Abstract
Root-knot nematodes (Meloidogyne spp.) are a significant problem in potato (Solanum tuberosum) production. There is no potato cultivar with Meloidogyne resistance, even though resistance genes have been identified in wild potato species and were introgressed into breeding lines. The objectives of this study were to generate stable transgenic potato lines in a cv. Russet Burbank background that carry an RNA interference (RNAi) transgene capable of silencing the 16D10 Meloidogyne effector gene, and test for resistance against some of the most important root-knot nematode species affecting potato, i.e., M. arenaria, M. chitwoodi, M. hapla, M. incognita, and M. javanica. At 35 days after inoculation (DAI), the number of egg masses per plant was significantly reduced by 65% to 97% (P < 0.05) in the RNAi line compared to wild type and empty vector controls. The largest reduction was observed in M. hapla, whereas the smallest reduction occurred in M. javanica. Likewise, the number of eggs per plant was significantly reduced by 66% to 87% in M. arenaria and M. hapla, respectively, compared to wild type and empty vector controls (P < 0.05). Plant-mediated RNAi silencing of the 16D10 effector gene resulted in significant resistance against all of the root-knot nematode species tested, whereas RMc1(blb), the only known Meloidogyne resistance gene in potato, did not have a broad resistance effect. Silencing of 16D10 did not interfere with the attraction of M. incognita second-stage juveniles to roots, nor did it reduce root invasion.
Keywords: Effector, host–parasitic relationship, Meloidogyne arenaria, M. chitwoodi, M. hapla, M. incognita, M. javanica, potato (Solanum tuberosum), resistance, RNA interference
Root-knot nematodes (Meloidogyne spp.) pose serious problems to potato (Solanum tuberosum) production in both temperate and tropical climates (Brodie et al., 1993; Sikora and Fernández, 2005). In addition to direct losses resulting from reduced tuber yields, Meloidogyne spp. can render potatoes unmarketable because of quality defects caused by gall formation on the tuber surface (Finley, 1981). Some species are quarantine pests and interfere with international trade. Even though M. chitwoodi is present in most major potato-producing areas, its detection in tubers can lead to the rejection of entire shipments (Ingham et al., 2000; Elling, 2013). To date, no commercially available potato cultivars with resistance to root-knot nematodes exist. Current nematode control strategies in potato heavily rely on synthetic nematicides. This practice is not only costly but also potentially harmful to the environment and faces increasing restrictions from regulatory agencies in many countries.
Knowledge about root-knot nematode resistance genes in potato is sparse (Gebhardt and Valkonen, 2001; Sanchez-Puerta and Masuelli, 2011). There is no known root-knot nematode resistance gene in the cultivated potato S. tuberosum, but several genes have been found in wild potato species (Brown et al., 1991a, 1995; Janssen et al., 1996; Brown et al., 2004; Williamson and Kumar, 2006). The best characterized root-knot nematode resistance gene in Solanum sect. Petota is RMc1(blb) from S. bulbocastanum, a gene that is effective against some races of M. chitwoodi (Brown et al., 2009). The resistance mechanism of RMc1(blb) is based on a hypersensitive response and involves calcium signaling (Davies et al., 2015). Recent studies suggest that M. chitwoodi resistance in different species of Solanum is based on the same gene, thereby limiting the diversity of available resistance (Brown et al., 2014).
Resistance based on plant-mediated RNA interference (RNAi) is emerging as a promising new disease control tactic. RNAi was first discovered in Caenorhabditis elegans but is now regarded as a widespread phenomenon in virtually all eukaryotes (Fire et al., 1998). RNAi is based on the ability of a cell to detect and degrade double-stranded RNA (dsRNA). Dicer, a ribosome III-like enzyme, catalyzes the cleavage of long dsRNA into small interfering RNA (siRNA) segments of about 21 nt in length. Double-stranded siRNA molecules are unwound and separated into single strands. While the passenger strand is degraded, the guide strand is loaded into a large complex, RISC (RNA-induced silencing complex). If the guide strand binds to a complementary region in the mRNA of a target gene, the respective gene is silenced (Hammond et al., 2001).
Endogenous nematode genes can be silenced by expressing dsRNA that is complementary to nematode genes in planta. Since root-knot nematodes are able to ingest molecules of up to 140 kDa (Zhang et al., 2012), there is an opportunity for oral delivery of dsRNA or siRNA into the nematode. During feeding, the nematodes take up plant cytoplasm containing dsRNA or siRNA that may then silence the respective target gene in the parasite (Lilley et al., 2012; Elling and Jones, 2014). For RNAi applications in an agricultural setting, it is essential to avoid off-target effects that could silence genes in the host plant or nontarget animals or humans. Nematode effector genes represent a very specific and therefore attractive target. Effector genes are expressed in the esophageal gland cells of plant–parasitic nematodes and form the basis for the molecular interactions between the parasite and its host (Mitchum et al., 2013). The 16D10 effector gene was initially cloned in M. incognita, but orthologs have since been identified in M. arenaria, M. chitwoodi, M. hapla, and M. javanica (Huang et al., 2006a, 2006b; Dinh et al., 2014a). The nucleotide sequence of 16D10 is 95% to 98% identical between M. arenaria, M. hapla, M. incognita, and M. javanica (Huang et al., 2006a). In contrast, the M. chitwoodi ortholog is only 70% identical to M. incognita (Dinh et al., 2014a). Previous studies have shown that plant-mediated silencing of the Meloidogyne effector gene 16D10 can lead to a dramatic increase in resistance (Huang et al., 2006a; Dinh et al., 2014a, 2014b). However, these reports were based either on Arabidopsis or made use of only a single species of Meloidogyne. The objective of this study was to select a high-performing stable transgenic 16D10 RNAi line with the genetic background of a commercial potato cultivar and then evaluate the resistance of this line against a broad range of Meloidogyne spp. found in temperate and tropical potato-producing regions.
