Abstract
Background
Clock genes regulate circadian rhythm and are involved in various physiological processes, including digestion. We therefore investigated the association between the CLOCK 3111T/C single nucleotide polymorphism and the Period3 (PER3) variable-number tandem-repeat polymorphism (either 4 or 5 repeats 54 nt in length) with morning gastric motility.
Methods
Lifestyle questionnaires and anthropometric measurements were performed with 173 female volunteers (mean age, 19.4 years). Gastric motility, evaluated by electrogastrography (EGG), blood pressure, and heart rate levels were measured at 8:30 a.m. after an overnight fast. For gastric motility, the spectral powers (% normal power) and dominant frequency (DF, peak of the power spectrum) of the EGG were evaluated. The CLOCK and PER3 polymorphisms were determined by polymerase chain reaction (PCR) restriction fragment length polymorphism analysis.
Results
Subjects with the CLOCK C allele (T/C or C/C genotypes: n = 59) showed a significantly lower DF (mean, 2.56 cpm) than those with the T/T genotype (n = 114, 2.81 cpm, P < 0.05). Subjects with the longer PER3 allele (PER3 4/5 or PER3 5/5 genotypes: n = 65) also showed a significantly lower DF (2.55 cpm) than those with the shorter PER3 4/4 genotype (n = 108, 2.83 cpm, P < 0.05). Furthermore, subjects with both the T/C or C/C and PER3 4/5 or PER3 5/5 genotypes showed a significantly lower DF (2.43 cpm, P < 0.05) than subjects with other combinations of the alleles (T/T and PER3 4/4 genotype, T/C or C/C and PER3 4/4 genotypes, and T/T and PER3 4/5 or PER3 5/5 genotypes).
Conclusions
These results suggest that minor polymorphisms of the circadian rhythm genes CLOCK and PER3 may be associated with poor morning gastric motility, and may have a combinatorial effect. The present findings may offer a new viewpoint on the role of circadian rhythm genes on the peripheral circadian systems, including the time-keeping function of the gut.
Introduction
The endogenous circadian clock regulates daily oscillations in various behavioral and physiological processes of the human body, including those associated with digestive activity [1]. In mammals, the core molecular clock is present within the hypothalamic suprachiasmatic nucleus (SCN) in the brain, and in nearly all peripheral tissues [1, 2]. The peripheral clocks are organ-specific and tend to be delayed by 3–9 hours when compared to the SCN [3]; therefore, the SCN neurons synchronize peripheral tissue clocks through both neuronal and hormonal pathways [4, 5].
The SCN and peripheral core molecular clocks consist of interconnected positive and negative transcription and translation feedback loops that oscillate every 24 hours [1, 2, 4]. In this system, the forward loop involves a set of transcriptional enhancers that induce the transcription of a set of repressors, and the negative loop feeds back to inhibit the forward loop [3–5]. The core clock genes (CLOCK, Bmal1, Period, Cry) are considered essential elements of this biological molecular system, and genetic variants such as the CLOCK 3111T/C single nucleotide polymorphism (SNP) or the Period3 (PER3) variable-number tandem-repeat (VNTR) polymorphism (either 4 or 5 repeats 54 nt in length) may contribute to the disruption of the circadian clock [6–8].
Peripheral tissue clocks also exist in the gastrointestinal tract (GIT), located in special intestinal cells, with unstable membrane potentials positioned between the longitudinal and circular muscle layers [9–12]. These gut clocks are responsible for the periodic activity of various segments and transit along the GIT [9]. The migrating motor complex starts in the stomach and moves along the gut causing peristaltic contractions when the electrical activity spikes are superimposed on the slow waves [9]. The rhythm of slow waves occurs in the stomach (3 cycles per minute [cpm]), duodenum (12 cpm), jejunum and ileum (7–10 cpm), and colon (12 cpm), and can be cooperatively controlled by the gut and brain SCN clocks [9].
With respect to the stomach, gastric motility shows circadian rhythm as well as mealtime variations in healthy subjects [13]. Similarly, it has been reported that ghrelin, an appetite regulating hormone produced by the stomach, is secreted in anticipation of regularly scheduled mealtime [14]. Indeed, the expression of ghrelin and PER is rhythmic in light-dark cycles and is synchronized to prior feeding, suggesting that the anticipatory activity, such as gastric motility before breakfast, depends on an endogenous circadian timing system [14]. This system called circadian food-entrainable oscillators (FEOs) is closely related to both brain and peripheral clocks, and is located in oxyntic gland cells of stomach [9, 14]. In healthy subjects, ghrelin concentration elevated 1–2 h before breakfast [15], and an increase in gastric motility occurs several hours before the meal [9]. These findings raise the possibility that attenuated gastric locomotor activity before breakfast may reflect the modulation of peripheral and/or central circadian timing system.
Accordingly, we have previously demonstrated the involvement of CLOCK on gut function; subjects with the CLOCK 3111T/C SNP showed slower gastric waves (mean, 2.45 cpm) compared to those with the T/T genotype (2.94 cpm) [16]. Given the existence of several circadian clock genes [6–8], the impact of other clock gene polymorphisms on gut function should further be investigated. We hypothesized that the CLOCK 3111T/C SNP and PER3 VNTR polymorphism may modulate gastric contraction rhythm. Therefore, the present study was designed to investigate sole and combinatorial involvement of these polymorphisms with morning gastric motility in healthy female volunteers.
Materials and Methods
Subjects
We recruited 173 female university students (19.4 ± 0.1 years) from our campus for the present study. All subjects were non-smokers, free of any symptoms or medical history of gastrointestinal, cardiovascular, or any diseases that affect gastric motility, and were not taking any medications. The study protocol was reviewed and approved by the Ethics Committee of the University of Hyogo, and was in accordance with the principles of the Declaration of Helsinki. All subjects provided written informed consent.
