Abstract
Virus-like particles (VLPs) consisting of the influenza A virus proteins haemagglutinin (HA) and matrix protein (M1) represent a new alternative approach for vaccine design against influenza virus. Influenza VLPs can be fast and easily produced in sufficient amounts in insect cells using the baculovirus expression system. Up to now, influenza VLPs have been produced in the Spodoptera frugiperda cell line Sf9. We compared VLP production in terms of yield and quality in two insect cell lines, namely Sf9 and the Trichoplusia ni cell line BTI-TN5B1-4 (High Five™). Additionally we compared VLP production with three different HAs and two different M1s from influenza H1 and H3 strains including one swine-origin pandemic H1N1 strain. Comparison of the two cell lines showed dramatic differences in baculovirus background as well as in yield and particle density. Taken together, we consider the establishment of the BTI-TN5B1-4 cell line advantageous as production cell line for influenza VLPs.
Keywords: Virus-like particles, Influenza, Influenza vaccine, Baculovirus, Insect cells
Introduction
The emergence of the swine-origin influenza H1N1 pandemic demonstrates quite plainly the potential threat of influenza viruses [1]. In addition, annual influenza epidemics are responsible for significant mortality and morbidity [2]. The most effective way to prevent disease resulting from influenza virus infections is vaccination. Egg derived production methods for influenza vaccines were developed in the 40s of the last century. These production methods have various disadvantages including limited production capacity due to insufficient egg supply in case of a pandemic and allergic reactions of vaccinees to egg proteins [3]. Cell culture derived influenza vaccines may overcome most of the disadvantages of egg derived vaccines but biosafety issues, e.g. biosafety level 3 production facilities for some pandemic strains, and difficulties with growth and yield of certain isolates may cause constraints during production.
Alternatives to egg and cell culture derived vaccine approaches, like recombinant haemagglutinin (HA) expressed in E. coli and insect cells, influenza virus-like particles (VLPs) expressed in insect cells, mammalian cells or plants, and influenza DNA vaccines have been established [4-8].
Influenza viruses belong to the Orthomyxoviridae family and possess a segmented genome of seven (influenza C virus) or eight (influenza A and B viruses) segments. Influenza A virions consist of a host cell derived membrane with incorporated multiple copies of HA and neuraminidase (NA). HA and NA are the major glycoproteins and antigens of influenza, HA is responsible for binding to cellular receptors of the host cell whilst NA mediates the release of newly synthesized particles from the mammalian host cell surface [9]. The matrix protein (M1) lines the inner surface of the membrane and stabilizes the particle [9]. Effective and successful influenza vaccines have to induce neutralizing antibodies against HA; these antibodies prevent the virus particles from attaching to the host cell receptors and therefore prevent infection.
VLPs have previously been produced in insect cells and have shown to be promising candidates for vaccination [10]. VLPs are structurally very similar to their correspondent pathogenic virus, and therefore, they are able to induce CD4+ cell proliferation which results in B-cell as well as in cytotoxic T-cell immune responses [10, 11]. The market entry of GlaxoSmithKline’s human papilloma virus vaccine (Cervarix™) finally demonstrated the immunological potency of insect cell derived VLP vaccines [12].
Various expression systems have been used to produce influenza VLPs including mammalian cells, insect cells and plants [6, 8, 11]. VLPs of influenza subtypes H1, H3, H5 and H9 have been generated in insect cells by co-expression of HA, NA, M1 and M2 or HA, NA and M1 or by co-expression of HA and M1, respectively [13-20]. Immunological studies with all of these subtypes have shown superior immune responses against influenza A when administered intranasally, intraperitoneally or intramuscularly [11, 13-18, 20]. Up to date all of these studies have been carried out with VLPs that had been produced in the Spodoptera frugiperda insect cell line Sf9. In this study we tested the Trichoplusia ni cell line BTI-TN5B1-4 (High Five™) and compared VLP production in terms of yield and quality to the Sf9 production platform.
Materials and Methods
Cells and Viruses
Spodoptera frugiperda Sf9 cells (ATCC# CRL-1711) were maintained as adherent cultures in Roux flasks in modified IPL-41 media (supplemented with lipid mixture (Sigma, St. Louis, USA) and Yeast Extract (Sigma)) containing 3% FCS at 27°C [21]. Sf9 cells dedicated to VLP production were cultivated in the same media in suspension in 500-ml shaker flasks at 100 rpm. Trichoplusia ni BTI-TN5B1-4 cells (ATCC# CRL-10859) were maintained in shaker flasks in serum-free modified IPL-41 medium at 27°C shaking at 100 rpm [21]. Influenza strains A/Puerto Rico/8/1934 (H1N1) and A/Hiroshima/52/2005 (H3N2) were grown on Vero cells (ATCC# CCL-81) in Dulbecco’s modified Eagles medium/Hams F12 medium.
Cloning and Recombinant Baculovirus (rBV) Generation
The haemagglutinin (HA) gene from swine-origin H1N1 pandemic influenza strain A/California/04/2009 (GenBank: GQ117044.1) was synthesized by Geneart (Regensburg, Germany). The synthesis vector pGA was digested by the restriction endonucleases EcoRV and NotI. The haemagglutinin fragment was ligated into a StuI and NotI digested pBacPAK8 (Clonetech, CA, USA) resulting in pBacPAK8-SF.
