Abstract
Targeted cancer therapies have produced substantial clinical responses but most tumors develop resistance to these drugs. Here we describe a pharmacogenomic platform that facilitates rapid discovery of drug combinations that can overcome resistance. We established cell culture models derived from biopsy samples of lung cancer patients whose disease had progressed while on treatment with EGFR or ALK tyrosine kinase inhibitors and then subjected these cells to genetic analyses and a pharmacological screen. Multiple effective drug combinations were identified. For example, the combination of ALK and MEK inhibitors was active in an ALK-positive resistant tumor that had developed a MAP2K1 activating mutation, and the combination of EGFR and FGFR inhibitors was active in an EGFR mutant resistant cancer with a novel mutation in FGFR3. Combined ALK and SRC inhibition was effective in several ALK-driven patient-derived models, a result not predicted by genetic analysis alone. With further refinements, this strategy could help direct therapeutic choices for individual patients.
Genotype-based selection of patients for the application of targeted therapies has had a significant impact on the treatment of cancers. Effective targeted therapies, such as tyrosine kinase inhibitors (TKIs), are widely used to treat patients harboring non-small cell lung cancers (NSCLCs) with activating mutations in EGFR or ALK translocations (1–5). However, acquired resistance to these inhibitors eventually develops through a variety of mechanisms, usually within 1–2 years [EGFR inhibitors reviewed in (6) and ALK inhibitors in (7–9)]. In particular, secondary resistance mutations can develop in the oncogene preventing target inhibition by the corresponding TKI (e.g., EGFR T790M or ALK L1196M). Alternatively, resistant cells can develop a compensatory signaling pathway, or “bypass track”, that reestablishes activation of key downstream proliferation and survival signals despite inhibition of the original oncogene [reviewed in (10)]. As more drugs are developed that effectively overcome secondary resistance mutations in the targeted genes, these bypass track mechanisms of resistance will likely continue to emerge in the clinical setting.
Current efforts to understand resistance often center on two different strategies. One approach is to model the development of resistance in vitro using sensitive cell line models that are exposed to a specific targeted therapy until resistance emerges. A second approach focuses on the genetic analyses of resistant biopsies to identify new genetic anomalies that could be driving resistance. However, both approaches have deficiencies. Although the in vitro derived resistant cells are amenable to functional studies, it is unknown which models are clinically relevant, and they could never be used to inform treatment decisions for individual patients. Furthermore, there are few genetically appropriate cell lines in existence that could be used to develop such resistant models (e.g., there are less than 10 existing EGFR mutant and less than 5 EML4-ALK cell lines). Thus, these lines may model only a subset of potential resistance mechanisms. In contrast, studying the genetics of resistant biopsies has the advantage that the discovered genetic alterations actually occurred clinically. These studies can facilitate the development of hypotheses about what is causing resistance, and even speculation as to how one might treat individual patients. However, since the tissue is nonviable, such hypotheses cannot be directly tested on the resistant tumor cells. Furthermore, many resistant cancers do not harbor genetic abnormalities that clearly point to a treatment strategy. In this study, we describe a discovery platform that integrates the genetics of cancers with acquired resistance with pharmacologic interrogation of cell lines systematically developed from those same resistant patient tumors. This permits the discovery and evaluation of therapeutic strategies for clinically relevant mechanisms of resistance.
Establishment of resistant cell lines from clinical specimens
The ability to develop cell lines directly from patient specimens has been facilitated by recent technological advances, including methodologies developed by Schlegel and colleagues utilizing irradiated feeder cells (11). As shown in Table S1, NSCLC cell lines were developed with a ~ 50% success rate from patient samples (effusions and biopsies), including a 38% success rate from biopsy samples. Of note, the majority of the failures were associated with low cancer cellularity in the sample (see below). For many of these samples, cell viability was established on feeder cells and then transitioned off those cells prior to characterization and screening. As shown in Table S2, the oncogenic mutation (EGFR or ALK) present in the patient tumor was reliably identified in the derived cell line.
To identify effective drug combinations in these patient-derived models of acquired resistance, we built upon previous work identifying bypass track mechanisms of acquired resistance (10). In this type of resistance, the original driver oncogene and a secondary bypass track redundantly maintain downstream signaling, such as the PI3K and MAPK pathways, to promote cell survival and proliferation. These cancers are resistant to single-agent inhibition of the primary driver oncogene, and are similarly resistant to single-agent inhibition of the acquired bypass track because, in either case, the untargeted pathway maintains downstream signaling. However, simultaneous inhibition of both pathways suppresses downstream signaling, resulting in growth arrest and cell death (Fig. S1A) (12–15). Thus, drugs targeting relevant bypass tracks are effective when administered in combination with an inhibitor of the primary driver oncogene, but relatively impotent when administered as single-agents (Fig. S1B). Based on this principle, to discover effective therapeutic strategies and gain insight into the underlying mechanisms of resistance, we performed a screen that combined the original TKI (targeting the driving oncogene) with each of the drugs in an established panel.
We assembled a panel of 76 targeted agents (Table S3) directed against a range of key regulators of cell proliferation and survival, including growth factor and development signaling pathways, apoptosis regulators, transcription and protein folding machinery, and DNA damage sensors (Table S4). This drug panel included inhibitors of previously identified bypass tracks as well as several additional clinical targets. The potency of each drug was tested across a 10,000-fold range both in the presence and absence of a fixed concentration of the primary TKI (Fig. S2A). Resultant changes in GI50 (the drug concentration necessary to obtain 50% less cells than in the untreated condition) and AUC (area under the dose-response curve) were determined following addition of the primary TKI.