Materials and Methods
Plant transformation:
Stable transgenic lines of potato ‘Russet Burbank’ were generated as described by Dinh et al. (2014a). Briefly, potato stem segments (about 1 cm in length) obtained from plants growing under axenic conditions were coincubated with Agrobacterium tumefaciens strain GV3101 carrying silencing vector pART27(16D10i-2) (Huang et al., 2006a) or empty vector pART27 as control. Incubation was done for 3 d in the dark at 19°C on CIM media (MS basal salts, 0.25 ppm folic acid, 0.05 ppm D-biotin, 2 ppm glycine, 0.5 ppm nicotinic acid, 0.5 ppm pyridoxine HCl, 0.4 ppm thiamine HCl, 0.01% myo-inositol, 3% d-sucrose, 1 ppm 6-benzylaminopurine, 2 ppm 1-naphthaleneacetic acid, 0.6% Daishin agar, pH 5.6). The stem segments were transferred every 2 wk for up to 3 mon to fresh 3C5ZR media (MS basal salts, 0.5 ppm nicotinic acid, 0.5 ppm pyridoxine HCl, 1 ppm thiamine HCl, 0.01% myo-inositol, 3% d-sucrose, 0.5 ppm indole-3-acetic acid, 3 ppm zeatin ribose, 0.5 g/liter timentin, 70 µg/ml kanamycin sulfate, 0.6% Daishin agar, pH 5.9), supplemented with 50 µg/ml kanamycin sulfate and 50 µg/ml timentin (Sheerman and Bevan, 1988; Brown et al., 1991b; Dinh et al., 2014a). Potato plantlets that regenerated on 3C5ZR media were maintained and propagated on propagation media (MS basal salts, 3% d-sucrose, 50 µg/ml kanamycin sulfate, 50 µg/ml timentin, 0.6% Daishin agar, pH 5.7). Wild type ‘Russet Burbank’ and advanced breeding line PA99N82-4 were maintained on propagation media without kanamycin sulfate and timentin. PA99N82-4 carries the RMc1(blb) resistance gene from S. bulbocastanum (Brown et al., 2009).
Southern and northern blotting:
DNA was extracted from potato leaves ground in liquid nitrogen with TPS buffer (100 mM Tris-HCl pH 8, 100 mM EDTA pH 8, 1 M KCl), precipitated with isopropanol, treated with RNase, and purified with chloroform/isopropanol precipitation. The DNA pellets were resuspended in 100 µl sterile water before being used for Southern blots. For each plant line, 15 µg DNA was digested with 50 U Xba I (New England Biolabs, Ipswich, MA) for 16 hr at 37°C, then separated on a 0.8% agarose gel at 70 V for 16 hr and transferred to a GeneScreen Plus nylon membrane (PerkinElmer, Boston, MA) in 10× saline sodium citrate (SSC) buffer (1.5 M NaCl, 0.15 M sodium citrate, pH 7). The membrane was UV cross-linked and hybridized for 16 hr at 42°C with a [α-32P] dATP labeled 16D10i-2 probe (Dinh et al., 2014b) in hybridization buffer (50% deionized formamide, 0.1 mg/ml salmon sperm DNA, 1% sodium dodecyl sulfate (SDS), 1 M NaCl, 10% dextran sulfate). Following hybridization, the membrane was washed twice with 2× SSC buffer for 5 min at 42 °C, three times with 2× SSC plus 1% SDS for 20 min at 65°C, and three times with 0.1× SSC plus 1% SDS for 20 min at 42°C. After washing, the membrane was exposed to X-ray film for 2 d at −70°C.
For northern blots, total RNA enriched for small RNA was extracted from 1 g potato leaves using the mirVana miRNA isolation kit (Ambion, Austin, TX) according to the manufacturer’s instructions. Twenty micrograms denatured total RNA of each potato line was separated on a 1% agarose gel at 120 V for 1 hr, transferred to a Nytran N nylon membrane (Sigma-Aldrich, St. Louis, MO) for 16 hr, UV cross-linked, and hybridized with [α-32P] dATP labeled 16D10 and U6 probes (Dinh et al., 2014b). After 16 hr of hybridization at 25°C in hybridization buffer (50% deionized formamide, 3× SSC, 0.1 mg/ml salmon sperm DNA, 1% SDS, 0.05 M phosphate buffer, 0.2% bovine serum albumin, 0.2% polyvinylpyrrolidone, 0.2% Ficoll), the membrane was washed three times for 20 min at 46°C with 2× SSC plus 0.2% SDS and exposed to X-ray film for 1 to 5 d at −70°C. Details about probes used for Southern and northern blots were described previously by Dinh et al. (2014a, 2014b).
Nematode culture and extraction:
Meloidogyne arenaria, M. chitwoodi isolate WAMC1, M. hapla, M. incognita isolate OP-50, and M. javanica were maintained on tomato (S. lycopersicum) ‘Rutgers’ grown in autoclaved sand under greenhouse conditions. Nematode eggs were extracted from roots 3 mon postinoculation. To release the eggs, roots were cut into 1- to 3-cm pieces and agitated in a 0.5% NaOCl solution following the method of Hussey and Barker (1973). The suspension was poured over nested sieves (850-, 75-, 25-µm pore size from top to bottom) and eggs collected on the 25-µm pore size sieve. Eggs were purified on a 70% sucrose gradient (Elling and Jones, 2014) before being used for infection assays. To obtain M. incognita infective second stage juveniles (J2) for attraction assays, eggs were hatched in modified Baermann pans (Dinh et al., 2014a).
Nematode infection assays:
Single nodes of each potato line were grown in propagation media for 30 d before being transferred to a sterilized sand/soil mixture (3 parts sand : 1 part soil). Plants were supplied with 20–10–20 N–P–K liquid fertilizer every 2 d for the duration of the experiment. All infection assays were set up in randomized complete block designs with host genotype as the main effect.
The initial infection assay screened for highly resistant potato lines. Five plants each of 22 transgenic 16D10i-2 lines and wild type ‘Russet Burbank’ were grown in separate 6-in clay pots and inoculated with 1,200 M. chitwoodi eggs per pot after the plants were acclimated to greenhouse conditions (16-hr light, 22°C, and 8-hr dark, 20°C) for 10 d. Roots were harvested at 55 days after inoculation (DAI), washed to remove soil particles, and processed for egg extraction as described above. The total number of eggs was estimated by counting three 100-µl aliquots of a 50-ml suspension per root system under a Stemi 2000C stereomicroscope (Zeiss, Jena, Germany). The reproductive efficiency R was calculated as the ratio between the initial inoculated number of eggs (Pi) and the final number of eggs at harvest (Pf).
