Abstract
There is extensive genomic diversity among Streptococcus pneumoniae isolates. Approximately half of the comprehensive set of genes in the species (the supragenome or pangenome) is present in all the isolates (core set), and the remaining is unevenly distributed among strains (distributed set). The Streptococcus pneumoniae Supragenome Hybridization (SpSGH) array provides coverage for an extensive set of genes and polymorphisms encountered within this species, capturing this genomic diversity. Further, the capture is quantitative. In this manner, the SpSGH array allows for both genomic and transcriptomic analyses of diverse S. pneumoniae isolates on a single platform. In this unit, we present the SpSGH array, and describe in detail its design and implementation for both genomic and transcriptomic analyses. The methodology can be applied to construction and modification of SpSGH array platforms, as well as applied to other bacterial species as long as multiple whole genome sequences are available that collectively capture the vast majority of the species supragenome.
Keywords: supragenome, pangenome, gene array, transcriptional profiling, gene expression, genome content, Streptococcus pneumoniae
Introduction
In many bacterial species, isolates differ from one another by extensive genomic variability (Ahmed et al., 2012; Davie et al., 2011; Boissy et al., 2011; Donati et al., 2010; Hiller et al., 2007; Hiller et al., 2010; Ehrlich et al., 2010; Hogg et al., 2007; Tettelin et al., 2005; Borneman et al., 2012; He et al., 2010; Conlan et al., 2012). This variability is observed as single nucleotide polymorphisms (allelic differences), as well as extensive differences in gene possession where a percentage of the genes are shared across all strains (core set), and the remainder are unevenly distributed across isolates (distributed/accessory/variable set). The comprehensive set of all the genes across all the strains of the species is referred to as the pangenome or supragenome (Ehrlich et al., 2010, Erdos et al., 2003, Tettelin et al., 2005) This variability in gene possession is an important factor in determining the broad array of phenotypes displayed by various isolates with respect to disease, as well as drug and vaccine resistance (Forbes et al., 2008; Coffey et al., 1991; Dowson et al., 1989; Engelmoer and Rozen, 2011; Prudhomme et al., 2006; Wyres et al., 2013).
We have developed supragenome hybridization (SGH) arrays to study the differences in gene content and gene expression for Haemophilus influenzae (Eutsey et al., 2013 and Janto et al., 2014) and Streptococcus pneumoniae (pneumococcus), two opportunistic pathogens that colonize the human nasopharynx. For both of these species, ~50% of the supragenome is core, and strain pairs often differ by ~20% of their genomic content (Boissy et al., 2011; Donati et al., 2010; Hiller et al., 2007; Hogg et al., 2007). Thus, in these species, when an array is designed to a reference strain, the analysis of a distantly related isolate will be hampered due to the loss of information associated with the lack of probes that can capture highly variable alleles or genes that were absent in the reference strain. The SGH array is designed to capture the diversity of the species by providing coverage of multiple alleles and most genes in the supragenome; and in doing so, allow for the analysis of diverse isolates on the same platform.
The H. influenzae SGH array (HiSGH) was designed based on a supragenome (pangenome) analysis of 24 clinical H. influenzae strains. The array contains 31,307 probes that collectively cover 2,890 H. influenzae genes, corresponding to greater than 85% of all non-rare genes (that is, those present in 10% or more of isolates). This array has been used to investigate the gene content of a library of isolates (Eutsey et al., 2013); as well as to measure transcriptomic differences between a wild-type (WT) strain and an associated deletion mutant (Janto et al., 2014 PLoS ONE). For genome content studies, the HiSGH array accuracy was shown to be ~ 98% by comparing whole genome sequence (WGS) of eight strains with their hybridization data obtained using the HiSGH array. Once tested, the array was used to investigate the gene content of 193 geographically and clinically diverse H. influenzae clinical strains (Eutsey et al., 2013). In transcriptomic studies, the HiSGH aray was used to compare transcripts levels between a WT strain and the cognate AI-2 sensing mutant. The strains were grown in multiple conditions using different media and sampling time points. Additionally, technical and biological replicates were analyzed for reproducibility. The results were highly reproducible, and the differentially expressed genes were confirmed by quantitative reverse transcription polymerase chain reaction (qRTPCR) (Janto et al., 2014-in press).
In this unit, we describe the development and testing of an SGH array, and use the SpSGH array as an example. Each section has a general description, followed by details from the SpSGH array. The unit is divided into three major sections: I) probe design; II) analysis of genomic content; and III) analysis of transcriptomic content. These are organized into the following basic protocols: 1) probe design for the SpSGH Array; 2) SpSGH Array to determine gene content; 3) Data Analysis of SpSGH Array to determine gene content; 4) SpSGH Array for gene expression profiling; and 5) Data Analysis of SpSGH Array for gene expression profiling. Development of an SGH array can assist in understanding the finer genomic and transcriptomic differences contributing to diverse phenotypes with respect to disease and carriage of historical, present day as well as emerging pneumococcal strains. In addition, it provides a holistic view to elucidate gene regulatory networks differentially regulated in selected in vitro conditions.
BASIC PROTOCOL 1: Probe Design for SpSGH Array
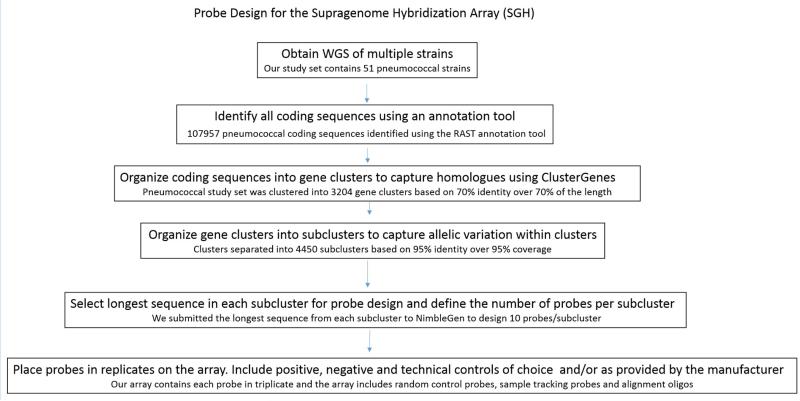
The goal of probe design is to generate DNA probes that recognize most genes in the supragenome, but which do not cross hybridize with paralogous genes. Probe design is a multistep process that requires: 1) preparation of sequence input by selection and annotation of WGS; 2) organization of genes into homologous clusters and then into allelic groups; and finally 3) selection of sequences for probe design and manufacturing. A schematic of SGH probe design is provided in Figure 1.
Figure 1.
Schematic of probe design for the SGH array. Specific information on the SpSGH array is indicated in smaller fonts.
Materials
WGS of multiple strains that capture the diversity of the set of interest Rapid Annotations with Subsystems Technology (RAST) (http://rast.nmpdr.org/) FASTA36 from the FASTA package (http://faculty.virginia.edu/wrpearson/fasta/fasta36/.) Scripts for gene clustering (available from the authors on request, or at https://github.com/jpearl01/)
Array manufacturer's probe design tool
1. Prepare Input: Select Strains and Annotate to Obtain CDSs
The coverage potential of the final probe set will depend on how well the input sequences capture the distributed gene content within the species. The goal is to capture as many genes as possible, by selecting not only a large number of strains with available high quality WGS, but also highly variable strains with respect to gene content, geographic isolation and clinical phenotypes. Boissy and colleagues describe in detail a model to predict the coverage of a supragenome/pangenome given a subset of strains (Boissy et al., 2011). For H. influenzae, S. pneumoniae and Staphylococcus aureus it was found that less than 50 strains cover >95% of the non-rare (ν< 0.1) genes.
After selecting the strains, annotate the WGSs to identify the CDSs. We recommend that all strains be annotated in parallel given that gene annotations vary significantly depending on the tool selected and the version of the algorithm at the time of submission. We used Rapid Annotations with Subsystems Technology (RAST), a fully-automated web service for annotating bacterial genomes, where the annotated genomes are made available in a GenBank format (Overbeek et al., 2014). RAST is available at http://rast.nmpdr.org/.