To evaluate the potential of our strategy we initially examined five previously established models of acquired resistance developed in vitro (i.e., by chronically exposing sensitive cells to TKI in vitro) with known resistance bypass tracks. In these models, the known mechanisms of resistance were identified by our approach with high specificity: For example, in a previously characterized EGFR mutant cell line with MET amplification (12), the MET inhibitors were the sole hits identified in the screen (Fig. S2B, C). In four tested cell lines [HCC827 GR6 (13), HN11 GR (16), SNU638 C1 (17) and H3122 PFR3 (7)] drugs that target known bypass tracks were among those producing the largest shifts in GI50 and AUC (Fig. S3A–D). In the fifth model [A431 GR (16)] the effect of IGFR1 inhibitors was less marked but recapitulated the previously observed combination effect (Fig. S3E). Thus, in these previously investigated models, unbiased screening of a 76 drug panel successfully identified inhibitors of the known bypass tracks. We therefore applied the approach to 55 models of acquired resistance with unknown mechanisms of resistance. Twenty of these models were derived directly from patients who had progressed on either an ALK inhibitor (n=9) or an EGFR inhibitor (n=11). The remaining lines were derived in vitro (Table S5). To compare the information yielded by genetic analysis to the pharmacologic interrogation, patient-derived models were also analyzed by next-generation sequencing to identify potential genetic causes of resistance (Table S6, S7, Database S1, S2).
Effective Drug Combinations in Patient-Derived Resistant NSCLC Models
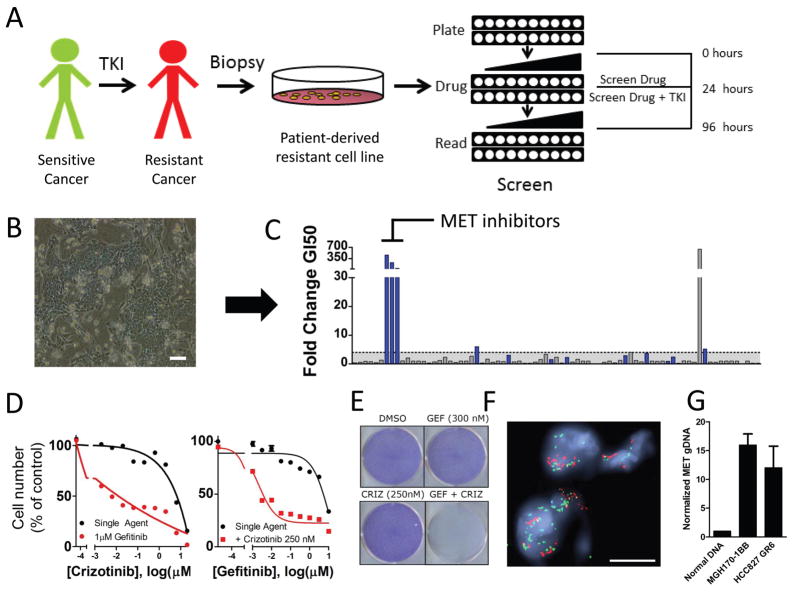
Each of the 55 models of acquired resistance was tested against the panel of 76 compounds in the presence or absence of the inhibitor of the primary target as described above (schema in Fig. 1A). For patient derived resistance models with gatekeeper resistance mutations in the driver oncogene (i.e, EGFR or ALK), next-generation inhibitors that overcome those mutations were used as the primary TKI in the combination screen. The results from the initial screening were analyzed to determine the specific thresholds of GI50 and AUCs changes most likely to yield strong effect on viability and maximize the potential for in vivo efficacy (see Materials and Methods and Database S2, S3 and S4). The process of screening and evaluating hits is demonstrated for the cell line MGH170-1BB in Figure 1A–C. These cells were derived from a patient with an EGFR mutant lung cancer who had become resistant to multiple lines of EGFR TKIs (Table S2, Fig. 1B). The screen clearly identified MET inhibitors as hits (Fig. 1C), and MET inhibitors effectively re-sensitized these resistant cells to EGFR inhibition (Fig. 1D left: screen format, right: dose response to gefitinib as single agent or in the presence of a fixed concentration of the MET inhibitor, crizotinib). The combination of EGFR and MET inhibitors was synergistic across a range of concentrations tested with on average 25% lower viability than predicted by the Bliss independence model for the 9 concentrations tested (see Table S8 for synergy calculations). Indeed, EGFR and MET inhibitor combination therapy was effective in eliminating resistant cells (Fig. 1E). Subsequent assessment of a paraffin-embedded biopsy from this patient’s cancer demonstrated clear evidence of MET amplification (Fig. 1G) and quantitative PCR performed on the corresponding MGH170-BB cell line confirmed MET amplification (Fig. 1H). Thus, the unbiased pharmacologic interrogation of the cells derived from the patient specimen unequivocally indicated the combination treatment supported by genetic analyses of the patient specimen.
Fig. 1. Screen schematic and proof of concept in a patient derived cell line.
A. Schematic of the screen workflow. Cell line models of acquired resistance were obtained directly from biopsies of patients after the development of acquired resistance to either EGFR inhibitor or ALK inhibitor in the clinic. Screen drugs were tested as single agent and in the presence of a single fixed concentration of the primary TKI across 10 concentrations encompassing a 10,000-fold dilution range. After 72 hours, cell viability was determined with CellTiter-Glo. B. Phase contrast microscopy of cell line MGH170-1BB, derived from an EGFR mutant lung cancer metastatic lesion with acquired resistance to EGFR inhibitors. Scale bar, 100 μm C. Representation of screen data for the MGH170-1BB cell line. The y-axis represents the fold-change GI50 that resulted with addition of gefitinib (GI50 single agent/GI50 combination). Each bar is the result for an individual drug. The bars are color-coded blue when the percent decrease in AUC from single agent to combination was greater than 10%. Drugs were defined as “hits” when the GI50 shift was > 4 and the AUC change > 10% (see Materials and Methods) D. Left: The MET inhibitor crizotinib was more potent in combination with 1 μM gefitinib (in red) than as single agent (in black). Right: Crizotinib (1 μM) resensitizes the MGH170-1BB cells to gefitinib. Error bars are mean +/− SEM. E. Long-term proliferation assay of MGH170-1BB cells that had been exposed to the indicated drug for 7 days. Cells were stained using crystal violet. F. FISH analysis of a biopsy sample from a metastatic bone lesion obtained after the patient had progressed while on treatment with erlotinib. The scale bar represents 10 μm. The MET gene is represented in red and the EGFR gene in green. G. Quantitative PCR analysis demonstrating overexpression of MET in MGH170-1BB in comparison to normal DNA. DNA from HCC827 GR6, which has MET amplification (13), is presented as a reference. Error bars are mean −/+ SEM. This experiment was repeated 3 times.