For subsequent infection assays, the best performing transgenic line D21 was tested for resistance against M. arenaria, M. hapla, M. incognita, and M. javanica, with empty vector line E34, wild type PA99N82-4, and ‘Russet Burbank’ controls. Plantlets were transferred to Ray Leach SC10U Cone-tainers (Stuewe and Sons, Tangent, OR) and acclimated to a growth chamber for 10 d before being inoculated with nematode eggs. The infection assay was performed under 16-hr light, 24°C, and 8-hr dark, 22°C conditions in a growth chamber. For each line, 20 plants were inoculated with 2,000 eggs per nematode species. At 35 DAI, 10 root systems were harvested to count egg masses and at 55 DAI an additional 10 root systems were harvested to count the number of eggs, respectively, for each plant line and nematode combination. At 35 DAI, roots were washed, fresh weight per root system determined, and roots were stained for 15 min with 0.15 g/liter phloxine B (Fisher Scientific, Fair Lawn, NJ) to visualize egg masses. At 55 DAI, root fresh weight was determined and eggs extracted as described above. The infection assay was conducted twice for each plant line and nematode combination.
Nematode attraction assays:
The attraction of M. incognita J2 to 16D10i-2 potato line D21 and PA99N82-4, empty vector E34, and wild type ‘Russet Burbank’ controls was investigated following the methods of Wang et al. (2009). Briefly, potato plantlets grown for 15 d from single nodes in propagation media were placed on microscope glass slides. The root systems were coated with 1 ml of 23% pluronic gel PF-127 (Sigma-Aldrich) containing 300 M. incognita J2. For each potato line, 15 replicates with one plantlet each were prepared on separate glass slides and then covered with cover slips. After exposure in a moist chamber for 6 hr at 25°C, the numbers of J2 touching the terminal 10 mm of each root tip were counted. Following, samples were kept in a moist chamber for an additional 4 d before the roots were stained with acid fuchsin (Acros Organics, Morris Plains, NJ). For staining, roots were soaked in 0.5% NaOCl for 3 min, washed under running tap water for 10 min, and boiled in acid fuchsin solution (0.35% acid fuchsin, 25% acetic acid) for 5 min (Byrd et al., 1983). Stained roots were kept in distilled water at 4°C until parasitic J2 inside roots were counted. Nematode attraction and invasion were visualized with a SteREO Discovery.V8 stereomicroscope, AxioCam ICc1 digital camera, and ZEN imaging software (Zeiss) at 6 h after inoculation and 4 DAI and staining, respectively.
Data analysis:
Average numbers of eggs and egg masses and SE were calculated using Microsoft Excel. A Student’s t-test (LSD) at alpha level 0.05 (SAS 9.2 software; SAS Institute, Cary, NC) was used to estimate statistical significant differences between treatments.
Results
Screening of 16D10i-2 stable transgenic potato lines for resistance against M. chitwoodi:
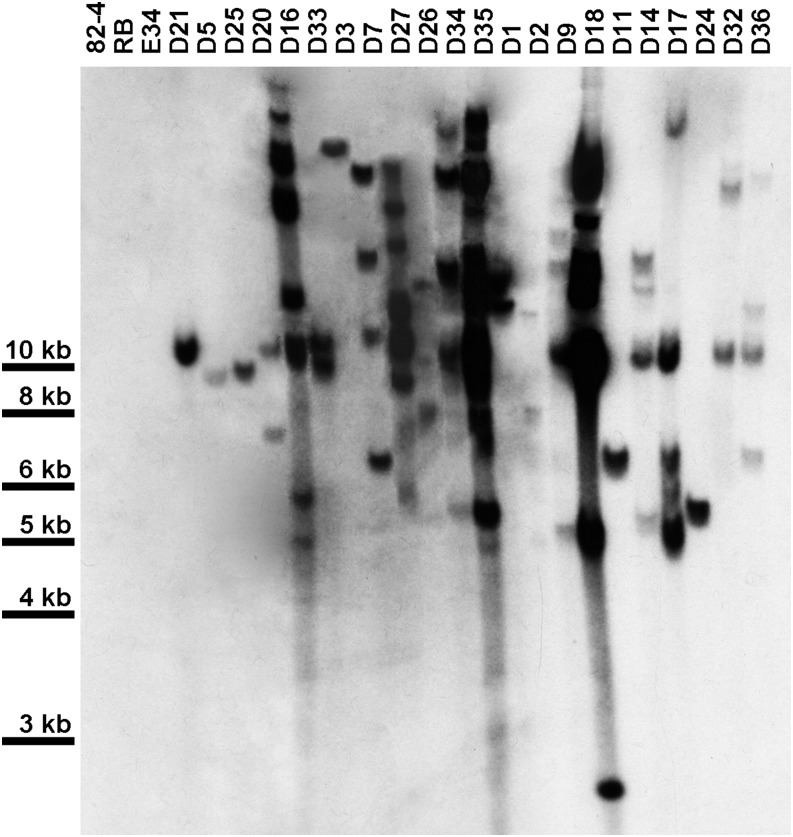
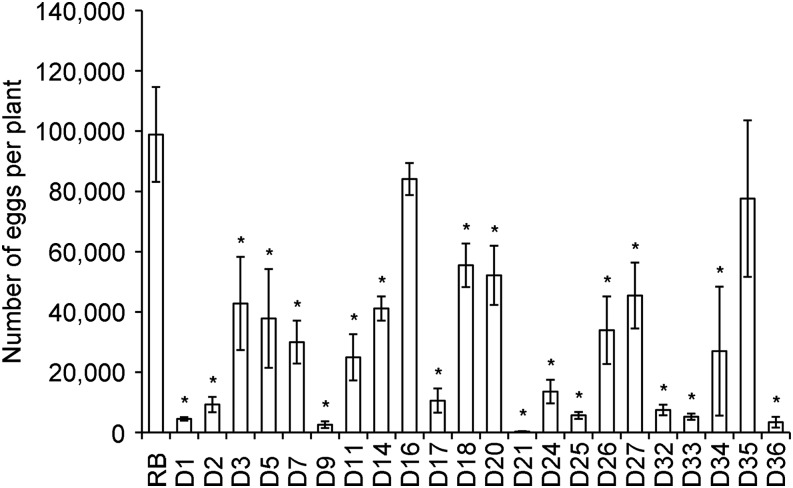
Silencing construct pART27(16D10i-2) was introduced into ‘Russet Burbank’ potato to generate stable transgenic RNAi lines. Twenty-two independent lines, none of which showed any overt phenotypical changes compared to wild type plants, were chosen for further analysis. Southern blots indicated that lines D3, D5, D11, D21, D24, and D25 carried single copies of the 16D10i-2 transgene. Lines D2, D16, and D20 had double insertions, and the remaining lines carried more than two copies of 16D10i-2 (Fig. 1). For the 22 transgenic RNAi lines and wild type control plants inoculated with M. chitwoodi eggs, the number of eggs at 55 DAI and Pf/Pi ratios revealed that lines D1, D9, D21, D25, D33, and D36 were most resistant against M. chitwoodi. The average number of eggs per plant at 55 DAI in the resistant lines ranged from 263 in D21 to 5,633 in D25, and had corresponding Pf/Pi ratios from 0.22 to 4.69. The reduction was significant (P < 0.05) compared to wild type plants, which yielded on average 98,882 eggs per plant and had a Pf/Pi ratio of 82.4 (Fig. 2; Table 1). D21 emerged as the most highly resistant line in this screening assay and was chosen for further analysis. Northern blots confirmed a strong expression of 16D10-specific small RNAs in line D21 (Fig. 3).
