Abstract
Autophagosome biogenesis requires two ubiquitin-like conjugation systems. One couples ubiquitin-like Atg8 to phosphatidylethanolamine, and the other couples ubiquitin-like Atg12 to Atg5. Atg12∽Atg5 then forms a heterodimer with Atg16. Membrane recruitment of the Atg12∽Atg5/Atg16 complex defines the Atg8 lipidation site. Lipidation requires a PI3P-containing precursor. How PI3P is sensed and used to coordinate the conjugation systems remained unclear. Here, we show that Atg21, a WD40 β-propeller, binds via PI3P to the preautophagosomal structure (PAS). Atg21 directly interacts with the coiled-coil domain of Atg16 and with Atg8. This latter interaction requires the conserved F5K6-motif in the N-terminal helical domain of Atg8, but not its AIM-binding site. Accordingly, the Atg8 AIM-binding site remains free to mediate interaction with its E2 enzyme Atg3. Atg21 thus defines PI3P-dependently the lipidation site by linking and organising the E3 ligase complex and Atg8 at the PAS.
Keywords: autophagy, PROPPIN, ubiquitin-like conjugation, WIPI
Introduction
Macroautophagy (hereafter autophagy) is an evolutionarily conserved process where cytosol and damaged or superfluous organelles are enclosed into double-membraned autophagosomes. After fusion of autophagosomes with the lysosome, their content is degraded and recycled (Farré et al, 2009; Backues & Klionsky, 2012; Hamasaki et al, 2013; Reggiori & Klionsky, 2013; Zavodszky et al, 2013). Autophagy affects ageing, cell death, removal of intracellular bacteria and antigen presentation. It is involved in cancer, Parkinson's, Huntington's and Crohn's disease (Choi et al, 2013). In S. cerevisiae, most of the autophagy (Atg) proteins colocalise at least transiently at the preautophagosomal structure (PAS), where the isolation membrane (phagophore) is assembled, elongated and sealed to an autophagosome. Autophagy subtypes selectively remove bulky cargos such as organelles or aggregates. The yeast cytoplasm-to-vacuole (Cvt) pathway, which targets proaminopeptidase I (pApeI) under vegetative conditions to the vacuole, is a prototype of selective autophagy.
Crucial PAS components are the regulatory serine/threonine protein kinase Atg1, the transmembrane protein Atg9, which is proposed to deliver membranes (Mari et al, 2010; Yamamoto et al, 2012) and a phosphatidylinositol 3-kinase complex containing Atg14. Autophagosome biogenesis further requires two ubiquitin-like conjugation systems. First, ubiquitin-like Atg8 is processed by the cysteine proteinase Atg4, then activated by the E1 enzyme Atg7, transferred to the E2 Atg3 and finally coupled to phosphatidylethanolamine (PE). Atg8-PE might act as tether during phagophore expansion (Nakatogawa et al, 2007; Xie et al, 2008; Weidberg et al, 2010, 2011; Nair et al, 2011). Secondly, ubiquitin-like Atg12 is activated by Atg7, transferred to the E2 Atg10 and covalently coupled to Atg5. The Atg12∽Atg5 conjugate then binds Atg16, which dimerises via its coiled-coil domain. Binding of dimeric Atg12∽Atg5/Atg16 to the PAS activates Atg3 analogous to an E3 ubiquitin-ligase complex (Hanada et al, 2007; Fujioka et al, 2010; Romanov et al, 2012; Noda et al, 2013; Sakoh-Nakatogawa et al, 2013). Atg16 is the key determinant of the lipidation site (Hanada et al, 2007; Fujita et al, 2008). Although no phosphoinositide-binding sites have been detected, yeast Atg5 and Atg16 require PI3P for PAS recruitment (Suzuki et al, 2007) and the PI3P-kinase inhibitor wortmannin affects mammalian Atg5 membrane association (Mizushima et al, 2001).
Autophagosome biogenesis requires PI3P at the PAS. Its presence on autophagosomal membranes is deciphered by members of the PROPPIN family. These β-propellers that bind phosphoinositides are a WD40 protein family conserved from yeast to human (Lemmon, 2008; Moravcevic et al, 2012). PROPPINs fold as seven-bladed β-propellers, with each blade containing four antiparallel β-strands (A to D, from in- to outside) (Fig 4A). We and others identified based on the crystal structure two lipid-binding sites at the circumference of the propeller; they act jointly and can bind both PI3P and PI(3,5)P(2) (Dove et al, 2004; Baskaran et al, 2012; Krick et al, 2012; Watanabe et al, 2012).
Figure 4.
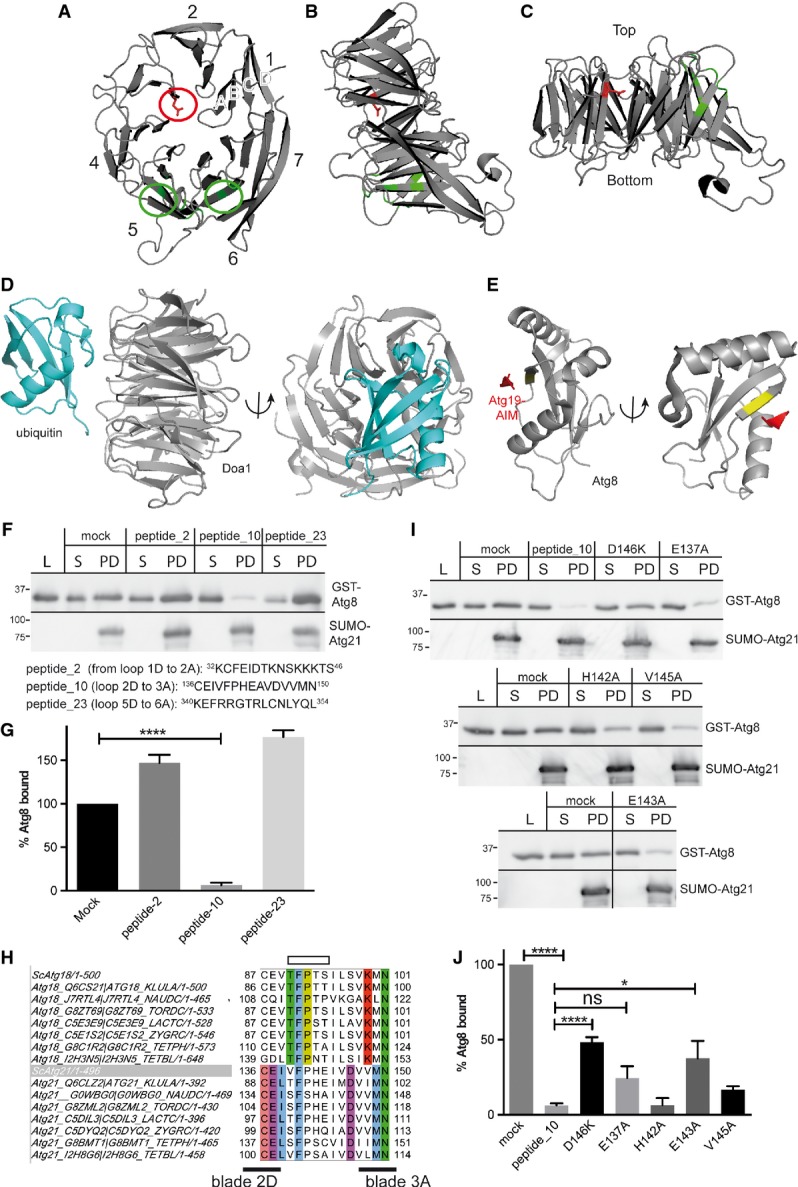
- A-C Cartoons based on the homologous K. lactis Hsv2 structure. The two phosphoinositide-binding sites are in green. They indicate the orientation upon membrane binding. In the top view (A), the position equivalent to D146 of Atg21 is marked red. (B) Side view, the membrane plane would be at the bottom. (C) The propeller top is typically smaller than the bottom.
- D Interaction of ubiquitin with the WD40 β-propeller of Doa1 (Pashkova et al, 2010).
- E Atg3 and the cargo receptor Atg19 contain a WxxL-like AIM-motif (Atg8 interacting motif), which mediates interaction with Atg8 (Noda et al, 2008). Orientation of Atg8 similar to ubiquitin in (D) indicates that the AIM of Atg19 (red) would be opposed to the Atg8 region interacting with the propeller. Part of the AIM-binding site of Atg8 is in yellow. Pictures were made with PyMOL (Schrodinger, 2010).
- F, G In competition experiments, GST-Atg8 was incubated with peptides and added to Sepharose-bound His6-SUMO-Atg21. After bead sedimentation, supernatant (S) and bound (PD) fractions were immunoblotted with GST (top) or His-tag antibodies (bottom). Samples of the GST-Atg8 peptide mixture (L) were included. (L) and (S) correspond to 8% of the (PD). Quantification of three experiments is shown in (G). Ratios of bound and lysates were normalised to the mock-treated sample. Peptide_10 corresponding to loop 2D to 3A affected binding.
- H-J Sequence alignment of loop 2D to 3A of Atg18 and Atg21 orthologues with Jalview (Waterhouse et al, 2009). Conserved residues are coloured; the binding site of Atg18 for Atg2 is indicated on top as open box. As in (F), effects of amino acid substitutions in peptide_10 on competition were analysed by immunoblots (I) and quantified (J). Molecular weight is in kDa.
The S. cerevisiae PROPPINs Atg18, Atg21 and Hsv2 are homologous, but differently affect autophagy subtypes. Atg18 binds PI3P-dependently to the PAS, where it mediates with Atg2 the retrieval of Atg9 from mature autophagosomes (Reggiori et al, 2004; Obara et al, 2008). Loops 2AB, 2CD and 2D3A in Atg18, localised opposite to the PIP-binding sites, are involved in Atg2 binding (Watanabe et al, 2012; Rieter et al, 2013). Interestingly, for Atg18 in P. pastoris, modulation of phosphoinositide binding by phosphorylation was reported (Tamura et al, 2013). Hsv2 partially affects piecemeal microautophagy of the nucleus, where non-essential parts of the nucleus are removed by autophagy (Krick et al, 2008a,b). Atg21 is essential for selective autophagy as the Cvt pathway or mitophagy; in its absence, unselective autophagy is retarded (Meiling-Wesse et al, 2004; Obara et al, 2008; Nair et al, 2010; Reggiori & Klionsky, 2013). We here dissect the molecular function of the yeast PROPPIN Atg21.
Results
Atg21 localises PI3P-dependently to the PAS
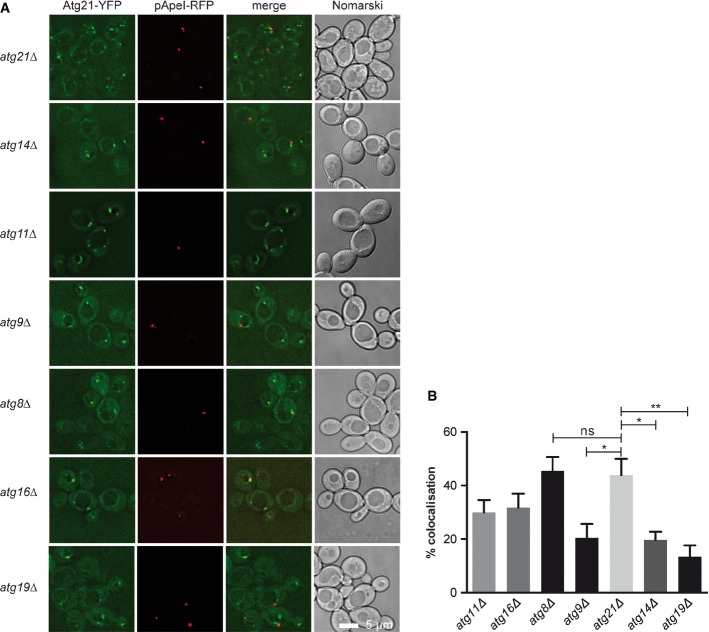
Atg21 was so far only detected at endosomes. Depletion of Atg21 from endosomes did not affect autophagy, leaving its role during autophagy elusive (Krick et al, 2008a; Obara et al, 2008). With a sensitive fluorescence microscope, we now detected in growing cells colocalisation of the PAS marker pApeI-RFP with one of the multiple Atg21-YFP punctae (Fig1A and B). Atg14 is crucial for the PAS PtdIns 3-kinase complex (Obara & Ohsumi, 2011). Diminished colocalisation in atg14Δ cells showed that Atg21 PAS binding depends on PI3P (Fig1A and B). Fluorescence microscopy (Fig 5A) further showed for the PI3P-binding-deficient cherry-Atg21FTTG only 0.0035 puncta/cell (1 punctum in 420 cells), while cherry-Atg21 formed 3.5 ± 0.21 puncta/cell. Atg21 PAS localisation was confirmed by reduced colocalisation in cells lacking the pApeI receptor (atg19Δ) and in cells with PAS assembly defects (atg9Δ) (Fig1A and B). Also, pApeI-YFP and cherry-Atg21 colocalised to 46% in atg21Δ and to 23% in atg14Δ cells. Cherry-Atg21 also colocalised in atg8Δ cells (Fig 6F and G) with the PAS markers Atg16-GFP (65%) and Atg5-YFP (78%).