2. Organize CDSs into Allelic Groups: Compare all CDSs to Each Other using FASTA36 and Parse Data into Clusters and Subclusters
Organize the gene sequences into clusters of related sequences, so that cluster-specific probes can be designed. The clustering requires the following steps: A) prepare the input; B) compare all sequences using FASTA36, C) parse the sequence comparison into clusters of homologous genes with (presumed) shared function, D) parse each cluster into subclusters to ensure probes will recognize all alleles, and E) submit selected sequences for probe selection and manufacturing.
A. Prepare the Input for FASTA36 Comparisons:
Organize the GenBank files from the RAST output into three files: i) a multi-fasta with all CDS as amino acid sequences, ii) a multi-fasta with CDS as nucleotide sequences, and iii) a multi-fasta with all the contigs. An in-house program for these functions is available from authors by request or can be downloaded from: https://github.com/jpearl01/prepare_supragenome_project
B. Compare all CDSs and Contigs using FASTA36
Use the programs within the FASTA Package (FASTA36) to compare all the sequences (Pearson and Lipman, 1988). Tfasty36 is used to compare the protein CDSs to the DNA CDSs database, calculating similarities with frameshifts to the forward and reverse orientations. Fasta36 is used to compare the DNA CDSs to the contigs, and capture any genes that may have been missed in the annotation process. The programs can be downloaded from http://faculty.virginia.edu/wrpearson/fasta/fasta36/.
Run the fasta36 programs using the following parameters:
Fasta36: fasta36 –E 1 –m 9 –n –Q –d 0 input ii (multi-fasta of all CDS as nucleic acids) input iii (multi-fasta of all contig sequences) > output name
Tfasty36: tfasty36 –E 1 –m 9 –p –Q –d 0 input i (multi-fasta of all CDS as amino acids) input ii (multi-fasta of all CDS as nucleic acids) > output name
C. Group the CDSs into Gene Clusters to Capture Similar Sequences
To parse the gene comparison into clusters we recommend our in-house Perl script – termed ClusterGenes - developed by Justin Hogg and originally presented by Hogg and colleagues (Hogg et al., 2007). A cluster is defined as a group of genes that share at least 70% identity, over 70% of their length, with one or more of the other genes in the group, and where at least one sequence in the cluster is equal to or longer than 120 amino acids. This script also organizes the cluster as either core or distributed. ClusterGenes is available from authors by request or can be downloaded from https://github.com/jpearl01/
D. Group the Gene Clusters into Subclusters to Capture Allelic Differences
Many of these clusters contain multiple allelic variants, such that if probes are designed to only one representative sequence from each cluster, they may not hybridize to all the alleles. To ensure that probes are designed that will collectively hybridize to all known alleles, each cluster should be further split into subclusters. Within a subcluster, all sequences are 95% identical over 95% of the length of the shorter sequence. For the subclustering step, apply the sequence comparison and parsing to each individual cluster using the same ClusterGenes script.
E. Submit the Longest Sequence in Each Subcluster for Probe Design and Manufacturing
The selected company that manufactures the probes will apply their in-house algorithms to design probes when given a user-defined set of sequences. Our probes were designed by Roche NimbleGen, and currently could be designed by the Agilent platform.
The company algorithms will ensure the design of probes of the desired length (60-200 bp), while avoiding homopolymers and low complexity regions. To this end, submit a multi-fasta file with the longest sequence in each subcluster for probe design and manufacturing. Regarding the number of probes per sequence, we suggest generating the maximum number that would fit one array while allowing each probe to be placed in triplicate. For the NimbleGen pneumococcal array, this meant that we could include up to 10 probes for each subcluster.
Control probes should also be included; two such sets are included in the SpSGH array. First, a set of 1000 random control probes (generated by NimbleGen) with the same length and GC characteristics as the experimental probes on the array and these can be used to estimate non-specific hybridization for background correction. Second, a set of alignment and tracking probes that serve for accurate positioning of the probe grid during image analysis, detection of erroneous mixing of samples, and gauging the uniformity of hybridization over the probe covered area of the array.
Example: Probe Design for the SpSGH Array
For design of the SpSGH array, 51 strains were selected (Table 1). The strains include multiple representatives of the major pathogenic lineages, and multiple serotypes and multi locus sequencing types (MLST). Furthermore, the chosen strains were isolated from subjects on multiple continents, and included representatives associated with nasopharyngeal carriage as well as disease. Together, the 51 genomes code for 107,957 CDSs. These were compared and organized into 3,204 pneumococcal gene clusters of which 1,597 are core and 1,607 are distributed. All clusters were further subdivided into subclusters and the longest sequences from each subcluster were used to design probes by the manufacturing company. Some subclusters were eliminated because no suitable probes could be designed and/or only suitable probes were predicted to cross react with multiple subclusters. 40,988 experimental probes were designed to 3,027 clusters subdivided into 4,450 subclusters (9.2 probes/subcluster), of which 2,344 are core and 2,106 are distributed. The final probes for the SpSGH are provided in Table S1.
Table 1.
List of strains used for design of the SpSGH
| Pneumococcal Strain | Serotype | MLST | Genome (bp) | #ORFs | Location of isolation | Carriage / disease | Status | Source | Accession Number | Technology | Coverage |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SP3 | 3 | 180 | 2033581 | 2177 | Pittsburgh, US | disease | draft | Ref. [4] | AAZZ00000000 | 454 GS20 | 18x |
| SP6 | 6 | 460 | 2162916 | 2325 | Pittsburgh, US | disease | draft | Ref. [4] | ABAA00000000 | 454 GS20 | 19x |
| SP9 | 9 | 1269 | 2117908 | 2241 | Pittsburgh, US | disease | draft | Ref. [4] | ABAB00000000 | 454 GS20 | 20x |
| SP11 | 11 | 62 | 2060705 | 2127 | Pittsburgh, US | disease | draft | Ref. [4] | ABAC00000000 | 454 GS20 | 17x |
| SP14 | 14 | 124 | 2148093 | 2624 | Pittsburgh, US | disease | draft | Ref. [4] | ABAD00000000 | 454 GS20 | 16x |