Experimental protocol
All subjects were asked to maintain their usual lifestyle and body weight for at least one month before the test. On the day before the test, the consumption of coffee, tea, spicy foods, and high-fat foods was prohibited. Subjects were tested in the morning, from 8:00 to 9:00 a.m., after an overnight fast beginning at 10:00 p.m. of the previous night. After measurement of body mass and percent body fat were using a bioelectrical impedance analyzer (InBody520, Biospace Co., Seoul, Korea), seated blood pressure levels were measured twice at 5 min intervals using a sphygmomanometer (HEM-7250-IT; OMRON, Co., Ltd., Kyoto, Japan). Subjects were then prepared for electrogastrography (EGG) and electrocardiogram (ECG) and allowed to rest for at least 15 min in a temperature-controlled (24–25°C) room in a sitting-up, 45° inclined position. After the resting period, EGG and ECG were continuously recorded for 20 min. The resting heart rate (HR) values were obtained from ECG data. The subjects were separated by partition screens and instructed to maintain their positions for the duration of data collection.
EGG measurement and spectral analysis procedure
To derive bipolar EGG signals, two active electrodes and one ground were positioned on the abdomen, as suggested by the American Motility Society Clinical GI Motility Testing Task Force [17]. The high cut-off frequency of the EGG amplifier was 0.15 Hz, and the low cut-off frequency was 0.016 Hz. These band-pass filters completely eliminated ECG and 60 Hz power source artifacts [16, 18]. The EGG signals were amplified (EGG Amplifier BBA-8321, Bio-tex, Kyoto, Japan) and digitized via a 13-bit analog-to-digital converter (DAQ AD135, Germany) at a sampling rate of 0.5 Hz. The acquired data were sequentially stored on a hard disk for later analysis. The root mean square value of the EGG was calculated as representing the average amplitude. After passing through the hamming-type data window, power spectral analysis by means of a fast Fourier transformation was performed on a consecutive 512 time point series of data obtained during the test. Signal acquisition, storage, and processing were performed on a personal computer. The computer programs for sampling and analysis were written in HTBasic (Trans Era version 9.0, Utah, USA).
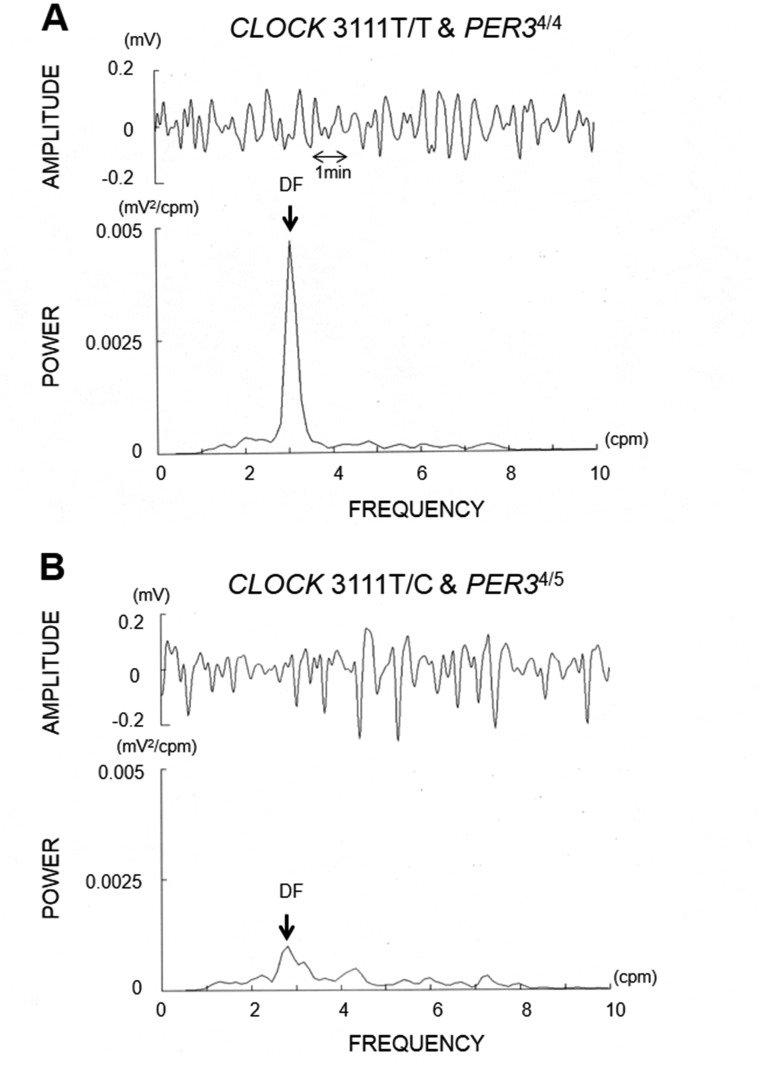
To determine gastric motility, the components of gastric slow wave, normal power (2–4 cpm), % normal power (normal power / total power derived from 1–9 cpm), and dominant frequency (DF) were calculated [16–18]. Raw gastric myoelectrical signals (amplitude) and the corresponding power spectrum are illustrated in Fig. 1.
Fig 1. EGG power spectrum analysis results.
Typical sets of raw EGG waves (top of each figure) and the corresponding power spectral data (bottom of each figure) obtained from a T/T and PER3 4/4 (A) and a T/C and PER3 4/5 genotype subject (B). By visual inspection, in the T/C and PER3 4/5 genotype subject, the DF of the waves was diminished and shifted toward the slower frequency compared to the T/T and PER3 4/4 genotype subject.
Dietary intake and breakfast frequency
Dietary intake and nutritional values were estimated from self-reported, weighed food records with photographs of all consumed food and drink taken using a camera-equipped cellular phone for two typical weekdays. These records were carefully checked by registered dietitians through an interview with each subject on the test day [16, 18]. Energy intake and nutritional values were calculated using computer-assisted procedures (Excel Eiyokun, ver. 6.0, Kenpakusya Co., Tokyo, Japan) based on the Japanese food composition table. A lifestyle questionnaire was also performed; breakfast frequency was divided into three categories to make clear comparisons among subjects who consumed breakfast daily, intermittently, or never.