In some instances, pharmacologic interrogation permitted evaluation of the functional relevance of uncharacterized genetic variants. For example, a previously undescribed FGFR3 variant was identified as a key contributor to resistance in the MGH156-1A cell line derived from a patient with acquired resistance to EGFR TKIs (Fig. S4A and Table S2). The screen and subsequent follow-up studies clearly indicated that FGFR inhibitors re-sensitized these cells to EGFR inhibitors. The combination also suppressed key signaling events known to regulate proliferation and survival (Fig. S4B–E). Genetic analyses of both the cell line and corresponding biopsy revealed an FGFR3 mutation, Y649C, located in the tyrosine kinase domain (Table S6). While this FGFR3 mutation has not been observed previously (http://www.cbioportal.org/public-portal), it is adjacent to a recurring activating mutation in the kinase domain. Thus, in this model combining genetic analysis of tumor material and pharmacologic evaluation of cells from the resistant tumor allows for the identification of actionable therapeutic strategies. Furthermore, this finding demonstrates that FGFR activation is a bona fide mechanism of acquired resistance to EGFR inhibition in this patient.
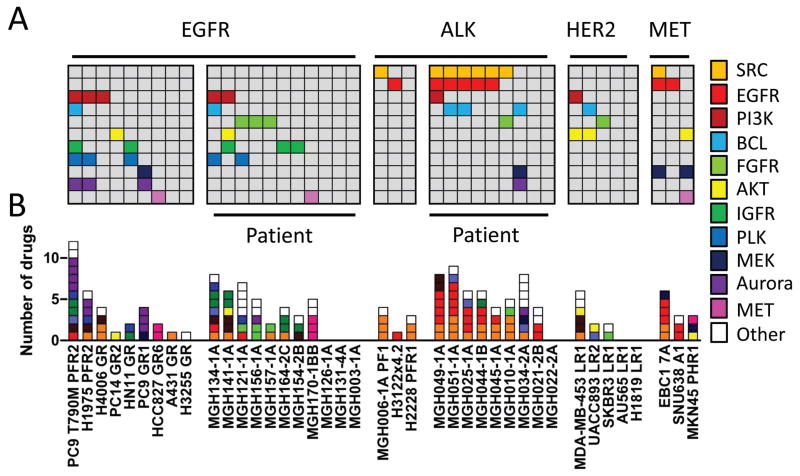
Among the 60 models screened, 201 hits were identified, for a mean of 3.4 hits per cell line (range 0 to 12). At least one hit was identified in 50 of 60 cell lines (Fig. S5 and 2A). Drugs known to have overlapping specificity were found to have overlapping activity across cell lines, demonstrating robustness of the dataset (see, for example, aurora, SRC and MET inhibitors in Figs. 2A, 2B and S5). Notably, EGFR inhibitors tended to be hits in both ALK- and MET-driven resistant lines, consistent with previously published reports (7, 17). Because re-activation of the PI3K pathway via activating PIK3CA mutations and bypass RTKs has commonly been observed in cancers with acquired resistance (18), it was not surprising that PI3K inhibitors were hits in a subset of resistant cell lines. Importantly, genetic analyses of the cell lines were insufficient to inform which cancers would be sensitive to this combination. Notably, PI3K inhibitors were not sufficient to re-sensitize to the original TKI in the majority of models tested (Figs. 2A, S5). Other unanticipated drug combinations were identified. In particular, aurora kinase inhibitors were active in combination with EGFR inhibition in a number of EGFR-mutant cell lines. Similarly, the polo-like kinase inhibitor (BI2536) was a hit in 5 EGFR-driven lines. The complete hit profile of each resistant cell line is presented in Fig. S6. In the in vitro models of acquired resistance (which have a paired sensitive, “parental” cell line from which the resistant cells were derived), we also sought to determine whether resistant models had developed increased sensitivity to any single-agent treatments compared to the parental cell lines (Fig. S7). This analysis revealed that, in the vast majority of cases, resistant models did not acquire sensitivity to single-agent therapies, further supporting the notion of developing combination therapies (Fig. S7).
Fig. 2. Representation of selected screen hits in independent resistant models.
A. The pattern of hits across cell lines harboring the indicated oncogene are shown. Each column represents a cell line, and each row represents a target inhibited by the following drugs: Afatinib (EGFR), AZD0530 (SRC), BYL719 (PI3K), ABT-263 (BCL), Dovitinib or BGJ-398 (FGFR), MK2206 (AKT), OSI906 (IGFR), BI2536 (PLK), AZD6244 (MEK), AZD1152-HQPA (Aurora kinase B), MGCD265 (MET). Each drug is color-coded as indicated. B. The number and profile of all hit drugs for each model. Each box represents a single drug, and the drugs are color-coded by target. The white boxes indicate a hit that corresponds to a drug that is not among the targets listed. For resistant lines derived from a single parental line, only one representative model is presented except in the case of PC9, for which PC9 GR1 and PC9 GR2 are both presented due to the presence of a T790M mutation in PC9 GR2 only.
