DOI] [PubMed] [Google Scholar]
- 25. Teuliere J, Faraldo MM, Deugnier MA, Shtutman M, Ben-Ze'ev A, Thiery JP, et al. Targeted activation of beta-catenin signaling in basal mammary epithelial cells affects mammary development and leads to hyperplasia. Development. 2005;132: 267–277. [DOI] [PubMed] [Google Scholar]
- 26. Polakis P. Wnt signaling and cancer. Genes Dev. 2000;14: 1837–1851. [PubMed] [Google Scholar]
- 27. Furuuchi K, Tada M, Yamada H, Kataoka A, Furuuchi N, Hamada J, et al. Somatic mutations of the APC gene in primary breast cancers. Am J Pathol. 2000;156: 1997–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jonsson M, Borg A, Nilbert M, Andersson T. Involvement of adenomatous polyposis coli (APC)/beta-catenin signalling in human breast cancer. Eur J Cancer. 2000;36: 242–248. [DOI] [PubMed] [Google Scholar]
- 29. Zhang J, Li Y, Liu Q, Lu W, Bu G. Wnt signaling activation and mammary gland hyperplasia in MMTV-LRP6 transgenic mice: implication for breast cancer tumorigenesis. Oncogene. 2010;29: 539–549. 10.1038/onc.2009.339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang ZC, Lin M, Wei LJ, Li C, Miron A, Lodeiro G, et al. Loss of heterozygosity and its correlation with expression profiles in subclasses of invasive breast cancers. Cancer Res. 2004;64: 64–71. [DOI] [PubMed] [Google Scholar]
- 31. Archbold HC, Yang YX, Chen L, Cadigan KM. How do they do Wnt they do?: regulation of transcription by the Wnt/beta-catenin pathway. Acta Physiol (Oxf). 2012;204: 74–109. 10.1111/j.1748-1716.2011.02293.x [DOI] [PubMed] [Google Scholar]
- 32. Vlad A, Rohrs S, Klein-Hitpass L, Muller O. The first five years of the Wnt targetome. Cell Signal. 2008;20: 795–802. [DOI] [PubMed] [Google Scholar]
- 33. Hoverter NP, Waterman ML. A Wnt-fall for gene regulation: repression. Sci Signal. 2008;1: pe43 10.1126/scisignal.139pe43 [DOI] [PubMed] [Google Scholar]
- 34. Ziegler S, Rohrs S, Tickenbrock L, Moroy T, Klein-Hitpass L, Vetter IR, et al. Novel target genes of the Wnt pathway and statistical insights into Wnt target promoter regulation. FEBS J. 2005;272: 1600–1615. [DOI] [PubMed] [Google Scholar]
- 35. Baljinnyam B, Klauzinska M, Saffo S, Callahan R, Rubin JS. Recombinant R-spondin2 and Wnt3a up- and down-regulate novel target genes in C57MG mouse mammary epithelial cells. PLoS One. 2012;7: e29455 10.1371/journal.pone.0029455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wan X, Liu J, Lu JF, Tzelepi V, Yang J, Starbuck MW, et al. Activation of beta-catenin signaling in androgen receptor-negative prostate cancer cells. Clin Cancer Res. 2012;18: 726–736. 10.1158/1078-0432.CCR-11-2521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Van der Flier LG, Sabates-Bellver J, Oving I, Haegebarth A, De Palo M, Anti M, et al. The Intestinal Wnt/TCF Signature. Gastroenterology. 2007;132: 628–632. [DOI] [PubMed] [Google Scholar]
- 38. Schwartz DR, Wu R, Kardia SL, Levin AM, Huang CC, Shedden KA, et al. Novel candidate targets of beta-catenin/T-cell factor signaling identified by gene expression profiling of ovarian endometrioid adenocarcinomas. Cancer Res. 2003;63: 2913–2922. [PubMed] [Google Scholar]
- 39. Lee HS, Park MH, Yang SJ, Park KC, Kim NS, Kim YS, et al. Novel candidate targets of Wnt/beta-catenin signaling in hepatoma cells. Life Sci. 2007;80: 690–698. [DOI] [PubMed] [Google Scholar]
- 40. Zirn B, Samans B, Wittmann S, Pietsch T, Leuschner I, Graf N, et al. Target genes of the WNT/beta-catenin pathway in Wilms tumors. Genes Chromosomes Cancer. 2006;45: 565–574. [DOI] [PubMed] [Google Scholar]
- 41. Maire V, Nemati F, Richardson M, Vincent-Salomon A, Tesson B, Rigaill G, et al. Polo-like kinase 1: a potential therapeutic option in combination with conventional chemotherapy for the management of patients with triple-negative breast cancer. Cancer Res. 2013;73: 813–823. 10.1158/0008-5472.CAN-12-2633 [DOI] [PubMed] [Google Scholar]