| SP18 | 6 | new | 2105593 | 2211 | Pittsburgh, US | disease | draft | Ref. [4] | ABAE00000000 | 454 GS20 | 19x |
| SP19 | 19 | 485 | 2136434 | 2301 | Pittsburgh, US | disease | draft | Ref. [4] | ABAF00000000 | 454 GS20 | 16x |
| SP23 | 23 | 37 | 2103479 | 2200 | Pittsburgh, US | disease | draft | Ref. [4] | ABAG00000000 | 454 GS20 | 15x |
| INV104B | 1 | 227 | 2142122 | 1941 | Oxford, UK | disease | complete | Ref. [4] | FQ312030 | Sanger | 9x |
| OXC141 | 3 | 180 | 2036967 | 1973 | Oxford, UK | carriage | complete | Ref. [4] | FQ312027 | Sanger | 9x |
| INV200 | 14 | 9 | 2093318 | 2045 | Oxford, UK | disease | complete | Ref. [4] | FQ312029 | Sanger / Illumina | 12x/85x |
| SpnATCC700669 | 23F | 81 | 2221315 | 2132 | Spain | carriage | complete | Ref. [19] | FM211187 | Sanger | 8x |
| Sp03_4156 | 3 | 180 | 2058353 | 1954 | The Netherlands | carriage | draft | Donati et al | FQ312045 | Sanger / 454 FLX | 5x/25x |
| Sp03_4183 | 3 | 180 | 1993183 | 1933 | The Netherlands | carriage | draft | Donati et al | FQ312043 | Sanger / 454 FLX | 6x/16x |
| Sp07_2838 | 3 | 180 | 1990038 | 1901 | Bolivia | carriage | draft | Donati et al | CACI01000000 | Sanger / 454 FLX | 5x/23x |
| Sp99_4038 | 3 | 180 | 2010908 | 1952 | Glasgow Reference Lab | disease | draft | Donati et al | FQ312041 | Sanger / 454 FLX | 7x/26x |
| Sp99_4039 | 3 | 180 | 2010104 | 1954 | Glasgow Reference Lab | disease | draft | Donati et al | FQ312044 | Sanger / 454 FLX | 4x/32x |
| Sp02_1198 | 3 | 180 | 1989367 | 1938 | Glasgow Reference Lab | disease | draft | Donati et al | CACH01000000 | Sanger / 454 FLX | 5x/28x |
| A45 | 3 | New | 2041833 | 1932 | Newmarket | disease | draft | Donati et al | CACG01000000 | Sanger / 454 FLX | 6x/15x |
| P1041 | 1 | 217 | 2166490 | 1905 | Ghana | disease | draft | Donati et al | CACE01000000 | Sanger / 454 FLX / Illumina |
5x/13x/96x |
| Sp03_2672 | 1 | 306 | 2144331 | 1904 | Glasgow Reference Lab | disease | draft | Donati et al | FQ312039 | Sanger / 454 FLX | 5x/25x |
| Sp03_3038 | 1 | 306 | 2164519 | 1936 | Glasgow Reference Lab | disease | draft | Donati et al | FQ312042 | Sanger / 454 FLX | 5x/20x |
| Sp06_1370 | 1 | 306 | 2012346 | 1874 | Glasgow Reference Lab | disease | draft | Donati et al | CACJ01000000 | Sanger / 454 FLX | 5x/19x |
| NCTC7465 | 1 | 615 | 2100988 | 1845 | Type strain, Rockefeller USA, 1948 |
disease | draft | Donati et al | CACF01000000 | Sanger / 454 FLX / Illumina |
5x/11x/86x |
| P1031 | 1 | 303 | 2111882 | 2073 | Ghana | disease | complete | Donati et al | CP000920 | Sanger / 454 FLX | Finished* |
| D39 | 2 | 595 | 2046115 | 1914 | US | disease | complete | Ref. [17] | CP000410 | Sanger | Finished* |
| TIGR4 | 4 | 205 | 2160842 | 2125 | Norway | disease | complete | Ref. [15] | AE005672 | Sanger | Finished* |
| 70585 | 5 | 289 | 2184682 | 2202 | Bangladesh | disease | complete | Donati et al | CP000918 | Sanger / 454 FLX | Finished* |
| JJA | 14 | 66 | 2120234 | 2123 | Brazil | disease | complete | Donati et al | CP000919 | Sanger / 454 FLX | Finished* |
| MLV-016 | 11A | 62 | 2247118 | 2159 | USA, Europe | carriage | draft | Donati et al | ABGH00000000 | Sanger / 454 FLX | 9x/28x |
| CDC0288-04 | 12F | 220 | 2051140 | 2105 | USA, UK | disease | draft | Donati et al | ABGF00000000 | Sanger / 454 FLX | 10x/31x |
| CDC3059-06 | 19A | 199 | 2293277 | 2379 | Iceland, UK, USA, others | disease | draft | Donati et al | ABGG00000000 | Sanger / 454 FLX | 9x/26x |
| Hungary19A-6 | 19A | 268 | 2245615 | 2155 | Hungary | disease | complete | Donati et al | CP000936 | Sanger / 454 FLX | Finished* |
| Taiwan19F-14 | 19F | 236 | 2112148 | 2044 | Taiwan | disease | complete | Donati et al | CP000921 | Sanger / 454 FLX | Finished* |
| CDC1873-00 | 6A | 376 | 2265195 | 2402 | USA | disease | draft | Donati et al | ABFS00000000 | Sanger / 454 FLX | 9x/20x |
| 670-6B | 6B | 90 | 2240045 | 2384 | Spain | disease | complete | Ref. [4] | CP002176 | Sanger | Finished* |
| CDC1087-00 | 7F | 191 | 2190853 | 2232 | Bra, Den, Fin, Neth, Nor, UK, Uru, USA |
disease | draft | Donati et al | ABFT00000000 | Sanger / 454 FLX | 9x/14x |
| SP195 | 9V | 156 | 2198294 | 2287 | Worldwide | disease | draft | Donati et al | ABGE00000000 | Sanger / 454 FLX | 10x/29x |
| G54 | 19F | 63 | 2078953 | 2115 | Italy | disease | complete | Ref. [4] | CP001015 | Sanger | Finished* |
| R6 | 2 | 595 | 2038615 | 2043 | Laboratory | laboratory | complete | Ref. [16] | AE007317 | Sanger | Finished* |
| ST13v1-CGSSp14BS292 | 14 | 13 | 2100368 | 2290 | Children's Hospital of Pittsburgh |
disease | draft | Hiller 2010 | ABWQ00000000.1 | 454 FLX | 21.5x |
| ST13v12-CGSSpBS293 | NT | 13 | 2065452 | 2242 | Children's Hospital of Pittsburgh |
disease | draft | Hiller 2010 | ABWU00000000.1 | 454 FLX | 23x |
| ST13v6-CGSSpBS457 | NT | 13 | 2053197 | 2225 | Children's Hospital of Pittsburgh |
disease | draft | Hiller 2010 | ABWB00000000.1 | 454 FLX | 27x |
| ST2011v4-CGSSpBS455 | NT | 2011 | 2086050 | 2182 | Children's Hospital of Pittsburgh |
disease | draft | Hiller 2010 | ADHN00000000.1 | 454 FLX Titanium | 28x |
| SV35-T23 | 23F | 81 | 2156885 | 2242 | AIDS clinic of St. Vincent's Medical Center, Richmond, New York, US |
disease | draft | Hiller,2011 | ADNN01000000 | 454 FLX | 26x |
| SV36-T3 | 3 | 81 | 2162633 | 2239 | AIDS clinic of St. Vincent's Medical Center, Richmond, New York, US |
disease | draft | Hiller,2011 | ADNO01000000 | 454 FLX | 26x |
| PN4595-T23 | 23F | 81 | 2169192 | 2259 | Lisbon, Portugal | carriage | draft | Hiller,2011 | ABXO01 | 454 FLX | 27.6x |
| CGSP14 | 14 | 15 | 2209198 | 2206 | China, Beijing Institute of Genomics |
disease | complete | Ref. [18] | CP001033 | Sanger | Finished* |
| CCRI 1974 | 14 | 124 | 2005075 | 2074 | McGill University, Canada | disease | draft | Ref. [20] | ABZC00000000 | 454 FLX | 20x |
| CCRI 1974M2 | 14 | 124 | 2003231 | 2069 | McGill University, Canada | disease | draft | Ref. [20] | ABZT00000000 | 454 FLX | 20x |
| AP200 | 11A | 62 | 2084139 | 2149 | University of Siena, Italy | disease | draft | Unpublished | CP002121 | 454 | 10x |
BASIC PROTOCOL 2: SpSGH Array to Determine Gene Content
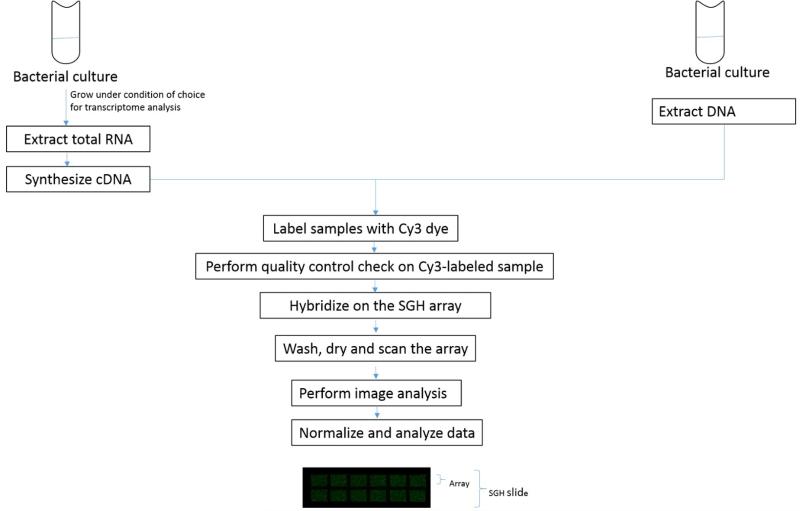
The SpSGH array can be used to determine the gene content of isolates by employing DNA-DNA hybridizations. This has been described for the H. influenzae SGH array (Eutsey et al., 2013) and is described here for the SpSGH. The process has five steps: 1) grow bacterial strains; 2) extract genomic DNA (gDNA); 3) label Cy3 gDNA; 4) hybridize gDNA on array, wash and scan 5) analyze the data. A schematic for these steps is represented in Figure 2.