Identification of mechanisms of resistance and combination therapies for ALK-positive lung cancers
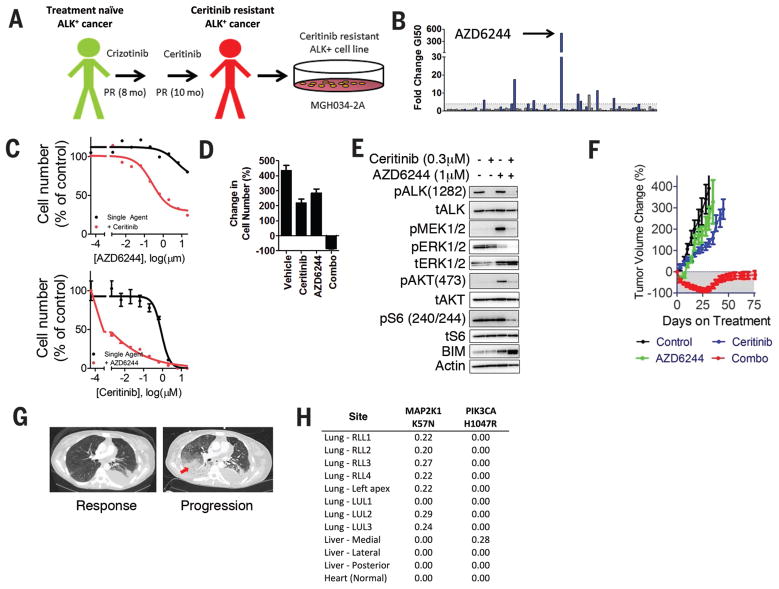
Assessment of the patient-derived ALK-positive models identified previously undescribed mechanisms of resistance. The MGH034-2A cell line was derived from a biopsy of a patient harboring an ALK-positive cancer that had become resistant to ceritinib (LDK378), a second-generation ALK inhibitor that was recently approved by the FDA (19, 20) (Fig. 3A). The MEK inhibitor, AZD6244, was a potent hit in combination with ceritinib (Fig. 3B, 3C top panel, synergistic effect with on average 45% less viability than predicted by Bliss (Table S8). Furthermore, AZD6244 treatment also led to marked re-sensitization to ceritinib in MGH034-2A (Fig. 3C, bottom panel). To our knowledge, there have been no previous reports demonstrating that MEK inhibitors re-sensitize resistant ALK-positive cancer cells to an ALK inhibitor. Furthermore, MEK inhibitor sensitization was not observed in any of the other ALK-positive patient-derived or laboratory-derived models examined in this study (Fig. S8A), illustrating the potential for the present approach to identify patient-specific efficacious combinations. Long-term viability assays revealed that the combination had a potent effect on cell viability, with a marked net decrease in cell number compared to the cell number before drug treatment (Fig. 3D). Accordingly, the combination was required to inhibit PI3K, MAPK and mTORC activity as well as to upregulate BIM and promote substantial apoptosis (14) (Fig. 3E, S8B). In vivo, neither single agent was effective, but the combination resulted in robust tumor regression (Fig. 3F). Importantly, next generation sequencing (NGS) analysis of the cell line revealed a MAP2K1 K57N mutation (Table S7), which has previously been reported as a MEK activating event in lung adenocarcinoma (21), though neither in conjunction with an activating RTK mutation nor in the setting of acquired resistance to any TKI. Notably, this cell line also harbored a JAK3 V722I variant, an activated allele of JAK3 (22). Despite this, the JAK3 specific inhibitor tofacitinib was not a screen hit (Fig. S5) and furthermore did not re-sensitize MGH034-2A cells or other ALK cell lines to ALK inhibition (Fig. S9A). Indeed, these cells did not express appreciable levels of JAK3 (Fig. S9B, C). This patient subsequently expired and NGS analysis of 10 resistant lesions acquired at autopsy demonstrated that the MAP2K1 K57N mutation was present in 7 of the 11 lesions (of note, a PIK3CA mutation was identified in one of the other lesions) (Fig. 3H). Importantly, the MAP2K1 K57N mutation was found in the lesions that were rapidly progressing and led to respiratory failure, which caused the patient’s death. The autopsy revealed that the JAK3 mutation was a germline variant, supporting the functional data that JAK3 activity was not driving resistance. These results suggest that a combination of MEK and ALK inhibitors may have provided a therapeutic benefit to this patient had these drugs been administered after the cancer had acquired resistance to ceritinib. Importantly, these results also suggest that functional assessment adds information to that provided by genetic analysis alone. Genetic profiling of the tumor alone, as if often performed in the clinic, would not have discriminated between targeting the MAP2K1 K57N mutation and the less consequential JAK3 V722I mutation.
Fig. 3. MEK activation is a mechanism of resistance to ceritinib.
A. Schematic of the derivation of model MGH034-2A. B. Representation of screen data for the MGH034-2A cell line. The y-axis represents the fold-change GI50 that resulted with addition of ceritinib (0.3 μM) (GI50 single agent/GI50 combination). The bars are color-coded blue when the percent decrease in AUC from single agent to combination was greater than 10%. C. Top: Primary screen data of the effect of ceritinib (0.3 μM) on AZD6244 effect in MGH034-2A. Bottom: A dose-response curve to ceritinib is shown in the presence and absence of a fixed concentration of the MEK inhibitor, AZD6244 (1 μM). D. Viability assay of MGH034-2A cells demonstrating the change in cell number after 6 days of treatment with vehicle, ceritinib (300 nM), AZD6244 (1 μM) or the combination of both drugs in comparison to the number of cells at the initiation of drug exposure. E. Western blot analysis of MGH034-2A. Cells were treated with vehicle, ceritinib (0.3 μM), AZD6244 (1 μM) or the combination of both drugs for 24 hours. Lysates were analyzed with antibodies to the indicated proteins. F. Subcutaneous xenografts of MGH034-2A grown in mice were used to determine in vivo efficacy by measuring change in tumor volume when treated as indicated. n=6 mice per group. G. Axial CT images of the chest demonstrate the patient’s disease burden after responding to ceritinib (5.5 weeks on treatment), and at the time of progression on ceritinib (after 9.5 months on treatment). The site of progression in the right lower lobe is indicated by an arrow. H. Table of allele frequencies for MAP2K1 and PIK3CA mutations discovered at autopsy in the patient.