Genetic analysis
DNA samples were obtained from whole blood using a DNA Quick II, genomic DNA separation kit (DS Pharma Biomedical Co., Ltd., Osaka, Japan). The polymorphisms of CLOCK and PER3 were determined by a polymerase chain reaction (PCR) restriction fragment length polymorphism analysis. The 3111T/C polymorphic region of CLOCK (rs1801260) was amplified using PCR with forward (5’- TCC AGC AGT TTC ATG AGA TGC-3’) and reverse (5’-GAG GTC ATT TCA TAG CTG AGC-3’) primers. The VNTR polymorphic region of PER3 (rs57875989) was amplified using PCR with forward (5'-TTC TAG CAG TGT GTT ACA GGC AAC-3') and reverse (5'-TCC TGA TGC TGC TGA ACC AGT TC-3') primers. PCR amplification was performed under the conditions recommended by the enzyme supplier. In brief, a 10-μL reaction contained 200 ng of genomic DNA, the reaction buffer supplied, 3.0 mM MgCl2, 0.2 mM dNTP, and 1.25 U of rTaq (TAKARA Bio Inc., Tokyo, Japan). For the 3111T/C polymorphic region of CLOCK, cycling parameters were an initial denaturation at 94°C for 3 minutes, then denaturation at 95°C for 30 seconds, annealing at 58°C for 30 seconds, and primer extension at 72°C for 60 seconds for five cycles, then denaturation at 95°C for 30 seconds, annealing at 55°C for 30 seconds, and primer extension at 72°C for 60 seconds for thirty cycles in a thermal cycler (GeneAmp PCR System 2700). The 3111T/C polymorphism was identified by restriction of the 221bp PCR-fragment with Bsp1286I; the C-allele is cut by Bsp1286I. In the VNTR polymorphic region of PER3, cycling parameters were an initial denaturation at 94°C for 3 minutes, then denaturation at 94°C for 30 seconds, annealing at 65°C for 30 seconds, and primer extension at 72°C for 30 seconds for thirty-five cycles, and post-extension at 72°C for 4 minutes in a thermal cycler. The resultant PCR products (335bp and/or 282bp) were separated on 1.5% agarose gels and visualized using ethidium bromide. All samples were checked by two independent investigators, and no samples showed differing results between the investigators.
Statistical analyses
The data were expressed as the mean ± standard errors (s.e.). Comparison between two groups (T/T vs. T/C or C/C genotypes, PER3 4/4 vs. PER3 4/5 or PER3 5/5 genotypes) was tested by unpaired Student’s t-test. Comparison among four groups (T/T and PER3 4/4 genotype, T/C or C/C and PER3 4/4 genotypes, T/T and PER3 4/5 or PER3 5/5 genotypes, and T/C or C/C and PER3 4/5 or PER3 5/5 genotypes) was tested by one way analysis of variance (ANOVA) using post hoc Tukey’s test. In ANOVA, we performed adjustments of the association by multiple regression analysis, adjusted for age, body mass index (BMI) and the four genotype groups. Comparison of breakfast frequency between genotype groups was analyzed by χ2-tests or Fisher exact tests, as appropriate. All analyses were performed using SPSS 20 for Windows (IBM Inc., Tokyo, Japan). P < 0.05 was considered significant.
Results
Distribution of genotypes
Table 1 shows subject characteristics by CLOCK and PER3 polymorphisms. The frequency of the CLOCK −3111 C allele was 0.18, and the longer PER3 allele was 0.21. The genotype frequencies of CLOCK T/T, T/C, and C/C were 0.66, 0.32, and 0.02, and PER3 4/4, PER3 4/5, and PER3 5/5 were 0.63, 0.32, and 0.05, respectively. These genotype frequencies were in Hardy-Weinberg equilibrium (P > 0.05). The CLOCK −3111 T/C and longer PER3 polymorphisms were independent of one other in terms of linkage disequilibrium.
Table 1. Subject characteristics according to genotype of CLOCK and PER3.
| Genotype | |||||||
|---|---|---|---|---|---|---|---|
| CLOCK 3111T/C | PER3 VNTR | ||||||
| Total | T/T | T/C or C/C | P value a | 4/4 | 4/5 or 5/5 | P value a | |
| Number | 173 | 114 | 59 | – | 108 | 65 | – |
| Age (years) | 19.4 ± 0.1 | 19.6 ± 0.2 | 18.9 ± 0.1 | 0.015 | 19.3 ± 0.2 | 19.5 ± 0.2 | 0.41 |
| Height (cm) | 158.5 ± 0.4 | 158.4 ± 0.5 | 158.7 ± 0.7 | 0.73 | 159.0 ± 0.5 | 157.8 ± 0.6 | 0.12 |
| Body mass (kg) | 51.0 ± 0.5 | 50.3 ± 0.5 | 52.3 ± 0.9 | 0.042 | 51.3 ± 0.7 | 50.5 ± 0.7 | 0.42 |
| Body mass Index (kg/m2) | 20.3 ± 0.2 | 20.0 ± 0.2 | 20.7 ± 0.3 | 0.046 | 20.3 ± 0.2 | 20.3 ± 0.2 | 1.00 |
| Body fat (%) | 26.1 ± 0.4 | 25.7 ± 0.5 | 26.8 ± 0.7 | 0.21 | 25.9 ± 0.5 | 26.5 ± 0.7 | 0.50 |
| Systolic blood pressure (mmHg) | 98.3 ± 0.6 | 98.2 ± 0.7 | 98.3 ± 0.9 | 0.92 | 98.3 ± 0.7 | 98.1 ± 0.9 | 0.87 |
| Diastolic blood pressure (mmHg) | 59.3 ± 0.5 | 60.0 ± 0.7 | 58.0 ± 0.8 | 0.067 | 59.9 ± 0.7 | 58.5 ± 0.8 | 0.19 |
| Heart rate (bpm) | 65.6 ± 0.7 | 67.1 ± 0.8 | 62.8 ± 1.0 | 0.002 | 65.9 ± 0.9 | 65.0 ± 0.9 | 0.53 |
| Body temperature (°C) | 36.11 ± 0.32 | 36.13 ± 0.04 | 36.08 ± 0.06 | 0.54 | 36.11 ± 0.04 | 36.12 ± 0.06 | 0.88 |
| % normal power (%) | 50.3 ± 1.3 | 51.6 ± 1.5 | 47.8 ± 2.2 | 0.15 | 51.1 ± 1.5 | 48.9 ± 2.2 | 0.39 |
| Dominant frequency (cpm) | 2.72 ± 0.06 | 2.81 ± 0.07 | 2.56 ± 0.12 | 0.049 | 2.83 ± 0.07 | 2.55 ± 0.11 | 0.021 |
| Enegy intake (kJ/day) | 6735 ± 122 | 6611 ± 146 | 6987 ± 209 | 0.14 | 6669 ± 163 | 6853 ± 172 | 0.47 |
| Sleep duration (hour) | 6.08 ± 0.76 | 6.06 ± 0.10 | 6.12 ± 0.13 | 0.68 | 6.11 ± 0.10 | 6.03 ± 0.11 | 0.66 |
| Breakfast frequency | |||||||
| Daily | 141 (81.5) | 91 (79.8) | 50 (84.7) | 89 (82.4) | 52 (80.0) | ||
| Intermettently | 26 (15.0) | 18 (15.8) | 8 (13.6) | 0.59 b | 14 (13.0) | 12 (18.5) | 0.38 b |
| Never | 6 (3.5) | 5 (4.4) | 1 (1.7) | 5 (4.6) | 1 (1.5) |
Values are expressed as subject numbers or mean ± s.e. or number and frequency (%).