Figure 2.
Schematic of processing of nucleic acid samples for the SGH array.
We tested the SpSGH array by comparing array results with WGS data for 5 strains. We also investigated genome content for 2 non-sequenced strains isolated from a patient with a polyclonal upper respiratory infection.
Materials
Bacterial strain of interest
Standard media for bacterial growth (Columbia broth is used for S. pneumoniae)
Pneumococcal cell lysis cocktail: lysozyme (15ml/mL), mutanolysin (30μg/mL), proteinase K (20mg/mL), 1x Tris EDTA Buffer
Chloroform:isoamylalcohol (24:1)
RNAseA (4mg/mL)
1% TAE agarose gel
NanoDrop 1000 UV spectrophotometer
Centrifuge that allows harvesting cells from a 15mL culture volume
Vacuum concentrator
SGH array
gDNA Cy-3 labeling reagents from the array manufacturer
Hybridization and washing reagents from the array manufacturer
Hybridization station from the array manufacturer
Fluorescent scanner
Thermocycler
Vortex
Strain and growth conditions
- Set up 15mL bacterial cultures in duplicates and allow growth to mid-log phase or stationary phase, in standard media.The goal is to determine gene content, thus the only condition for growth is that which provides sufficient DNA. For S. pneumoniae we use Columbia broth and continue culture until an OD600 of 0.5 is achieved, as these conditions provide for high cell numbers yet limited Lyt-A-mediated autolysis. For H. influenzae we use overnight cultures grown in supplemented BHI broth.
- Harvest the bacterial cells by centrifugation at 5000xg for 10mins, and freeze the pelleted cultures at −80°C so that the subsequent steps can be performed at a later time point, if desired.We recommend frozen pellets be used within two weeks of freezing.
Thaw the pelleted cells by incubating at room temperature for 15 minutes. Lyse the bacterial cells. To achieve S. pneumoniae cell lysis, resuspend pellets in a 220μL cocktail of lysozyme (15mg/ml), mutanolysin (30μg/mL), and proteinase K (20mg/mL, Qiagen) in 1X Tris-EDTA buffer for 15 minutes at room temperature, with intermittent vortexing every 2 minutes.
gDNA extraction
-
4
Perform a standard 24:1 chloroform/isoamyl alcohol method for gDNA extraction and store samples in 1X TE buffer (Sambrook and Russell, 2001).
-
5
Measure the concentration and purity of the gDNA using the ratio of absorbance at 260 and 280 nm on a UV spectrophotometer (NanoDrop 1000, Thermo Scientific), where pure gDNA has an A260/280 ratio of 1.8.
-
6
Confirm the purity of the gDNA by running ~1 μg of DNA on a 1% TAE agarose gel. If the purity is low, the gDNA should be treated with RNaseA (4mg/mL) and/or Proteinase K (20mg/mL), then re-precipitated and re-analyzed to ensure purity of the DNA.
Cy3 gDNA labeling and quality control
-
7
Label gDNA samples with Cy3 dye. This step utilizes a nucleotide synthesis reaction which incorporates Cy3 labeled random nonamers into double stranded DNA using the NimbleGen One Color DNA Labeling Kit (NimbleGen Arrays User's Guide, Gene Expression Arrays version 6.0). To this end, heat the gDNA sample to 98°C for 10 minutes in the presence of the Cy3 labeled random nonamers, and rapidly cool in an ice water bath. For DNA polymerization, add the dNTPs and Klenow fragment to the reaction and incubate for 2 hours at 37°C, as described in the kit instructions.
-
8
Use isopropanol precipitation of the labeled gDNAs to eliminate any unincorporated nucleotides and primers from the labeling reaction.
-
9
Dry the DNA pellet in a vacuum concentrator (SpeedVac) and protect from light. Rehydrate the sample in nuclease-free water.
-
10Measure the concentration and quality using 260/280 absorbance ratio on a spectrophotometer.The final concentration and volume of Cy3-labeled gDNA for the next step, depends on the array design and manufacturer. A NimbleGen array, with 12 hybridization regions of 135K each (that is, 135,000 probe capacity), requires 2 μg of labeled gDNA per region.
Hybridization, washing and slide scanning
SGH slides allow for parallel processing of multiple samples per slide, such that each sample is loaded onto a separate array on the slide. Our NimbleGen slide contains 12 arrays. Cross-reaction of different samples is monitored using sample tracking controls (STC) provided by the manufacturer. Each array has a different STC. For the NimbleGen 12 X 135K array, 2 μg of Cy3-labeled DNA is lyophilized in a SpeedVac and resuspended in sample tracking solution. On each slide, probes specific to the STC are placed as repeating sets of 20 along the perimeter of each array and bordering their corners. The STCs assist later during imaging where, by performing a sample tracking control analysis and visually verifying the outlines of each array, the user can confirm that samples have not mixed with each other.
-
11
Prior to hybridization, mix the sample with the components of the Hybridization Kit (NimbleGen Hybridization buffer, component A, and alignment oligomer) and incubate at 95°C for 5 minutes.
-
12
The array manufacturer supplies a proprietary mixing device that is designed to align and adhere to the surface of the array. Place the adherence assembly (mixer device + SpSGH slide) on the hybridization station, and load 6μl of each sample into a fill port.
-
13
Set the hybridization station at 42°C and incubate the slides for 18 hours.
-
14After the incubation, disassemble the slide from the mixer and wash to remove unbound sample. Washing and drying involve a series of wash buffers from the NimbleGen Hybridization Wash Buffer kit and the Microarray slide dryer.For best results, perform the steps leading from washing to scanning without any pauses.
-
15
Measure the fluorescence using a fluorescent scanner with suitable resolution; we used the Molecular Devices Axon GenePix 4200AL for the one-color array scanning of the SpSGH slide. Process the images using the NimbleScan, or equivalent imaging software, to measure the intensity and the relative position of each fluorescent signal.
BASIC PROTOCOL 3: Data Analysis of SpSGH Array to Determine Gene Content
The analysis of gene content involves multiple steps: a) conversion of the fluorescent signals into quantitative intensity values and determination of data integrity; b) normalization of the values across all arrays on the same slide, or among slides; c) determination of the threshold for presence/absence of a gene to establish genome content for the strains of interest.
Materials
Slide scanning software provided by the array manufacturer (such as NimbleScan)
Generating quantitative intensity values and assessing data integrity
Following the slide scan, use the array software to burst the single multiplex image of the slide into separate array images based on the format of the slide. Each image will correspond to one strain/sample.
Align each of the separated images to the design file that contains information on the placement of the probes on the array. A grid setup in the software assists in aligning the images correctly. During this step, perform the sample tracking control check that verifies absence of cross-contamination among samples by indicating which sample tracking controls (STC) are present in each array. Only one STC should be present per sample. The same analysis step also generates an experimental metrics report, consisting of a spreadsheet reporting signal density, signal range and uniformity.
Normalize the data across regions on the array as per the user's choice. Normalization uses a Robust Multichip Average (RMA) algorithm and quantile normalization (also available from the chip manufacturer, in this case via the NimbleScan software). In the raw data, an intensity value is available for each probe. The NimbleScan normalization process will combine the multiple probes for each subcluster using a median polish, generating a table that lists each subcluster and an associated intensity value for each probe set. At the end of this step, the user will have a tab-delineated sheet with one value integrating the multiple probes per subcluster (i.e per allele) (~9/ alleles for the SpSGH array), with triplicates for each value (since all probes were placed on the array in triplicate). An example sheet can be visualized in Table S2.
Determining present/absent genes
-
4To determine which genes are present in the sample, select a hybridization value threshold that separates genes present versus genes absent.We recommend this threshold be 1.5 times the median background value.