SRC signaling mediates acquired resistance in ALK-positive NSCLC
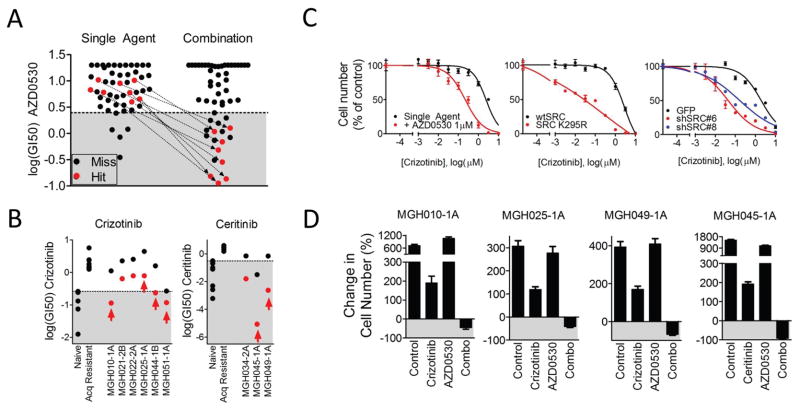
Multiple SRC family kinase inhibitors were consistently effective across several patient-derived ALK-positive resistant NSCLC models (Fig. 2). In particular, AZD0530 (saracatinib) was a hit in 6 of 9 patient-derived ALK lines tested (Fig. 2A). Models in which AZD0530 was a screen hit had unremarkable sensitivity to single agent AZD0530 indicating that, as in other cases, these cell lines have not switched to an entirely different dependency. On the other hand these resistant ALK-positive cell lines were highly sensitive to AZD0530 in the presence of ALK inhibitors (Fig. 4A). Drug synergism between AZD0530 and ALK inhibitors was also observed (average of 20% less viability than expected across all concentrations for five models re-tested in triplicate with maximum differences ranging from 18 to 45% over Bliss (Table S8). Two other drugs (dasatinib and KIN001-113) that potently inhibit SFKs (23, 24) were often hits in models in which AZD0530 was a hit (Figs. 2B, S5). However, due to the more favorable specificity profile of AZD0530 (25), we used this drug in our subsequent studies. Each model in which AZD0530 was a hit (as indicated by arrows in Fig. 4A) was significantly sensitized to ALK inhibition by AZD0530 (Fig. 4B). Notably, other ALK driven models also demonstrated shifts in sensitivity with AZD0530 pointing to the possibility of broad involvement of SRC kinases in ALK inhibitor response. Interestingly, AZD0530 was not a hit in any of the mutant EGFR or HER2 amplified cancers and in only 1 of 9 MET amplified cancers (Fig. S5).
Fig. 4. SRC inhibition restores sensitivity to ALK inhibitor in multiple models.
A. Representation of the GI50 of AZD0530 in each screened model as a single-agent or in combination with the primary TKI. Models that were hits are color-coded red. The GI50s of cell lines in which AZD0530 scored as hits are connected by an arrow. The shaded area represents the GI50 values among the top 10% sensitive models for single agent values among all lines screened. B. GI50 of each ALK+ patient-derived model of acquired resistance to either crizotinib or ceritinib. Control cell line models of sensitivity (MGH006-1A, H3122, SU-DHL-1, KARPAS299, NB-1) and acquired resistance (MGH006-1A PFR1, MGH006-1A PFR2, H2228 PFR1, H3122 PFR1, H3122 PFR3, H3122 x4.2) to crizotinib are presented as standards for comparison. Models of sensitivity (H3122, H2228, MGH051-1B, H3122 PFR2, MGH021-2cl4, MGH006-1A, MGH026-1A, MGH039-1A) and acquired resistance (MGH021-5, H3122 LDKR1, H3122 LDKR2, H3122 LDKR2, H3122 LDRK4) to ceritinib are presented as standards for comparison. The GI50 of each model is presented as single-agent (black) and in combination with AZD0530 (1 μM) (red). The mean GI50 of the three experiments is presented. Arrows indicate hits identified by the screen. C. Dose-response curves to crizotinib in model MGH010-1A (crizotinib resistant) are presented. The left panel demonstrates the dose-response of single-agent crizotinib (black) in the absence or presence of AZD0530 (1 μM) (red). The middle panel presents the effect of crizotinib in cells with lentiviral overexpression of either wild-type SRC (black) or kinase-dead SRC (K295R, red). The right panel demonstrates the effect of lentiviral expression of GFP (black), or either of two SRC-targeted shRNAs (blue and red). D. Six-day viability assay of 4 ALK lines: MGH010-1A, MGH025-1A, MGH049-1A, MGH045-1A. Each panel presents percentage change in cell number after treatment with vehicle, ALK inhibitor (crizotinib 1 μM or ceritinib 300 nM), AZD0530 (1 μM) or the combination compared to cell number at the initiation of treatment.
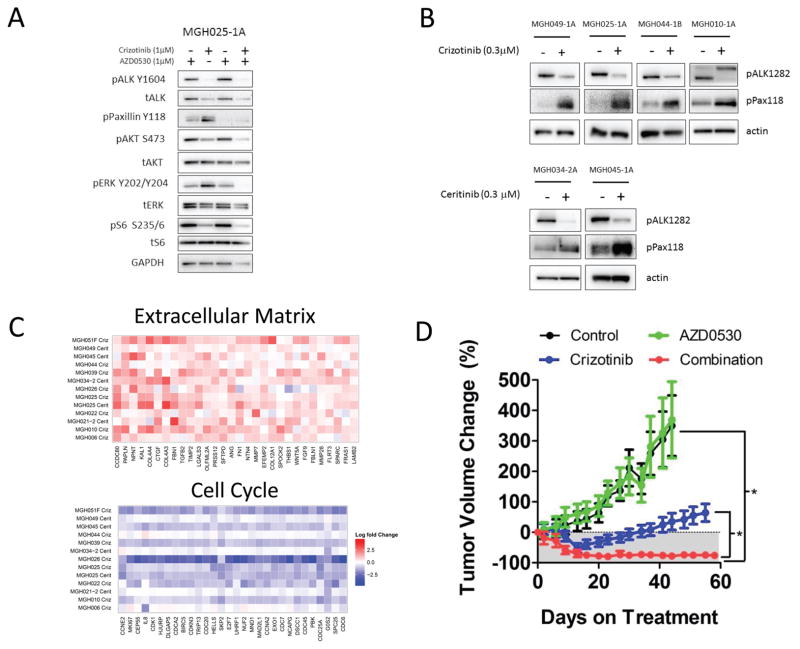
We next aimed to determine the relevant target of AZD0530. Overexpression of the kinase-dead SRC K295R (26), as well as knockdown of SRC alone with either of two shRNAs effectively recapitulated the effect of AZD0530, demonstrating that among AZD0530 targets, including multiple SFKs, SRC inhibition is sufficient to re-sensitize cells to ALK inhibition (Fig. 4C). We observed that multiple ALK driven models were sensitive to both SRC and EGFR inhibitors when combined to an ALK inhibitor. However, the activity of AZD0530 does not appear to be driven by EGFR inhibition directly or indirectly since AZD0530 did not inhibit EGFR activation in the ALK-positive MGH025-1A cells, which were sensitized by AZD0530 (Fig. S10A). Furthermore, some cell lines, such as MGH010-1A, were sensitized by AZD0530, but not EGFR inhibitors (Figs. 2A, S10B). We next examined the effect of combined ALK and SRC inhibition on three resistant ALK-positive models derived from patient biopsies: MGH010-1A and MGH025-A (resistant to crizotinib, no ALK resistance mutations) and MGH049-1A (resistant to ceritinib, no ALK resistance mutations (27)). In all three models, cells grew at 6 days when treated with either drug as single agent, but combination treatment resulted in loss of cell viability compared to pre-treatment cell number (Fig. 4D) and robust apoptotic cell death (S11A). Consistent with these results, the ALK TKI failed to fully inhibit downstream signaling (AKT, MAPK or S6K) except in the presence of AZD0530 in each of these resistance models (Fig. 5A and S11B).