Fig. 1.
Southern blot showing copy numbers of the 16D10i-2 RNAi transgene in transformed potato lines and controls. For each plant line, 15 µg DNA was digested with 50 U Xba I and separated on a 0.8% agarose gel. 82-4, PA99N82-4 advanced breeding line; RB, wild type ‘Russet Burbank’; E34, empty vector control; D1 to D36, 16D10i-2 transgenic lines.
Fig. 2.
Number of Meloidogyne chitwoodi ‘WAMC1’ eggs per plant in wild type and transgenic RNAi potato lines at 55 days after inoculation. RB, wild type ‘Russet Burbank’. Each bar represents the mean of five plants per independent line with SE. Asterisk indicates statistically significant differences compared to RB controls using a Student’s t-test (P < 0.05).
Table 1.
Reproductive efficiency of Meloidogyne chitwoodi isolate ‘WAMC1’ on wild type and 16D10i-2 RNAi transgenic ‘Russet Burbank’ potato lines at 55 days after inoculation.
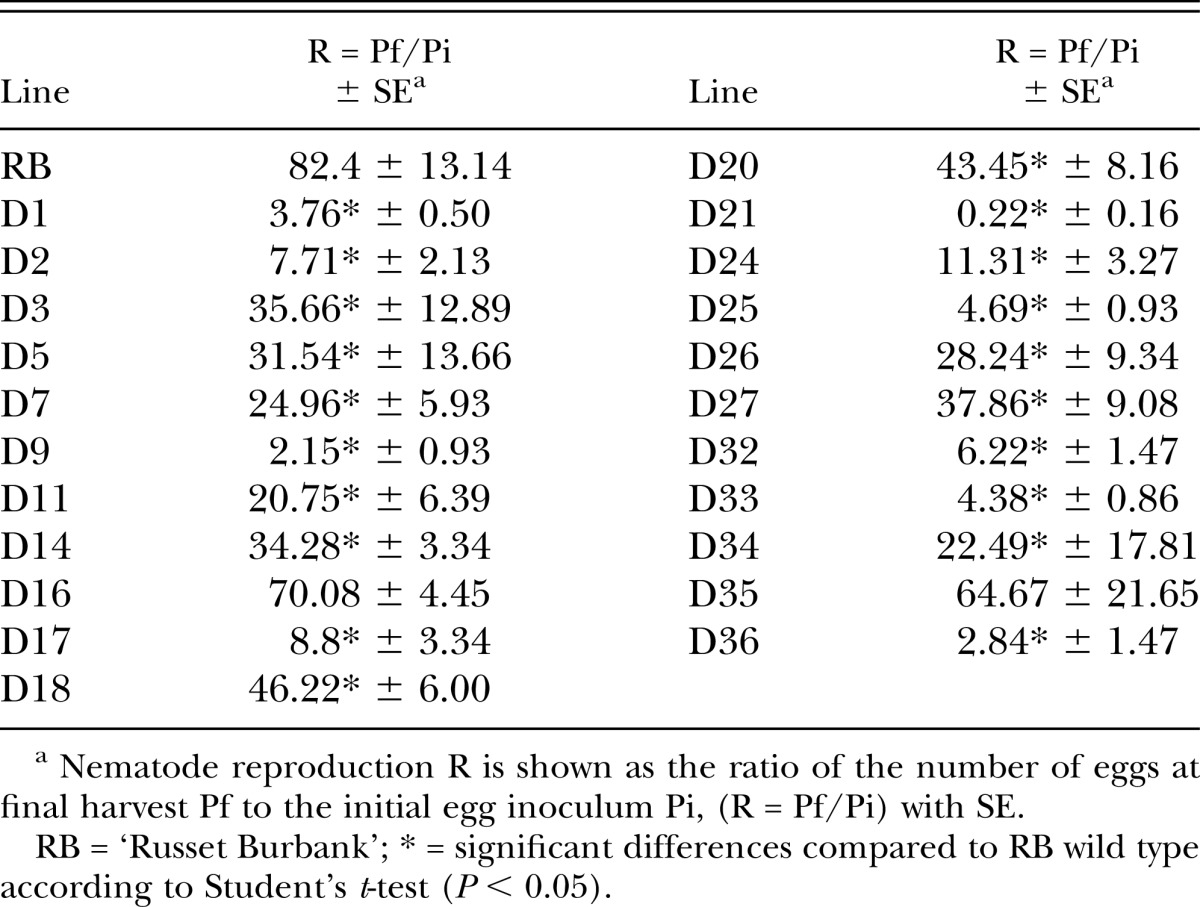
Fig. 3.