-
5Convert the normalized data into a log2 scale and determine the inter-slide consistency using the Student's t distribution analysis. These standard statistics can be applied from any program of choice; we use a python-based script.A gene is considered present if the signal for any of its subclusters is above the threshold and the p-value from the Student's t test is less than 0.05. Conversely, a gene is considered absent if the signal for all its subclusters is below the threshold or if the p-value from the Student's t test is above 0.05 for all subclusters above threshold. Importantly, binding to subsets of subclusters is not used to investigate polymorphisms given that hybridization can occur even when small variations exist between the probe and the allele.
Example: Genomic Content of Seven Pneumococcal Isolates Using the SpSGH Array
The gene possession profile of 5 pneumococcal isolates was interrogated for presence/absence of 3,027 clusters (out of 3,204 total) using the SpSGH array. The presence/absence profile was compared to WGS to calculate the sensitivity (determined by the number of false positives) and specificity (determined by the number of false negatives) of the SpSGH array output relative to WGS. The results are described in Table 2, where false positives varied from 3-5 genes/genome and false negatives between 21-37 genes/genome, suggesting that >98% of genes were accurately predicted by the SpSGH array.
Table 2.
Validation of the SpSGH using whole genome sequence. The supragenome analysis used for SpSGH design contained 3204 gene clusters, of which 3027 are represented on the SpSGH. The last two rows depict data from unsequenced genomes isolated during a polyclonal infection. The SpSGH results demonstrate they are almost identical to other strains isolated from the same patient (ST13v3 is compared to ST13v1 and ST2011v5 to ST2011v4)
| strain | # CLUSTERS | # CLUSTERS w/PROBE ON CHIP | # CLUSTERS DETECTED BY CHIP | % CORR. PREDICTED | # FALSE POSITIVE | # FALSE NEGATIVE |
|---|---|---|---|---|---|---|
| ST13v1 | 2028 | 1918 | 1902 | 99.2 | 5 | 21 |
| SP3 | 1996 | 1890 | 1857 | 98.3 | 4 | 37 |
| SP14 | 2119 | 2002 | 1973 | 98.6 | 4 | 33 |
| SV35 | 2059 | 1950 | 1927 | 98.8 | 3 | 26 |
| SP23 | 2044 | 1933 | 1909 | 98.8 | 4 | 28 |
| ST13v3 | 2028 | 1918 | 1902 | 99.2 | 5 | 21 |
| ST2011v5 | 2009 | 1903 | 1868 | 98.2 | 8 | 43 |
An additional 2 pneumococcal genomes, ST13v3 and ST2011v5 (Hiller et al., 2010) were assayed using the SpSGH array. These strains were isolated from a young child with a polyclonal infection, previously referred to as patient 19 (Hiller et al., 2010). MLST and serotype analyses suggested these strains were similar to a pair of strains isolated from patient 19 at different clinical visits, strains ST13v1 (ST13v1-CGSSp14BS292) and ST2011v4 (ST2011v4-CGSSpBS455), respectively. The SpSGH analysis confirms this prediction, demonstrating that these strains were isolated from the same patient at separate time points consistent with chronic colonization (Table 2).
BASIC PROTOCOL 4: SpSGH Array for Gene Expression Profiling
The SpSGH array data is quantitative in nature, thus it can be used for gene expression profiling. To this end, cDNA, instead of gDNA is analyzed.
Profiling of gene expression requires the following steps: 1) strain growth; 2) RNA extraction, 3) conversion of RNA to cDNA; 4) Cy3 labeling of the cDNA; 5) cDNA hybridization, washing, and scanning; and 6) data analyses. Steps 1, 4-5 are very similar to those described for gDNA, such that this section will focus on the differences only. As an example, we used the SpSGH array to investigate the relative levels of transcripts for two pneumococcal strains relative to housekeeping controls. To measure accuracy, the results were compared with data for 54 genes using the nCounter Analysis System by nanoString Technologies (Geiss et al., 2008). Figure 2 provides a schematic for these steps.
Materials
Bacterial strain of interest
Standard media for bacterial growth (Columbia broth is used for S. pneumoniae)
Centrifuge that allows harvesting cells from a 15mL culture volume
RNAProtect Bacterial Reagent (Qiagen)
RNeasy Mini Kit (Qiagen) for RNA extraction
Pneumococcal cell lysis cocktail: lysozyme (15ml/mL), mutanolysin (30μg/mL), proteinase K (20mg/mL), 1x Tris EDTA Buffer
Lysis buffer RLTplus (Qiagen)
DNAse, 2units/μL (Turbo DNAse, Ambion)
gDNA eliminator column (Qiagen)
Agilent Bioanalyzer and RNA 6000 Nano Kit
SuperScript III First-Strand Synthesis SuperMix kit (Invitrogen)
SuperScript Double-Stranded cDNA Synthesis kit (Invitrogen)
RNaseA (4mg/mL)
Phenol:chloroform:isoamylalcohol (25:24:1)
Ammonium acetate, glycogen and ethanol
Vacuum concentrator (SpeedVac)
Nuclease-free water
NanoDrop1000 spectrophotometer
SGH array
gDNA Cy3 labeling reagents from the array manufacturer
Hybridization and washing reagents from the array manufacturer
Hybridization station from the array manufacturer
Fluorescent scanner
Thermocycler
Vortex
Strains and growth conditions
- Grow bacteria under in vitro condition(s) of interest. cDNA profiles can be compared across multiple types of samples. For example, the same strain under different growth conditions, or a wild type (WT) strain versus its cognate mutant strain.For the pneumococcal work presented here, we selected S. pneumoniae strains PN4595-T23 (ABXO01) and 3063-00 (AGQG01). The former is one of the 51 genomes used in the probe design, while the latter was not included in the probe design. PN4595-T23 is a member of the Pneumococcal Molecular Epidemiology Network clone 1 lineage (PMEN1) and 3063-00 is related to the Taiwan19F, thus both represent isolates from widespread and multidrug-resistant lineages. These strains are grown in 15mL Columbia broth in a 50mL tube to an OD600 of 0.5. This mid-log phase OD is chosen as it yields high cell numbers while the Lyt-A mediated autolysis is absent.
Harvest bacterial cells by centrifugation at 5000xg for 10 minutes and immediately resuspend the pellet in RNAProtect Bacterial Reagent (Qiagen) to stabilize RNA before storing at -80°C.
RNA extraction and quality check
It is critical that all tubes and water used for sample preparation are DNase/RNase-free.
-
3
Use the RNeasy Mini Kit (Qiagen) to extract total bacterial RNA. This process can be divided into 3 steps: cell lysis, RNA extraction, and elimination of any residual DNA. To achieve pneumococcal cell lysis, resuspend the cell pellets in a cocktail of lysozyme (15 mg/ml), mutanolysin (30 μg/mL), and proteinase K (20 mg/mL) in 1X Tris-EDTA buffer for 15 minutes at room temperature, with intermittent vortexing every 2 minutes to aid the lysis process.
-
4
Add lysis buffer RLTplus (Qiagen) to the preparation. Apply the lysate to the gDNA eliminator column (Qiagen) to remove genomic DNA, next apply to an RNeasy column for RNA isolation.
-
5
Treat the eluted RNA with 2 units/μL of DNAse (TurboDNase, Ambion) for 1.5h at 37°C.
-
6Assess the RNA integrity by running samples on the Agilent Bioanalyzer using an RNA 6000 Nano Kit.These RNA chips consist of micro-channels that separate nucleic acid fragments based on their electrophoretic mobility (Agilent Technologies, Inc.). Intact peaks corresponding to 16SrRNA and 23SrRNA and high RIN number in electropherograms are measures of RNA integrity.
-
7
Confirm the RNA purity using polymerase chain reaction (PCR) for the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) housekeeping gene, such that no amplification would be observed in pure RNA samples, while amplicons would be observed in the pure genomic DNA control.
-
8For an additional quality check, measure sample absorbance using a spectrophotometer.High quality RNA has an A260/280 ratio of ≥ 2.0.
cDNA preparation and quality check
All RNA, cDNA and reagents should be maintained on ice.