The matrix protein (M1) gene from strain A/Udorn/307/1972 (H3N2) (GenBank: DQ508932.1) was synthesized by Sloning (Puchheim, Germany). The synthesis vector pPCR-M1 was digested by the restriction endonucleases XhoI and NotI. The M1 fragment was ligated into an XhoI and NotI digested pBacPAK8 resulting in pBacPAK8-UM.
The gen sequences of A/Hiroshima/52/2005 HA (GenBank: CY031336.1), A/Puerto Rico/8/1934 HA (GenBank: EF467821.1) and A/Puerto Rico/8/1934 M1 (GenBank: EF467824.1) were obtained by TRIZOL™ extraction (Invitrogen, Carlsbad, USA) and reverse transcription from Vero cell infection supernatant. The sequence of the A/Hiroshima/52/2005 HA gene was amplified by PCR by the use of primers 5′atgatgatggatatcAGCAAAAGCAGGGGATAATTCTATTAAC and 3′atgatgatggcggccgcAGT AGAAACAAGGGTGTTTTTAATTAATGCAC, the product was digested by EcoRV and NotI restriction endonucleases and ligated into StuI and NotI digested pBacPAK8 resulting in pBacPAK8-HIR. The sequence of the A/Puerto Rico/8/1934 M1 gene was amplified by PCR by the use of primers 5′gatgatTCTAGAATGAGTCTTCTAACCGAGGTC and 3′gatgatGCGGCCGCTCACTTGAACCGTTGCATCTG, the product was digested by XbaI and NotI restriction endonucleases and ligated into XbaI and NotI digested pBacPAK8 resulting in pBacPAK8-PM. The sequence of the A/Puerto Rico/8/1934 HA gene was amplified by PCR by the use of primers 5′gatgatTCTAGAATGAAGGCAAACCTACTGGTC and 3′gatgatGCGGCCGCTCAGATGCATATTCTGCACTG, the product was digested by XbaI and NotI restriction endonucleases and ligated into XbaI and NotI digested pBacPAK8 resulting in pBacPAK8-PR. The green fluorescent protein (GFP) gene was ligated into an HindIII and XhoI digested pBac-5 (Merck, Darmstadt, Germany) resulting in pBac-5-GFP. Recombinant baculoviruses were generated by cotransfection of pBacPAK8-SF pBacPAK8-UM, pBacPAK8-HIR, pBacPAK8-PM, pBacPAK8-PR or pBac-5-GFP together with Baculogold AcNPV linearized DNA (Pharmingen, San Diego, USA) into Sf9 cells by Cellfectin (Invitrogen, Carlsbad, USA) according to the manufacturer’s instructions. For abbreviations of the different HA and M1 proteins please consult Table 1.
Table 1.
Abbreviations for cell lines, HA and M1 proteins
| Abbreviation | Full-length designation |
|---|---|
| Hi5 | BTI-TN5B1-4 cell derived material |
| Sf9 | Sf9 cell derived material |
| SF | HA from A/California/04/2009 (H1N1) |
| HIR | HA from A/Hiroshima/52/2005 (H3N2) |
| PR | HA from A/Puerto Rico/8/1934 (H1N1) |
| PM | M1 from A/Puerto Rico/8/1934 (H1N1) |
| UM | M1 from A/Udorn/307/1972 (H3N2) |
Production, Purification and Characterization of VLPs
BTI-TN5B1-4 or Sf9 cells were co-infected with rBV expressing HA (SF, PR or HIR) and M1 (PM or UM) or GFP (negative control) at a multiplicity of infection (MOI) of 10 and a cell density of 1 × 106 cells/ml in 500 ml shaker flasks (Table 1). Cells were harvested 72 h post infection and separated from supernatant by low speed centrifugation for 10 min at 2,000×g at room temperature. The supernatant was tested for VLP content by haemagglutination assay and for baculovirus titer by plaque assay on Sf9 cells [22]. VLPs were pelleted by ultracentrifugation at 136,000×g at 20°C for 90 min. Pellets were resuspended in phosphate-buffered saline (PBS) for one hour at room temperature. VLPs were further purified by discontinuous sucrose density gradient centrifugation (20%–30%–40%–50%–60%, 100 mM NaCl, 10 mM Tris–HCl and 1 mM EDTA) at 190,000×g at 4°C for 16 h. Gradients were split into fractions which were analysed by SDS-PAGE followed by Western Blot or coomassie staining.
SDS-PAGE and Blots
Sucrose gradient fractions and expression supernatants were examined by SDS-PAGE, coomassie staining and Western Blot [23]. Western Blot analysis was performed using anti-M1 (#290, Institute of Virology, Bratislava), anti-H1 antibodies (#18L/4, Institute of Virology, Bratislava), anti-viral protein 39 (vp39) antibodies (p10C6), anti-glycoprotein 64 (gp64) antibodies (AcV5) and a secondary anti-mouse IgG alkaline phosphatase labelled goat antibody (Sigma, #A1047) [24]. For Western Blot analysis regarding H3 HA a polyclonal anti-A/Panama/2007 H3N2 HA sheep serum (NIBSC, London, UK) was used in combination with an alkaline phosphatase labelled anti-sheep IgG antibody (Sigma, #A5187). Detection was carried out using 5-bromo-4-chloro-3-indolyl phosphate as substrate. Quantification by Western Blot was performed by standard curves with H1 and H3 haemagglutinin of known concentration and visual estimation.