a Unpaired Student's t-test (T/T vs. T/C or C/C, PER3 4/4 vs. PER3 4/5 or 5/5).
b χ2-tests or Fisher's exact test (T/T vs. T/C or C/C, PER3 4/4 vs. PER3 4/5 or 5/5).
The association of CLOCK or PER3 polymorphisms with biological parameters
To examine the association between the CLOCK C allele and biological parameters, subjects were divided into two groups either with (T/C or C/C genotypes) or without (T/T genotype) the C allele. Likewise, to examine the association between the longer PER3 allele and biological function, subjects were divided into two groups either with (PER3 4/5 or PER3 5/5 genotypes) or without (PER3 4/4 genotype) the longer allele. Carriers of the CLOCK C allele showed significantly higher body mass and BMI compared to subjects with the T/T genotype. Additionally, resting HR and the DF were significantly lower in C allele carriers than in T/T genotype subjects, in confirmation with our previous report [16]. Carriers of the longer PER3 allele (PER3 4/5 or PER3 5/5 genotypes) had a significantly lower DF compared with subjects with the PER3 4/4 genotype. No significant differences were found in breakfast frequency and other parameters between the groups (Table 1).
Combinatorial effect of CLOCK and PER3 polymorphisms on biological parameters
Subjects were classified into four combination groups of genotypes based on two gene polymorphisms; T/T and PER3 4/4 genotype (frequency, 0.42), T/C or C/C and PER3 4/4 genotypes (0.20), T/T and PER3 4/5 or PER3 5/5 genotypes (0.24), and T/C or C/C and PER3 4/5 or PER3 5/5 genotypes (0.14), respectively, to examine the combinatorial effect of each allele on biological parameters (Table 2).
Table 2. Comparison of biological parameters and breakfast frequency among four genotype groups.
| Genotype | ||||||
|---|---|---|---|---|---|---|
| Total | T/T | T/C or C/C | T/T | T/C or C/C | P value a | |
| 4/4 | 4/4 | 4/5 or 5/5 | 4/5 or 5/5 | |||
| Number | 173 | 73 | 35 | 41 | 24 | – |
| Age (years) | 19.4 ± 0.1 | 19.5 ± 0.2 | 18.7 ± 0.2 | 19.7 ± 0.3 | 19.2 ± 0.2 | 0.065 |
| Height (cm) | 158.5 ± 0.4 | 158.7 ± 0.6 | 159.7 ± 1.0 | 158.1 ± 0.8 | 157.3 ± 0.9 | 0.29 |
| Body mass (kg) | 51.0 ± 0.5 | 50.1 ± 0.7 | 53.7 ± 1.3b | 50.6 ± 0.9 | 50.4 ± 1.1 | 0.061 |
| Body mass Index (kg/m2) | 20.3 ± 0.2 | 19.9 ± 0.3 | 21.0 ± 0.4 | 20.2 ± 0.3 | 20.3 ± 0.4 | 0.11 |
| Body fat (%) | 26.1 ± 0.4 | 25.3 ± 0.6 | 27.1 ± 1.0 | 26.5 ± 0.9 | 26.4 ± 1.0 | 0.39 |
| Systolic blood pressure (mmHg) | 98.3 ± 0.6 | 98.3 ± 1.0 | 98.3 ± 1.0 | 98.0 ± 1.1 | 98.4 ± 1.8 | 1.00 |
| Diastolic blood pressure (mmHg) | 59.3 ± 0.5 | 60.6 ± 0.9 | 58.3 ± 1.0 | 59.0 ± 1.0 | 57.6 ± 1.3 | 0.17 |
| Heart rate (bpm) | 65.6 ± 0.7 | 67.1 ± 1.2 | 63.4 ± 1.3 | 67.0 ± 1.1 | 61.8 ± 1.5 b | 0.017 |
| Body temperature (°C) | 36.11 ± 0.32 | 36.12 ± 0.05 | 36.09 ± 0.07 | 36.14 ± 0.07 | 36.07 ± 0.11 | 0.92 |
| % normal power (%) | 50.3 ± 1.3 | 53.7 ± 1.7 | 45.6 ± 2.9 | 47.7 ± 2.8 | 50.9 ± 3.3 | 0.069 |
| Dominant frequency (cpm) | 2.72 ± 0.06 | 2.92 ± 0.07 | 2.65 ± 0.15 | 2.62 ± 0.13 | 2.43 ± 0.19 b | 0.030 |
| Enegy intake (kJ/day) | 6735 ± 122 | 6514 ± 200 | 7073 ± 300 | 6764 ± 209 | 6887 ± 280 | 0.37 |
| Sleep duration (hour) | 6.08 ± 0.76 | 6.06 ± 0.13 | 6.19 ± 0.17 | 6.04 ± 0.14 | 6.03 ± 0.20 | 0.91 |
| Breakfast frequency | ||||||
| Daily | 141 (81.5) | 60 (82.2) | 29 (82.9) | 31 (75.6) | 21 (87.5) | |
| Intermettently | 26 (15.0) | 9 (12.3) | 5 (14.3) | 9 (22.0) | 3 (12.5) | 0.69 c |
| Never | 6 (3.5) | 4 (5.5) | 1 (2.8) | 1 (2.4) | 0 (0) |
Values are expressed as subject numbers or mean ± s.e. or number and frequency (%).
a ANOVA using post hoc Turkey's test.
b P < 0.05 vs. T/T and PER3 4/4
c χ2-tests or Fisher's exact test.