- 42. Maire V, Baldeyron C, Richardson M, Tesson B, Vincent-Salomon A, Gravier E, et al. TTK/hMPS1 Is an Attractive Therapeutic Target for Triple-Negative Breast Cancer. PLoS One. 2013;8: e63712 10.1371/journal.pone.0063712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. van Veelen W, Le NH, Helvensteijn W, Blonden L, Theeuwes M, Bakker ER, et al. beta-catenin tyrosine 654 phosphorylation increases Wnt signalling and intestinal tumorigenesis. Gut. 2011;60: 1204–1212. 10.1136/gut.2010.233460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Creyghton MP, Roel G, Eichhorn PJ, Hijmans EM, Maurer I, Destree O, et al. PR72, a novel regulator of Wnt signaling required for Naked cuticle function. Genes Dev. 2005;19: 376–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Goel S, Chin EN, Fakhraldeen SA, Berry SM, Beebe DJ, Alexander CM. Both Lrp5 and Lrp6 receptors are required to respond to physiological Wnt ligands in mammary epithelial cells (and fibroblasts). J Biol Chem. 2012;287: 16454–16466. 10.1074/jbc.M112.362137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu BY, Soloviev I, Huang X, Chang P, Ernst JA, Polakis P, et al. Mammary tumor regression elicited by Wnt signaling inhibitor requires IGFBP5. Cancer Res. 2012;72: 1568–1578. 10.1158/0008-5472.CAN-11-3668 [DOI] [PubMed] [Google Scholar]
- 47. Bilir B, Kucuk O, Moreno CS. Wnt signaling blockage inhibits cell proliferation and migration, and induces apoptosis in triple-negative breast cancer cells. J Transl Med. 2013;11: 280 10.1186/1479-5876-11-280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Matsuda Y, Schlange T, Oakeley EJ, Boulay A, Hynes NE. WNT signaling enhances breast cancer cell motility and blockade of the WNT pathway by sFRP1 suppresses MDA-MB-231 xenograft growth. Breast Cancer Res. 2009;11: R32 10.1186/bcr2317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33: e175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31: e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Smyth GK. Limma: linear models for microarray data In: Bioinformatics and Computational Biology Solutions using R and Bioconductor, Gentleman R., Carey V., Dudoit S., Irizarry R., Huber W. (eds.), Springer, New York: 2005;397–420. [Google Scholar]
- 52. Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. 1995;57: 289–300. [Google Scholar]
- 53. Backes C, Keller A, Kuentzer J, Kneissl B, Comtesse N, Elnakady YA, et al. GeneTrail—advanced gene set enrichment analysis. Nucleic Acids Res. 2007;35: W186–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Takahashi M, Fujita M, Furukawa Y, Hamamoto R, Shimokawa T, Miwa N, et al. Isolation of a novel human gene, APCDD1, as a direct target of the beta-Catenin/T-cell factor 4 complex with probable involvement in colorectal carcinogenesis. Cancer Res. 2002;62: 5651–5656. [PubMed] [Google Scholar]
- 55. Shimomura Y, Agalliu D, Vonica A, Luria V, Wajid M, Baumer A, et al. APCDD1 is a novel Wnt inhibitor mutated in hereditary hypotrichosis simplex. Nature. 2010;464: 1043–1047. 10.1038/nature08875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Railo A, Pajunen A, Itaranta P, Naillat F, Vuoristo J, Kilpelainen P, et al. Genomic response to Wnt signalling is highly context-dependent—evidence from DNA microarray and chromatin immunoprecipitation screens of Wnt/TCF targets. Exp Cell Res. 2009;315: 2690–2704. 10.1016/j.yexcr.2009.06.021 [DOI] [PubMed] [Google Scholar]
- 57. Mokry M, Hatzis P, Schuijers J, Lansu N, Ruzius FP, Clevers H, et al. Integrated genome-wide analysis of transcription factor occupancy, RNA polymerase II binding and steady-state RNA levels identify differentially regulated functional gene classes. Nucleic Acids Res. 2012;40: 148–158. 10.1093/nar/gkr720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Herbst A, Jurinovic V, Krebs S, Thieme SE, Blum H, Goke B, et al. Comprehensive analysis of beta-catenin target genes in colorectal carcinoma cell lines with deregulated Wnt/beta-catenin signaling. BMC Genomics. 