-
9
Use SuperScript III First-Strand Synthesis SuperMix kit (Invitrogen) for synthesis of the first strand of cDNA. For this, start with 5 μg of good quality total RNA as a template, add the random hexamers primers supplied in the kit, and heat at 70°C for 10 minutes. Next, add First Strand Buffer, DTT and dNTPs. These aid in the removal of any secondary structures in the RNA template. Add Superscript III reverse transcriptase as the last component and incubate the sample at 42 °C for 1 hour to synthesize an RNA:DNA hybrid.
-
10
Use SuperScript Double-Stranded cDNA Synthesis kit (Invitrogen) for second strand cDNA synthesis. Incubate the samples with kit components (DEPC water, 5x Second Strand Buffer, dNTPs, DNA Ligase, DNA Polymerase I and RNase H) at 16°C for 2 hours.
-
11
Eliminate residual template RNA by treating the reaction with RNaseA (4mg/mL) and extract using phenol:chloroform:isoamylalcohol (25:24:1). Precipitate the cDNA in the upper aqueous layer using ammonium acetate, glycogen and ethanol, followed by concentrating the pellet in a vacuum concentrator (SpeedVac) until a gel-like consistency is reached.
-
12Rehydrate the cDNA samples with nuclease-free water and assess their quality and quantity (we use the NanoDrop1000 spectrophotometer, A260/280 ≥ 1.8). If desired, check the cDNA samples by running on an agarose gel or Agilent Bioanalyzer.It is important to ensure that degraded/poor quality samples are not carried through further steps.
Cy3 cDNA labeling, hybridization, washing, and scanning
-
13
These steps are as described above in Basic Protocol 2.
BASIC PROTOCOL 5: Data Analysis of SpSGH Array for Gene Expression Profiling
The data analysis for transcriptional profiling can be subdivided into the following steps: A) selection of the relevant probes for the analysis; B) normalization within and among arrays; C) comparison of transcription levels between/among sample sets.
Materials
Slide scanning software provided by the array manufacturer (such as NimbleScan) NCBI-BLAST
Software of choice for transcriptomic analysis (e.g. CyberT, http://cybert.microarray.ics.uci.edu/; SAM http://statweb.stanford.edu/~tibs/SAM/faq.html; TM4/MeV, http://www.tm4.org/mev.html)
If the probe set of choice in selected by hybridization, also refer to Basic Protocol 2.
Selection of the relevant probes for the analysis
The advantage of using the SpSGH array is that the same platform can be used to assay any isolate from the species. However, for each isolate, the relevant probe set must be selected. For this quantitative analysis, a distribution of the intensity values per subcluster should fit a normal distribution. If the analysis accounts for all the subclusters, the majority of probes will not hybridized and will have very low fluorescent values. Thus, the relevant subclusters should be singled out.
Perform subcluster selection using one of two methods described below. If the genome of the test strain is known, the relevant subclusters can be selected in silico (i). If the genome sequence is not known, the relevant clusters can be selected using hybridization of gDNA on the SpSGH (ii).
i. Clusters selection using WGS
Compare the representative sequence for each subcluster to the WGS of selected strain using BLAST (basic local alignment search tool) from NCBI (Altschul et al. 1990). Include the top hit for each query sequence into the relevant subcluster set. We recommend downloading BLASTn onto a Linux computer and running the program locally with the following command line:
Prepare database for blastn
makeblastdb –in [multi-fasta file with the longest sequence for all subclusters where gene is labeled with the subcluster ID (i.e sequences submitted for probe design)] –dbtype nucl
Run blastn
blastn –evalue 1e-20 –query [multi-fasta with the CDSs for the WGS of selected strain] –db [multi-fasta file with the longest sequence for all subclusters where gene is labeled with the subcluster ID (i.e file submitted for probe design)] > output
Parse the output to select the top hit. We use a BioPerl script to parse the output, such that only the cluster in the top hit is included in the relevant set of subclusters. For any cluster, all subclusters are included.
ii. Cluster Selection using SpSGH array
Hybridize the gDNA to the SpSGH array, as described in Basic Protocol 2 above. Finalize Basic Protocol 2. Select only the clusters with a signal intensity above threshold and include all subclusters for each of these positive clusters. This represents the subcluster set relevant for your strain of interest.
Data Normalization
The analysis involves multiple steps: A) conversion of the fluorescent signal into quantitative intensity values and integrity check of the data; B) normalization of the values across all arrays on the same slide, or between arrays.
-
2
Use the array manufacturer's software to convert fluorescence intensity to quantitative value for each probe. The user can follow the description in “Part II: data analyses” above that describes how to: burst the single multiplex image of the slide into separate array images; check for cross-contamination among samples; acquire the signal density, signal range and uniformity; normalize the data across regions on the array; and generate a tab-delineated sheet with one value integrating the multiple probes per subcluster, each in triplicate. An example sheet can be visualized in Table S2.
Comparison of transcription between sample sets
-
3
This analysis will differ depending on the samples being compared, and standard array methods can be employed (e.g. CyberT: http://cybert.microarray.ics.uci.edu/; SAM: http://statweb.stanford.edu/~tibs/SAM/faq.html; TM4/MeV: http://www.tm4.org/mev.html). Janto and colleagues provide a detailed analysis comparing wildtype and deletion mutant strains over multiple conditions and time points using the HiSGH (Janto et al PLoS One, 2014). In their analysis, a web-based microarray analysis tool, Cyber T (Kayala and Baldi, 2012) is used to obtain Bayesian corrected p-values, Bonferroni corrected p-values and Benjamini-Hochberg values. These data are then combined and filtered in the following order: 1) SAM FDR <10%, 2) Bayesian p-values < .05, 3) Benjamini-Hochberg FDR < 10%, 4) Bonferroni corrected p-value < .05. This pipeline, with progressively more stringent statistical parameters, generates a robust set of differentially regulated genes for transcriptomic analysis.
Example: Transcriptional analysis of two pneumococcal Isolates using the SpSGH Array
The following section presents an example analysis that reflects on reproducibility of the SpSGH by comparing transcriptional values within a single genome using the SpSGH array and an alternative transcriptomic profiling technique, the nanoString technology.
Cluster Selection
For the analysis of strains PN4595-T23 and 3063-00, the relevant clusters were selected using in silico analysis. 1,929 and 1,886 total clusters were analyzed for PN4595-T23 and 3063-00, respectively.
Reproducibility of the SpSGH Array
To assess the SpSGH array reproducibility we compared: 1) the signal intensity values across the triplicate probe sets within the same array (Figure 3); 2) the signal intensity values across biological replicates on the same slide (Figure 4A); and 3) the signal intensity values across biological replicates on two slides, hybridized and analyzed independently (Figure 4B).
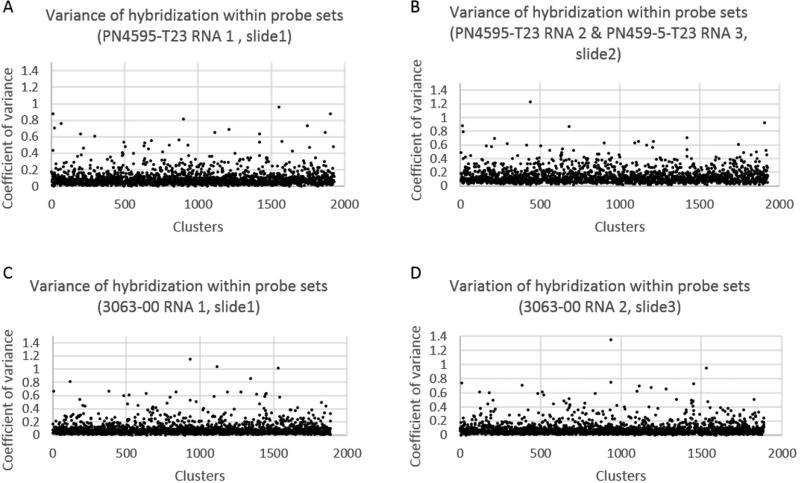
Figure 3.