Blood pressure
Average systolic and diastolic BP of the four groups was shown in Table 2. There was no significant difference among the four groups.
Resting heart rate
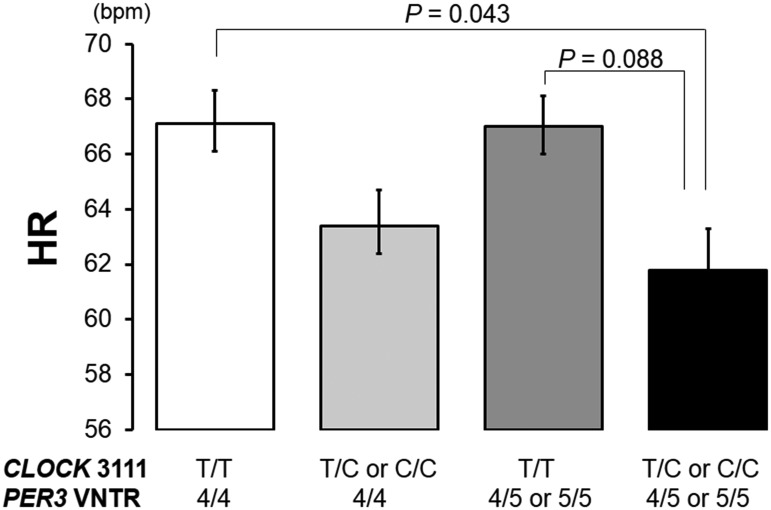
Average resting HR of the four groups was shown in Table 2. There were significant differences in the resting HR among the four groups (ANOVA, F = 3.49, P = 0.017). As shown in Fig. 2, subjects with the T/C or C/C and PER3 4/5 or PER3 5/5 genotypes had a significantly lower resting HR compared with subjects with the T/T and PER3 4/4 genotype (P = 0.043), indicating a combinatorial effect of the CLOCK −3111 C and longer PER3 alleles on decreased resting HR in the morning. Additionally, the resting HR of subjects with the T/C or C/C and PER3 4/5 or PER3 5/5 genotypes tended to be lower than in subjects with T/T and PER3 4/5 or PER3 5/5 genotypes (P = 0.088). After adjustment for age (β = 0.09, P = 0.26) and BMI (β = −0.05, P = 0.48), the difference among four genotype groups remained significant (R = 0.25, β = −0.21, P = 0.007).
Fig 2. The resting HR of the four combined polymorphic types.
Subjects with both T/C or C/C and PER3 4/5 or PER3 5/5 genotypes displayed the lowest resting HR values among the four groups. Additionally, there was a significant difference in the resting HR between the T/T and PER3 4/4 genotype and T/C or C/C and PER3 4/5 or PER3 5/5 genotypes (ANOVA followed by post-hoc Tukey’s test).
EGG parameters
Average % normal power of the four groups was shown in Table 2. The % normal power of subjects with the T/T and PER3 4/4 genotype tended to be greater than in subjects with the other genotypes (ANOVA, F = 2.41, P = 0.069).
DF
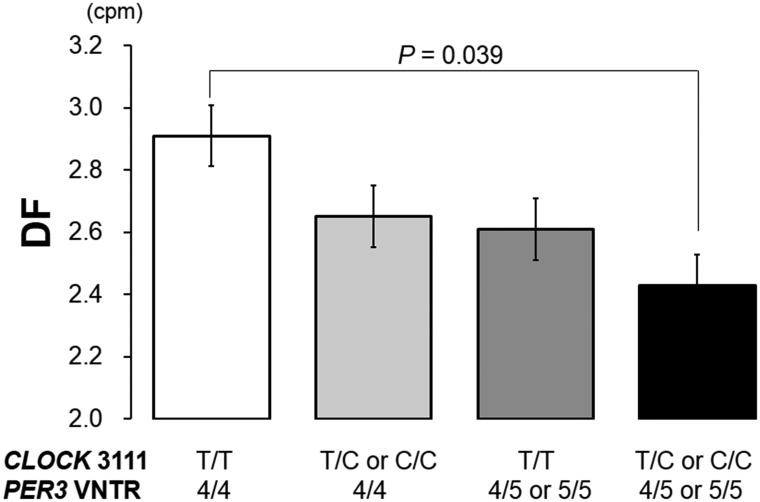
Fig. 1 represents typical sets of raw EGG waves (top of each figure) and the corresponding power spectral data (bottom of each figure) obtained from a T/T and PER3 4/4 (A) and a T/C and PER3 4/5 (B) subject. By visual inspection, in the T/C and PER3 4/5 subject, the DF of the waves was diminished and shifted toward the slower frequency compared to the T/T and PER3 4/4 subject. The group data indicated that there were significant differences in the DF among the four groups (ANOVA, F = 3.06, P = 0.030) (Table 2); combination of the CLOCK −3111 C and longer PER3 alleles had a stronger impact on the DF than that of either polymorphism. As shown in Fig. 3, subjects with the T/C or C/C and PER3 4/5 or PER3 5/5 genotypes displayed the lowest DF values among the four groups, suggesting the slowest gastric motility. Additionally, a significant difference in the DF was found between the T/T and PER3 4/4 genotype and the T/C or C/C and PER3 4/5 or PER3 5/5 genotypes (P = 0.039) (Fig. 3). After adjustment for age (β = 0.07, P = 0.35) and BMI (β = –0.05, P = 0.51), the difference among four genotype groups remained significant (R = 0.23, β = –0.19, P = 0.013). These results imply a combinatorial effect of the CLOCK −3111 C and longer PER3 alleles on morning gastric motility in the fasting condition.
Fig 3. The DF of the four combined polymorphic types.
