Abstract
Beneficial microbes hold great promise for the treatment of a wide range of immune and inflammatory disorders. In this issue of The EMBO Journal, Lightfoot and colleagues report how the food-grade bacterium Lactobacillus acidophilus helps the immune system to limit experimental colitis in mice through interaction between SIGNR3 and surface layer protein A (SlpA) in L. acidophilus. These results pave the way for future development of novel therapies for inflammatory diseases, including inflammatory bowel disease.
See also: YL Lightfoot et al (April 2015)
Research in the field of host–microbial interactions has historically focused on the diverse immune responses to pathogenic infection afflicting humans and, reciprocally, on how the pathogen subverts the myriad of immune mechanisms in place to protect the host. However, the vast majority of the host–microbial interactions are of a symbiotic nature. The human epithelia are inhabited by an estimated 100 trillion commensals (the so-called microbiota) that outnumber host cells by a factor greater than ten. The human microbiota includes mostly bacteria and profoundly influences health and disease. The last decade has shed light to the essential contribution of the gut microbiota to human health, which ranges from energy homeostasis, nutrient uptake, and vitamin synthesis functions, to immune homeostasis and protection against gut inflammation and infection (Dorrestein et al, 2014), and to more unexpected behavioral and even emotional functions. Importantly, the therapeutic potential of manipulating the gut microbiota using probiotics, genetically engineered commensals, and microbiota transplantation has burgeoned (Buffie & Pamer, 2013). In addition, the impact of the gut microbiota on immunity at body sites other than the intestine was documented with early reports showing that germ-free (GF) mice are impaired in oral tolerance to foodborne allergens (Buffie & Pamer, 2013). Therefore, the prominent role of the microbiota in immunity and inflammatory disease has awakened keen interest to investigate the cellular and molecular mechanisms, by which commensal gene products and their corresponding host sensors influence the immune system and tolerance mechanisms.
Microbial-associated molecular patterns (MAMPs) are conserved bacterial moieties in commensal bacteria that modulate multiple aspects of the immune system. Classical examples of MAMPs are lipopolysaccharide (LPS), peptidoglycan (PG), polysaccharide A (PSA), lipoteichoic acids (LTA), lipoproteins (LP), and microbial RNA and DNA (Thaiss et al, 2014). In the case of inflammatory bowel disease (IBD), PSA promotes peripheral tolerance through the induction of regulatory T cells, which efficiently mitigate the disease and contribute to microbial coexistence and colonic health (Buffie & Pamer, 2013). Important for the establishment of this type of peripheral tolerance is the presence of host “sensors,” such as pattern-recognition receptors (PRR), which recognize and transduce the signals and effects induced by MAMPs. Epithelium-associated, enteric macrophages, dendritic cells (DC), T cells and B cells, differentially express three major groups of PRRs: the cell surface Toll-like receptors, the intracellular nucleotide-binding oligomerization domain receptors, and the C-type lectin receptors (CLRs). Studies in mice with deficiencies in innate immune pathways dictated by these PRR families demonstrated that some of their members play critical roles in the interface with microbiota (Thaiss et al, 2014). The context of the interaction between a PRR and its MAMP ligand determines whether the activation and function of the immune cells are beneficial or harmful to the host (Dorrestein et al, 2014; Thaiss et al, 2014).
A major challenge to elucidate how microbiota can be employed as a treatment or prevention of numerous diseases, including IBD, is to characterize the composition and properties of individual MAMPs and to identify their respective PRRs expressed by intestinal innate immune cells. Yet, the strong potential for variation at the functional level for each MAMP according to bacterial specie, and the redundant role among different MAMPs, represents a difficult obstacle to fully delineate the signaling pathways that establish the beneficial effects provided by commensal bacteria.
In this issue of The EMBO Journal, Lightfoot et al (2015) provide an elegant demonstration of how, by applying genetic tools to generate a beneficial strain of Lactobacillus acidophilus, the impact of individual microbial MAMPs can be assessed on the host immune response and development of IBD. Previous work by this group established that L. acidophilus directly stimulates DCs to activate T cells, inducing an immunomodulatory effect heavily influenced by microbial surface-exposed (S-layer) molecules, such as proteins encoded by the slpA, slpB, and slpX genes (Lightfoot & Mohamadzadeh, 2013). From these studies, it was suggested that the SlpA protein interaction with the human CLR, DC-specific ICAM-3-grabbing non-integrin (DC-SIGN/CD209), significantly contributed to the potential regulation role for L. acidophilus (Lightfoot & Mohamadzadeh, 2013). In Lightfoot et al (2015), irrefutable evidence is presented for the in vivo role of the SlpA-induced protective immune regulation. Using the L. acidophilus strain expressing SlpA (SlpA+SlpB−SlpX−LTA−), the authors now demonstrate this protein (i) promotes intestinal regulation at steady-state conditions; (ii) maintains intestinal barrier function and prevents dysbiosis in pathogenic T-cell induced colitis; and (iii) binds specifically to one of the murine homologs of DC-SIGN, SIGNR3, to induce regulatory signals that ensure a strong protection against dextran sulfate sodium (DSS)-induced colitis (Fig1).