Comparison of probe specificity within each array based on coefficient of variance of hybridization of RNA samples to probe set. The RNA samples for each strain are numbered based on independent experiments. (A) PN4595-T23 RNA 1 on slide 1, (B) PN4595-T23 RNA 2 and PN459-5-T23 RNA 3 on slide 2, (C) 3063-00 RNA1 on slide 1, (D) 3063-00 RNA2 on slide 1.
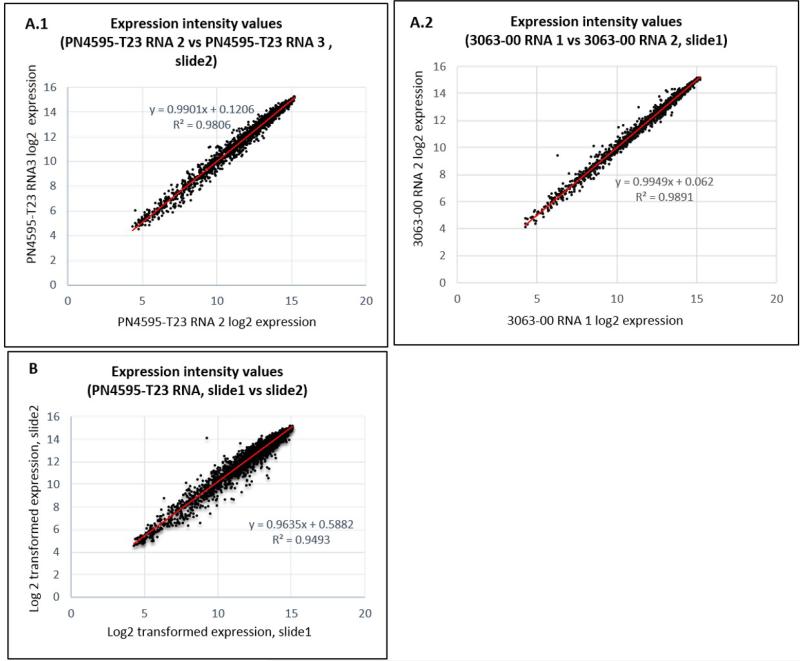
Figure 4.
Comparison of hybridization values for biological RNA replicates. (A) within the same slide. A.1. slide, A.2. slide; and (B) across slides.
The robustness of the array can be measured by the reproducibility of triplicate sets within each array. Each subcluster is represented by up to 10 unique probes, the probe values are condensed into a final hybridization value per subcluster. Given the triplicate probe sets, there are 3 final hybridization values per array. The values across the triplicate probe sets were analyzed using coefficient of variance, where a standard deviation is calculated for each set of triplicate probe sets and reported as a percentage of the average signal for that probe set. We find that over 94% of the probe sets have a coefficient of variance below 0.3 (Figure 3).
Each slide is manufactured with multiple arrays present on the same slide allowing multiple samples to be processed together. We compared the final hybridization values for biological replicates hybridized on the same slide Figure 4A (A.1 and A.2); as well as biological replicates hybridized on separate slides and processed independently (Figure 4B). In both cases, the arrays showed good reproducibility as illustrated by an R2 of 0.980 and 0.949, respectively.
Validation
The quantitative value of the SpSGH array was assessed by comparing the intensity values for RNA extracted from wildtype strains relative to two housekeeping genes: gyrase B (gyrB) and methionyl-tRNA synthetase (metG). We compared these fold changes to those obtained using another transcriptomics technology, the nCounter Analysis System (nanoString Technologies). The nCounter system directly captures mRNA with a sequence-specific DNA probe and quantifies the signal by single molecule imaging of unique transcripts (thus without any amplification step). The method is highly quantitative and reproducible, thus serves as a good method to verify the results from SpSGH array. The probe sets for the SpSGH array and the nCounter system were designed independently. Finally, all the RNA analyses, across SpSGH arrays and between the array and nCounter, were derived from different biological replicates (where cells were grown and RNA extracted independently).
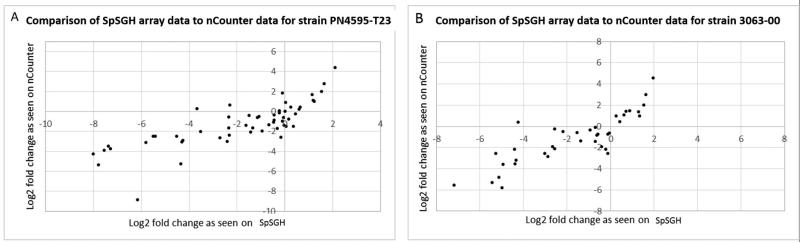
In both the SpSGH array and nCounter analyses, all values were normalized against gyrB and metG using the geometric mean for their signal intensity. Next, the value of signal intensity for each cluster was divided by the geometric mean, yielding a relative intensity value which was converted to log2. Finally the results from each method were plotted against each other (Figure 5). The comparison between the nCounter and SGH arrays reveals similar trends, suggesting that like the HiSGH array, the SpSGH arrays can also be used for transcriptomic studies.
Figure 5.
Validation of the SpSGH array, by comparing relative expression using SpSGH array and the nCounter from NanoString technologies. (A) PN4595-T23 . (B) 3063-00.
COMMENTARY
Background Information
There can be extensive differences in allelic content and gene possession among strains of a single bacterial species. Our goal was to design a gene chip that quantitatively captured the genetic diversity in a bacterial population. The SpSGH array described in this unit: 1) captures >90% of non-rare genes allowing genomic analysis of any S. pneumoniae isolate, and 2) is quantitative, thereby allowing for gene expression profiling of S. pneumoniae strains under in vitro conditions. The methodology described can be applied to the construction and modification of an S.pneumoniae SGH array, as well as applied to other bacterial species as long as multiple WGSs are available.
Critical Parameters and Troubleshooting
This unit describes the design of a SGH array and its implementation for genomic and transcriptomic analyses. In the design step, it is important to ensure that the probe set capture the genetic diversity of the population of interest. The coverage of the probe set will depend on the number and the phylogenetic distance of the whole genome sequences in the input set. The final probes must capture the differences in gene possession as well as the allelic variations. For probe selection, the user may select any pangenome analysis tool. We describe in detail methods to organize the genomic content of any number of strains into clusters of highly similar genes for probe design. The genomic analysis and/or transcriptomic profiling require multiple steps from cell growth and nucleic acid extraction to nucleic acid labeling, hybridization and washing. It is imperative that every step be carefully monitored by performing quality control of the output and adding additional control probes.
Anticipated Results
The SGH array can be used to analyze DNA and reveal the genetic content of an isolate, or to analyze cDNA and reveal the gene expression profile of an isolate.
Time Considerations
Once whole genome sequences are selected for the pangenome analysis, the clustering and selection of sequences for probe design can be finalized within 1-2 weeks. The rate- limiting step is the comparison of each sequence to all other sequences, which depends on the number of sequences in the set and the processing power available. If no problems are encountered, the processing of DNA or cDNA to genomic content or gene expression respectively can be achieved in 1 week. The number of samples that can be processed in parallel depends on the design of the chip.