Figure 1.
- Cells expressing Atg21-YFP and the PAS marker pApeI-RFP were grown to log-phase and visualised in fluorescence microscopy. Images were deconvoluted with SoftWoRx (Applied Precision).
- To dissect the requirement of other Atg proteins for Atg21 PAS localisation, the colocalisation rate as the percentage of perivacuolar pApeI-RFP PAS dots overlapping with Atg21-YFP was determined. Images from at least three cultures were taken, and for each image, puncta/cells were counted. Error bars are SEM. Statistical relevance was determined by unpaired two-tailed t-test. ns, not significant P > 0.05; *P < 0.05; **P < 0.01.
Figure 5.
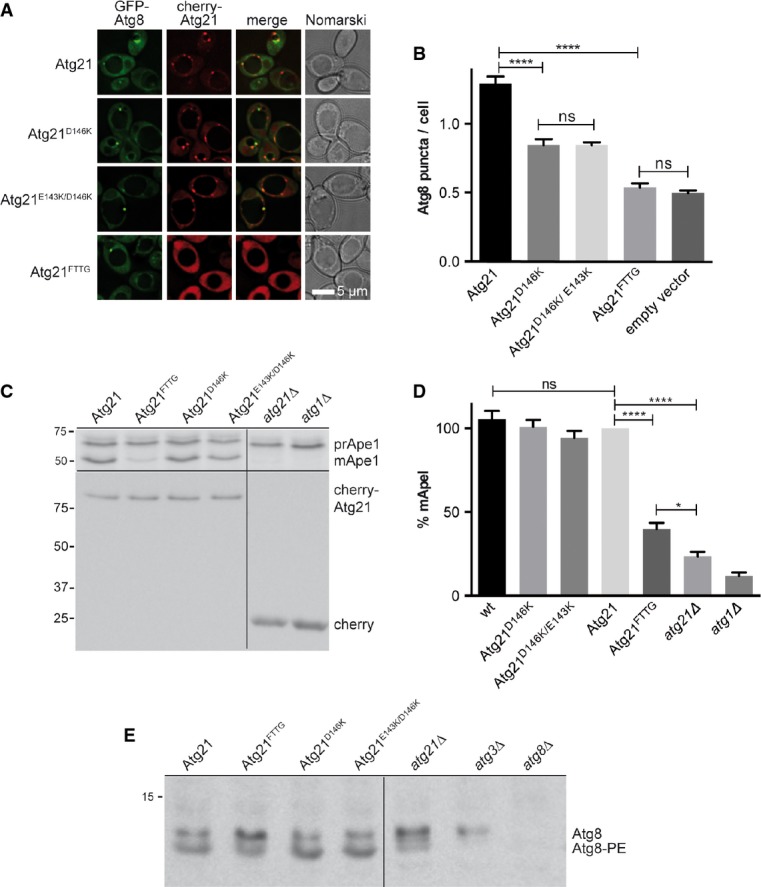
- A, B Fluorescence microscopy of growing (OD600 1.5) atg1Δ atg8Δ atg21Δ cells expressing cherry-Atg21 and GFP-Atg8. Cherry-Atg21D146K significantly reduced the number of Atg8 puncta/cell (B). The PI3P-binding-deficient Atg21FTTG reduced the Atg8 puncta/cell comparable to the absence of Atg21.
- C-E pApeI maturation (C, D) and Atg8 lipidation (E) are affected by Atg21FTTG. Percentages of mApeI were calculated and those for Atg21 set to 100%.
Atg21 recruits Atg8 to the PAS by direct interaction with the Atg8 FK-motif
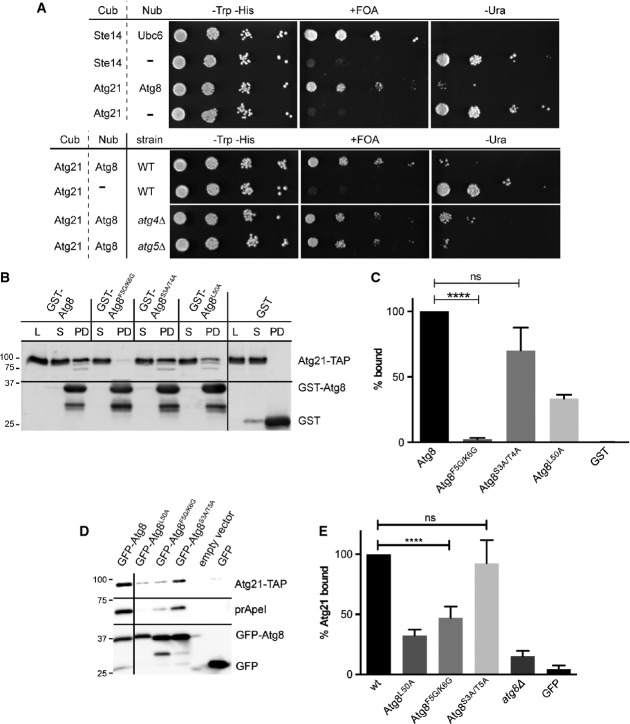
In growing atg21Δ cells, Atg8 PAS recruitment is impaired (Meiling-Wesse et al, 2004; Stromhaug et al, 2004), and we thus speculated that Atg21 might interact with Atg8. Since cell lysis often affects Atg21 membrane association, we used the split-ubiquitin system in intact cells. The split-ubiquitin system is similar to the 2-hybrid system but better suited for membrane-associated proteins. Briefly, the amino terminal half of ubiquitin (Nub) is fused to the bait and its carboxy-terminal domain (Cub) to the prey (Müller & Johnsson, 2008). Protein interaction restores ubiquitin leading to degradation of a proteolytically sensitive Ura3 variant at the carboxy-terminal ubiquitin domain. Interaction thus decreases growth without uracil and allows growth with 5-fluoroorotic acid (FOA). The split-ubiquitin system indeed indicated interaction of Atg21 with Atg8 comparable to the known interaction between Ste14 and Ubc6 (Fig2A). The interaction was unaffected in atg4Δ and atg5Δ cells, suggesting that Atg21 can interact with unprocessed and unlipidated Atg8 (Fig2A). To further confirm this interaction, E. coli-expressed GST-Atg8 on glutathione beads was incubated with extracts of S. cerevisiae cells expressing Atg21-TAP. Atg21-TAP effectively bound GST-Atg8, but not GST (Fig2B and C). Which Atg8 region mediates interaction with Atg21? Leucine 50 in the Atg8 carboxy-terminal ubiquitin-like domain is part of the AIM (Atg8 interacting motif)-binding site crucial for interaction with the WxxL-like motif (AIM) in proteins such as Atg1, Atg3 and Atg19 (Noda et al, 2010; Alemu et al, 2012; Nakatogawa et al, 2012; Shaid et al, 2013). The additional N-terminal helical domain (NHD) of Atg8 is flexible in solution (Schwarten et al, 2010), and a conserved FK-motif (residues 5 and 6) mediates further interactions (Krick et al, 2010). Interestingly, replacement of the FK-motif with glycine almost blocked Atg21-TAP binding from yeast extracts to GST-Atg8 beads (Fig2B and C). While mutagenesis of the non-conserved NHD residues 3 and 4 (ST) of Atg8 did not affect Atg21-TAP binding, GST-Atg8L50A showed reduced binding. We further coexpressed GFP-Atg8 and Atg21-TAP in atg1Δ pep4Δ cells and immunoprecipitated GFP-Atg8 with a GFP-binding protein on beads. GFP-Atg8F5G/K6G and GFP-Atg8L50A coprecipitated less Atg21-TAP than GFP-Atg8 (Fig2D and E).
Figure 2.
- In the split-ubiquitin system, the N-terminal ubiquitin domain (Nub) is fused to the bait and its C-terminal part (Cub) to the prey. Protein interaction restores ubiquitin and induces degradation of a destabilised Ura3 variant, which was attached to the Cub. Cells were diluted in tenfold steps and spotted on CM –Trp –His (growth control), CM –Trp –His +FOA (growth implies protein interaction) and CM –Trp –His –Ura (growth indicates no interaction). Images were taken after three days at 30°C. Ste14-Cub/Nub-Ubc6: positive control; Ste14-Cub/pRS314: negative control.
- GST-Atg8 variants on beads were incubated with extracts from atg21Δ pep4Δ cells expressing Atg21-TAP. Bound fraction (PD), lysate (L) and supernatant (S) corresponding to 4% of the PD were included. Immunoblots were probed with TAP (top) or GST antibodies (bottom). GST samples were diluted 1:10. Molecular weight markers are in kDa.
- Experiments from (B) were quantified as detailed in Materials and Methods. The ratio of bound Atg21-TAP and related lysate was calculated. The ratio for GST-Atg8 was set to 100%. SEM is from three experiments. ns, P > 0.05; ****P < 0.0001 (unpaired two-tailed t-test).
- Extracts of atg1Δ pep4Δ cells expressing GFP-Atg8 and Atg21-TAP were incubated with GFP-binding protein on beads. The co-immunoprecipitates were immunobloted with TAP (top), pApeI (middle) and GFP antibodies (lower row).
- Quantification of (D). The bound fraction for GFP-Atg8 was set to 100%. SEM is from four experiments. ns, P > 0.05; ****P < 0.0001 (unpaired two-tailed t-test).
Since crude extracts were used, the Atg21–Atg8 interaction could depend on further components. As shown in Fig3A and B, the Atg12∽Atg5/Atg16 complex, the Atg21 homologue Atg18 and the cargo receptor Atg19 do not mediate interaction of Atg21 with Atg8. To prove a direct interaction, we used recombinant E. coli proteins. We generated a SUMO-Atg21 fusion, which was isolated with Ni2+ beads via a His6-tag (Supplementary Fig S1). Its ability to pull down pApeI and cherry-Atg19 from yeast extracts showed its functionality (Supplementary Fig S2). Binding of E. coli-expressed GST-Atg8 to SUMO-Atg21 carrying beads (FigC and D) and of SUMO-Atg21 to GST-Atg8 on beads (Supplementary Fig S3) confirmed direct interaction. Interaction between the recombinant proteins strongly required the FK-motif, but not the ST-motif in the Atg8-NHD. In contrast to the experiments with yeast extracts, GST-Atg8L50A bound almost normally to SUMO-Atg21 (Fig3C and D). This suggests that direct interaction between Atg21 and Atg8 mainly depends on the FK-motif of the Atg8-NHD, but not the AIM-binding site of Atg8 via leucine 50. According to the binding of Atg19 and pApeI (Supplementary Fig S2), we expect that from yeast extracts, additional components stabilize the interaction of Atg21 with Atg8 via Atg8 leucine 50.
Figure 3.