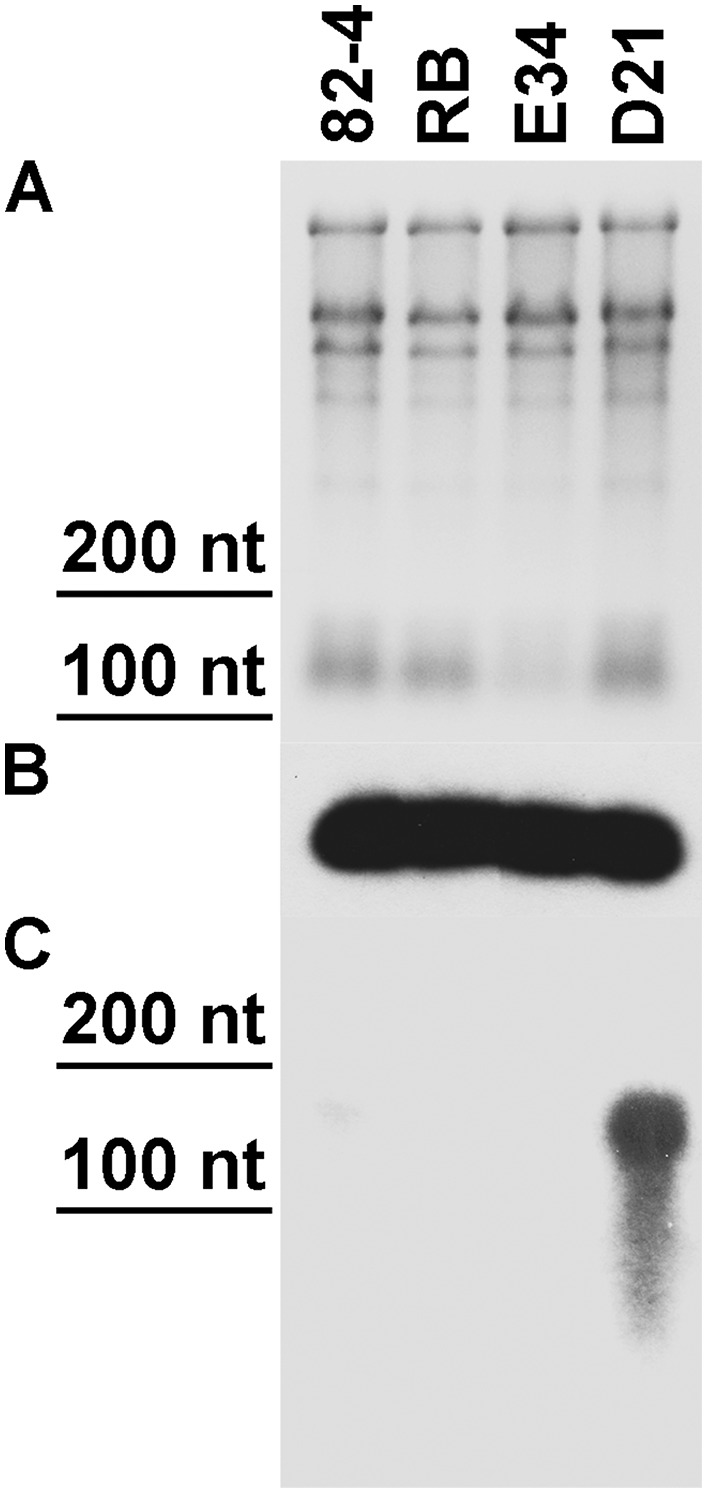
Northern blot for 16D10i-2 RNAi transgene. A. Total RNA loading control. B. U6 small nuclear RNA (snRNA) loading control. C. 16D10i-2-specific small RNA (smRNA) using probe 16D10 (Dinh et al., 2014a). 82-4, PA99N82-4 advanced breeding line; RB, wild type ‘Russet Burbank’; E34, empty vector control; D21, 16D10i-2 transgenic line D21.
Broad Meloidogyne resistance in stable transgenic 16D10i-2 potato line D21:
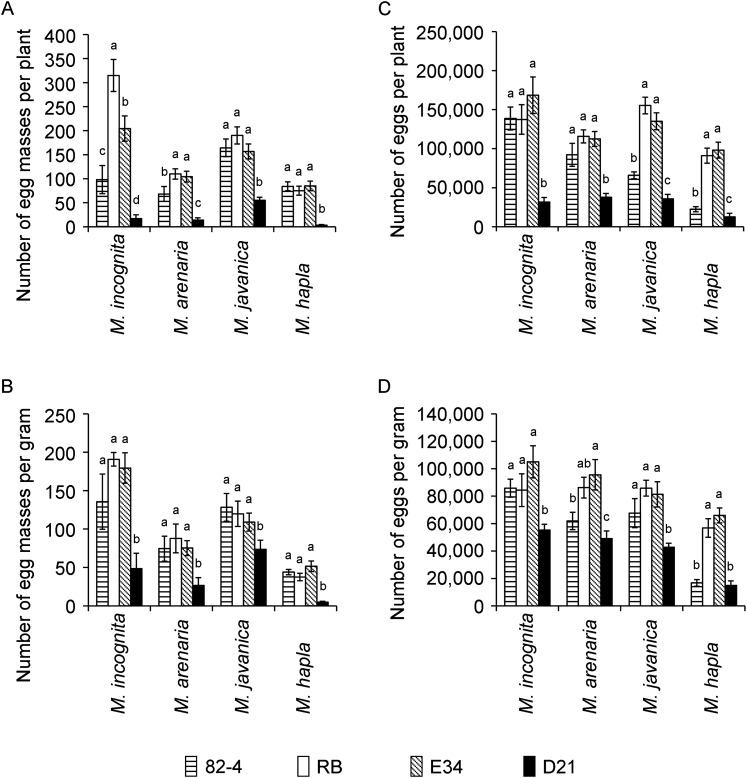
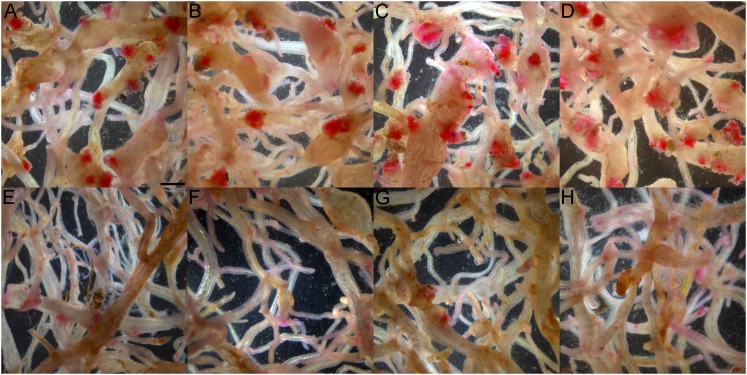
When challenged with M. arenaria, M. hapla, M. incognita, or M. javanica, line D21 showed strong resistance regardless of root-knot nematode species. At 35 DAI, the number of egg masses per plant was significantly reduced (P < 0.05) in D21 compared to wild type ‘Russet Burbank’, empty vector, and PA99N82-4 control plants by 65% to 97% (Figs. 4,5). The greatest reduction in the number of egg masses was achieved against M. hapla, whereas M. javanica showed the smallest reduction. Comparable levels of resistance were observed when the number of egg masses was analyzed on a per gram root basis (Fig. 4A,B). PA99N82-4 control plants, which carried the RMc1(blb) resistance gene, had significantly fewer egg masses for M. arenaria and M. incognita on a per plant basis, but not when the data were adjusted for root fresh weight (P < 0.05). At 55 DAI, the number of eggs per plant remained significantly lower in line D21 for all root-knot nematodes tested (P < 0.05). Compared to wild type and empty vector controls, egg production in line D21 was reduced from 66% in M. arenaria to 87% in M. hapla on a per plant basis (Fig. 4C). Similar levels of resistance were achieved when the number of eggs were expressed on a per gram root basis (Fig. 4D). Line PA99N82-4 significantly lowered the number of eggs on a per plant and per gram root basis for M. hapla, but not for the other root-knot nematodes tested.
Fig. 4.
Reproductive success of Meloidogyne spp. in potato lines with and without 16D10i-2 RNAi transgene. Each bar represents the mean of 10 plants per independent line and time point with SE. Letters indicate statistically significant differences using a Student’s t-test (P < 0.05). 82-4, PA99N82-4 advanced breeding line; RB, wild type ‘Russet Burbank’; E34, empty vector control; D21, 16D10i-2 transgenic line D21. A and B. Reproductive success of nematodes as number of egg masses per plant and per gram root fresh weight at 35 days after inoculation (DAI). C and D. Reproductive success of nematodes as number of eggs per plant and per gram root fresh weight at 55 DAI.