Subjects with both T/C or C/C and PER3 4/5 or PER3 5/5 genotypes displayed the lowest DF values among the four groups, suggesting the slowest gastric motility. Additionally, there was a significant difference in the DF between the T/T and PER3 4/4 genotype and T/C or C/C and PER3 4/5 or PER3 5/5 genotypes (ANOVA followed by post-hoc Tukey’s test).
No significant differences were found among the four groups for energy intake, breakfast frequency, or sleep duration (Table 2). These variables did not have any association with the measured biological parameters (data not shown).
Discussion
Genetic and metabolic circadian oscillators are coupled to synchronize between the light-dark/feeding-fasting cycles and human internal biochemical processes [19]. Therefore, genetic variations in the circadian rhythm genes may influence not only circadian time-keeping but also the energetic cycles in humans [19]. The present study examined the association of the CLOCK 3111T/C SNP and PER3 VNTR polymorphism with morning gastric motility in humans. The novelty of this study is that 1) the subjects carrying the minor allele at CLOCK or PER3 possessed significantly lower DF values, suggesting poor gastric motility compared to non-carrier subjects, and that 2) the subjects carrying both minor alleles at CLOCK and PER3 showed the lowest DF value of the groups, suggesting this combined polymorphism may be linked to a greater suppression of gastric motility in the morning.
It has been thought that the CLOCK 3111T/C SNP is associated with higher plasma ghrelin concentrations, altered eating behaviors, higher total energy intake, and resistance to weight loss [20–22], implying a genetic susceptibility to obesity [7]. Our subjects with the CLOCK C allele also had a modest but significantly higher BMI than subjects with the T/T genotype. Interestingly, a recent report [23] using actimetry observed that CLOCK C allele carriers were less active and started their activities later in the morning. In the present study, CLOCK C allele carriers showed decreased HR, slightly lower diastolic BP, and poor gastric motility in the morning. Such attenuated cardiovascular function together with the gastric function that characterizes C allele carriers may be attributable to behavior or performance after wake-up.
Intriguingly, not only sole (CLOCK) but combinatorial effect of CLOCK and PER3 VNTR miner alleles on the resting HR were observed in the present study. In an animal study [24], cardiomyocyte-specific circadian clock mutant mice showed bradycardia and attenuation of HR diurnal variations, suggesting that peripheral CLOCK gene in heart may directly influence myocardial contractile function. A grovel understanding of the role of circadian clock genes in cardiovascular function is still lacking in humans; however, there are a few reports regarding the association between CLOCK or PER3 polymorphism and the resting HR. As to the CLOCK 3111T/C SNP, we recently reported that the CLOCK C allele carriers had a lower resting HR in the morning [16]. In agreement of the present results, a previous research suggested that the PER3 VNTR polymorphism was not associated with resting HR at any time point [25]. By interacting with CLOCK; however, PER3 could have a greater impact on cardiovascular function or parasympathetic-sympathetic nervous system balance in the morning.
The role of PER3 in regulating metabolism and adiposity has been described in animal studies [26, 27], but is not yet clear in humans [28]. We found no relationship between the PER3 polymorphism and BMI, suggesting this polymorphism may have only a relatively modest effect on metabolic function. On the other hand, previous findings regarding the role of PER3 on habitual and behavioral circadian rhythm in humans have been controversial. The shorter 4-repeat [29–31] or longer 5-repeat [32] alleles of PER3 have been associated with diurnal preference. Earlier wake/bed time [33] and favorable sleep homeostasis [34, 35] were observed in the longer 5-repeat allele carriers. However, some studies reported no correlation between the PER3 polymorphism and habitual sleep patterns or morningness-eveningness phenotypes in healthy volunteers [34–37] and university students [38].
In the present study, although usual sleep duration as well as breakfast frequency did not differ across genotype groups, the longer 5-repeat allele (PER3 4/5 or PER3 5/5 genotypes) showed slower morning gastric motility. Interestingly, recent investigations have emphasized that the PER3 5/5 genotype has a detrimental impact on sleep deprivation compared to the PER3 4/4 genotype [34–37]. Subjects with the PER3 5/5 genotype showed good quality of sleep; however, under sleep deprivation conditions, demonstrated poor cognitive capacity [34–36, 39], declined time-on-task performance [37], and greater sleepiness [34]. It is possible that sleep deficiency-related vulnerability in attention may be enhanced by particular genotypes under sleep deficient conditions. These findings raise the possibility that the diminished morning gastric motility observed in the subjects with the longer 5-repeat allele (PER3 4/5 or PER3 5/5 genotypes) may be a physiological vulnerability in response to short sleep time. It has been reported that Japanese women’s sleep duration is the shortest in the world [40, 41]; participants of the present study were also short-time sleepers (approximately six hours, see Table 1, 2). Being in such a chronically vigilant state may selectively influence gut motility in the longer 5-repeat allele carriers.
In addition to short-sleep, Chellappa et al. [42, 43] provided intriguing evidence that the exposure to blue-enriched light may attenuate sleep-wake regulation in subjects with the PER3 5/5 genotype. In short, the PER3 5/5 genotype exhibited a more pronounced alert response to a two-hour exposure to blue light in the evening. In modern society, there are many opportunities to be exposed to blue-enriched light via smart phones, mobile computers, and large-screen televisions. We hypothesize that the combination of light sensitivity and vulnerability to short-sleep may have a serious effect on the biological circadian system, particularly in PER3 5/5 individuals. To what extent this polymorphism may influence gut function warrants future studies under controlled laboratory conditions.
The present study has some limitations. First, this study had selection bias; all subjects were female university students, although the homogeneity of the studied population was a strength of the study. Further studies including different populations should be performed. Second, the signals recorded by EGG correlated to some degree with gastric emptying, and hence with gastric motility, but these signals may not be exactly correlated to actual gastric motility; therefore, the data should be interpreted carefully. Finally, ghrelin, following the circadian rhythm, is known to act as a stimulant on gastric motility, but was not measured. Future experiment for understanding the diurnal variations of gastric motility, gut hormones and related biological parameters is needed. Notwithstanding, the present study provides the first evidence of the sole and combinatorial effect of the CLOCK 3111T/C SNP and PER3 VNTR polymorphism on morning gastric motility.