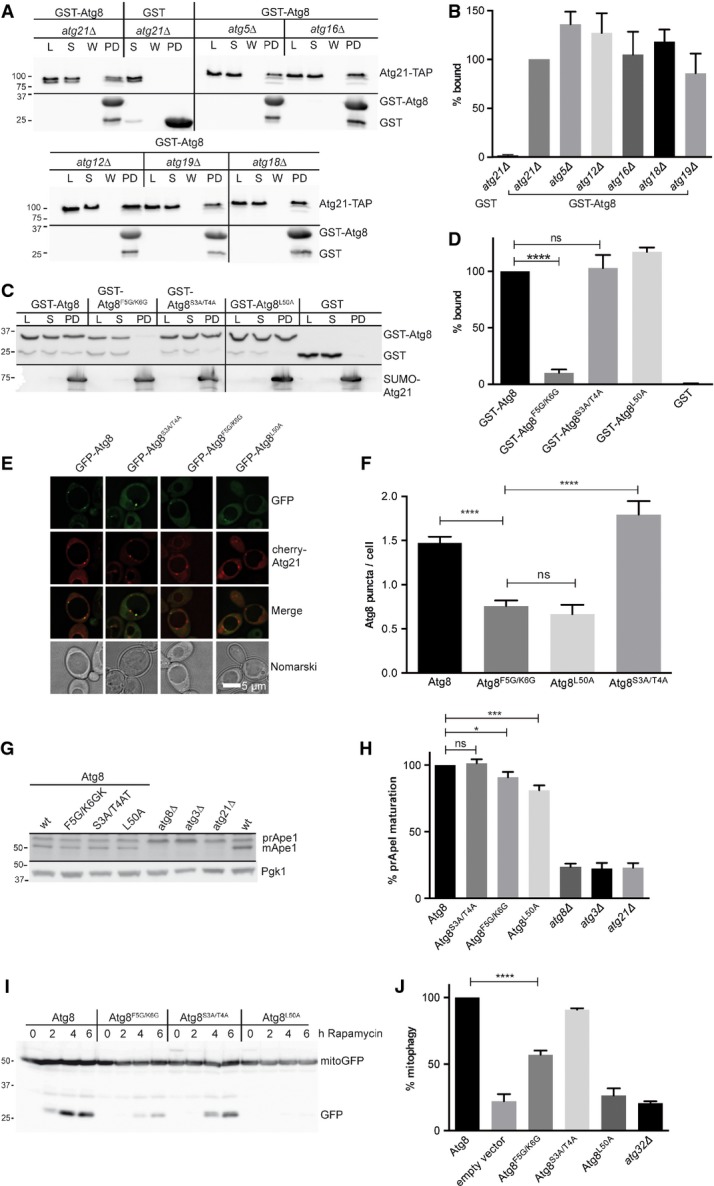
- A To test whether the interaction of Atg21 with Atg8 depends on other proteins, GST-Atg8 on beads was incubated with extracts from BY4741 deletion strains. PD: bound fraction; lysate (L) and supernatant (S). Immunoblots were with TAP (top) or GST antibodies (lower row).
- B Quantification of (A). Again, the ratio of bound Atg21-TAP and related lysate was normalised to wild-type.
- C Sepharose-bound SUMO-Atg21 was incubated with recombinant GST-Atg8 variants. Samples of the load (L), supernatant (S), wash (W) and bound fraction (PD) are immunobloted with GST (top) or His-tag antibodies (bottom).
- D Quantification of (C). Normalised ratios of bound and lysates are shown. ns, P > 0.05; ****P < 0.0001 (unpaired two-tailed t-test).
- E, F Logarithmic (OD600 1.5) atg8Δ atg1Δ cells expressing GFP-Atg8F5G/K6G showed significantly reduced PAS puncta/cell. Fluorescence microscopy (E) and quantification (F) are shown. ns, P > 0.05; ****P < 0.0001 (unpaired two-tailed t-test).
- G, H Expression of Atg8F5G/K6G affected pApeI maturation. Cell extracts are immunoblotted with anti-pApeI (top) and anti-Pgk1 (3-phosphoglycerate kinase). Percentage of mature ApeI was calculated and set to 100% for Atg8.
- I, J atg8Δ cells expressing Atg8 variants and the mitochondrial marker mito-GFP were grown on lactate medium, and mitophagy was induced with rapamycin. Samples were taken and after immunoblotting with GFP antibodies the level of free GFP after 6 h was quantified (J). The GFP level corresponds to the mitophagy rate and was set to 100% for Atg8. Molecular weights are in kDa.
Disturbed interaction of Atg8 with Atg21 should affect PAS recruitment of Atg8. Indeed, in growing cells, the number of GFP-Atg8F5G/K6G puncta per cell was significantly reduced (Fig3E and F). This reduction is comparable to that of GFP-Atg8L50A, which has defects in interactions with WxxL-containing Atg proteins (Noda et al, 2010). In line with the normal PAS recruitment of Atg21 in atg8Δ cells (Fig1B), the Atg8 variants had little effect on Atg21 localisation (Atg8: 3.45 ± 0.14 Atg21 puncta/cell; Atg8F5G/K6G: 3.51 ± 0.13; Atg8S3A/T4A: 3.69 ± 0.17; Atg8L50A: 3.9 ± 0.16).
Expression of Atg8F5G/K6G affected pApeI maturation slightly but significantly (Fig3G and H). Atg21 is required for selective autophagic subtypes. Indeed, the effect of Atg8F5G/K6G on selective rapamycin-induced mitophagy was severe (Fig3I and J).
To analyse the importance of PI3P binding by Atg21, we included PI3P-binding-defective Atg21-FTTG (Fig 5A and B). Cherry-Atg21FTTG almost completely failed to form puncta confirming that Atg21 PAS recruitment depends on PI3P. Compared to Atg21, cherry-Atg21FTTG significantly reduced the number of Atg8 puncta/cell similar to atg21Δ cells. This shows that the PI3P binding of Atg21 is required for normal formation of Atg8 puncta. Previous studies suggested an almost complete lack of Atg8 puncta in atg21Δ cells. The detection of residual Atg8 puncta in atg21Δ cells is due to our sensitive microscope. Additionally, we determined colocalisation of GFP-Atg8 with pApeI-RFP (Supplementary Fig S4) in cells expressing Atg21 or Atg21FTTG. The significant (P < 0.001) difference further confirms requirement of Atg21 PI3P binding for efficient PAS recruitment of Atg8. As expected, Atg21FTTG expression caused defects in the Cvt pathway and Atg8 lipidation (Fig 5C–E).
Loop 2D to 3A on the top side of Atg21 contributes to the interaction with Atg8
Yeast PROPPINs are non-velcro closed β-propellers. The four antiparallel β-sheets of each blade are denoted A to D, from the in- to the outside (Fig4A–C). The propeller top is typically smaller than the bottom and exposes loops connecting D with A strands of the next blade (Stirnimann et al, 2010; Krick et al, 2012). Interestingly, a functionally diverse set of WD40 β-propellers, among them components of ubiquitin E3 ligases, binds ubiquitin via their top region (Fig4D) (Pashkova et al, 2010). To identify the Atg21 domain involved in interaction with ubiquitin-like Atg8, we used synthetic 15 amino acid peptides corresponding to the Atg21 loops of the top. For longer loops, peptides overlapping for five amino acids were synthesised. SUMO-Atg21 on beads was incubated with GST-Atg8 and these peptides. The peptide CEIVFPHEIVDVVMN (Atg21 amino acids 136–150, connecting strand 2D with 3A), but no other peptide caused a significant reduction in GST-Atg8 binding to SUMO-Atg21 (Fig4F and G). Interestingly, in Atg18, the beginning of this loop has been identified to mediate interaction with Atg2 (Fig4H), (Rieter et al, 2013). Sequence alignment showed that D146 of this loop is conserved among Atg21, but not Atg18 orthologues from different yeasts (Fig4H). D146 is at the beginning of blade A at the entry of the central β-propeller channel, a site often involved in protein interactions (Stirnimann et al, 2010), (Fig4A–C). We next replaced D146 in peptides by alanine or lysine. The D146K peptide showed a significantly reduced competition in GST-Atg8 binding (Fig4I and J). In parallel, other residues of the peptide were consecutively replaced by alanine. Due to problems with synthesis, the P141A peptide was omitted. Only replacement of the non-conserved E143 caused some loss of competition (Fig4I and J). To verify the relevance of these residues, we coexpressed cherry-Atg21 variants and GFP-Atg8 in atg8Δ atg21Δ atg1Δ cells. Lack of the serine/threonine kinase Atg1 leads to Atg8 accumulation at the PAS. Cells expressing Atg21 had 1.29 Atg8 puncta/cell, Atg21D146K reduced this to 0.845 (P < 0.0001), Atg21FTTG yielded 0.54, and atg21Δ yielded 0.497. Additional replacement of E143 with K had no significant effect (Fig5A and B). We conclude that D146K of Atg21 is significantly involved in recruiting Atg8 to the PAS, but that additional interactions must exist. Maturation of pApeI (Fig5C and D) and Atg8 lipidation (Fig5E) was not obviously reduced by Atg21D146K. Most likely, the residual PAS recruitment of Atg8 is sufficient for these processes.
Atg21 interacts with Atg16
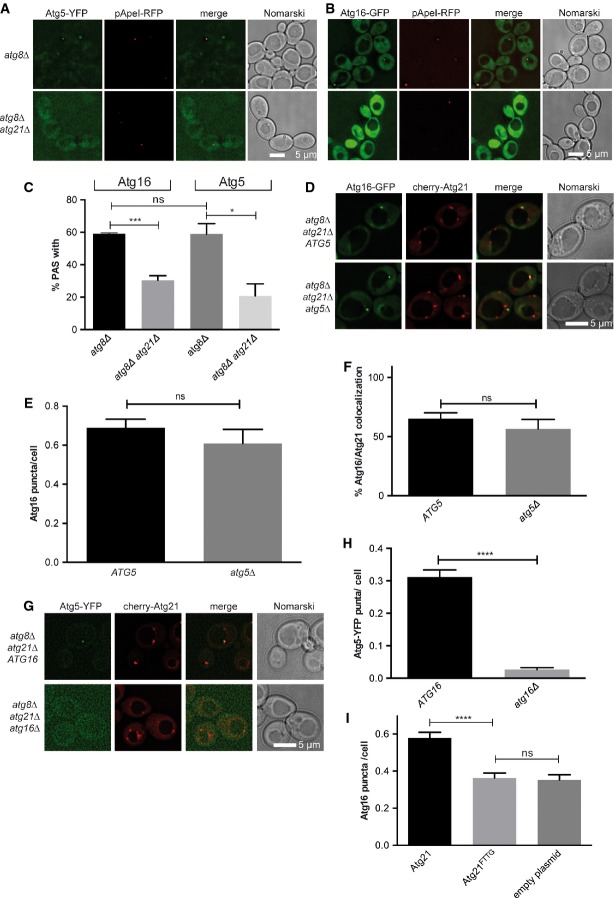
In atg8Δ cells, the otherwise faint PAS pool of Atg5 and Atg16 is enhanced (Stromhaug et al, 2004; Suzuki et al, 2007). We analysed Atg5-YFP and Atg16-GFP in atg8Δ and atg8Δ atg21Δ cells. As published, in growing atg21Δ atg8Δ cells, colocalisation of Atg5-YFP with pApeI-RFP was reduced (Fig6A and C), while Atg12∽Atg5 conjugation was unaffected (Stromhaug et al, 2004). We now found that also PAS recruitment of Atg16-GFP requires Atg21 (Fig6B and C). Does Atg21 recruit Atg16 or the Atg12-Atg5 conjugate? In growing atg8Δ atg21Δ cells of our background, the number of Atg16-GFP puncta/cell (Fig6D and E) and the colocalisation rate of Atg16-GFP with cherry-Atg21 (Fig6D and F) did not significantly change in the absence of Atg5. In contrast, ATG16 deletion decreased the number of Atg5 puncta/cell drastically (Fig6G and H). Since Atg12∽Atg5 conjugation occurs in atg16Δ cells (Mizushima et al, 1999), our data suggest that Atg21 specifically recruits Atg16. Compared to Atg21, the number of Atg16-GFP puncta/cell was significantly reduced with the PI3P-binding-deficient mutant Atg21FTTG (Fig6I). This shows that PAS recruitment of Atg16 depends on the ability of Atg21 to bind PI3P.
Figure 6.
- A-C Fluorescence microscopy of growing cells expressing the PAS marker pApeI-RFP and Atg5-YFP (A) or Atg16-GFP (B). The percentage of pApeI-RFP-positive cells showing a perivacuolar Atg5-YFP or Atg16-GFP puncta was determined (C).
- D-F Fluorescence microscopy of growing atg8Δ atg21Δ and atg5Δ atg8Δ atg21Δ cells expressing Atg16-GFP and cherry-Atg21. Quantification of (D) showed that lack of Atg5 did not affect the number of Atg16-GFP puncta/cell (E) and the colocalisation rate of Atg16-GFP and cherry-Atg21 (F).
- G, H Fluorescence microscopy of atg8Δ atg21Δ and atg8Δ atg21Δ atg16Δ cells expressing Atg5-YFP and cherry-Atg21 showed that lack of Atg16 significantly reduced the number of Atg5-YFP puncta/cell (H).