2014;15: 74 10.1186/1471-2164-15-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yan D, Wiesmann M, Rohan M, Chan V, Jefferson AB, Guo L, et al. Elevated expression of axin2 and hnkd mRNA provides evidence that Wnt/beta-catenin signaling is activated in human colon tumors. Proc Natl Acad Sci U S A. 2001;98: 14973–14978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Jho EH, Zhang T, Domon C, Joo CK, Freund JN, Costantini F. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol. 2002;22: 1172–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lustig B, Jerchow B, Sachs M, Weiler S, Pietsch T, Karsten U, et al. Negative feedback loop of Wnt signaling through upregulation of conductin/axin2 in colorectal and liver tumors. Mol Cell Biol. 2002;22: 1184–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jackson A, Vayssiere B, Garcia T, Newell W, Baron R, Roman-Roman S, et al. Gene array analysis of Wnt-regulated genes in C3H10T1/2 cells. Bone. 2005;36: 585–598. [DOI] [PubMed] [Google Scholar]
- 63. Tomimaru Y, Koga H, Yano H, de la Monte S, Wands JR, Kim M. Upregulation of T-cell factor-4 isoform-responsive target genes in hepatocellular carcinoma. Liver Int. 2013;33: 1100–1112. 10.1111/liv.12188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kim JS, Crooks H, Dracheva T, Nishanian TG, Singh B, Jen J, et al. Oncogenic beta-catenin is required for bone morphogenetic protein 4 expression in human cancer cells. Cancer Res. 2002;62: 2744–2748. [PubMed] [Google Scholar]
- 65. Kamiya N, Kaartinen VM, Mishina Y. Loss-of-function of ACVR1 in osteoblasts increases bone mass and activates canonical Wnt signaling through suppression of Wnt inhibitors SOST and DKK1. Biochem Biophys Res Commun. 2011;414: 326–330. 10.1016/j.bbrc.2011.09.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rieger ME, Sims AH, Coats ER, Clarke RB, Briegel KJ. The embryonic transcription cofactor LBH is a direct target of the Wnt signaling pathway in epithelial development and in aggressive basal subtype breast cancers. Mol Cell Biol. 2010;30: 4267–4279. 10.1128/MCB.01418-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Angonin D, Van Raay TJ. Nkd1 functions as a passive antagonist of Wnt signaling. PLoS One. 2013;8: e74666 10.1371/journal.pone.0074666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Waaler J, Machon O, von Kries JP, Wilson SR, Lundenes E, Wedlich D, et al. Novel synthetic antagonists of canonical Wnt signaling inhibit colorectal cancer cell growth. Cancer Res. 2011;71: 197–205. 10.1158/0008-5472.CAN-10-1282 [DOI] [PubMed] [Google Scholar]
- 69. Van Raay TJ, Fortino NJ, Miller BW, Ma H, Lau G, Li C, et al. Naked1 antagonizes Wnt signaling by preventing nuclear accumulation of beta-catenin. PLoS One. 2011;6: e18650 10.1371/journal.pone.0018650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Qiu W, Chen L, Kassem M. Activation of non-canonical Wnt/JNK pathway by Wnt3a is associated with differentiation fate determination of human bone marrow stromal (mesenchymal) stem cells. Biochem Biophys Res Commun. 2011;413: 98–104. 10.1016/j.bbrc.2011.08.061 [DOI] [PubMed] [Google Scholar]
- 71. Shin H, Kwack MH, Shin SH, Oh JW, Kang BM, Kim AA, et al. Identification of transcriptional targets of Wnt/beta-catenin signaling in dermal papilla cells of human scalp hair follicles: EP2 is a novel transcriptional target of Wnt3a. J Dermatol Sci. 2010;58: 91–96. 10.1016/j.jdermsci.2010.02.011 [DOI] [PubMed] [Google Scholar]
- 72. Takahashi N, Yamaguchi K, Ikenoue T, Fujii T, Furukawa Y. Identification of two Wnt-responsive elements in the intron of RING finger protein 43 (RNF43) gene. PLoS One. 2014;9: e86582 10.1371/journal.pone.0086582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hao HX, Xie Y, Zhang Y, Charlat O, Oster E, Avello M, et al. ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature. 2012;485: 195–200. 10.1038/nature11019 [DOI] [PubMed] [Google Scholar]
- 74. Zhou Y, Lan J, Wang W, Shi Q, Lan Y, Cheng Z, et al. ZNRF3 acts as a tumour suppressor by the Wnt signalling pathway in human gastric adenocarcinoma. J Mol Histol. 2013;44: 555–563. 10.1007/s10735-013-9504-9 [DOI] [PubMed] [Google Scholar]