In conclusion, our results suggest that minor polymorphic genotypes of the circadian rhythm genes CLOCK and PER3 may be associated with poor morning gastric motility, and may also have a combinatorial effect. These findings may offer a new viewpoint on the role of circadian rhythm genes on the peripheral circadian system, including the time-keeping function of the gut.
Acknowledgments
We are grateful to all subjects of the study for their cooperation.
Data Availability
All relevant data are within the paper.
Funding Statement
This study was supported by a Grant-in-Aid for Science Research from the Ministry of Education and Science of Japan (24500988). URL of funder's website: http://www.jsps.go.jp/english/e-grants/index.html. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. King DP, Takahashi JS. Molecular genetics of circadian rhythms in mammals. Annu Rev Neurosci. 2000;23: 713–742. [DOI] [PubMed] [Google Scholar]
- 2. Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330 (6009): 1349–1354. 10.1126/science.1195027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zylka MJ, Shearman LP, Weaver DR, Reppert SM. Three period homologs in mammals: differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron. 1998;20(6): 1103–1110. [DOI] [PubMed] [Google Scholar]
- 4. Lowrey PL, Takahashi JS. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet. 2004;5: 407–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ramsey KM, Affinati AH, Peek CB, Marcheva B, Hong HK, Bass J. Circadian measurements of sirtuin biology. Methods Mol Biol. 2013;1077: 285–302. 10.1007/978-1-62703-637-5_19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pendergast JS, Niswender KD, Yamazaki S. Tissue-specific function of Period3 in circadian rhythmicity. PLoS One. 2012;7(1): e30254 10.1371/journal.pone.0030254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Marcheva B, Ramsey KM, Peek CB, Affinati A, Maury E, Bass J. Circadian clocks and metabolism. Handb Exp Pharmacol. 2013;(217): 127–155. 10.1007/978-3-642-25950-0_6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Etain B, Milhiet V, Bellivier F, Leboyer M. Genetics of circadian rhythms and mood spectrum disorders. Eur Neuropsychopharmacol. 2011;Suppl 4: S676–S682. 10.1016/j.euroneuro.2011.07.007 [DOI] [PubMed] [Google Scholar]
- 9. Konturek PC, Brzozowski T, Konturek SJ. Gut clock: implication of circadian rhythms in the gastrointestinal tract. J Physiol Pharmacol. 2011;62(2): 139–150. [PubMed] [Google Scholar]
- 10. Hoogerwerf WA, Hellmich HL, Cornélissen G, Halberg F, Shahinian VB, Bostwick J, et al. Clock gene expression in the murine gastrointestinal tract: endogenous rhythmicity and effects of a feeding regimen. Gastroenterology. 2007;133(4): 1250–1260. [DOI] [PubMed] [Google Scholar]
- 11. Sladek M, Rybova M, Jindrakova Z, Zemanová Z, Polidarová L, Mrnka L, et al. Insight into the circadian clock within rat colonic epithelial cells. Gastroenterology. 2007;133(4): 1240–1249. [DOI] [PubMed] [Google Scholar]
- 12. Hoogerwerf WA. Role of clock genes in gastrointestinal motility. Am J Physiol Gastrointest Liver Physiol. 2010;299(3): G549–G555. 10.1152/ajpgi.00147.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Suzuki A, Asahina M, Ishikawa C, Asahina KM, Honma K, Fukutake T, et al. Impaired circadian rhythm of gastric myoelectrical activity in patients with multiple system atrophy. Clin Auton Res. 2005;15(6): 368–372. [DOI] [PubMed] [Google Scholar]
- 14. LeSauter J, Hoque N, Weintraub M, Pfaff DW, Silver R. Stomach ghrelin-secreting cells as food-entrainable circadian clocks. Proc Natl Acad Sci USA. 2009;106(32): 13582–13587. 10.1073/pnas.0906426106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cummings DE, Purnell JQ, Frayo RS, Schmidova K, Wisse BE, Weigle DS. A preprandial rise in plasma ghrelin levels suggests a role in meal initiation in humans. Diabetes. 2001;50(8): 1714–1719. [DOI] [PubMed] [Google Scholar]
- 16. Yamaguchi M, Kotani K, Sakane N, Tsuzaki K, Takagi A, Wakisaka S, et al. The CLOCK 3111T/C SNP is associated with morning gastric motility in healthy young women. Physiol Behav. 2012;107(1): 87–91. 10.1016/j.physbeh.2012.06.006 [DOI] [PubMed] [Google Scholar]
- 17. Parkman HP, Hasler WL, Barnett JL, Eaker EY; American Motility Society Clinical GI Motility Testing Task Force. Electrogastropraphy: a document prepared by the gastric section of the American Motility Society Clinical GI Motility Testing Task Force. Neurogastroenterol Motil. 2003;15(2): 89–102. [DOI] [PubMed] [Google Scholar]
- 18. Wakisaka S, Nagai H, Mura E, Matsumoto T, Moritani T, Nagai N. The effects of carbonated water upon gastric and cardiac activities and fullness in healthy young women. J Nutr Sci Vitaminol (Tokyo). 2012;58(5): 333–338. [DOI] [PubMed] [Google Scholar]
- 19. Bass J, Takahashi JS. Circadian rhythms: Redox redux. Nature. 2011;469(7331): 476–478. 10.1038/469476a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Garaulet M, Corbalán MD, Madrid JA, Morales E, Baraza JC, Lee YC, et al. CLOCK gene is implicated in weight reduction in obese patients participating in a dietary programme based on the Mediterranean diet. Int J Obes (Lond). 2010;34(3): 516–523. 10.1038/ijo.2009.255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Garaulet M, Lee YC, Shen J, Parnell LD, Arnett DK, Tsai MY, et al. Genetic variants in human CLOCK associate with total energy intake and cytokine sleep factors in overweight subjects (GOLDN population). Eur J Hum Genet. 2010;18(3): 364–369. 10.1038/ejhg.2009.176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Garaulet M, Sánchez-Moreno C, Smith CE, Lee YC, Nicolás F, Ordovás JM. Ghrelin, sleep reduction and evening preference: relationships to CLOCK 3111 T/C SNP and weight loss. PLoS One. 2011;6(2): e17435 10.1371/journal.pone.0017435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bandín C, Martinez-Nicolas A, Ordovás JM, Ros Lucas JA, Castell P, Silvente T, et al. Differences in circadian rhythmicity in CLOCK 3111T/C genetic variants in moderate obese women as assessed by thermometry, actimetry and body position.Int J Obes (Lond). 2013;37(8): 1044–1050. 10.1038/ijo.2012.180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bray MS, Shaw CA, Moore MW, Garcia RA, Zanquetta MM, Durgan DJ, et al. Disruption of the circadian clock within the cardiomyocyte influences myocardial contractile function, metabolism, and gene expression. Am J Physiol Heart Circ Physiol. 2008;294(2): H1036–H1047. [DOI] [PubMed] [Google Scholar]