- I Quantification of fluorescence microscopy of atg8Δ atg16Δ atg2Δ cells expressing Atg16-GFP and Atg21-TAP variants. Expression of PI3P-binding-deficient Atg21FTTG-TAP reduced Atg16 puncta/cell similar to lack of Atg21. This shows the importance of PI3P binding of Atg21 in recruiting Atg16 to the PAS.
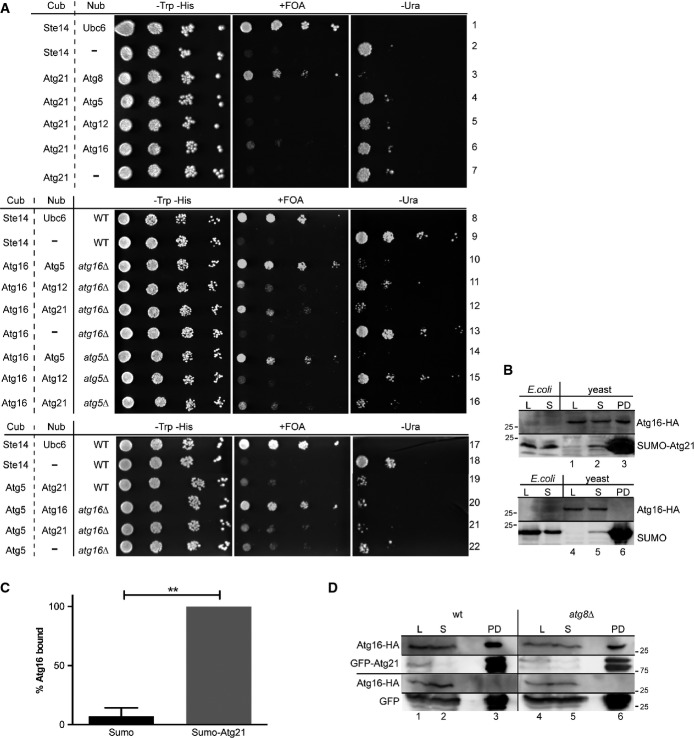
Atg16 interacts with the Atg12-Atg5 conjugate via Atg5, and this transfers Atg12 in an open conformation able to bind and activate Atg3 (Romanov et al, 2012; Noda et al, 2013). Accordingly, 2-hybrid analyses indicated interaction of Atg16 with Atg12 in wild-type, but not in atg5Δ cells (Mizushima et al, 1999). Indeed, with the split-ubiquitin system, we found interaction between Atg16 and Atg5 (Fig7A, row 10,14). The interaction between Atg16 and Atg12 was weaker (Fig7A, row 11) and absent in atg5Δ cells (row 15). Nub-Atg21 showed strong interaction with Atg16-Cub (row 12), which was only slightly reduced in atg5Δ cells (row 16), suggesting interaction of Atg21 with Atg16 without Atg12∽Atg5. Also, Atg21-Cub showed some interaction with Atg16 (row 6), but not with Atg5 (row 4) or Atg12 (row 5).
Figure 7.
- A In a split-ubiquitin assay, tenfold serial dilutions of cells expressing Cub (bait) and Nub (prey) constructs were spotted on plates with CM –Trp –His (growth control), CM –Trp –His +FOA (growth indicates interaction) and CM –Trp –His –Ura (growth indicates no interaction). Ste14-Cub/Nub-Ubc6: positive control, Ste14-Cub/pRS314: negative control.
- B, C Recombinant His6-SUMO-Atg21 and as control His6-SUMO were purified on Ni2+-NTA beads and incubated with extract of growing cells chromosomally expressing Atg16-HA. Samples from the E. coli lysates (E. coli L), cleared lysate (E. coli S), yeast lysates (yeast L), unbound fraction (yeast S) and the purified proteins (PD) were immunoblotted with antibodies against HA and His6. Quantification of bound Atg16 is shown in (C). Error bars are SEM. **P < 0.01(unpaired two-tailed t-test).
- D Lysates of growing cells expressing Atg16-HA from the chromosome and GFP-Atg21 or GFP from plasmids were immunoprecipitated with beads carrying GFP-binding protein. Samples from the cell lysate (L), the supernatant (S) and the purified proteins (PD) were analysed by immunoblotting using HA and GFP antibodies.
To biochemically confirm the interaction of Atg21 with Atg16, we incubated recombinant SUMO-Atg21 on beads with extracts from yeast cells chromosomally expressing Atg16-HA from its endogenous promoter. Atg16-HA bound to SUMO-Atg21, but not to SUMO (Fig7B, lane 3, 6, and C). We further used extracts from yeast cells expressing Atg16-HA and GFP-Atg21 for co-immunoprecipitations with GFP-binding proteins on beads. Again, Atg16-HA precipitated with GFP-Atg21, but not with GFP (Fig7D, lane 3). Precipitation of Atg16-HA with GFP-Atg21 was also possible in the absence of Atg8 (Fig7D, lane 6), consistent with PAS recruitment of cherry-Atg21 and Atg16-GFP in atg8Δ cells (Fig6D and F), (Suzuki et al, 2007). Together, our data support that Atg21 recruits PI3P-dependently Atg12∽Atg5/Atg16 via Atg16 to the PAS.
Atg21 directly interacts with D101 and E102 of the Atg16 coiled-coil domain
To confirm direct interaction of Atg21 with Atg16, we isolated recombinant proteins from E. coli. GST-Atg16, but not GST effectively bound SUMO-Atg21 (Fig8A and B). Atg16 has beside an Atg5 binding a coiled-coil domain, which forms a homodimer (Fujioka et al, 2010). Recombinant GST-Atg161-57 only containing the Atg5-binding domain did not bind SUMO-Atg21 (Supplementary Fig S5). However, GST-Atg1658–150 containing the coiled-coil domain bound similar to full-length Atg16. We confirmed these results by co-immunoprecipitation of yeast extracts expressing GFP-Atg21 and Atg16-HA truncated versions (Supplementary Fig S6). Fujioka et al (2010) identified based on the structure two residues (D101 and E102) in the coiled-coil domain whose replacement by alanine did not affect dimerisation, but autophagy and the Cvt pathway (Fig8C). We speculated that these residues might mediate interaction of Atg16 with Atg21. Using GFP-binding beads, GFP-Atg21 efficiently coprecipitated Atg16-HA, while Atg16D101A/E102A-HA showed almost no interaction (Fig8D and E). Analysis of single mutations uncovered that D101 is more important for interaction of Atg16 with Atg21 than E102 (Fig8D and E). Co-immunoprecipitation of Atg16D101A/E102A-GFP with Atg16D101A/E102A-HA (Fig8F) confirmed published data (Fujioka et al, 2010) that these residues are not required for Atg16 dimerisation. We then evaluated the relevance of these residues by fluorescence microscopy. As above, we used growing atg8Δ atg16Δ cells to better detect Atg16-GFP PAS puncta. Atg16D101A, Atg16E102A and Atg16D101A/E102A were predominantly cytosolic, and only Atg16E102A formed some faint dots (Fig8G and H). Atg16D101A/E102A did not complement the pApeI maturation defect (Fujioka et al, 2010) and the incomplete Atg8 lipidation in growing atg16Δ cells (Fig8I). Together, our data show that D101 and E102 of Atg16 mediate the direct interaction with Atg21. Remarkably, crystallisation of Atg16 was only possible after replacing D101 and E102 with alanine (Fujioka et al, 2010). Atg21 PAS recruitment was unaffected in atg8Δ cells, but reduced in atg16Δ cells (Fig1A and B), suggesting that binding of Atg21 to Atg16 enhances its association with the PAS. To detect whether the interaction of Atg21 with Atg8 depends on the Atg12∽Atg5/Atg16 complex, we probed binding of Atg21-TAP from extracts of atg16Δ, atg5Δ and atg12Δ cells to GST-Atg8 beads (Fig3A and B). Efficient binding suggests that Atg21 can interact with Atg8 without Atg12∽Atg5/Atg16.
Figure 8.
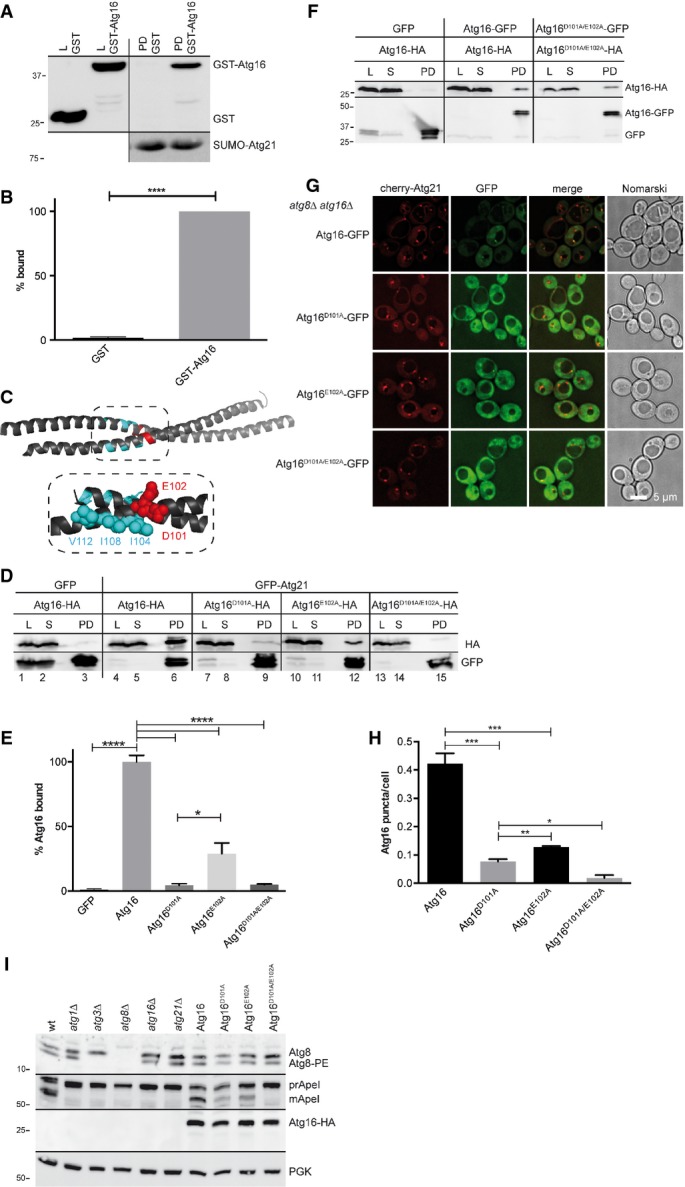
- A, B Recombinant His6-SUMO-Atg21 on beads was incubated with purified recombinant GST or GST-Atg16. Samples from purified GST-Atg16 and GST (L) and bound proteins (PD) were immunoblotted with antibodies against GST and His6. Quantification is shown in (B).
- C Ribbon diagram of the dimeric Atg16 coiled-coil domain made with PyMOL. D101 and E102 are not involved in dimerisation of Atg16, but for unknown reasons for normal autophagy (Fujioka et al, 2010). V112, I108 and I104 (cyan) mediate after Atg8 lipidation the coat formation of Atg12˜Atg5/Atg16 (Kaufmann et al, 2014).
- D, E To test the relevance of D101 and E102 for the interaction of Atg16 with Atg21, extracts of growing atg16Δ cells expressing GFP-Atg21 or GFP as control and Atg16-HA versions were immunoprecipitated with GFP-binding protein on beads. Samples of the lysate (L), supernatant (S) and bound proteins (PD) were blotted using HA and GFP antibodies. Replacement of E102 and especially of D101 with alanine impaired the interaction of Atg16 with Atg21 (E).
- F Lysates of growing atg16Δ cells expressing Atg16-GFP variants were mixed with lysate of growing atg16Δ cells expressing Atg16-HA variants. Co-immunoprecipitation using GFP-binding protein on beads in addition to published data (Fujioka et al, 2010) confirmed that D101 and E102 are not required for Atg16 self-interaction. Lysate (L), supernatant (S) and bound proteins (PD) were analysed by immunoblotting using HA and GFP antibodies.
- G, H Growing atg8Δ atg16Δ cells expressing cherry-Atg21 and Atg16-GFP variants were analysed by fluorescence microscopy (G) and the number of Atg16 puncta/cell determined (H).
- I Immunoblot analysis of the Cvt pathway and Atg8 lipidation showed in cells expressing Atg16D101A/E102A defects comparable to atg21Δ cells.