- 75. Valenta T, Hausmann G, Basler K. The many faces and functions of beta-catenin. EMBO J. 2012;31: 2714–2736. 10.1038/emboj.2012.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Staal FJ, Weerkamp F, Baert MR, van den Burg CM, van Noort M, de Haas EF, et al. Wnt target genes identified by DNA microarrays in immature CD34+ thymocytes regulate proliferation and cell adhesion. J Immunol. 2004;172: 1099–1108. [DOI] [PubMed] [Google Scholar]
- 77. Reichling T, Goss KH, Carson DJ, Holdcraft RW, Ley-Ebert C, Witte D, et al. Transcriptional profiles of intestinal tumors in Apc(Min) mice are unique from those of embryonic intestine and identify novel gene targets dysregulated in human colorectal tumors. Cancer Res. 2005;65: 166–176. [PubMed] [Google Scholar]
- 78. Rohrs S, Kutzner N, Vlad A, Grunwald T, Ziegler S, Muller O. Chronological expression of Wnt target genes Ccnd1, Myc, Cdkn1a, Tfrc, Plf1 and Ramp3. Cell Biol Int. 2009;33: 501–508. 10.1016/j.cellbi.2009.01.016 [DOI] [PubMed] [Google Scholar]
- 79. Corradetti MN, Inoki K, Guan KL. The stress-inducted proteins RTP801 and RTP801L are negative regulators of the mammalian target of rapamycin pathway. J Biol Chem. 2005;280: 9769–9772. [DOI] [PubMed] [Google Scholar]
- 80. Shimobayashi M, Hall MN. Making new contacts: the mTOR network in metabolism and signalling crosstalk. Nat Rev Mol Cell Biol. 2014;15: 155–162. 10.1038/nrm3757 [DOI] [PubMed] [Google Scholar]
- 81. Yang M, Zhong WW, Srivastava N, Slavin A, Yang J, Hoey T, et al. G protein-coupled lysophosphatidic acid receptors stimulate proliferation of colon cancer cells through the {beta}-catenin pathway. Proc Natl Acad Sci U S A. 2005;102: 6027–6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Oderup C, Lajevic M, Butcher EC. Canonical and noncanonical wnt proteins program dendritic cell responses for tolerance. J Immunol. 2013;190: 6126–6134. 10.4049/jimmunol.1203002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Tang W, Dodge M, Gundapaneni D, Michnoff C, Roth M, Lum L. A genome-wide RNAi screen for Wnt/beta-catenin pathway components identifies unexpected roles for TCF transcription factors in cancer. Proc Natl Acad Sci U S A. 2008;105: 9697–9702. 10.1073/pnas.0804709105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Miyazaki M, Esser KA. REDD2 is enriched in skeletal muscle and inhibits mTOR signaling in response to leucine and stretch. Am J Physiol Cell Physiol. 2009;296: C583–592. 10.1152/ajpcell.00464.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Pecina-Slaus N. Wnt signal transduction pathway and apoptosis: a review. Cancer Cell Int. 2010;10: 22 10.1186/1475-2867-10-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Niehrs C, Acebron SP. Mitotic and mitogenic Wnt signalling. EMBO J. 2012;31: 2705–2713. 10.1038/emboj.2012.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Schaale K, Neumann J, Schneider D, Ehlers S, Reiling N. Wnt signaling in macrophages: augmenting and inhibiting mycobacteria-induced inflammatory responses. Eur J Cell Biol. 2011;90: 553–559. 10.1016/j.ejcb.2010.11.004 [DOI] [PubMed] [Google Scholar]
- 88. Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24: 2899–2908. [DOI] [PubMed] [Google Scholar]
- 89. Katoh M. STAT3-induced WNT5A signaling loop in embryonic stem cells, adult normal tissues, chronic persistent inflammation, rheumatoid arthritis and cancer (Review). Int J Mol Med. 2007;19: 273–278. [PubMed] [Google Scholar]
- 90. Guo X, Wang XF. Signaling cross-talk between TGF-beta/BMP and other pathways. Cell Res. 2009;19: 71–88. 10.1038/cr.2008.302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ingham PW. Hedgehog signaling. Cold Spring Harb Perspect Biol. 2012;4: a011221 10.1101/cshperspect.a011221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Laplante M, Sabatini DM. mTOR Signaling. Cold Spring Harb Perspect Biol. 2012;4: a011593 10.1101/cshperspect.a011593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Silva CM. Role of STATs as downstream signal transducers in Src family kinase-mediated tumorigenesis. Oncogene. 2004;23: 8017–8023. [DOI] [PubMed] [Google Scholar]