Transmission Electron Microscopy
Sucrose gradient fractions were used for electron microscopy. VLPs were adsorbed for 1 min on copper grids directly from different sucrose gradient fractions and washed with PBS containing 1% bovine serum albumin (BSA). Negative staining was performed using uranyl acetate (pH 4.5) [25]. Samples were examined on a transmission electron microscope at various magnifications.
Haemagglutination Assay
VLPs were serially 2-fold diluted in PBS and incubated at room temperature for two hours with 50 μl of 1.25% human red blood cells (RBC, blood group 0). The haemagglutination extent was estimated visually. The haemagglutination titer designated as haemagglutination units (HAU) was determined by the reciprocal of the highest dilution without complete sedimentation of RBC.
Results
Influenza A VLP Yields in Two Different Insect Cell Lines
Influenza A VLPs were generated by co-infection of Sf9 or BTI-TN5B1-4 cells with recombinant baculovirus (rBV) expressing HA (SF = HA from A/California/04/2009 (H1N1), PR = HA from A/Puerto Rico/8/1934 (H1N1) or HIR = HA from A/Hiroshima/52/2005 (H3N2)) and M1 (PM = M1 from A/Puerto Rico/8/1934 (H1N1) or UM = M1 from A/Udorn/307/1972 (H3N2)) (abbreviations see Table 1). VLPs were detected and quantified in two different ways. The expression supernatants were analysed for haemagglutination activity by haemagglutination assay and additionally the HA content of ultracentrifuged expression supernatant was analysed by Western Blot. Haemagglutination assays showed 16 HAU for all supernatants from both cell lines.
Analysis of the HA content in pellets after ultracentrifugation is shown in Fig. 1. Differences between cell lines are clearly visible; ultracentrifugation pellets from BTI-TN5B1-4 expression supernatants showed a higher HA content in all samples. Differences are most significant for HA from A/California/04/2009 (ranges from ~3/6 μg/ml in BTI-TN5B1-4 supernatants to ~0.4/0.4 μg/ml in Sf9 supernatants) and A/Puerto Rico/8/1934 (ranges from ~15 μg/ml in BTI-TN5B1-4 supernatants to ~3 μg/ml in Sf9 supernatants) but are also observed with HA for A/Hiroshima/52/2005 (ranges from ~6/4.5 μg/ml in BTI-TN5B1-4 supernatants to ~3/3 μg/ml in Sf9 supernatants) (Fig. 1; Table 2). Also the choice of M1 (from H1N1 or H3N2) seems to influence HA expression; HA expression levels are slightly higher when co-expressed with the homologues M1 protein (Fig. 1; Table 2).
Fig. 1.
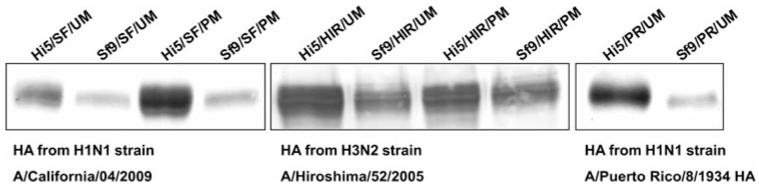
HA content of ultracentrifugation pellets from expression supernatants. A/California/04/2009 and A/Puerto Rico/8/1934 blots were developed using monoclonal anti-HA mouse antibodies in combination with secondary anti-mouse IgG alkaline phosphatase labelled antibodies. The A/Hiroshima/52/2005 blot was developed using anti-A/Panama/2007 H3N2 HA polyclonal sheep serum and a secondary anti-sheep IgG alkaline phosphatase labelled antibody. For abbreviations of the different HA and M1 proteins as well as cell lines please consult Table 1
Table 2.