Discussion
Atg12∽Atg5/Atg16 has two distinct functions at the phagophore. First, a small amount catalyses Atg8 lipidation; then, larger amounts assemble with Atg8-PE into a scaffold at the outside of the phagophore (Kaufmann et al, 2014). This is consistent with the proposed role of Atg16 in determining the Atg8 lipidation site (Hanada et al, 2007; Fujita et al, 2008). Our data now support the following refined model of autophagosome biogenesis (Fig9). Atg21 binds via PI3P to the phagophore and translates the presence of this phosphoinositide into assembly of the Atg8 lipidation system including its E3 ligase complex. To this end, Atg21 recruits Atg12∽Atg5/Atg16 via direct interaction with D101 and E102 of the Atg16 coiled-coil domain (Figs7 and 8). These residues are on the side of the coiled-coil domain, which is not involved in Atg16 dimerisation.
Figure 9.
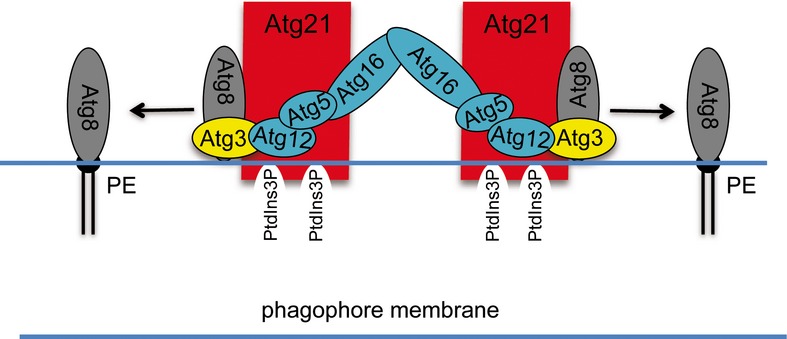
Efficient Atg8 lipidation requires spatial and temporal coordination of Atg8 and its lipidation machinery
In our proposed model, Atg21 binds via PI3P to the phagophore and interacts directly with D101 and E102 of the coiled-coil domain of Atg16. Atg21 thus recruits the dimeric E3-like Atg12∽Atg5/Atg16 complex to the membrane and correctly positions Atg12 to allow activation of the E2-like Atg3. Atg21 additionally binds with the help of D146 on the top site of its β-propeller to Atg8 and thus positions the Atg8 carboxyterminus close to phosphatidylethanolamine (PE). Atg8 for its part interacts with the F5K6-motif of its NHD-domain with Atg21. Additionally, the Atg8 AIM-binding site interacts with the AIM-motif of Atg3. Together, Atg21 is the key component, which determines PI3P-dependently the lipidation site of Atg8.
Mammalian PROPPINs are termed ‘WIPI’ (WD repeat proteins interacting with phosphoinositides). Like their yeast orthologues, WIPIs act during autophagosome biogenesis. While our manuscript was under revision, an elegant study unravelled that WIPI2b via the bottom side of its propeller directly interacts with Atg16L1 and thus defines PI3P-dependently the lipidation site of the Atg8 orthologue LC3 (Dooley et al, 2014). Compared to yeast Atg16, mammalian Atg16L1 is extended at the carboxyterminus. Well in agreement with our results, WIPI2b interacts with two adjacent acidic amino acids of Atg16L1, located after its self-interacting domain. The interaction of Atg16L1 with the bottom side of the WIPI2b propeller strengthens our model, which proposes binding of Atg8 to the top side of the Atg21 propeller.
We further found that the Atg21 PAS frequency is partially reduced without Atg16 (Fig1A and B). This synergistic effect on PAS binding is most likely caused by the bridging of two Atg21 molecules by the Atg12∽Atg5/Atg16 heterodimer and probably also by direct membrane association with Atg12∽Atg5/Atg16 (Romanov et al, 2012). D146 on the top side of the Atg21 propeller contributes to Atg8 binding (Figs4 and 5). This recruits Atg8 to the PAS and positions its carboxyterminus near the PE-containing membrane. For the direct binding of Atg8 to Atg21, its NHD-domain, especially the conserved F5K6-motif, but not its AIM-binding region was required (Fig3). In analogy to the orientation of ubiquitin bound to β-propellers of the F box family of SCF ubiquitin E3 ligase adaptors (Fig4D) (Pashkova et al, 2010), we propose that the AIM-binding domain of Atg8 is exposed away from the propeller (Fig4E). This leaves the AIM-binding domain unoccupied for binding to Atg3, which has a WxxL motif within its handle domain (Yamaguchi et al, 2010; Metlagel et al, 2013). In this scenario, Atg21 organises both the ubiquitin-like Atg8 and the E3-like Atg12∽Atg5/Atg16 complex, allowing coordinated interaction of Atg12 with Atg3 (Noda et al, 2013; Sakoh-Nakatogawa et al, 2013), which leads to Atg3 activation and efficient Atg8 lipidation. We found that interaction of Atg21 with Atg16 occurs in the absence of Atg8 and also interaction of Atg21 with Atg8 does not require Atg16. Atg21 is thus crucial for determining PI3P-dependently (Figs5 and 6) the Atg8 lipidation site and for spatial arrangement of the multi-subunit E3 ligase complex.
After lipidation, Atg8-PE is bound to the Atg12∽Atg5/Atg16 complex via a non-canonical AIM of Atg12 (Kaufmann et al, 2014). Together, they form a rigid scaffold restricted to the convex side of the phagophore. Formation of this scaffold requires interaction with Atg12∽Atg5/Atg16 heterodimers via I104, I108 and V112 within the coiled-coil domain of Atg16 (Fig7D). Remarkably, these three hydrophobic residues are in close proximity to D101 and E102, which we identified to be required for interaction of Atg16 with Atg21 (Figs7 and 8). This suggests that interaction of Atg16 with Atg21 and scaffold formation via these two sites of the Atg16 coiled-coil domain might be mutually exclusive. Well in agreement, cells expressing Atg16I104D/I108D/V112D showed normal Atg8 lipidation, but a severe autophagy defect (Kaufmann et al, 2014). At the end of autophagosome biogenesis, the Atg12∽Atg5/Atg16 scaffold disassembles, and at the outside of autophagosomes, Atg8 is released and PI3P hydrolysed. Inside autophagosomes, Atg8-PE and PI3P stay intact and reach the vacuole. Since we could not detect cherry-Atg21 in autophagic bodies of pep4Δ cells, Atg21 similar to Atg12∽Atg5/Atg16 may be mainly on the outside of phagophores.
As outlined, Atg8 is part of several spatially and temporally distinct protein complexes. The FK-motif of Atg8 has also been implicated in interaction with the Cdc48-adaptor Shp1 (Krick et al, 2010). Since Atg21 also interacts with unlipidated Atg8 (Fig2A), while Shp1 preferentially interacted with lipidated Atg8, we expect that Shp1 acts later than Atg21.
Atg8 lipidation and pApeI maturation defects in growing atg21Δ cells show that Atg21-dependent PAS recruitment of Atg8 and of Atg12∽Atg5/Atg16 is especially important for selective autophagy. Upon starvation, the autophagy rate of atg21Δ cells is only partially reduced (Meiling-Wesse et al, 2004; Nair et al, 2010). We expect that this is due to starvation-induced changes at the PAS and especially increased Atg8 expression (Reggiori & Klionsky, 2013).
Atg18 is part of several protein complexes at the PAS, endosomes and the vacuolar membrane with distinct functions (Dove et al, 2004; Reggiori et al, 2004; Efe et al, 2007; Jin et al, 2008; Graef et al, 2013; Suzuki et al, 2013). In analogy, the dual localisation of Atg21 to endosomes and the PAS most likely depends on interaction with a different set of proteins. Further work will be necessary to dissect, if further interactions of Atg21, Atg8 or the E3-complex with Atg proteins restrict this complex to the PAS. Interaction of Atg21 with Atg8 depends on the same loop 2D to 3A, which mediates interaction of Atg18 with Atg2. Indeed, this propeller region is typically involved in protein interactions (Stirnimann et al, 2010). Interestingly, D146 is only conserved among Atg21 yeast homologues, while Atg18 homologues typically have no negatively charged residue at this position.
WIPIs are implicated in severe diseases such as cancer and specific types of neurodegeneration (Polson et al, 2010; Bakula et al, 2013; Saitsu et al, 2013). Together with the involvement of Atg16L1 in Crohn's disease and its role in bacteria-induced autophagy (xenophagy), our study might also influence our understanding of the medical role of autophagy (Zavodszky et al, 2013).
Materials and Methods
Strains
atg8Δ atg1Δ and atg8Δ atg1Δ atg21Δ cells were made by the deletion of ATG1 in WCG4-α atg8Δ (Lang et al, 1998) or atg8Δ atg21Δ using a PCR product of primers atg1_ko_fw and atg1_ko_rev and pFa6a-natNT2 or pFa6a-hphNT1 (Janke et al, 2004).
Strains cAtg16-HA and cAtg16-HA atg8Δ were made using primer Atg16-HA-S2/-S3 to amplify the 6xHA natNT2 cassette from pYM17 (Janke et al, 2004) and homologues recombination in WCG4-α (Thumm et al, 1994) and WCG4-α atg8Δ, respectively. Plasmid KS-PRA1ΔEN::HIS3 (Thumm et al, 1994) was cut with SacI/ApaI, and the insert was transformed in WCG4-α atg21Δ (Barth et al, 2002) to create WCG4-α atg21Δ pep4Δ. atg14Δ was made by the deletion of ATG14 in WCG4-α using a product of atg14_ko_fwd and atg14_ko_rev and pFa6a-natNT2 (Janke et al, 2004).
atg16Δ, atg8Δ atg16Δ and atg21Δ atg16Δ cells were made in WCG4-α or WCG4-α atg8Δ (Lang et al, 1998) or WCG4-α atg21Δ (Barth et al, 2002) using a product of Atg16_S1 and Atg16_S2 and pFa6a-natNT2. atg16Δ atg8Δ atg21Δ cells were made in atg16Δ atg8Δ with a PCR product of Atg21_KO_S1 and Atg21_KO_S2 and pFa6a-hphNT1. atg8Δ atg21Δ was made by the deletion of ATG21 WCG4-α atg8Δ using a product of Atg21_KO_S1 and Atg21_KO_S2 and pFa6a-natNT2. atg8Δ atg21Δ atg5Δ cells were made in atg8Δ atg21Δ (above) using a product of Atg5_HIS forward and Atg5_HIS reverse and pFa6a-HIS3MX6. atg5Δ was made in WCG4-α with a product of S-APG5-kan and AS-APG5-kan and pUG6 (Güldener et al, 1996). atg8Δ pep4Δ was made in WCG4-α atg8Δ with a PCR product of pep4KO_S1 and pep4KP_S2 and pFa6a-natNT2. Other WCG4-α deletion strains: atg1Δ (Straub et al, 1997), atg3Δ (Schlumpberger et al, 1997), atg9Δ (Lang et al, 2000) and atg11Δ (Krick et al, 2008b).
In SEY6210 (Harding et al, 1996), ATG5 and ATG16 were deleted using a product of atg5_ko_fwd and atg5_ko_rev or atg16_ko_fwd and atg16_ko_rev and pFa6a-natNT2. SEY6210 atg4Δ is from Krick et al (2010). BY4741 strains were from Euroscarf (Frankfurt/Main, Germany).