Fig. 5.
Egg masses of Meloidogyne spp. in roots of potato lines with and without 16D10i-2 RNAi transgene. Potato plants were inoculated with nematode eggs and developing egg masses from mature females were stained 35 days after inoculation with phloxine B. A and B. Egg masses of M. incognita and M. javanica, respectively, in roots of 82-4, PA99N82-4 advanced breeding line. C and D. Egg masses of M. incognita in roots of RB, wild type ‘Russet Burbank’ and E34, empty vector control plants, respectively. E–H. Egg masses of M. incognita, M. arenaria, M. javanica, and M. hapla, respectively, in D21, 16D10i-2 transgenic line D21. Scale bar = 1 mm.
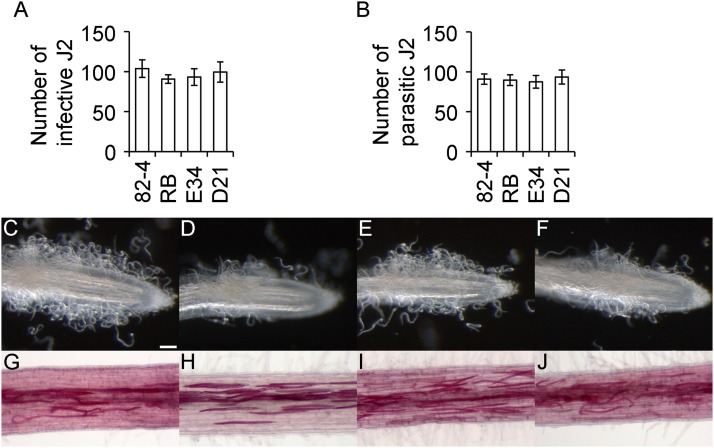
Plant-mediated 16D10i-2 resistance does not affect Meloidogyne attraction to host roots:
To test whether 16D10i-2 alters the nematodes’ ability to find and invade roots, an attraction assay was performed. There was no statistically significant difference between D21 and PA99N82-4, ‘Russet Burbank’, and empty vector controls when the number of M. incognita infective J2 or parasitic J2 were counted at 6 hr after inoculation or 4 DAI, respectively. No overt phenotypical differences were observed between the roots of either line (Fig. 6).
Fig. 6.
Attraction and invasion of Meloidogyne incognita J2 to potato roots with and without 16D10i-2 RNAi transgene. A. Number of infective J2 touching the 10-mm terminal end of potato roots with and without 16D10i-2 RNAi transgene at 6 hr after inoculation. B. Number of parasitic J2 invading potato roots with and without 16D10i-2 RNAi transgene at 4 days after inoculation (DAI). Each bar represents the mean of 15 root tips per independent line with SE. Student’s t-test did not indicate significant differences (P > 0.05). 82-4, PA99N82-4 advanced breeding line; RB, wild type ‘Russet Burbank’; E34, empty vector control; D21, 16D10i-2 transgenic line D21. C–F. Attraction of M. incognita infective J2 to representative root tips at 6 hr after inoculation in potato lines 82-4, RB, E34, and D21, respectively. G–J. Invasion of M. incognita parasitic J2 in representative roots at 4 DAI after acid fuchsin staining in potato lines 82-4, RB, E34, and D21, respectively. Scale bar = 100 µm.