Comparison of Sf9 and BTI-TN5B1-4 cells as production platforms for influenza A VLPs
| HAU/50 μl | HA protein yield in μg/ml | Baculovirus titer pfu/ml | Main fractions in sucrose density centrifugation | |
|---|---|---|---|---|
| Hi5 mock infection | 0 | – | 6.0 × 106 | – |
| Sf9 mock infection | 0 | – | 2.9 × 108 | – |
| Sf9 SF + UM | 16 | ~0.4 | 1.5 × 108 | 6, 7, 8 |
| Hi5 SF + UM | 16 | ~3 | 2.8 × 106 | 7, 8 |
| Sf9 SF + PM | 16 | ~0.4 | 1.2 × 108 | 6, 7 |
| Hi5 SF + PM | 16 | ~6 | 2.3 × 106 | 8, 9, 10 |
| Sf9 HIR + UM | 16 | ~3 | 2.2 × 108 | 6, 7, 8 |
| Hi5 HIR + UM | 16 | ~6 | 2.5 × 106 | 7, 8, 9 |
| Sf9 HIR + PM | 16 | ~3 | 3.1 × 108 | 7, 8, 9 |
| Hi5 HIR + PM | 16 | ~4.5 | 5.8 × 106 | 8, 9 |
| Sf9 PR + PM | 16 | ~3 | 3.0 × 108 | – |
| Hi5 PR + PM | 16 | ~15 | 8.8 × 106 | – |
For abbreviations of the different HA and M1 proteins as well as cell lines please consult Table 1
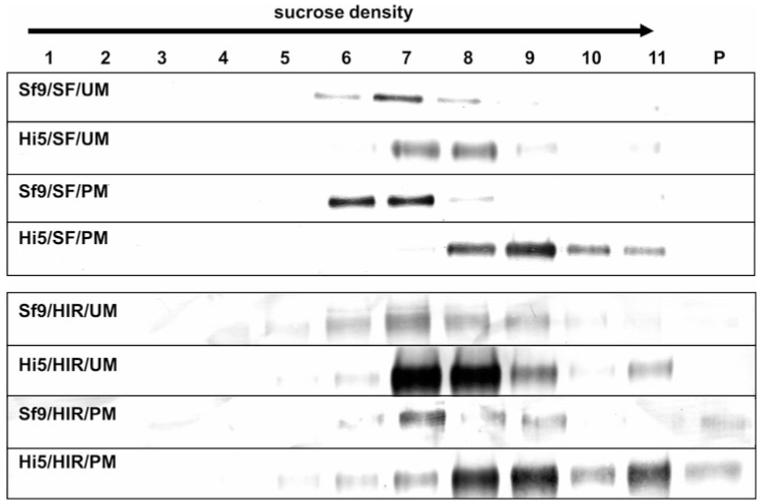
Migration Pattern of VLPs in Sucrose Density Gradient Centrifugation
VLPs from both cell lines produced by co-expression of HA (SF or PR) and M1 (PM or UM) were analysed in context of their migration pattern in sucrose density gradient centrifugation. VLPs were harvested from supernatants by ultracentrifugation and subjected to a sucrose density gradient. After centrifugation, the gradient was split into 11 fractions and the pellet and analysed by Western Blot analysis. Migration patterns of VLPs were found to be cell line and therefore yield dependent. VLPs produced in BTI-TN5B1-4 cells were located dominantly in fractions 7, 8 and 9 (40–50% sucrose) whereas VLPs derived from Sf9 cells were dominantly found in fractions 6, 7 and 8 (35–45%) (Table 2; Fig. 2).
Fig. 2.
Migration patterns of H1N1 and H3N2 VLPs in sucrose density gradients (20–60%). 1 represents the fraction with the lowest, 11 the fraction with the highest sucrose density and P indicates the pellet. A/California/04/2009 blots were developed using monoclonal anti-HA mouse antibodies in combination with secondary anti-mouse IgG alkaline phosphatase labelled antibodies. A/Hiroshima/52/2005 blots were developed using anti-A/Panama/2007 H3N2 HA polyclonal sheep serum and a secondary anti-sheep IgG alkaline phosphatase labelled antibody. For abbreviations of the different HA and M1 proteins as well as cell lines please consult Table 1. BTI-TN5B1-4 cell derived VLPs migrated into fractions with higher sucrose densities than Sf9 derived VLPs. Furthermore M1 proteins influence the migration pattern
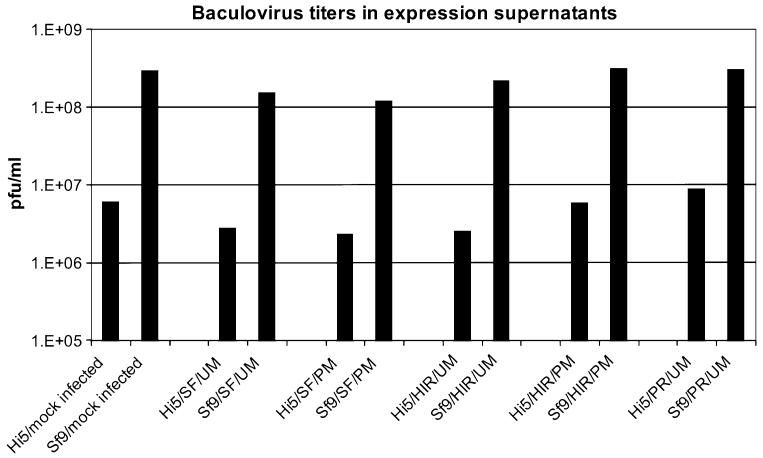
Baculovirus Background
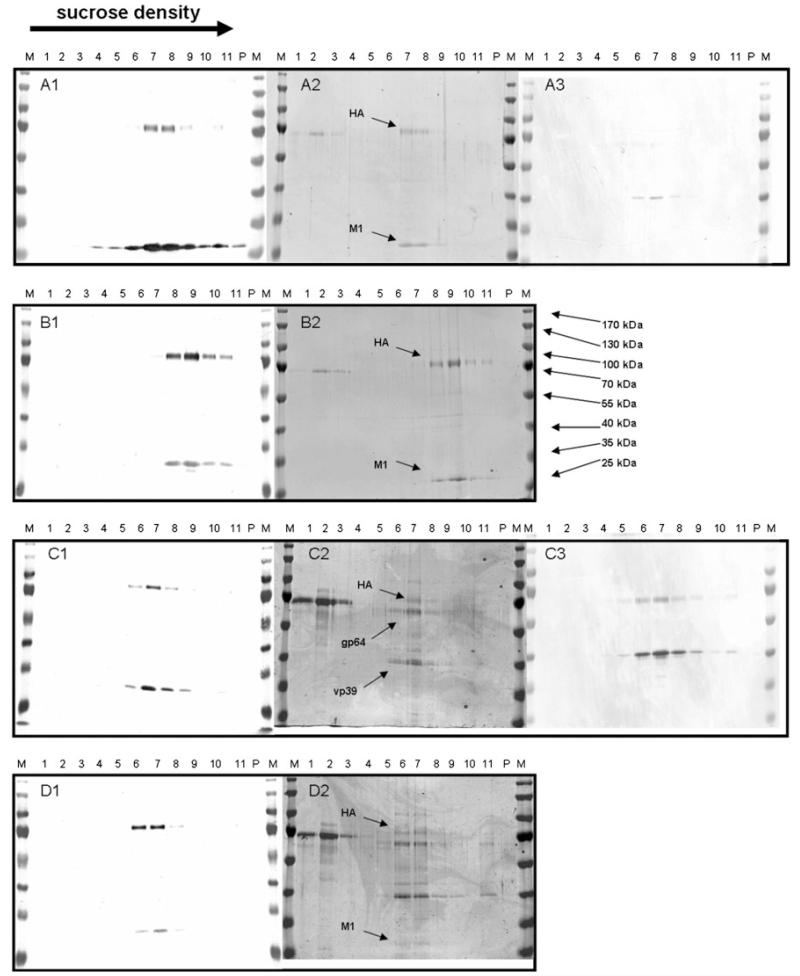