Plasmids
GST-Atg8 mutants are from Krick et al (2010). To generate pRS315-GFP-Atg8, GFP-Atg8 (Suzuki et al, 2001) was digested with NotI/SalI and the insert ligated into pRS315. Mutagenesis of pRS315-GFP-Atg8 used the Quikchange II Site-Directed Mutagenesis kit (Agilent Technologies) with primer GFPATG8 S3A/T4A for/rev, GFPATG8 F5G/K6G for/rev and nATG8 L50A for/rev. pRS316-Atg5-YFP is from Suzuki et al (2001).To generate pRS313-Atg8, ATG8 was amplified using EcoR1-Atg8_fwd and Xho1-Atg8_rev and chromosomal DNA. pRS313 and the PCR product were cut with EcoR1 and Xho1 and ligated. Mutagenesis of pRS313-Atg8 used the Quikchange II Site-Directed Mutagenesis kit (Agilent Technologies) and the primer 313ATG8 ST f/r, 313ATG8 FK f /r and nATG8 L50A for/rev. pMET25-mCherry was constructed by replacing GFP in pUG36 (Niedenthal et al, 1996) by mCherry. mCherry was amplified with MET25-Cherry for/rev and Nab-NLS-Cherry (Krick et al, 2008b). PCR product and pUG36 were cut with XbaI and ligated. pMET25-Cherry-Atg21 was constructed by isolating the Atg21 insert with BamHI/XhoI from pUG36-Atg21 (Krick et al, 2008a) and ligation into BamHI/XhoI cut pUG36. Mutagenesis of pMET25-cherry-Atg21 used overlapping extension PCR with primers Atg21_D146K_fwd and _rev, atg21 mut f and atg21 mut r (for FTTG), or Atg21_ E143K_D146K_fwd and rev with the outer primers GST-Atg21 f/r and BamHI/XhoI as cloning sites into pMET25-Cherry. pRS316-Atg21-YFP is from Meiling-Wesse et al (2004).
pRS316-Atg21-TAP or pRS316-Atg21 FTTG-TAP were made by homologous recombination in yeast. The TAP cassette was amplified using Atg21-TAP f/r and chromosomal DNA. pRS316-Atg21-3HA-eGFP or pRS316-Atg21 FTTG-3HA-eGFP (Krick et al, 2006) was linearised using KpnI/SalI, and the HA-eGFP cassette was replaced by the TAP cassette. pYES2-GFP-Atg8 and pYES2-GFP-Atg8 L50A are from Amar et al (2006). For pYES2-GFP-Atg8F5GK6G and pYES2-GFP-Atg8S3AT4A, the inserts GFP-Atg8F5GK6G and GFP-Atg8S3AT4A were amplified with GFP-Atg8for and GFP-Atg8rev using pRS316-GFP-Atg8F5GK6G or pRS316-GFP-Atg8S3AT4A as templates. The PCR products were cut with BamHI and XhoI and ligated into pYES2.
pRS425-Atg21-TAP was constructed by isolating Atg21-TAP with HindIII/NaeI from pRS315-Atg21-TAP and ligation into the HindIII/NaeI sites of pRS425.
For expression in E. coli, Atg21 was C-terminally fused to His6-SUMO (SUMO: Smt3, the yeast SUMO homologue). MET25-Cub-RURA3 was from F. Reggiori (Utrecht, the Netherlands) and N. Johnsson (Universitaet Ulm, Ulm, Germany). pRS314-PCUP1-Nub was from Wittke et al (1999). CUP1-Nub-ATG8 was from Krick et al (2010). For pRS313-MET25-ATG21-Cub-RURA3, ATG21 was amplified with Atg21-Cub for2 and Atg21-Cub rev2. pRS313-MET25-Cub-RURA3 was cut with ClaI and SphI, and the PCR product and the linearised vector were used for homologous recombination in yeast.
To generate pRS313-MET25-ATG16-Cub-RURA3, ATG16 was amplified with Atg16Cub_ClaI_fwd and Atg16Cub_SalI-rev, cut with ClaI and SalI and ligated in pRS313-MET25-Cub-RURA3. For pRS313-MET25-ATG5-Cub-RURA3, ATG5 was amplified with Atg5Cub_StuI_fwd and Atg5Cub_SalI_rev and cut with StuI and SalI. pRS313-MET25-Cub-RURA3 was cut with ClaI, blunted with Klenow DNA-polymerase and cut with SalI. Both were ligated to pRS313-MET25-ATG5-Cub-RURA3. For pRS314-CUP1-Nub-ATG16, ATG16 was amplified with Nub-Atg16_BamHI_fwd and Nub-Atg16_XhoI_rev, cut with BamHI and XhoI and ligated into pRS314-CUP1-Nui. For pRS314-CUP1-Nui-ATG12, ATG12 was amplified with Nui-Atg12_StuI_fwd and Nui-Atg12_XhoI_rev and cut with StuI and XhoI. pRS314-CUP1-Nui was cut with BamHI, blunted with Klenow polymerase and cut with XhoI. Both were ligated to pRS314-CUP1-Nub-ATG12.
For pRS314-CUP1-Nub-ATG5, ATG5 was amplified with Nui-Atg5 for and Nui-Atg5 rev, cut with BamHI and XhoI and ligated into pRS314-CUP1-Nui.
For pRS313-CUP1-Atg16-HA, ATG16-HA was amplified with Atg16Cub_ClaI_fwd and Atg16HA_XhoI_rev using genomic DNA from cells chromosomally expressing cAtg16-HA. Similarly, for pRS313-CUP1-ATG161-57-HA ATG161-57-HA was amplified with the same primer and genomic DNA from cAtg161-57-HA cells. For pRS313-CUP1-ATG1658-150-HA, ATG1658-150-HA was amplified with Atg16HA_58-150_ClaI_fwd and Atg16HA_XhoI_rev using genomic DNA of cAtg16-HA cells. DNA fragments were cut with ClaI and XhoI and ligated into pRS313-CUP1.
pRS313-CUP1-Atg16D101A-HA, pRS313-CUP1-Atg16E102A-HA, and pRS313-CUP1-Atg16D101A/E102A-HA were constructed using the Quikchange II Site-Directed Mutagenesis kit (Agilent Technologies). Point mutations were introduced using primers: Atg16_D101A_fwd_rev; Atg16_E102A_fwd_rev; Atg16_DE_fwd_rev; Atg16_E97A_fwd_rev.
For pGEX-4T3-GST-Atg16, ATG16 was amplified with GST-Atg16_BamHI_fwd and GST-Atg16_XhoI_rev using genomic DNA. Similarly, ATG161-57 was amplified with GST-Atg16_BamHI_fwd and GST-Atg161-57_XhoI_rev, and ATG1658-150 was amplified with GST-Atg1658-150_BamHI_fwd and GST-Atg16_XhoI_rev. DNA fragments were digested with BamHI and XhoI and ligated into pGEX-4T3. Point mutations were introduced as above.
cAtg16-GFP (Invitrogen; Carlsbad, CA, USA) was taken to amplify Atg16-GFP with its endogenous promoter using primer cAtg16-GFP XbaI and cAtg16-GFP XhoI. PCR product and pRS313 were cut with XbaI and XhoI and ligated into pRS313-Atg16-GFP. pRS313-Atg16D101A-GFP, pRS313-Atg16E102A-GFP and pRS313-Atg16D101A/E102A-GFP were generated as above using pRS313-Atg16-GFP as template.
pUG36-GFP-Atg21 is from Krick et al (2008a); pRS315-pApeI-RFP and pRS313-pApeI-RFP are from Meiling-Wesse et al (2005).
Antibodies
Anti-pApe I is from Barth and Thumm (2001); anti-Atg8 antibody is from Meiling-Wesse et al (2004). Anti-GFP antibodies were from Roche; anti-Pgk1 and anti-carboxypeptidase Y were from Invitrogen (Carlsbad, Kalifornien, USA), and anti-HA was from Santa Cruz Biotechnology, Inc. (Dallas, Texas, USA.). HisProbe™-HRP was from Thermo SCIENTIFIC (Waltham, MA 02454; USA); TAP-tag antibody was from GenScript USA Inc. (Piscataway, NJ 08854 USA); and RFP antibody [5F8] was from ChromoTek (Planegg-Martinsried, Germany).
Horseradish peroxidase (HRPO)-conjugated goat anti-rabbit was from Medac, Hamburg, Germany; HRPO-conjugated goat anti-mouse and anti-rat were from Dianova, Hamburg, Germany.
Split-ubiquitin assay
1 OD600 of cells was diluted 1:10, 1:100, 1:1,000 and 1:10,000 on selection plates, on plates with 50 μg/ml uracil and 1 mg/ml 5-FOA, and on plates without uracil. A total of 250 μM methionine and 100 μM CuSO4 were added to induce expression (Laser et al, 2000). Plates were grown 2–3 days at 30°C.
Isolation of GST-Atg8 and GST-Atg16
Plasmids were transformed into E. coli BL21 containing the lysozyme-expressing plasmid pLysS (K.D. Entian, Frankfurt, Germany). Cells containing both plasmids were grown in LB with ampicillin (75 μg/ml) and chloramphenicol (25 μg/mL) to an OD600 of 0.5–0.8, induced with isopropyl-β-D-thiogalactopyranoside (0.1 mM, 4–5 h at 30°C), harvested, snap-frozen and stored at −80°C. Cells were resuspended in ice-cold PBS with protease inhibitor mixture (Sigma Aldrich, St. Louis, MO, USA), 10 mM MgCl2, Benzonase (Sigma Aldrich, St. Louis, MO, USA) and 1% Triton X-100. Supernatants from lysed cells (15 min, 8,000 g at 4°C) were applied to glutathione–Sepharose 4B beads (30–50 min at 4°C). Elution followed the manufacturer's protocol. Protein concentration was determined using the Bradford method, and protein purity was verified by SDS–PAGE and Coomassie staining.
Isolation of His6-SUMO-Atg21
Plasmids were transformed into E. coli BL21 as above. Cells were grown in LB with kanamycin (50 μg/ml) and chloramphenicol (25 μg/ml) to an OD600 of 0.5–0.8, induced with isopropyl-β-D-thiogalactopyranoside (0.1 mM, 4–5 h at 30°C), harvested, snap-frozen and stored at −80°C. Cells were resuspended in ice-cold PBS containing protease inhibitor mixture (Sigma Aldrich, St. Louis, MO, USA), 2 mM MgCl2, Benzonase (Sigma Aldrich, St. Louis, MO, USA) and 1% Triton X-100. Supernatants from lysed cells (15 min, 8,000 g at 4°C) were applied to Ni-NTA Sepharose (Quiagen, Hilden, Germany) for 30–50 min at 4°C. Elution followed the manufacturer's protocol.
Fluorescence microscopy
Cells were grown to logarithmic phase (1.5 OD600) in selection medium. Cells expressing pMET25-mCherry-Atg21 were grown with 0.3 mM methionine. A DeltaVision Spectris (Applied Precision) fluorescence microscope with a 100× objective and FITC and TRITC filter sets (excitation wavelengths of 340–380 and 465–495 and emission wavelengths of 435–485 and 515–555, respectively) or with GFP (excitation wavelengths 475/28 and emission wavelengths 525/50) and mCherry (excitation wavelengths 575/25 and emission wavelengths 632/60) filter set was used. Images were deconvoluted using WoRx (Applied Precision) software and processed with Adobe CS6.
Pull-down experiments with yeast extracts
atg21Δ pep4Δ cells expressing pRS316-Atg21-TAP were grown to logarithmic phase. 40 OD600 were glass-bead-lysed in cold PBS, 0.5% Triton X-100 and protease inhibitors. 2 OD600 of cleared lysate was removed (L), and the rest incubated for 4 h at 4°C with equal amounts of GST fusions on beads. After sedimentation, 2 OD600 were removed (S). Beads were washed 4× with PBS, 0.5% Triton X-100 and protease inhibitors. Beads were resuspended in 50 μl Laemmli (PD). For immunoblots, 10 μl was used. Lysate (L) and the supernatant (S) corresponding to 4% of the eluate were loaded. For immunoblotting of GST, samples were diluted 1:10.
50 OD600 cAtg16-HA logarithmic cells were lysed. For immunoblots, 2.5 OD600 of cleared lysate was removed (L). The rest was shaken with beads carrying His6-SUMO or His6-SUMO-Atg21 for 4 h at 4°C. 2.5 OD600 of the supernatant was removed (S) and the beads washed 4× with lysis buffer. Proteins were eluted with 50 μl Laemmli with 50 mM DTT (PD). 10 μl was immunobloted. Lysate (L) and supernatant (S) corresponded to 4% of the eluate.
Co-immunoprecipitations with GFP-Atg8
atg1Δ pep4Δ cells expressing pYES2-GFP-Atg8, pYES2-GFP-Atg8L50A, pYES2-GFP-Atg8F5GK6G, GFP-Atg8S3AT4A or pYES2 together with pRS425-Atg21-TAP were grown to mid-log-phase with 2% galactose. 250 OD600 of cells were washed with cold 10 mM NaN3 and resuspended in SP-buffer (1.4 M sorbitol, 50 mM potassium phosphate buffer pH 7.5, 10 mM NaN3, 40 mM β-mercaptoethanol) with 0.8 mg zymolyase 100T at 30°C for 30 min. The spheroplasts were washed in SP-buffer and lysed in PBS pH 7.4, 0.2 M sorbitol, 5 mM MgCl2, 1 mM PMSF and protease inhibitors using a glass homogenizer. Triton X-100 was added to 1%. After sedimentation of debris, the supernatant (input) was incubated with equilibrated beads carrying GFP-binding protein (GFP-Trap_A; ChromoTek, Planegg-Martinsried, Germany) for 2 h at 4°C with mixing. The beads were washed 3× with lysis buffer with 0.5% Triton X-100. Proteins were eluted with 50 μl Laemmli. 15 μl was immunoblotted.