Fig. 5. ALK inhibition and SRC signaling.
A. Western blot analysis of MGH025-1A. Cells were treated with vehicle, crizotinib (1 μM), AZD0530 (1 μM) or the combination of both drugs for 24 hours. Lysates were analyzed with antibodies to the indicated proteins. B. Western blot analysis of patient-derived resistant ALK models treated for 24 hours with crizotinib (300 nM) or ceritinib (300 nM). Lysates were prepared and blotted with the indicated antibodies. C. Fold-change in gene expression (Log2) upon treatment with the indicated ALK inhibitor for 24 hours. Top: Upregulated genes annotated with the GO term “extracellular matrix”. Bottom: Downregulated genes annotated with the GO term “cell cycle” (top 30 genes only). D. MGH025-1A subcutaneous xenografts grown in mice were treated as indicated: Vehicle (n=4 mice), crizotinib 25 mg/kg daily (n=6 mice), AZD0530 50 mg/kg daily (n=5 mice), or the combination of both drugs (n=6 mice). Error bars are mean −/+ SEM. Asterisks indicate P<0.0001 by Dunn’s Multiple Comparison Test.
In each of the patient-derived ALK models in which AZD0530 was effective (including MGH034-2A, which narrowly failed to meet our threshold for hit call for AZD0530), ALK inhibition resulted in robust upregulation of SRC activity as measured by the phosphorylation of the SRC substrate Paxillin (Fig. 5B). Thus, ALK inhibition may lead to upregulation of SRC signaling, perhaps via release of a negative regulatory signal normally coordinating ALK and SRC activities. In contrast, we did not consistently observe an increase in SRC activity as measured by p-Paxillin in EGFR mutant cancers following EGFR inhibitor treatment (Fig. S11C), consistent with the absence of efficacy noted with AZD0530 in EGFR mutant cancer. Furthermore, in the ALK driven models, SRC signaling was also upregulated by inhibition of signaling pathways downstream of ALK. Interestingly, although the downstream pathways regulated by ALK in individual models vary, the pathways regulated by ALK tended to be the one suppressing SRC signaling. For example, when ALK inhibition primarily impacted PI3K signaling but not MEK activity, PI3K inhibition upregulated SRC signaling (Fig S12A). Moreover, when ALK inhibition suppressed both MAPK and PI3K signaling, SRC signaling was robustly upregulated by either PI3K or MAPK signaling (Fig S12B). Overall these results are compatible with a model in which ALK activity suppresses SRC activity broadly in the setting of ALK-driven cancers.
To further characterize the effect of ALK inhibition on these models, we performed gene expression analysis on each of the ALK-positive patient-derived models in the presence or absence of an ALK inhibitor for 24 hours. The gene ontologies most enriched within genes whose expression was induced by ALK inhibition were extracellular matrix and basal membrane (Benjamini-Hochberg corrected p values 1.75E-04 and 2.31E-04) (Fig. 5C, Databases S6–S8). As SRC signaling is known to be a focal point of integrin-mediated signaling and the transduction of extracellular signals, these results further support the finding that SRC activity is increased upon inhibition of ALK signaling in ALK-positive lung cancers.
Finally, we tested the efficacy of the combination of ALK TKIs and AZD0530 in vivo using mouse xenograft models. In MGH025-1A (derived from an ALK-positive patient who had become resistant to crizotinib), treatment with single-agent crizotinib resulted in tumor progression after 34 days. However, combining AZD0530 and crizotinib resulted in a sustained, profound response for over 60 days (Fig. 5D). Notably, when AZD0530 was added to the treatment of the xenografts that had progressed on crizotinib, the tumors regressed (Fig. S13A). To test the specificity of AZD0530 for resistant models that demonstrated synergy in the screen, we tested it in the HCC827 GR6 line, which harbors a MET bypass track and was not a hit for AZD0530. In this model the combination of AZD0530 with gefitinib was ineffective in comparison to gefitinib plus crizotinib (which is a potent MET inhibitor) (Fig. S13B). Thus, the effect of AZD0530 appears particular to the models in which combination efficacy was found in the screen.
Since we observed impressive activity of the SFK inhibitors in large proportion of patient-derived ALK-positive resistant models, we also determined if the combination of ALK inhibitor with AZD0530 might delay the emergence of acquired resistance in a relatively sensitive model. We examined cell line MGH045-1A, a model established from a patient tumor resistant to crizotinib due to the acquisition of a mutation in the ALK kinase domain gatekeeper residue (L1196M) (Table S2) (27). Ceritinib, which can overcome the L1196M mutation, was used as the primary TKI in the screen of this cell line, and AZD0530 was a hit (Fig. 2A). The cell line is relatively sensitive to the next-generation ALK inhibitor ceritinib, which can effectively suppress L1196M (27). Over 6 days of treatment in vitro, single-agent ceritinib effectively inhibited growth, but the combination of ceritinib and AZD0530 resulted in near-complete obliteration of cell viability (Fig. 4D). Accordingly, both ALK inhibition and AZD0530 were required to completely suppress key downstream signaling events (Fig. S13C). In vivo, single-agent ceritinib slowed tumor growth as previously described (27), but the combination resulted in a more sustained response (Fig. S13D). This reinforces the notion that initial treatment combining a SRC and an ALK inhibitor could help induce a more sustained response in patients with ALK-positive lung cancer.