Baculovirus titers of Sf9 and BTI-TN5B1-4 expression supernatants were assessed by plaque assay in Sf9 cells. Baculovirus titers of Sf9 supernatants ranged from 1.2 × 108 to 3 × 108 whereas titers of BTI-TN5B1-4 supernatants were significantly lower and ranged from 2.5 × 106 to 8.8 × 106 (Table 2; Fig. 3). The difference in baculovirus titers and, therefore, baculovirus protein content is also reflected by results from coomassie stainings and Western Blot analysis against baculoviral proteins vp39 and gp64 (Fig. 4). Coomassie stainings from VLP gradients derived from BTI-TN5B1-4 cells showed a high ratio of HA and M1 protein whereas stainings from Sf9 derived material showed predominantly baculovirus proteins and BSA residues from foetal calf serum (FCS) (Fig. 4). Western Blots from VLP gradients against vp39 and gp64, the major baculovirus structural proteins, confirmed the result (Fig. 4). Blots of material from BTI-TN5B1-4 showed only slight bands for vp39 and no bands for gp64 at all, whereas blots from Sf9 derived material showed massive bands for both proteins (Fig. 4, A3 and C3).
Fig. 3.
Baculovirus titers in VLP expression supernatants from BTI-TN5B1-4 and Sf9 cells. Bars represent arithmetic mean titers from one series of expressions. Mock infection was carried out with baculovirus expressing GFP. For abbreviations of the different HA and M1 proteins as well as cell lines please consult Table 1
Fig. 4.
Western Blot analysis and coomassie stainings of VLPs from co-expressions of A/California/04/2009 HA with two different M1 proteins derived from BTI-TN5B1-4 (a, b) and Sf9 cells (c, d). A1 to D2 shows Western Blots and coomassie stainings of migration patterns of VLPs in sucrose density gradients. M indicates the marker, 1 indicates the fraction with the lowest sucrose density, 11 indicates the fraction with the highest sucrose density and P indicates the pellet. Western Blots in A1, B1, C1 and D1 were developed using monoclonal anti-HA and anti-M1 mouse antibodies in combination with secondary anti-mouse IgG alkaline phosphatase labelled antibodies and show co-migration of HA and M1 proteins. A2, B2, C2 and D2 show coomassie stainings, HA, M1 and baculoviral proteins gp64 and vp39 are indicated by arrows. A3 and C3 show Western Blots developed with anti-gp64 and anti-vp64 monoclonal mouse antibodies in combination with an alkaline phosphatase labelled secondary anti-mouse antibody
Integrity of VLPs
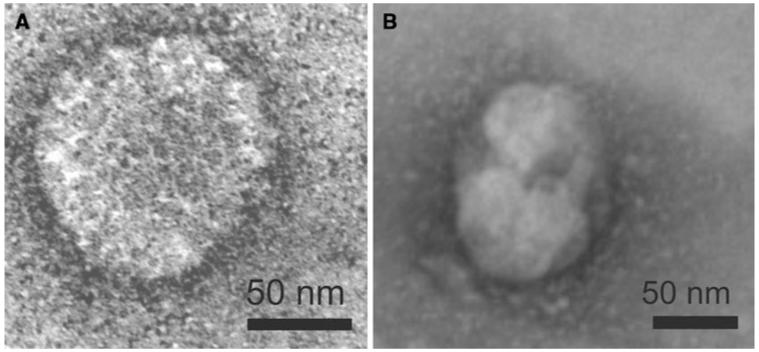
Co-migration of HA and M1 proteins in sucrose density gradients and migration into zones of higher sucrose density, as shown in Figs. 2 and 4 (A1–A4) are one hint for the integrity of influenza VLPs. As shown in Fig. 4 the HA protein was detected as approximately 70 kDa protein and the M1 protein was detected as 29 kDa protein. The same results could be observed for HA from A/Puerto Rico/8/34 (H1N1) and HA from A/Hiroshima/52/2005 (H3N2) (data not shown). In order to confirm the structure of insect cell derived VLPs, electron microscopy was performed for VLPs comprising A/California/04/2009 HA and A/Udorn/307/1972 M1. Particles from different fractions from Sf9 or BTI-TN5B1-4 gradients were stained with uranyl acetate and used for transmission electron microscopy (Fig. 5) [25]. Both cell lines produced spheric and partially polymorphic VLPs of 80–120 nm diameter which is consistent with the morphology of wild-type influenza virions [26].