Pull-down experiments with recombinant proteins
2 μM GST, GST-Atg8 or variants were incubated in binding buffer (BB: 20 mM sodium phosphate, pH 7.2; 0.2% Triton X-100; 1 mM EDTA; 20 mM imidazole and protease inhibitors) for 2 h at 4°C with 2 μM SUMO-Atg21 on Ni-NTA agarose. Samples of the BB containing the GST fusion were taken (L). After sedimentation, samples of the supernatant were taken (S) and the rest removed. Beads were washed 4× with BB and resuspended in 50 μl Laemmli (PD). (L) and (S) samples correspond to 8% of the (PD) samples. Of 10 μl was immunoblotted. For the His6-SUMO-Atg21 pull down with recombinant GST-Atg16, 10 μM of the GST fusions was used.
For peptide competition, 2 μM GST-Atg8 in BB was preincubated for 1 h at 4°C with 600 μM (300-fold excess) of the peptide and then incubated for 50 min at 4°C with 2 μM SUMO-Atg21 on Ni-NTA agarose. Samples were processed as above.
0.5 μM isolated SUMO-Atg21 was preincubated with BSA in BB2 (20 mM sodium phosphate, pH 7.2; 0.5% Triton X-100; 1 mM EDTA; 20 mM imidazole; 0.4% BSA; and protease inhibitors) for 30 min at 4°C. As input control, 15% of the SUMO-Atg21 eluate were loaded. The preincubated SUMO-Atg21 solution was incubated with 10 μM GST-Atg8 on glutathione–Sepharose 4B for 2 h at 4°C. After sedimentation, samples of the supernatant were taken (S) and the rest removed. Beads were washed 4× with 20 mM sodium phosphate, pH 7.2; 0.5% Triton X-100; 1 mM EDTA; and protease inhibitors and resuspended in 50 μl Laemmli (PD).
Immunoprecipitation of GFP fusion proteins
cAtg16-HA and cAtg16-HA atg8Δ cells expressing pUG36-GFP-Atg21 or pUG36-GFP were grown to mid-log without methionine. Similarly, atg16Δ cells expressing pUG36-GFP and Atg16-HA or pUG36-GFP-Atg21 and Atg16-HA or its variants were grown to mid-log without methionine. The expression level of Atg161–57-HA and Atg1658–150-HA was adjusted with 50 μM or 25 μM CuSO4 to the selective medium for ∽12 h, respectively. 350 OD600 were harvested, washed with 10 mM NaN3 and resuspended in SP-buffer (1.4 M sorbitol, 50 mM potassium phosphate buffer pH 7.5, 10 mM NaN3, 40 mM β-mercaptoethanol) containing 1.5 mg zymolyase T100 at 30°C for 40 min. Spheroblasts were washed with SP-buffer and lysed in buffer (PBS pH 7.4, 0.2 M sorbitol, 5 mM MgCl2, 1 mM PMSF, protease inhibitors) with a homogeniser. Triton X-100 was added to the lysate to 1%. For immunoblots, 12 OD600 of cleared lysate were removed (L). The rest was incubated with equilibrated beads carrying GFP-binding protein (GFP-Trap_A; ChromoTek, Planegg-Martinsried, Germany) for 2 h at 4°C under mixing. After sedimentation, 12 OD600 of the supernatant were removed (S). Beads were washed 4× with lysis buffer. Proteins were eluted with 50 μl Laemmli (PD). 10 μl was blotted. Lysate (L) and the supernatant (S) corresponded to ∽2% of the eluate (PD) and for chromosomally expressing cAtg16 strains to ∽3.5% of the eluate (PD).
To confirm self-interaction of Atg16-HA, 100 OD600 of atg16Δ cells expressing Atg16-HA wild-type, Atg16D101A/E102A-HA or Atg16E97A-HA were glass-bead-lysed (PBS pH 7.4, 5 mM MgCl2, 1 mM PMSF, protease inhibitors). The cleared lysates were mixed with the lysate of atg16Δ cells expressing GFP, or Atg16-GFP variants, Atg16D101A/E102A-GFP or Atg16E97A-GFP and incubated with GFP-TRAP beads as described. Lysate (L) and the supernatant (S) corresponded to 3.5% of the eluate (PD).
Mitophagy assay and Atg8 lipidation
Cells expressing the mito marker mito-GFP were grown in lactate medium and mitophagy induced with 0.2 μg/ml rapamycin (Welter et al, 2013).
Analysis of Atg8 lipidation is described in Suzuki et al (2001).
Quantification and statistical analyses
Western blots were quantified with a LAS3000 imaging system and the AIDA software. From at least triplicate experiments, SEM was calculated. We used Graph Pad Prism 6 and unpaired two-tailed t-tests to calculate the statistical relevance. The figures are labelled as follows: not significant (ns) for P > 0.05; * for P < 0.05; ** for P < 0.01; *** for P < 0.001; and **** for P < 0.0001.
For fluorescence microscopy, images from at least three cultures were taken. For each image, puncta/cells were counted. Error bars are SEM.
Acknowledgments
We thank F. Reggiori, N. Johnsson and Z. Elazar for plasmids and antibodies. This work was funded by the Deutsche Forschungsgemeinschaft in the Collaborative Research Centre 860 ‘Integrative Structural Biology of Dynamic Macromolecular Assemblies’.
Author contributions
LJ, MM and PR planned and performed experiments; PS performed experiments; RK planned, supervised and performed experiments; MT planned and supervised experiments and wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary Information
Review Process File
References
- Alemu EA, Lamark T, Torgersen KM, Birgisdottir AB, Larsen KB, Jain A, Olsvik H, Øvervatn A, Kirkin V, Johansen T. ATG8 family proteins act as scaffolds for assembly of the ULK complex: sequence requirements for LC3-interacting region (LIR) motifs. J Biol Chem. 2012;287:39275–39290. doi: 10.1074/jbc.M112.378109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amar N, Lustig G, Ichimura Y, Ohsumi Y, Elazar Z. Two newly identified sites in the ubiquitin-like protein Atg8 are essential for autophagy. EMBO Rep. 2006;7:635–642. doi: 10.1038/sj.embor.7400698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backues SK, Klionsky DJ. Atg11: a Rab-dependent, coiled-coil membrane protein that acts as a tether for autophagy. Autophagy. 2012;8:1275–1278. doi: 10.4161/auto.21153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakula D, Takacs Z, Proikas-Cezanne T. WIPI β-propellers in autophagy-related diseases and longevity. Biochem Soc Trans. 2013;41:962–967. doi: 10.1042/BST20130039. [DOI] [PubMed] [Google Scholar]
- Barth H, Thumm M. A genomic screen identifies AUT8 as a novel gene essential for autophagy in the yeast Saccharomyces cerevisiae. Gene. 2001;274:151–156. doi: 10.1016/s0378-1119(01)00614-x. [DOI] [PubMed] [Google Scholar]
- Barth H, Meiling-Wesse K, Epple UD, Thumm M. Mai1p is essential for maturation of proaminopeptidase I but not for autophagy. FEBS Lett. 2002;512:173–179. doi: 10.1016/s0014-5793(02)02252-4. [DOI] [PubMed] [Google Scholar]
- Baskaran S, Ragusa MJ, Boura E, Hurley JH. Two-site recognition of phosphatidylinositol 3-phosphate by PROPPINs in autophagy. Mol Cell. 2012;47:339–348. doi: 10.1016/j.molcel.2012.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi AMK, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368:651–662. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- Dooley HC, Razi M, Polson HEJ, Girardin SE, Wilson MI, Tooze SA. WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Mol Cell. 2014;55:238–252. doi: 10.1016/j.molcel.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove S, Piper R, McEwen R, Yu J, King M, Hughes D, Thuring J, Holmes A, Cooke F, Michell R, Parker P, Lemmon M. Svp1p defines a family of phosphatidylinositol 3,5-bisphosphate effectors. EMBO J. 2004;23:1922–1933. doi: 10.1038/sj.emboj.7600203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efe J, Botelho R, Emr S. Atg18 regulates organelle morphology and Fab1 kinase activity independent of its membrane recruitment by phosphatidylinositol 3,5-bisphosphate. Mol Biol Cell. 2007;18:4232–4244. doi: 10.1091/mbc.E07-04-0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré J-C, Krick R, Subramani S, Thumm M. Turnover of organelles by autophagy in yeast. Curr Opin Cell Biol. 2009;21:522–530. doi: 10.1016/j.ceb.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka Y, Noda NN, Nakatogawa H, Ohsumi Y, Inagaki F. Dimeric coiled-coil structure of Saccharomyces cerevisiae Atg16 and its functional significance in autophagy. J Biol Chem. 2010;285:1508–1515. doi: 10.1074/jbc.M109.053520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008;19:2092–2100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graef M, Friedman JR, Graham C, Babu M, Nunnari J. ER exit sites are physical and functional core autophagosome biogenesis components. Mol Biol Cell. 2013;24:2918–2931. doi: 10.1091/mbc.E13-07-0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güldener U, Heck S, Fielder T, Beinhauer J, Hegemann JH. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki M, Shibutani ST, Yoshimori T. Up-to-date membrane biogenesis in the autophagosome formation. Curr Opin Cell Biol. 2013;25:455–460. doi: 10.1016/j.ceb.2013.03.004. [DOI] [PubMed] [Google Scholar]
- Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F, Ohsumi Y. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem. 2007;282:37298–37302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- Harding TM, Hefner-Gravink A, Thumm M, Klionsky DJ. Genetic and phenotypic overlap between autophagy and the cytoplasm to vacuole protein targeting pathway. J Biol Chem. 1996;271:17621–17624. doi: 10.1074/jbc.271.30.17621. [DOI] [PubMed] [Google Scholar]
- Janke C, Magiera M, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, Knop M. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- Jin N, Chow CY, Liu L, Zolov SN, Bronson R, Davisson M, Petersen JL, Zhang Y, Park S, Duex JE, Goldowitz D, Meisler MH, Weisman LS. VAC14 nucleates a protein complex essential for the acute interconversion of PI3P and PI(3,5)P(2) in yeast and mouse. EMBO J. 2008;27:3221–3234. doi: 10.1038/emboj.2008.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann A, Beier V, Franquelim HG, Wollert T. Molecular mechanism of autophagic membrane-scaffold assembly and disassembly. Cell. 2014;156:469–481. doi: 10.1016/j.cell.2013.12.022. [DOI] [PubMed] [Google Scholar]
- Krick R, Tolstrup J, Appelles A, Henke S, Thumm M. The relevance of the phosphatidylinositolphosphat-binding motif FRRGT of Atg18 and Atg21 for the Cvt pathway and autophagy. FEBS Lett. 2006;580:4632–4638. doi: 10.1016/j.febslet.2006.07.041. [DOI] [PubMed] [Google Scholar]
- Krick R, Henke S, Tolstrup J, Thumm M. Dissecting the localization and function of Atg18, Atg21 and Ygr223c. Autophagy. 2008a;4:896–910. doi: 10.4161/auto.6801. [DOI] [PubMed] [Google Scholar]
- Krick R, Muehe Y, Prick T, Bremer S, Schlotterhose P, Eskelinen E-L, Millen J, Goldfarb DS, Thumm M. Piecemeal microautophagy of the nucleus requires the core macroautophagy genes. Mol Biol Cell. 2008b;19:4492–4505. doi: 10.1091/mbc.E08-04-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krick R, Bremer S, Welter E, Schlotterhose P, Muehe Y, Eskelinen E-L, Thumm M. Cdc48/p97 and Shp1/p47 regulate autophagosome biogenesis in concert with ubiquitin-like Atg8. J Cell Biol. 2010;190:965–973. doi: 10.1083/jcb.201002075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krick R, Busse RA, Scacioc A, Stephan M, Janshoff A, Thumm M, Kühnel K. Structural and functional characterization of the two phosphoinositide binding sites of PROPPINs, a β-propeller protein family. Proc Natl Acad Sci USA. 2012;109:E2042–E2049. doi: 10.1073/pnas.1205128109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang T, Schaeffeler E, Bernreuther D, Bredschneider M, Wolf DH, Thumm M. Aut2p and Aut7p, two novel microtubule-associated proteins are essential for delivery of autophagic vesicles to the vacuole. EMBO J. 1998;17:3597–3607. doi: 10.1093/emboj/17.13.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang T, Reiche S, Straub M, Bredschneider M, Thumm M. Autophagy and the cvt pathway both depend on AUT9. J Bacteriol. 2000;182:2125–2133. doi: 10.1128/jb.182.8.2125-2133.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laser H, Bongards C, Schuller J, Heck S, Johnsson N, Lehming N. A new screen for protein interactions reveals that the Saccharomyces cerevisiae high mobility group proteins Nhp6A/B are involved in the regulation of the GAL1 promoter. Proc Natl Acad Sci USA. 2000;97:13732–13737. doi: 10.1073/pnas.250400997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- Mari M, Griffith J, Rieter E, Krishnappa L, Klionsky DJ, Reggiori F. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J Cell Biol. 2010;190:1005–1022. doi: 10.1083/jcb.200912089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiling-Wesse K, Barth H, Voss C, Eskelinen E-L, Epple UD, Thumm M. Atg21 is required for effective recruitment of Atg8 to the preautophagosomal structure during the Cvt pathway. J Biol Chem. 2004;279:37741–37750. doi: 10.1074/jbc.M401066200. [DOI] [PubMed] [Google Scholar]