Supplementary Material
Acknowledgments
The authors thank Drs. Aaron Mitchell and Wenjie Xu for the assistance with the nCounter nanoString technology. We are also grateful to the generous financial support from Allegheny Health Network. This work was supported by NIH grants AI080935 (GDE), DC 02148 (GDE), DC02148 – 16S1 (GDE), and DC011322 (NLH). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Literature Cited
- Ahmed A, Earl J, Retchless A, Hillier SL, Rabe LK, Cherpes TL, Powell E, Janto B, Eutsey R, Hiller NL, Boissy R, Dahlgren ME, Hall BG, Costerton JW, Post JC, Hu FZ, Ehrlich GD. Comparative Genomic Analyses of 17 Clinical Isolates of Gardnerella vaginalis Provide Evidence of Multiple Genetically Isolated Clades Consistent with Subspeciation into Genovars. J Bacteriol. 2012;194:3922–3937. doi: 10.1128/JB.00056-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 5. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Baldi P, Long AD. A Bayesian Framework for the Analysis of Microarray Expression Data: Regularized t-Test and Statistical Inferences of Gene Changes. Bioinformatics. 2001;17:509–519. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- Boissy R, Ahmed A, Janto B, Earl J, Hall BG, Hogg JS, Pusch GD, Hiller LN, Powell E, Hayes J, Yu S, Kathju S, Stoodley P, Post JC, Ehrlich GD, Hu FZ. Comparative supragenomic analyses among the pathogens Staphylococcus aureus, Streptococcus pneumoniae, and Haemophilus influenzae using a modification of the finite supragenome model. BMC Genomics. 2011;12:187. doi: 10.1186/1471-2164-12-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borneman AR, McCarthy JM, Chambers PJ, Bartowsky EJ. Comparative analysis of the Oenococcus oeni pan genome reveals genetic diversity in industrially-relevant pathways. BMC Genomics. 2012;13:373. doi: 10.1186/1471-2164-13-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey TJ, Daniels M, Enright MC, Spratt BG. Serotype 14 variants of the Spanish penicillin-resistant serotype 9V clone of Streptococcus pneumoniae arose by large recombinational replacements of the cpsA-pbp1a region. Microbiology. 1999;145:2023–31. doi: 10.1099/13500872-145-8-2023. [DOI] [PubMed] [Google Scholar]
- Conlan S, Mijares LA, Becker J, Blakesley RR, Bouffard GG, Brooks S, Coleman HL, Gupta J, Gurson N, Park M, Schmidt B, Thomas PJ, Young A, Otto M, Kong HH, Murray PR, Segre JA. Staphylococcus epidermidis pan-genome sequence analysis reveals diversity of skin commensal and hospital infection-associated isolates. Genome Biol. 2012;13:R64. doi: 10.1186/gb-2012-13-7-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davie JJ, Earl J, de Vries SP, Ahmed A, Hu FZ, Bootsma HJ, Stol K, Hermans PW, Wadowsky RM, Ehrlich GD, Hays JP, Campagnari AA. Comparative analysis and supragenome modeling of twelve Moraxella catarrhalis clinical isolates. BMC Genomics. 2011;12:70. doi: 10.1186/1471-2164-12-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donati C, Hiller NL, Tettelin H, Muzzi A, Croucher NJ, Angiuoli SV, Oggioni M, Dunning Hotopp JC, Hu FZ, Riley DR, Covacci A, Mitchell TJ, Bentley SD, Kilian M, Ehrlich GD, Rappuoli R, Moxon ER, Masignani V. Structure and dynamics of the pan-genome of Streptococcus pneumoniae and closely related species. Genome Biol. 2010;11:R107. doi: 10.1186/gb-2010-11-10-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowson CG, Hutchison A, Brannigan JA, George RC, Hansman D, Liñares J, Tomasz A, Smith JM, Spratt BG. Horizontal transfer of penicillin-binding protein genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Proc Natl Acad Sci U S A. 1989;86:8842–8846. doi: 10.1073/pnas.86.22.8842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelmoer DJP, Rozen DE. Competence increases survival during stress in Streptococcus pneumoniae. Evolution. 2011;65:3475–3485. doi: 10.1111/j.1558-5646.2011.01402.x. [DOI] [PubMed] [Google Scholar]
- Erdos G, Sayeed S, Antalis P, Hu FZ, Hayes J, Goodwin J, Dopico R, Post JC, Ehrlich GD. Development and characterization of a pooled Haemophilus influenzae genomic library for the evaluation of gene expression changes associated with mucosal biofilm formation in otitis media. Int J Pediatr Otorhinolaryngol. 2003;67:749–55. doi: 10.1016/s0165-5876(03)00093-4. [DOI] [PubMed] [Google Scholar]
- Eutsey RA, Hiller NL, Earl JP, Janto BA, Dahlgren ME, Ahmed A, Powell E, Schultz MP, Gilsdorf JR, Zhang L, Smith A, Murphy TF, Sethi S, Shen K, Post JC, Hu FZ, Ehrlich GD. Design and validation of a supragenome array for determination of the genomic content of Haemophilus influenzae isolates. BMC Genomics. 17. 2013;14:484. doi: 10.1186/1471-2164-14-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes ML, Horsey E, Hiller NL, Buchinsky FJ, Hayes JD, Compliment JM, Hillman T, Ezzo S, Shen K, Keefe R, Barbadora K, Post JC, Hu FZ, Ehrlich GD. Strain-specific virulence phenotypes of Streptococcus pneumoniae assessed using the Chinchilla laniger model of otitis media. PLoS One. 2008;3:e1969. doi: 10.1371/journal.pone.0001969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, James JJ, Maysuria M, Mitton JD, Oliveri P, Osborn JL, Peng T, Ratcliffe AL, Webster PJ, Davidson EH, Hood L, Dimitrov K. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26(3):317–25. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- He M, Sebaihia M, Lawley TD, Tabler RA, Dawson LF, Martin MJ, Holt KE, Seth-Smith HM, Quail MA, Rance R, Brooks K, Churcher C, Harris D, Bentley SD, Burrows C, Clark L, Corton C, Murray V, Rose G, Thurston S, van Tonder A, Walker D, Wren BW, Dougan G, Parkhill J. Evolutionary dynamics of Clostridium difficile over short and long time scales. Proc Natl Acad Sci U S A. 2010;107:7527–7532. doi: 10.1073/pnas.0914322107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller NL, Janto B, Hogg JS, Boissy R, Yu S, Powell E, Keefe R, Ehrlich NE, Shen K, Hayes J, Barbadora K, Klimke W, Dernovoy D, Tatusova T, Parkhill J, Bentley SD, Post JC, Ehrlich GD, Hu FZ. Comparative genomic analyses of seventeen Streptococcus pneumoniae strains: insights into the pneumococcal supragenome. J Bacteriol. 2007;189:8186–8195. doi: 10.1128/JB.00690-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg JS, Hu FZ, Janto B, Boissy R, Hayes J, Keefe R, Post JC, Ehrlich GD. Characterization and modeling of the Haemophilus influenzae core and supragenomes based on the complete genomic sequences of Rd and 12 clinical nontypeable strains. Genome Biol. 2007;8:R103. doi: 10.1186/gb-2007-8-6-r103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janto B, Hiller NL, Eutsey R, Dahlgren ME, Earl J, Powell E, Ahmed A, Hu FZ, Ehrlich GD. Development and Validation of an Haemophilus influenzae Supragenome Hybridization (SGH) Array for Transcriptomic Analyses. PLoS ONE. 2014;9(10):e105493. doi: 10.1371/journal.pone.0105493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayala MA, Baldi P. Cyber-T web server: differential analysis of high-throughput data. Nucleic Acids Research. 2012;40:W553–W559. doi: 10.1093/nar/gks420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NimbleGen Arrays User’s Guide Gene Expression Arrays version 6.0 [Google Scholar]
- Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST) Nucl. Acids Res. 2013;42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988;85:2444–8. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudhomme M, Attaiech L, Sanchez G, Martin B, Claverys JP. Antibiotic stress induces genetic transformability in the human pathogen Streptococcus pneumoniae. Science. 7. 2006;313:89–92. doi: 10.1126/science.1127912. [DOI] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34(2):374–8. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Vol. 1. CSHL Press, Science; 2001. [Google Scholar]
- Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, Deboy RT, Davidsen TM, Mora M, Scarselli M, YRos MI, Peterson JD, Hauser CR, Sundaram JP, Nelson WC, Madupu R, Brinkac LM, Dodson RJ, Rosovitz MJ, Sullivan SA, Daugherty SC, Haft DH, Selengut J, Gwinn ML, Zhou L, Zafar N. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc Natl Acad Sci U S A. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyres KL, van Tonder A, Lambertsen LM, Hakenbeck R, Parkhill J, Bentley SD, Brueggemann AB. Evidence of antimicrobial resistance-conferring genetic elements among pneumococci isolated prior to 1974. BMC Genomics. 24. 2013;14:500. doi: 10.1186/1471-2164-14-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.