Fig. 5.
Transmission electron micrographs of negatively stained VLPs from A/California/04/2009 HA and A/Udorn/307/1972 M1 co-expressions derived from Sf9 (a) and BTI-TN5B1-4 cells (b)
Discussion
In this report we compared influenza A VLP production in two insect cell lines, Sf9 and BTI-TN5B1-4, in matters of yield and quality. We co-expressed HA and M1 of several influenza A strains, including HA from a swine-origin pandemic H1N1 isolate, and assessed VLP contents of expression supernatants by haemagglutination assay and HA quantity of ultracentrifugation pellets from supernatants by Western Blot analysis. Immunological characteristics of swine-origin pandemic H1N1 VLPs produced in BTI-TN5B1-4 cells were published elsewhere [27]. Supernatants from all HA-M1 combinations in both cell lines showed constant haemagglutination activities of 16 HAU (Table 2). In contrast, the HA content of ultracentrifugation pellets was markedly different between cell lines. HA content in BTI-TN5B1-4 derived material was 2–15 times higher than inSf9 derived material (Table 2; Fig. 1). Additionally, supernatants showed slightly higher HA contents when HA was co-expressed together with homologues M1 protein (H1 with M1 from H1N1 strain, H3 with M1 from H3N2 strain) (Fig. 1). We detected a total higher HA protein content with no influence on the haemagglutination titer, suggesting there might be a difference in size, shape or density of the particles (Table 2; Fig. 1). For further investigation, VLPs derived from both cell systems were purified and compared for their appearance in electron microscopy and for their migration pattern in sucrose gradient density centrifugation. Analysis by electron microscopy showed that VLPs from both cell systems have similar spherical, partially polymorphic size and shape (Fig. 5). In contrast, VLPs derived from different cell lines show discriminative migration patterns in sucrose gradients. Particles derived from BTI-TN5B1-4 cells migrated to higher sucrose densities than VLPs derived from Sf9 cells (Figs. 2 and 4).
We assume that two preparations of resembling VLPs that accumulate at different densities in a gradient have different numbers of HA molecules integrated in their membranes. This theory could explain the higher amount of HA protein found in the BTI-TN5B1-4 cell culture supernatants. The question that comes up is whether a higher density and thereby a higher HA content of the particles is an advantage in terms of immunology. This question can only be answered by additional immunological analysis and is subject to further investigations.
An important aspect regarding insect cell produced influenza A VLPs is contamination of the vaccine formulation with baculovirus or baculovirus DNA [20]. Baculovirus particles are able to transduce a variety of mammalian cells including human cells with high efficiencies [28-33]. Moreover, baculovirus transduction in vivo provokes expression of various kinds of cytokines and may lead to unwanted adjuvant effects and strong immune responses [28, 34, 35]. Potentially, baculovirus DNA may integrate into the genome of the host cell and consequently may cause DNA damage [28]. We were able to show that by using BTI-TN5B1-4 cells instead of Sf9 cells as production platform for influenza VLPs, the baculovirus background can be reduced about approximately 2 logs from ~108 to ~106 pfu/ml expression supernatant (Fig. 3). The much lower concentration of baculovirus in the BTI-TN5B1-4 expression supernatants simplifies the following purification procedure and results in a much lower baculovirus background of the final preparation.
Until now, production of insect cell derived influenza VLP vaccines has been carried out in Sf9 cells [13-20]. We conclude from our results that BTI-TN5B1-4 cells are an advantageous production system for influenza VLPs as it decreases the baculovirus background and therefore DNA contamination. Furthermore, BTI-TN5B1-4 cells produce homogenous VLPs which carry more HA than their Sf9 derived counterparts. Additionally, Cervarix™, a human papilloma virus vaccine that is approved by the European Medicines Agency (EMEA) is produced in Hi-5 Rix4446 cells, which are derived from a BTI-TN5B1-4 cell subclone [12].
Although it would possibly be necessary to maintain a second cell line for rBV propagation due to the lower replication efficiency of recombinant AcNPV in BTI-TN5B1-4 cells, we recommend to further proof the qualification of BTI-TN5B1-4 cells as production platform and take them into account for influenza VLP vaccine production.
Contributor Information
Florian Krammer, Vienna Institute of BioTechnology, Department of Biotechnology, University of Natural Resources and Applied Life Sciences, 1190 Vienna, Austria.