Analysis of the discovered mutations identified by the 1,000-gene NGS panel in the ALK-positive models failed to identify mutations in SRC family kinases and other known regulators of SRC activity (Table S7). Thus, the pharmacologic approach identified a drug combination that would not have readily been predicted by genomic analyses alone.
Discussion
In summary, we have developed cell culture models of acquired resistance to EGFR and ALK inhibition derived directly from patient specimens to rapidly identify combinations that can overcome resistance. These initial studies demonstrated success in developing NSCLC models in 50% of collected specimens. However, we believe that success rates could be further improved by using biopsies acquired for specifically for cell line generation. Currently, the biopsies were prioritized for standard pathological analysis, and cell lines were generated from any remaining tissue. As a result, the quality of the specimens was less than ideal. Indeed, in the majority (24/39) of the “failures,” the samples we analyzed contained fewer than 20% cancer cells. Despite these obstacles, cell line models were successfully developed in approximately half of the cases. Thus, if biopsies were isolated primarily for this purpose, under the auspices of an IRB-approved protocol, we believe that this methodology could potentially be explored as a diagnostic approach to guide treatment decisions. We also anticipate that this approach will be generalizable to other solid and liquid tumor malignancies.
The robustness of the approach presented here is demonstrated by the success rate of in vivo studies. All five tested models (MGH034-2A (Fig. 3), MGH045-1A (Fig. S13), MGH025-1A (Fig. 5D), PC9 PFR2 (Fig. S14) and PC9 GR1 (Fig. S15) demonstrated substantial regression in vivo with the discovered active combination. Importantly, this functional assessment of patient derived samples can provide insights not provided by genetic analysis. For example, the effect of SRC inhibition in resistance ALK-positive cancers is not readily predicted by genetic analyses as no mutation was identified in SFKs or their regulators. In addition, our results illustrate how functional assessment of patient-derived cells can complement genetic profiling. For example, FGFR inhibitors were effective in a model with a previously uncharacterized FGFR3 mutation (Fig. S4). In the absence of functional data, the biological consequence of the mutation would have been uncertain.
By interrogating patient-derived models of resistance with this pharmacologic platform, we have discovered several previously undescribed combinations in EGFR mutant and ALK-positive lung cancers that were validated in follow-up studies and in vivo. We speculate that a similar approach could be explored in the future as a diagnostic test to identify therapeutic strategies for individual patients [under the auspices of an Institutional Review Board (IRB)-approved protocol]. In the current study, we screened the cells after they became fully established cell lines, which often took 2–6 months, a time frame that would make this approach less than ideal as a routine diagnostic test. Nevertheless, the robustness of the results from the current program lays the groundwork for performing screens on viable cells obtained within weeks of a biopsy using newer technologies that would permit screening of the cancer cells while still in the presence of the stroma that is also present in the biopsy. Indeed, it is possible that such functional screens performed on cells derived from a biopsy of a particular patient’s resistant cancer, might inform the choice of experimental therapies that are most likely to be effective in a given patient, advancing toward a future of truly personalized cancer therapy.
Supplementary Material
Acknowledgments
We thank LHP for direction, re-direction, and support. We thank SRV for continual critical discussion and support. We thank W. Michaud for providing feeder cells and training for the development of patient-derived cell lines. We thank N. Gray for providing several kinase inhibitors. This study was funded by support from the NIH R01CA137008 (JAE), R01CA164273 (ATS and JAE), 1U54HG006097-01 (CHB), the Wellcome Trust (086357 and 102696 CHB), the National Cancer Institute Lung SPORE P50CA090578 (ACS, AJI, and JAE), the Department of Defense (LVS and JAE) Conquer Cancer Foundation Young Investigator Award (ASC), Uniting Against Lung Cancer (ASC and ATS), Free to Breathe (ASC), Lungevity (LVS, JAE), and Be a Piece of the Solution. J.A.E. is a paid consultant for Novartis, Sanofi-Aventis, AstraZeneca, Chugai, Amgen, Genentech, GSK, Merck, and Pfizer and he holds equity in Gatekeeper Pharmaceuticals. J.A.E. also receives research support from Novartis. J.F.G. is a paid consultant for Boehringer Ingelheim. A.T.S. is a paid consultant for Pfizer, Novartis, Ariad, Chugai, Genentech, Roche, and Ignyta. A.J.I. is a Senior Advisory Board member and holds equity in Enzymatics Inc. For gene expression analyses, the raw data are deposited in ArrayExpress (accession number is E-MTAB-783). The normalized data are available at: http://www.cancerrxgene.org/downloads/.