- 94. Kasper M, Jaks V, Fiaschi M, Toftgard R. Hedgehog signalling in breast cancer. Carcinogenesis. 2009;30: 903–911. 10.1093/carcin/bgp048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. So EY, Ouchi T. The application of Toll like receptors for cancer therapy. Int J Biol Sci. 2010;6: 675–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Moses H, Barcellos-Hoff MH. TGF-beta biology in mammary development and breast cancer. Cold Spring Harb Perspect Biol. 2011;3: a003277 10.1101/cshperspect.a003277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Chen S, McLean S, Carter DE, Leask A. The gene expression profile induced by Wnt 3a in NIH 3T3 fibroblasts. J Cell Commun Signal. 2007;1: 175–183. 10.1007/s12079-007-0015-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Labbe E, Lock L, Letamendia A, Gorska AE, Gryfe R, Gallinger S, et al. Transcriptional cooperation between the transforming growth factor-beta and Wnt pathways in mammary and intestinal tumorigenesis. Cancer Res. 2007;67: 75–84. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLS)
(XLS)
(XLSX)
(XLS)
(XLSX)
MDA-MB-468 (A) or HCC38 cells (B) were transiently transfected with WRE or MRE and pRL-TK plasmids. The transcriptional activity of β-catenin/Tcf was evaluated after Wnt3a or vehicle stimulation (3, 6, 9 or 12 hours). The error bars show the standard deviation of the mean and asterisks indicate a significant P value in Student’s t test (* P<0.05, i.e. higher luciferase activity versus control condition).
(TIF)
Gene expression was evaluated in TNBC cells in the presence (red dots) or the absence (blue dots) of Wnt3a ligand and results are expressed as log2 values. The fold change between treated and control cells is indicated when significant (P < 0.05). ns: not significant.
(TIF)
Gene expression was evaluated in TNBC cells in the presence (red dots) or the absence (blue dots) of Wnt3a ligand and results are expressed as log2 values. The fold change between treated and control cells is indicated when significant (P < 0.05). ns: not significant.
(TIF)
Cells were co-transfected with pRK5-SK-β-cateninΔGSK or pRK5-SK and WRE or MRE and pRL-TK plasmids. The transcriptional activity of β-catenin/Tcf was evaluated 6, 12, and 24 hours after transfection. The error bars show the standard deviation of the mean and asterisks indicate a significant P value in Student’s t test (* P<0.05, i.e. higher luciferase activity versus control condition).
(TIF)
mRNA expression of 4 markers of proliferation are shown in TNBC, HER2+, luminal B (LB), luminal A (LA) samples as well as in normal breast tissues (norm). RNA quantifications were logarithmic (log2) transformed and illustrated by boxplots.
(TIF)
To identify potentially up-regulated Wnt target genes that could reflect the chronic activation of the Wnt pathway in human cancer, we selected the Wnt target genes that were up-regulated at both the earliest time point (6h) and the latest time point (24h) after the stimulation of HCC38 cells with Wnt3a. Of the 133 genes up-regulated in HCC38 cells at both time points, 72 were more strongly expressed in TNBC than in LA tumors. The genes are ordered by their P value in the t-test for the TNBC subgroup. Rows: genes; columns: tumor samples. Red: more strongly expressed genes; blue: more poorly expressed genes.
(TIF)
The abundance of mRNA of the 17 Wnt target genes is shown for TNBC, HER2+, luminal B (LB), luminal A (LA) samples as well as normal breast tissues (norm). The values were log2 transformed and are illustrated by boxplots.
(TIF)
Identical data that are in Fig 4 but with the names of the genes indicated.
(PDF)
(DOCX)
Data Availability Statement
All the transcriptomic raw and normalized data are available from the Gene Expression Omnibus database. Experiment performed in Wnt3a stimulated cell line (GEO accession number: GSE65238). Breast tumors of our cohort (GEO accession number: GSE65216).