- Meiling-Wesse K, Epple UD, Krick R, Barth H, Appelles A, Voss C, Eskelinen E-L, Thumm M. Trs85 (Gsg1), a component of the TRAPP complexes, is required for the organization of the preautophagosomal structure during selective autophagy via the Cvt pathway. J Biol Chem. 2005;280:33669–33678. doi: 10.1074/jbc.M501701200. [DOI] [PubMed] [Google Scholar]
- Metlagel Z, Otomo C, Takaesu G, Otomo T. Structural basis of ATG3 recognition by the autophagic ubiquitin-like protein ATG12. Proc Natl Acad Sci USA. 2013;110:18844–18849. doi: 10.1073/pnas.1314755110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Noda T, Ohsumi Y. Apg16p is required for the function of the Apg12p-Apg5p conjugate in the yeast autophagy pathway. EMBO J. 1999;18:3888–3896. doi: 10.1093/emboj/18.14.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moravcevic K, Oxley CL, Lemmon MA. Conditional peripheral membrane proteins: facing up to limited specificity. Structure. 2012;20:15–27. doi: 10.1016/j.str.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J, Johnsson N. Split-ubiquitin and the split-protein sensors: chessman for the endgame. ChemBioChem. 2008;9:2029–2038. doi: 10.1002/cbic.200800190. [DOI] [PubMed] [Google Scholar]
- Nair U, Cao Y, Xie Z, Klionsky DJ. Roles of the lipid-binding motifs of Atg18 and Atg21 in the cytoplasm to vacuole targeting pathway and autophagy. J Biol Chem. 2010;285:11476–11488. doi: 10.1074/jbc.M109.080374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair U, Jotwani A, Geng J, Gammoh N, Richerson D, Yen W-L, Griffith J, Nag S, Wang K, Moss T, Baba M, McNew JA, Jiang X, Reggiori F, Melia TJ, Klionsky DJ. SNARE proteins are required for macroautophagy. Cell. 2011;146:290–302. doi: 10.1016/j.cell.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–178. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Ohbayashi S, Sakoh-Nakatogawa M, Kakuta S, Suzuki SW, Kirisako H, Kondo-Kakuta C, Noda NN, Yamamoto H, Ohsumi Y. The autophagy-related protein kinase Atg1 interacts with the ubiquitin-like protein Atg8 via the Atg8 family interacting motif to facilitate autophagosome formation. J Biol Chem. 2012;287:28503–28507. doi: 10.1074/jbc.C112.387514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedenthal R, Riles L, Johnston M, Hegemann J. Green fluorescent protein as a marker for gene expression and subcellular localization in budding yeast. Yeast. 1996;12:773–786. doi: 10.1002/(SICI)1097-0061(19960630)12:8%3C773::AID-YEA972%3E3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Noda NN, Kumeta H, Nakatogawa H, Satoo K, Adachi W, Ishii J, Fujioka Y, Ohsumi Y, Inagaki F. Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells. 2008;13:1211–1218. doi: 10.1111/j.1365-2443.2008.01238.x. [DOI] [PubMed] [Google Scholar]
- Noda NN, Ohsumi Y, Inagaki F. Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. 2010;584:1379–1385. doi: 10.1016/j.febslet.2010.01.018. [DOI] [PubMed] [Google Scholar]
- Noda NN, Fujioka Y, Hanada T, Ohsumi Y, Inagaki F. Structure of the Atg12-Atg5 conjugate reveals a platform for stimulating Atg8-PE conjugation. EMBO Rep. 2013;14:206–211. doi: 10.1038/embor.2012.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obara K, Sekito T, Niimi K, Ohsumi Y. The Atg18-Atg2 complex is recruited to autophagic membranes via phosphatidylinositol 3-phosphate and exerts an essential function. J Biol Chem. 2008;283:23972–23980. doi: 10.1074/jbc.M803180200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obara K, Ohsumi Y. Atg14: a key player in orchestrating autophagy. Int J Cell Biol. 2011;2011:713435. doi: 10.1155/2011/713435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashkova N, Gakhar L, Winistorfer SC, Yu L, Ramaswamy S, Piper RC. WD40 repeat propellers define a ubiquitin-binding domain that regulates turnover of F box proteins. Mol Cell. 2010;40:433–443. doi: 10.1016/j.molcel.2010.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polson HEJ, De Lartigue J, Rigden DJ, Reedijk M, Urbé S, Clague MJ, Tooze SA. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy. 2010;6:506–522. doi: 10.4161/auto.6.4.11863. [DOI] [PubMed] [Google Scholar]
- Reggiori F, Tucker K, Stromhaug P, Klionsky D. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell. 2004;6:79–90. doi: 10.1016/s1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- Reggiori F, Klionsky DJ. Autophagic processes in yeast: mechanism, machinery and regulation. Genetics. 2013;194:341–361. doi: 10.1534/genetics.112.149013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieter E, Vinke F, Bakula D, Cebollero E, Ungermann C, Proikas-Cezanne T, Reggiori F. Atg18 function in autophagy is regulated by specific sites within its β-propeller. J Cell Sci. 2013;126:593–604. doi: 10.1242/jcs.115725. [DOI] [PubMed] [Google Scholar]
- Romanov J, Walczak M, Ibiricu I, Schüchner S, Ogris E, Kraft C, Martens S. Mechanism and functions of membrane binding by the Atg5-Atg12/Atg16 complex during autophagosome formation. EMBO J. 2012;31:4304–4317. doi: 10.1038/emboj.2012.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitsu H, Nishimura T, Muramatsu K, Kodera H, Kumada S, Sugai K, Kasai-Yoshida E, Sawaura N, Nishida H, Hoshino A, Ryujin F, Yoshioka S, Nishiyama K, Kondo Y, Tsurusaki Y, Nakashima M, Miyake N, Arakawa H, Kato M, Mizushima N, et al. De novo mutations in the autophagy gene WDR45 cause static encephalopathy of childhood with neurodegeneration in adulthood. Nat Genet. 2013;45:445–449. doi: 10.1038/ng.2562. -449e441. [DOI] [PubMed] [Google Scholar]
- Sakoh-Nakatogawa M, Matoba K, Asai E, Kirisako H, Ishii J, Noda NN, Inagaki F, Nakatogawa H, Ohsumi Y. Atg12-Atg5 conjugate enhances E2 activity of Atg3 by rearranging its catalytic site. Nat Struct Mol Biol. 2013;20:433–439. doi: 10.1038/nsmb.2527. [DOI] [PubMed] [Google Scholar]
- Schlumpberger M, Schaeffeler E, Straub M, Bredschneider M, Wolf DH, Thumm M. AUT1, a gene essential for autophagocytosis in the yeast Saccharomyces cerevisiae. J Bacteriol. 1997;179:1068–1076. doi: 10.1128/jb.179.4.1068-1076.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrodinger LLC. The PyMOL Molecular Graphics System. Portland, OR: Schrodinger; 2010. [Google Scholar]
- Schwarten M, Stoldt M, Mohrlüder J, Willbold D. Solution structure of Atg8 reveals conformational polymorphism of the N-terminal domain. Biochem Biophys Res Commun. 2010;395:426–431. doi: 10.1016/j.bbrc.2010.04.043. [DOI] [PubMed] [Google Scholar]
- Shaid S, Brandts CH, Serve H, Dikic I. Ubiquitination and selective autophagy. Cell Death Differ. 2013;20:21–30. doi: 10.1038/cdd.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnimann CU, Petsalaki E, Russell RB, Müller CW. WD40 proteins propel cellular networks. Trends Biochem Sci. 2010;35:565–574. doi: 10.1016/j.tibs.2010.04.003. [DOI] [PubMed] [Google Scholar]
- Straub M, Bredschneider M, Thumm M. AUT3, a serine/threonine kinase gene, is essential for autophagocytosis in Saccharomyces cerevisiae. J Bacteriol. 1997;179:3875–3883. doi: 10.1128/jb.179.12.3875-3883.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stromhaug P, Reggiori F, Guan J, Wang C, Klionsky D. Atg21 is a phosphoinositide binding protein required for efficient lipidation and localization of Atg8 during uptake of aminopeptidase I by selective autophagy. Mol Biol Cell. 2004;15:3553–3566. doi: 10.1091/mbc.E04-02-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Akioka M, Kondo-Kakuta C, Yamamoto H, Ohsumi Y. Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. J Cell Sci. 2013;126:2534–2544. doi: 10.1242/jcs.122960. [DOI] [PubMed] [Google Scholar]
- Tamura N, Oku M, Ito M, Noda NN, Inagaki F, Sakai Y. Atg18 phosphoregulation controls organellar dynamics by modulating its phosphoinositide-binding activity. J Cell Biol. 2013;202:685–698. doi: 10.1083/jcb.201302067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumm M, Egner R, Koch B, Schlumpberger M, Straub M, Veenhuis M, Wolf DH. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 1994;349:275–280. doi: 10.1016/0014-5793(94)00672-5. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Kobayashi T, Yamamoto H, Hoshida H, Akada R, Inagaki F, Ohsumi Y, Noda NN. Structure-based analyses reveal distinct binding sites for Atg2 and phosphoinositides in Atg18. J Biol Chem. 2012;287:31681–31690. doi: 10.1074/jbc.M112.397570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidberg H, Shvets E, Shpilka T, Shimron F, Shinder V, Elazar Z. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 2010;29:1792–1802. doi: 10.1038/emboj.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidberg H, Shpilka T, Shvets E, Abada A, Shimron F, Elazar Z. LC3 and GATE-16 N termini mediate membrane fusion processes required for autophagosome biogenesis. Dev Cell. 2011;20:444–454. doi: 10.1016/j.devcel.2011.02.006. [DOI] [PubMed] [Google Scholar]
- Welter E, Montino M, Reinhold R, Schlotterhose P, Krick R, Dudek J, Rehling P, Thumm M. Uth1 is a mitochondrial inner membrane protein dispensable for post-log-phase and rapamycin-induced mitophagy. FEBS J. 2013;280:4970–4982. doi: 10.1111/febs.12468. [DOI] [PubMed] [Google Scholar]
- Wittke S, Lewke N, Muller S, Johnsson N. Probing the molecular environment of membrane proteins in vivo. Mol Biol Cell. 1999;10:2519–2530. doi: 10.1091/mbc.10.8.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Nair U, Klionsky DJ. Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell. 2008;19:3290–3298. doi: 10.1091/mbc.E07-12-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Noda NN, Nakatogawa H, Kumeta H, Ohsumi Y, Inagaki F. Autophagy-related protein 8 (Atg8) family interacting motif in Atg3 mediates the Atg3-Atg8 interaction and is crucial for the cytoplasm-to-vacuole targeting pathway. J Biol Chem. 2010;285:29599–29607. doi: 10.1074/jbc.M110.113670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Kakuta S, Watanabe TM, Kitamura A, Sekito T, Kondo-Kakuta C, Ichikawa R, Kinjo M, Ohsumi Y. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol. 2012;198:219–233. doi: 10.1083/jcb.201202061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavodszky E, Vicinanza M, Rubinsztein DC. Biology and trafficking of ATG9 and ATG16L1, two proteins that regulate autophagosome formation. FEBS Lett. 2013;587:1988–1996. doi: 10.1016/j.febslet.2013.04.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Review Process File