Discussion
This study demonstrates that plant-mediated RNAi resistance can protect potato against important root-knot nematode pathogens. Stable transgenic lines of potato expressing the RNAi transgene 16D10i-2 showed significant resistance against M. arenaria, M. chitwoodi, M. hapla, M. incognita, and M. javanica. The 16D10 gene is widely conserved in Meloidogyne spp., but sequence divergence can reach about 30% between some species. Importantly, all orthologs found so far carry a highly conserved region of about 21 nt that is targeted by the 16D10i-2 silencing construct (Dinh et al., 2014a). Thus, statistically significant increases in resistance compared to wild type and empty vector controls could be achieved for all root-knot nematode species tested here. The number of RNAi transgene insertions into the host genome was not a reliable indicator of the resistance level. This finding confirms previous results and underscores the dominating role of position effects (Barrell et al., 2013; Dinh et al., 2014b). Furthermore, one or more copies of promoters used to control exogenous silencing genes targeted at nematodes can be affected by epigenetic silencing in the plant. This phenomenon has recently been demonstrated for the Cauliflower mosaic virus 35S promoter (Kyndt et al., 2013), a ubiquitous promoter that also was used to control 16D10i-2 in this study.
Breeding for resistance against pathogens is a high priority in integrated pest management strategies. Unfortunately, the number of known root-knot nematode resistance genes in potato is extremely limited (Gebhardt and Valkonen, 2001; Williamson and Kumar, 2006). Previous studies indicated that moving root-knot nematode resistance genes across species barriers within the Solanaceae can lead to resistance in the recipient plant (Goggin et al., 2006). However, this approach is limited by the fact that the respective resistance gene might not be effective against root-knot nematodes that are relevant in the recipient plant species (Brown et al., 1997). As an alternative, this study demonstrates that targeting effector genes that are highly conserved between a wide range of Meloidogyne spp. may be a promising control strategy and should lead to significant increases in nematode resistance.
Even though RNAi-based resistance is able to suppress the reproductive success of the nematode, this approach does not lend itself to shielding the plant from the primary infection. For a plant-mediated RNAi strategy to work, the nematode must take up plant cytoplasm containing small RNA or dsRNA complementary to an endogenous gene. Therefore, only feeding nematode life stages are vulnerable to acquiring the silencing trigger from the host. Nonfeeding life stages, such as the infective J2 are free to invade host roots, however. In M. chitwoodi, the 16D10 transcript is highly upregulated in infective J2, and in M. incognita antiserum confirmed the accumulation of 16D10 peptide in the subventral gland cells (Huang et al., 2006b; Dinh et al., 2014a). In this study, no difference was detected in the ability of M. incognita J2 to locate or invade roots of potato plants expressing or lacking the silencing gene 16D10i-2. This observation suggests that 16D10i-2-mediated resistance mechanisms occur after nematodes have invaded host roots and become sedentary. Importantly, the 16D10i-2 silencing effect can be transmitted epigenetically to nematode offspring and provide resistance even in non-RNAi plants (Dinh et al., 2014b).
In summary, this study demonstrates that RNAi-induced silencing of a nematode effector gene can lead to a dramatic reduction of a broad range of Meloidogyne spp. in stable transgenic lines of ‘Russet Burbank’ potato, thereby providing an attractive new tool for molecular breeding strategies against root-knot nematodes in this important crop.
Literature Cited
- Barrell PJ, Meiyalaghan S, Jacobs JME, Conner AJ. Applications of biotechnology and genomics in potato improvement. Plant Biotechnology Journal. 2013;11:907–920. doi: 10.1111/pbi.12099. [DOI] [PubMed] [Google Scholar]
- Brodie BB, Evans K, Franco J. 1993. Nematode parasites of potatoes. Pp. 87–132 in K. Evans, D. L. Trudgill, J. M. Webster, eds. Plant parasitic nematodes in temperate agriculture. Wallingford, UK: CAB International.
- Brown CR, Mojtahedi H, Santo GS. Resistance to Columbia root-knot nematode in Solanum spp. and in hybrids of S. hougasii with tetraploid cultivated potato. American Journal of Potato Research. 1991a;68:445–452. [Google Scholar]
- Brown CR, Mojtahedi H, Santo GS. Introgression of resistance to Columbia and northern root-knot nematodes from Solanum bulbocastanum into cultivated potato. Euphytica. 1995;83:71–78. [Google Scholar]
- Brown CR, Mojtahedi H, Bamberg J. Evaluation of Solanum fendleri as a source of resistance to Meloidogyne chitwoodi. American Journal of Potato Research. 2004;81:415–419. [Google Scholar]
- Brown CR, Mojtahedi H, Santo GS, Williamson VM. Effect of the Mi gene in tomato on reproductive factors of Meloidogyne chitwoodi and M. hapla. Journal of Nematology. 1997;29:416–419. [PMC free article] [PubMed] [Google Scholar]
- Brown CR, Mojtahedi H, Zhang LH, Riga E. Independent resistant reactions expressed in root and tuber of potato breeding lines with introgressed resistance to Meloidogyne chitwoodi. Phytopathology. 2009;99:1085–1089. doi: 10.1094/PHYTO-99-9-1085. [DOI] [PubMed] [Google Scholar]
- Brown CR, Yang CP, Kwiatkowski S, Adiwilaga KD. Insert copy number, chromosome-number, pollen stainability, and crossability of Agrobacterium-transformed diploid potato. American Potato Journal. 1991b;68:317–330. [Google Scholar]
- Brown CR, Zhang L, Mojtahedi H. Tracking the RMc1 gene for resistance to race 1 of Columbia root-knot nematode (Meloidogyne chitwoodi) in three Mexican wild potato species with different ploidies. American Journal of Potato Research. 2014;91:180–185. [Google Scholar]
- Byrd DW, Kirkpatrick T, Barker KR. An improved technique for clearing and staining plant tissue for detection of nematodes. Journal of Nematology. 1983;15:142–143. [PMC free article] [PubMed] [Google Scholar]
- Davies LJ, Brown CR, Elling AA. Calcium is involved in the RMc1(blb)-mediated hypersensitive response against Meloidogyne chitwoodi in potato. Plant Cell Reports. 2015;34:167–177. doi: 10.1007/s00299-014-1697-1. [DOI] [PubMed] [Google Scholar]
- Dinh PTY, Brown CR, Elling AA. RNA interference of effector gene Mc16D10L confers resistance against Meloidogyne chitwoodi in Arabidopsis and potato. Phytopathology. 2014a;104:1098–1106. doi: 10.1094/PHYTO-03-14-0063-R. [DOI] [PubMed] [Google Scholar]
- Dinh PTY, Zhang L, Brown CR, Elling AA. Plant-mediated RNA interference of effector gene Mc16D10L confers resistance against Meloidogyne chitwoodi in diverse genetic backgrounds of potato and reduces pathogenicity of nematode offspring. Nematology. 2014b;16:669–682. [Google Scholar]
- Elling AA. Major emerging problems with minor Meloidogyne species. Phytopathology. 2013;103:1092–1102. doi: 10.1094/PHYTO-01-13-0019-RVW. [DOI] [PubMed] [Google Scholar]
- Elling AA, Jones JT. 2014. Functional characterization of nematode effectors in plants. Pp. 113–124 in P. Birch, J. Jones, J. I. B. Bos, eds. Plant-pathogen interactions: Methods and protocols. Methods in Molecular Biology vol. 1127. Berlin, Germany: Springer Verlag.