Figure 1.
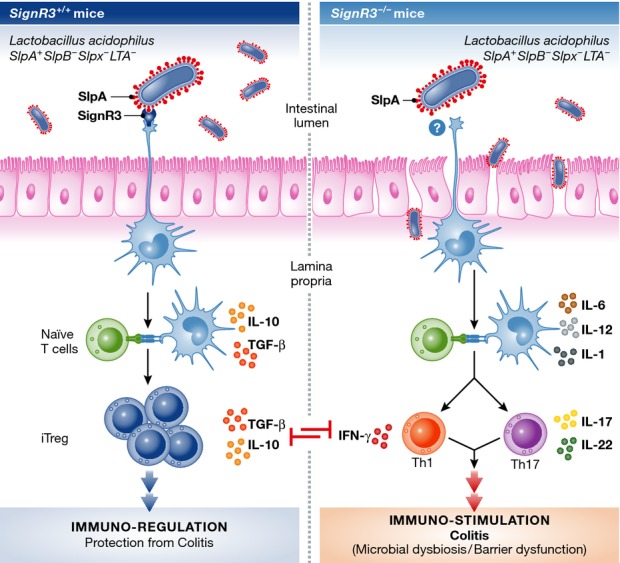
Immune modulation established by the SlpA-SIGNR3 interaction in murine experimental colitis
(Left) Epithelium-associated, enteric antigen-presenting cells (e.g. dendritic cells) recognize the Lactobacillus acidophilus strain expressing SlpA (SlpA+SlpB−SlpX−LTA−) through the C-type lectin receptor SIGNR3, which promotes intestinal regulation during experimental colitis. This includes the generation of an anti-inflammatory environment favoring the induction of regulatory T cells, which regulate other pro-inflammatory T-cell responses and protect against colitis, as reflected in the optimal maintenance of the intestinal barrier and prevention dysbiosis. (Right) Epithelium-associated, enteric antigen-presenting cells lacking SIGNR3 fail to recognize and bind to the L. acidophilus strain expressing SlpA. As a consequence, a pro-inflammatory environment becomes uncontrolled and characterized by the induction of T-cell responses that exacerbate experimental colitis and ultimately leads to permeability of the intestinal barrier and dysbiosis of the gut microbiota. The illustration was inspired and adapted from Lightfoot and Mohamadzadeh (2013).
An obvious implication for these observations is the therapeutic value for the SlpA-DC-SIGN interaction in humans. Human DC-SIGN is a type II transmembrane CLR that is characterized by one single extracellular CRD that recognizes fucosylated and mannosylated glycans, such as the blood-type Lewis antigens and high-mannose structures. This glycan specificity confers DC-SIGN the capacity to bind a wide range of ligands, both pathogen-derived (e.g. Mycobacterium tuberculosis) and self-glycoproteins (Garcia-Vallejo & van Kooyk, 2013). More recently, DC-SIGN was also shown to recognize sialylated immunoglobulins and mediate their therapeutic effects (Garcia-Vallejo & van Kooyk, 2013). As demonstrated in Lightfoot et al (2015), an alternative in vivo approach to study the role of DC-SIGN is to analyze the resistance or susceptibility of mice deficient for its closest mouse homologs (i.e. SIGNR1, SIGNR3) in experimental colitis. In the context of TLR-4 deficiency, mice lacking SIGNR1 display a reduced susceptibility to experimental colitis relative to severity of disease observed in wild-type animals, suggesting a pro-inflammatory role in response to LPS (Saunders et al, 2010). By contrast, mice lacking SIGNR3 exhibit susceptibility to DSS-induced colitis, as demonstrated by increased weight loss and aggravated colitis symptoms. Although no direct evidence was provided in this study, the authors hypothesized that SIGNR3 recognition of fungi present in the microbiota leads to protection against exacerbation of DSS-induced colitis, mainly through the control of TNFα production in the colon (Eriksson et al, 2013). Taking into account recent reports about the critical involvement of other CLRs to the development of murine colitis (Saba et al, 2009; Muller et al, 2010; Iliev et al, 2012; Hutter et al, 2014), we envisage that further understanding of CLR biology in the interface with microbiota will come from the continuing search for novel receptors and their MAMP ligands, studying signaling pathways and sequence motifs involved in CLR signaling cross talk and function upon interaction with different microbiota strains and their products, and elucidating the CLR impact in innate and adaptive immune responses in IBD, other autoimmune disorders, and chronic inflammatory disease.