Theresa Schinko, Vienna Institute of BioTechnology, Department of Biotechnology, University of Natural Resources and Applied Life Sciences, 1190 Vienna, Austria.
Dieter Palmberger, Vienna Institute of BioTechnology, Department of Biotechnology, University of Natural Resources and Applied Life Sciences, 1190 Vienna, Austria.
Christopher Tauer, Vienna Institute of BioTechnology, Department of Biotechnology, University of Natural Resources and Applied Life Sciences, 1190 Vienna, Austria.
Paul Messner, Vienna Institute of Biotechnology, Department of Nanobiotechnology, University of Natural Resources and Applied Life Sciences, 1190 Vienna, Austria.
Reingard Grabherr, Vienna Institute of BioTechnology, Department of Biotechnology, University of Natural Resources and Applied Life Sciences, 1190 Vienna, Austria.
References
- 1.Michaelis M, Doerr H, Cinatl JJ. Novel swine-origin influenza A virus in humans: another pandemic knocking at the door. Medical Microbiology and Immunology. 2009;198(3):175–183. doi: 10.1007/s00430-009-0118-5. [DOI] [PubMed] [Google Scholar]
- 2.Nicholson K, Wood J, Zambon M. Influenza. Lancet. 2003;362:1733–1745. doi: 10.1016/S0140-6736(03)14854-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Erlewyn-Lajeunesse M, Brathwaite N, Lucas J, Warner J. Recommendations for the administration of influenza vaccine in children allergic to egg. BMJ. 2009;339:b3680. doi: 10.1136/bmj.b3680. [DOI] [PubMed] [Google Scholar]
- 4.Biesova Z, Miller M, Schneerson R, Shiloach J, Green K, Robbins J, et al. Preparation, characterization, and immunogenicity in mice of a recombinant influenza H5 hemagglutinin vaccine against the avian H5N1 A/Vietnam/1203/2004 influenza virus. Vaccine. 2009;27:6234–6238. doi: 10.1016/j.vaccine.2009.07.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cox M. Progress on baculovirus-derived influenza vaccines. Current Opinion in Molecular Therapeutics. 2008;10:56–61. [PubMed] [Google Scholar]
- 6.D’Aoust M, Lavoie P, Couture M, Trépanier S, Guay J, Dargis M, et al. Influenza virus-like particles produced by transient expression in Nicotiana benthamiana induce a protective immune response against a lethal viral challenge in mice. Plant Biotechnology Journal. 2008;6:930–940. doi: 10.1111/j.1467-7652.2008.00384.x. [DOI] [PubMed] [Google Scholar]
- 7.Zheng L, Wang F, Yang Z, Chen J, Chang H, Chen Z. A single immunization with HA DNA vaccine by electroporation induces early protection against H5N1 avian influenza virus challenge in mice. BMC Infectious Diseases. 2009;9:17. doi: 10.1186/1471-2334-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szécsi J, Boson B, Johnsson P, Dupeyrot-Lacas P, Matrosovich M, Klenk H, et al. Induction of neutralising antibodies by virus-like particles harbouring surface proteins from highly pathogenic H5N1 and H7N1 influenza viruses. Virology Journal. 2006;3:70. doi: 10.1186/1743-422X-3-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nayak D, Balogun R, Yamada H, Zhou Z, Barman S. Influenza virus morphogenesis and budding. Virus Research. 2009;143(2):147–161. doi: 10.1016/j.virusres.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roy P, Noad R. Virus-like particles as a vaccine delivery system: myths and facts. Human Vaccines. 2008;4:5–12. doi: 10.4161/hv.4.1.5559. [DOI] [PubMed] [Google Scholar]
- 11.Kang S, Song J, Quan F, Compans R. Influenza vaccines based on virus-like particles. Virus Research. 2009;143:140–146. doi: 10.1016/j.virusres.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schiller J, Castellsagué X, Villa L, Hildesheim A. An update of prophylactic human papillomavirus L1 virus-like particle vaccine clinical trial results. Vaccine. 2008;26(Suppl 10):K53–K61. doi: 10.1016/j.vaccine.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bright R, Carter D, Daniluk S, Toapanta F, Ahmad A, Gavrilov V, et al. Influenza virus-like particles elicit broader immune responses than whole virion inactivated influenza virus or recombinant hemagglutinin. Vaccine. 2007;25:3871–3878. doi: 10.1016/j.vaccine.2007.01.106. [DOI] [PubMed] [Google Scholar]
- 14.Mahmood K, Bright R, Mytle N, Carter D, Crevar C, Achenbach J, et al. H5N1 VLP vaccine induced protection in ferrets against lethal challenge with highly pathogenic H5N1 influenza viruses. Vaccine. 2008;26:5393–5399. doi: 10.1016/j.vaccine.2008.07.084. [DOI] [PubMed] [Google Scholar]