- Finley AM. Histopathology of Meloidogyne chitwoodi (Golden et al.) on Russet Burbank potato. Journal of Nematology. 1981;13:486–491. [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Gebhardt C, Valkonen JPT. Organization of genes controlling disease resistance in the potato genome. Annual Review of Phytopathology. 2001;39:79–102. doi: 10.1146/annurev.phyto.39.1.79. [DOI] [PubMed] [Google Scholar]
- Goggin FL, Jia L, Shah G, Hebert S, Williamson VM, Ullman DE. Heterologous expression of the Mi-1.2 gene from tomato confers resistance against nematodes but not aphids in eggplant. Molecular Plant-Microbe Interactions. 2006;19:383–388. doi: 10.1094/MPMI-19-0383. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Caudy AA, Hannon GJ. Post-transcriptional gene silencing by double-stranded RNA. Nature Reviews Genetics. 2001;2:110–119. doi: 10.1038/35052556. [DOI] [PubMed] [Google Scholar]
- Huang G, Allen R, Davis EL, Baum TJ, Hussey RS. Engineering broad root-knot resistance in transgenic plants by RNAi silencing of a conserved and essential root-knot nematode parasitism gene. Proceedings of the National Academy of Sciences USA. 2006a;103:14302–14306. doi: 10.1073/pnas.0604698103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Dong R, Allen R, Davis EL, Baum TJ, Hussey RS. A root-knot nematode secretory peptide functions as a ligand for a plant transcription factor. Molecular Plant-Microbe Interactions. 2006b;19:463–470. doi: 10.1094/MPMI-19-0463. [DOI] [PubMed] [Google Scholar]
- Hussey RS, Barker KR. A comparison of methods of collecting inocula of Meloidogyne spp., including a new technique. Plant Disease Reporter. 1973;57:1025–1028. [Google Scholar]
- Ingham RE, Hamm PB, Williams RE, Swanson WH. Control of Meloidogyne chitwoodi in potato with fumigant and non-fumigant nematicides. Journal of Nematology (Suppl.) 2000;32:556–565. [PMC free article] [PubMed] [Google Scholar]
- Janssen GJW, von Norel A, Verkeerk-Bakker B, Janssen R. Resistance to Meloidogyne chitwoodi, M. fallax, and M. hapla in wild tuber-bearing Solanum spp. Euphytica. 1996;92:287–294. [Google Scholar]
- Kyndt T, Ji H, Vanholme B, Gheysen G. Transcriptional silencing of RNAi constructs against nematode genes in Arabidopsis. Nematology. 2013;15:519–528. [Google Scholar]
- Lilley CJ, Davies LJ, Urwin PE. RNA interference in plant parasitic nematodes: A summary of the current status. Parasitology. 2012;139:630–640. doi: 10.1017/S0031182011002071. [DOI] [PubMed] [Google Scholar]
- Mitchum MG, Hussey RS, Baum TJ, Wang X, Elling AA, Wubben M, Davis EL. Nematode effector proteins: An emerging paradigm of parasitism. New Phytologist. 2013;199:879–894. doi: 10.1111/nph.12323. [DOI] [PubMed] [Google Scholar]
- Sanchez-Puerta MV, Masuelli RW. Evolution of nematode-resistant Mi-1 gene homologs in three species of Solanum. Molecular Genetics and Genomics. 2011;285:207–218. doi: 10.1007/s00438-010-0596-6. [DOI] [PubMed] [Google Scholar]
- Sheerman S, Bevan MW. A rapid transformation method for Solanum tuberosum using binary Agrobacterium tumefaciens vectors. Plant Cell Reports. 1988;7:13–16. doi: 10.1007/BF00272967. [DOI] [PubMed] [Google Scholar]
- Sikora RA, Fernández J. 2005. Nematode parasites of vegetables. Pp. 319–392 in M. Luc, R. A. Sikora, J. Bridge, eds. Plant parasitic nematodes in subtropical and tropical agriculture. Wallingford, UK: CAB International.
- Wang C, Lower S, Williamson VM. Application of pluronic gel to the study of root-knot nematode behaviour. Nematology. 2009;11:453–464. [Google Scholar]
- Williamson VM, Kumar A. Nematode resistance in plants: The battle underground. Trends in Genetics. 2006;22:396–403. doi: 10.1016/j.tig.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Zhang F, Peng D, Ye X, Yu Z, Hu Z, Ruan L, Sun M. 2012. In vitro uptake of 140 kDa Bacillus thuringiensis nematicidal crystal proteins by the second stage juvenile of Meloidogyne hapla. PLoS ONE 7(6):e38534.