- 25. Viola AU, James LM, Archer SN, Dijk DJ. PER3 polymorphism and cardiac autonomic control: effects of sleep debt and circadian phase. Am J Physiol Heart Circ Physiol. 2008;295(5): H2156–H2163. 10.1152/ajpheart.00662.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dallmann R, Weaver DR. Altered body mass regulation in male mPeriod mutant mice on high-fat diet. Chronobiol Int. 2010;27(6): 1317–1328. 10.3109/07420528.2010.489166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Costa MJ, So AY, Kaasik K, Krueger KC, Pillsbury ML, Fu YH, et al. Circadian rhythm gene period 3 is an inhibitor of the adipocyte cell fate. J Biol Chem. 2011;286(11): 9063–9070. 10.1074/jbc.M110.164558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Otway DT, Mäntele S, Bretschneider S, Wright J, Trayhurn P, Skene DJ, et al. Rhythmic diurnal gene expression in human adipose tissue from individuals who are lean, overweight, and type 2 diabetic. Diabetes. 2011;60(5): 1577–1581. 10.2337/db10-1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Archer SN, Robilliard DL, Skene DJ, Smits M, Williams A, Arendt J, et al. A length polymorphism in the circadian clock gene Per3 is linked to delayed sleep phase syndrome and extreme diurnal preference. Sleep. 2003;26(4): 413–415. [DOI] [PubMed] [Google Scholar]
- 30. Jones KH, Ellis J, von Schantz M, Skene DJ, Dijk DJ, Archer SN. Age-related change in the association between a polymorphism in the PER3 gene and preferred timing of sleep and waking activities. J Sleep Res. 2007;16(1): 12–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Parsons MJ, Lester KJ, Barclay NL, Archer SN, Nolan PM, Eley TC, et al. Polymorphisms in the circadian expressed genes PER3 and ARNTL2 are associated with diurnal preference and GNβ3 with sleep measures. J Sleep Res. 2014;23(5): 595–604. 10.1111/jsr.12144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pereira DS, Tufik S, Louzada FM, Benedito-Silva AA, Lopez AR, Lemos NA, et al. Association of the length polymorphism in the human Per3 gene with the delayed sleep-phase syndrome: does latitude have an influence upon it? Sleep. 2005;28(1): 29–32. [PubMed] [Google Scholar]
- 33. Lázár AS, Slak A, Lo JC, Santhi N, von Schantz M, Archer SN, et al. Sleep, diurnal preference, health, and psychological well-being: a prospective single-allelic-variation study. Chronobiol Int. 2012;29(2): 131–146. 10.3109/07420528.2011.641193 [DOI] [PubMed] [Google Scholar]
- 34. Goel N, Banks S, Mignot E, Dinges DF. PER3 polymorphism predicts cumulative sleep homeostatic but not neurobehavioral changes to chronic partial sleep deprivation. PLoS One. 2009;4(6): e5874 10.1371/journal.pone.0005874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Viola AU, Archer SN, James LM, Groeger JA, Lo JC, Skene DJ, et al. PER3 polymorphism predicts sleep structure and waking performance. Curr Biol. 2007;17(7): 613–618. [DOI] [PubMed] [Google Scholar]
- 36. Maire M, Reichert CF, Gabel V, Viola AU, Strobel W, Krebs J, et al. Sleep ability mediates individual differences in the vulnerability to sleep loss: evidence from a PER3 polymorphism. Cortex. 2014;52: 47–59. 10.1016/j.cortex.2013.11.008 [DOI] [PubMed] [Google Scholar]
- 37. Maire M, Reichert CF, Gabel V, Viola AU, Krebs J, Strobel W, et al. Time-on-task decrement in vigilance is modulated by inter-individual vulnerability to homeostatic sleep pressure manipulation. Front Behav Neurosci. 2014;8: 59 10.3389/fnbeh.2014.00059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Osland TM, Bjorvatn BR, Steen VM, Pallesen Sl. Association study of a variable-number tandem repeat polymorphism in the clock gene PERIOD3 and chronotype in Norwegian university students. Chronobiol Int. 2011;28(9): 764–770. 10.3109/07420528.2011.607375 [DOI] [PubMed] [Google Scholar]
- 39. Dijk DJ, Archer SN. PERIOD3, circadian phenotypes, and sleep homeostasis. Sleep Med Rev. 2010;14(3): 151–160. 10.1016/j.smrv.2009.07.002 [DOI] [PubMed] [Google Scholar]
- 40.Organization for Economic Co-operation and Development Balancing paid work, unpaid work and leisure. (Accessed 2015/02/06) https://www.oecd.org/gender/data/balancingpaidworkunpaidworkandleisure.htm.
- 41. Steptoe A, Peacey V, Wardle J. Sleep duration and health in young adults. Arch Intern Med. 2006;166(16): 1689–1692. [DOI] [PubMed] [Google Scholar]
- 42. Chellappa SL, Viola AU, Schmidt C, Bachmann V, Gabel V, Maire M, et al. Light modulation of human sleep depends on a polymorphism in the clock gene Period3. Behav Brain Res. 2014;271: 23–29. 10.1016/j.bbr.2014.05.050 [DOI] [PubMed] [Google Scholar]
- 43. Chellappa SL, Viola AU, Schmidt C, Bachmann V, Gabel V, Maire M, et al. Human melatonin and alerting response to blue-enriched light depend on a polymorphism in the clock gene PER3. J Clin Endocrinol Metab. 2012;97(3): E433–E437. 10.1210/jc.2011-2391 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.