The use of genetic tools to derive novel beneficial strains of commensal bacteria will be crucial to identify the microbial source of therapeutic molecules and to characterize the molecular mechanisms responsible for the modulation of the immune system. In this regard, the pioneering efforts to generate beneficial strains of L. acidophilus pave the way for the development of cost-effective therapeutic strategies to reprogram mucosal immunity in order to treat disease, without any obvious side effects upon consumption or permanent suppression of naturally occurring inflammation, as this food-grade bacterium does not permanently colonize the gut and can be used as needed (Mohamadzadeh & Owen, 2011; Lightfoot & Mohamadzadeh, 2013). All things consider, when one ponders about how the gut environment hosts the presence of foreign organisms that outnumber host cells by a factor greater than ten, the SlpA-DC-SIGN interaction reminds us that a symbiotic pact has been signed and shaped by coevolution of the host and microbiota.
Acknowledgments
We thank Claire Lastrucci for the adaptation of the figure. The authors received no specific funding for this work. The Neyrolles laboratory is supported by CNRS, the University of Toulouse, the European Union (H2020 Program and ERA-NET), Agence Nationale de la Recherche (ANR), Fondation pour la Recherche Médicale (FRM), Institut Mérieux, and the Bettencourt-Schueller Foundation.
References
- Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol. 2013;13:790–801. doi: 10.1038/nri3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrestein PC, Mazmanian SK, Knight R. Finding the missing links among metabolites, microbes, and the host. Immunity. 2014;40:824–832. doi: 10.1016/j.immuni.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson M, Johannssen T, von Smolinski D, Gruber AD, Seeberger PH, Lepenies B. The C-type lectin receptor SIGNR3 binds to fungi present in commensal microbiota and influences immune regulation in experimental colitis. Front Immunol. 2013;4:196. doi: 10.3389/fimmu.2013.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Vallejo JJ, van Kooyk Y. The physiological role of DC-SIGN: a tale of mice and men. Trends Immunol. 2013;34:482–486. doi: 10.1016/j.it.2013.03.001. [DOI] [PubMed] [Google Scholar]
- Hutter J, Eriksson M, Johannssen T, Klopfleisch R, von Smolinski D, Gruber AD, Seeberger PH, Lepenies B. Role of the C-type lectin receptors MCL and DCIR in experimental colitis. PLoS ONE. 2014;9:e103281. doi: 10.1371/journal.pone.0103281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, Brown J, Becker CA, Fleshner PR, Dubinsky M, Rotter JI, Wang HL, McGovern DP, Brown GD, Underhill DM. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. 2012;336:1314–1317. doi: 10.1126/science.1221789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightfoot YL, Mohamadzadeh M. Tailoring gut immune responses with lipoteichoic acid-deficient Lactobacillus acidophilus. Front Immunol. 2013;4:25. doi: 10.3389/fimmu.2013.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightfoot YL, Selle K, Yang T, Goh YJ, Sahay B, Zadeh M, Owen JL, Colliou N, Li E, Johannssen T, Lepenies B, Klaenhammer TR, Mohamadzadeh M. SIGNR3-dependent immune regulation by Lactobacillus acidophilus surface layer protein A in colitis. EMBO J. 2015;34:881–895. doi: 10.15252/embj.201490296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamadzadeh M, Owen JL. Reprogramming intestinal immunity is the answer to induced pathogenic inflammation. Immunotherapy. 2011;3:1415–1417. doi: 10.2217/imt.11.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S, Schaffer T, Flogerzi B, Seibold-Schmid B, Schnider J, Takahashi K, Darfeuille-Michaud A, Vazeille E, Schoepfer AM, Seibold F. Mannan-binding lectin deficiency results in unusual antibody production and excessive experimental colitis in response to mannose-expressing mild gut pathogens. Gut. 2010;59:1493–1500. doi: 10.1136/gut.2010.208348. [DOI] [PubMed] [Google Scholar]
- Saba K, Denda-Nagai K, Irimura T. A C-type lectin MGL1/CD301a plays an anti-inflammatory role in murine experimental colitis. Am J Pathol. 2009;174:144–152. doi: 10.2353/ajpath.2009.080235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders SP, Barlow JL, Walsh CM, Bellsoi A, Smith P, McKenzie AN, Fallon PG. C-type lectin SIGN-R1 has a role in experimental colitis and responsiveness to lipopolysaccharide. J Immunol. 2010;184:2627–2637. doi: 10.4049/jimmunol.0901970. [DOI] [PubMed] [Google Scholar]
- Thaiss CA, Levy M, Suez J, Elinav E. The interplay between the innate immune system and the microbiota. Curr Opin Immunol. 2014;26:41–48. doi: 10.1016/j.coi.2013.10.016. [DOI] [PubMed] [Google Scholar]