References and Notes
- 1.Inoue A, et al. Prospective phase II study of gefitinib for chemotherapy-naive patients with advanced non-small-cell lung cancer with epidermal growth factor receptor gene mutations. J Clin Oncol. 2006 Jul 20;24:3340. doi: 10.1200/JCO.2005.05.4692. [DOI] [PubMed] [Google Scholar]
- 2.Kwak EL, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010 Oct 28;363:1693. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosell R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012 Mar;13:239. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 4.Sequist LV, et al. First-line gefitinib in patients with advanced non-small-cell lung cancer harboring somatic EGFR mutations. J Clin Oncol. 2008 May 20;26:2442. doi: 10.1200/JCO.2007.14.8494. [DOI] [PubMed] [Google Scholar]
- 5.Shaw AT, et al. Effect of crizotinib on overall survival in patients with advanced non-small-cell lung cancer harbouring ALK gene rearrangement: a retrospective analysis. Lancet Oncol. 2011 Oct;12:1004. doi: 10.1016/S1470-2045(11)70232-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chong CR, Janne PA. The quest to overcome resistance to EGFR-targeted therapies in cancer. Nat Med. 2013 Nov;19:1389. doi: 10.1038/nm.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katayama R, et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung Cancers. Sci Transl Med. 2012 Feb 8;4:120ra17. doi: 10.1126/scitranslmed.3003316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamaguchi N, et al. Dual ALK and EGFR inhibition targets a mechanism of acquired resistance to the tyrosine kinase inhibitor crizotinib in ALK rearranged lung cancer. Lung Cancer. 2014 Jan;83:37. doi: 10.1016/j.lungcan.2013.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanizaki J, et al. Activation of HER family signaling as a mechanism of acquired resistance to ALK inhibitors in EML4-ALK-positive non-small cell lung cancer. Clin Cancer Res. 2012 Nov 15;18:6219. doi: 10.1158/1078-0432.CCR-12-0392. [DOI] [PubMed] [Google Scholar]
- 10.Niederst MJ, Engelman JA. Bypass mechanisms of resistance to receptor tyrosine kinase inhibition in lung cancer. Sci Signal. 2013 Sep 24;6:re6. doi: 10.1126/scisignal.2004652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu X, et al. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am J Pathol. 2012 Feb;180:599. doi: 10.1016/j.ajpath.2011.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engelman JA, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008 Dec;14:1351. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engelman JA, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007 May 18;316:1039. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 14.Faber AC, et al. Differential induction of apoptosis in HER2 and EGFR addicted cancers following PI3K inhibition. Proc Natl Acad Sci U S A. 2009 Nov 17;106:19503. doi: 10.1073/pnas.0905056106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sos ML, et al. Identifying genotype-dependent efficacy of single and combined PI3K- and MAPK-pathway inhibition in cancer. Proc Natl Acad Sci U S A. 2009 Oct 27;106:18351. doi: 10.1073/pnas.0907325106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guix M, et al. Acquired resistance to EGFR tyrosine kinase inhibitors in cancer cells is mediated by loss of IGF-binding proteins. J Clin Invest. 2008 Jul;118:2609. doi: 10.1172/JCI34588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qi J, et al. Multiple Mutations and Bypass Mechanisms Can Contribute to Development of Acquired Resistance to MET Inhibitors. Cancer Res. 2011 Jan 25; doi: 10.1158/0008-5472.CAN-10-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sequist LV, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011 Mar 23;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marsilje TH, et al. Synthesis, Structure-Activity Relationships, and in Vivo Efficacy of the Novel Potent and Selective Anaplastic Lymphoma Kinase (ALK) Inhibitor 5-Chloro-N2-(2-isopropoxy-5-methyl-4-(piperidin-4-yl)phenyl)-N4-(2-(isopropylsulf onyl)phenyl)pyrimidine-2,4-diamine (LDK378) Currently in Phase 1 and Phase 2 Clinical Trials. J Med Chem. 2013 Jun 26; doi: 10.1021/jm400402q. [DOI] [PubMed] [Google Scholar]
- 20.Shaw AT, et al. Ceritinib in ALK-rearranged non-small-cell lung cancer. N Engl J Med. 2014 Mar 27;370:1189. doi: 10.1056/NEJMoa1311107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marks JL, et al. Novel MEK1 mutation identified by mutational analysis of epidermal growth factor receptor signaling pathway genes in lung adenocarcinoma. Cancer Res. 2008 Jul 15;68:5524. doi: 10.1158/0008-5472.CAN-08-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walters DK, et al. Activating alleles of JAK3 in acute megakaryoblastic leukemia. Cancer Cell. 2006 Jul;10:65. doi: 10.1016/j.ccr.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 23.Karaman MW, et al. A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol. 2008 Jan;26:127. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- 24.McDermott U, Pusapati RV, Christensen JG, Gray NS, Settleman J. Acquired resistance of non-small cell lung cancer cells to MET kinase inhibition is mediated by a switch to epidermal growth factor receptor dependency. Cancer Res. 2010 Feb 15;70:1625. doi: 10.1158/0008-5472.CAN-09-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hennequin LF, et al. N-(5-chloro-1,3-benzodioxol-4-yl)-7-[2-(4-methylpiperazin-1-yl)ethoxy]-5- (tetrahydro-2H-pyran-4-yloxy)quinazolin-4-amine, a novel, highly selective, orally available, dual-specific c-Src/Abl kinase inhibitor. J Med Chem. 2006 Nov 2;49:6465. doi: 10.1021/jm060434q. [DOI] [PubMed] [Google Scholar]
- 26.Bagrodia S, Chackalaparampil I, Kmiecik TE, Shalloway D. Altered tyrosine 527 phosphorylation and mitotic activation of p60c-src. Nature. 1991 Jan 10;349:172. doi: 10.1038/349172a0. [DOI] [PubMed] [Google Scholar]
- 27.Friboulet L, et al. The ALK inhibitor ceritinib overcomes crizotinib resistance in non-small cell lung cancer. Cancer Discov. 2014 Mar 27; doi: 10.1158/2159-8290.CD-13-0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ebi H, et al. Receptor tyrosine kinases exert dominant control over PI3K signaling in human KRAS mutant colorectal cancers. J Clin Invest. 2011 Nov;121:4311. doi: 10.1172/JCI57909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katayama R, et al. Therapeutic strategies to overcome crizotinib resistance in non-small cell lung cancers harboring the fusion oncogene EML4-ALK. Proc Natl Acad Sci U S A. 2011 May 3;108:7535. doi: 10.1073/pnas.1019559108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004 Feb 12;20:307. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 31.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical applications in genetics and molecular biology. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 32.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 33.Zhao M, Sun J, Zhao Z. TSGene: a web resource for tumor suppressor genes. Nucleic Acids Research. 2013;41:D970. doi: 10.1093/nar/gks937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McDermott U, et al. Genomic alterations of anaplastic lymphoma kinase may sensitize tumors to anaplastic lymphoma kinase inhibitors. Cancer Res. 2008 May 1;68:3389. doi: 10.1158/0008-5472.CAN-07-6186. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.