- 15.Ross T, Mahmood K, Crevar C, Schneider-Ohrum K, Heaton P, Bright R. A trivalent virus-like particle vaccine elicits protective immune responses against seasonal influenza strains in mice and ferrets. PLoS One. 2009;4:e6032. doi: 10.1371/journal.pone.0006032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perrone L, Ahmad A, Veguilla V, Lu X, Smith G, Katz J, et al. Intranasal vaccination with 1918 influenza virus-like particles protects mice and ferrets from lethal 1918 and H5N1 influenza virus challenge. Journal of Virology. 2009;83:5726–5734. doi: 10.1128/JVI.00207-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quan F, Huang C, Compans R, Kang S. Virus-like particle vaccine induces protective immunity against homologous and heterologous strains of influenza virus. Journal of Virology. 2007;81:3514–3524. doi: 10.1128/JVI.02052-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galarza J, Latham T, Cupo A. Virus-like particle (VLP) vaccine conferred complete protection against a lethal influenza virus challenge. Viral Immunology. 2005;18:244–251. doi: 10.1089/vim.2005.18.244. [DOI] [PubMed] [Google Scholar]
- 19.Latham T, Galarza J. Formation of wild-type and chimeric influenza virus-like particles following simultaneous expression of only four structural proteins. Journal of Virology. 2001;75:6154–6165. doi: 10.1128/JVI.75.13.6154-6165.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pushko P, Tumpey T, Bu F, Knell J, Robinson R, Smith G. Influenza virus-like particles comprised of the HA, NA, and M1 proteins of H9N2 influenza virus induce protective immune responses in BALB/c mice. Vaccine. 2005;23:5751–5759. doi: 10.1016/j.vaccine.2005.07.098. [DOI] [PubMed] [Google Scholar]
- 21.Wickham T, Davis T, Granados R, Shuler M, Wood H. Screening of insect cell lines for the production of recombinant proteins and infectious virus in the baculovirus expression system. Biotechnology Progress. 1992;8:391–396. doi: 10.1021/bp00017a003. [DOI] [PubMed] [Google Scholar]
- 22.King L, Hitchman R, Possee R. Recombinant baculovirus isolation. Methods in Molecular Biology. 2007;388:77–94. doi: 10.1007/978-1-59745-457-5_4. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli U. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Mäkelä A, Tuusa J, Volkman L, Oker-Blom C. Occlusion-derived baculovirus: interaction with human cells and evaluation of the envelope protein P74 as a surface display platform. Journal of Biotechnology. 2008;135:145–156. doi: 10.1016/j.jbiotec.2008.03.014. [DOI] [PubMed] [Google Scholar]
- 25.Sarkar N, Manthey W, Sheffield J. The morphology of murine oncornaviruses following different methods of preparation for electron microscopy. Cancer Research. 1975;35:740–749. [PubMed] [Google Scholar]
- 26.Harris A, Cardone G, Winkler D, Heymann J, Brecher M, White J, et al. Influenza virus pleiomorphy characterized by cryoelectron tomography. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:19123–19127. doi: 10.1073/pnas.0607614103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krammer F, Nakowitsch S, Messner P, Palmberger D, Ferko B, Grabherr R. Swine-origin pandemic H1N1 influenza virus-like particles produced in insect cells induce hemagglutination inhibiting antibodies in BALB/c mice. Biotechnology Journal. 2010;5:17–23. doi: 10.1002/biot.200900267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu Y. Baculoviral vectors for gene delivery: a review. Current Gene Therapy. 2008;8:54–65. doi: 10.2174/156652308783688509. [DOI] [PubMed] [Google Scholar]
- 29.Kost T, Condreay J. Recombinant baculoviruses as expression vectors for insect and mammalian cells. Current Opinion in Biotechnology. 1999;10:428–433. doi: 10.1016/s0958-1669(99)00005-1. [DOI] [PubMed] [Google Scholar]
- 30.Kost T, Condreay J. Recombinant baculoviruses as mammalian cell gene-delivery vectors. Trends in Biotechnology. 2002;20:173–180. doi: 10.1016/s0167-7799(01)01911-4. [DOI] [PubMed] [Google Scholar]
- 31.Nakowitsch S, Kittel C, Ernst W, Egorov A, Grabherr R. Optimization of baculovirus transduction on Free-Style293 cells for the generation of influenza B/Lee/40. Molecular Biotechnology. 2006;34:157–164. doi: 10.1385/mb:34:2:157. [DOI] [PubMed] [Google Scholar]
- 32.Ernst W, Schinko T, Spenger A, Oker-Blom C, Grabherr R. Improving baculovirus transduction of mammalian cells by surface display of a RGD-motif. Journal of Biotechnology. 2006;126:237–240. doi: 10.1016/j.jbiotec.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 33.Spenger A, Ernst W, Condreay J, Kost T, Grabherr R. Influence of promoter choice and trichostatin A treatment on expression of baculovirus delivered genes in mammalian cells. Protein Expression and Purification. 2004;38:17–23. doi: 10.1016/j.pep.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 34.Abe T, Takahashi H, Hamazaki H, Miyano-Kurosaki N, Matsuura Y, Takaku H. Baculovirus induces an innate immune response and confers protection from lethal influenza virus infection in mice. Journal of Immunology. 2003;171:1133–1139. doi: 10.4049/jimmunol.171.3.1133. [DOI] [PubMed] [Google Scholar]
- 35.Abe T, Hemmi H, Miyamoto H, Moriishi K, Tamura S, Takaku H, et al. Involvement of the Toll-like receptor 9 signaling pathway in the induction of innate immunity by baculovirus. Journal of Virology. 2005;79:2847–2858. doi: 10.1128/JVI.79.5.2847-2858.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]