Abstract
Nuclear receptors (NRs) are transcription regulators involved in an array of diverse physiological functions including key roles in endocrine and metabolic function. The aim of this study was to identify nuclear receptors in the fully sequenced genome of the gastropod snail, Biomphalaria glabrata, intermediate host for Schistosoma mansoni and compare these to known vertebrate NRs, with a view to assessing the snail's potential as a invertebrate model organism for endocrine function, both as a prospective new test organism and to elucidate the fundamental genetic and mechanistic causes of disease. For comparative purposes, the genome of a second gastropod, the owl limpet, Lottia gigantea was also investigated for nuclear receptors. Thirty-nine and thirty-three putative NRs were identified from the B. glabrata and L. gigantea genomes respectively, based on the presence of a conserved DNA-binding domain and/or ligand-binding domain. Nuclear receptor transcript expression was confirmed and sequences were subjected to a comparative phylogenetic analysis, which demonstrated that these molluscs have representatives of all the major NR subfamilies (1-6). Many of the identified NRs are conserved between vertebrates and invertebrates, however differences exist, most notably, the absence of receptors of Group 3C, which includes some of the vertebrate endocrine hormone targets. The mollusc genomes also contain NR homologues that are present in insects and nematodes but not in vertebrates, such as Group 1J (HR48/DAF12/HR96). The identification of many shared receptors between humans and molluscs indicates the potential for molluscs as model organisms; however the absence of several steroid hormone receptors indicates snail endocrine systems are fundamentally different.
Introduction
The tropical freshwater snail Biomphalaria glabrata is an intermediate host for several digenean trematode parasitic worms, including Schistosoma mansoni, the causative agent of human intestinal schistosomiasis. Human schistosomiasis is the most widespread trematode infection affecting around 200 million people, leading to a chronic debilitating disease and up to 200,000 deaths per year, across 75 developing countries [1]. Due to its medical significance as an intermediate host, B. glabrata has been the focus for much research, including several gene discovery projects [2–4]. Tools have been developed for investigating genomic and transcriptomic attributes of this species, including a BAC library for genome sequencing [5]; a 5K cDNA microarray [6]; a 1.2K oligo microarray [7] and the means to selectively silence gene expression in the snail (RNAi: [8,9]). This background of research has culminated in the sequencing the B. glabrata genome [10] (http://129.24.144.93/blast_bg/2index.html). The progress made in developing these resources has also provided the potential for the snail to become a new model organism for other purposes, including testing and identification of endocrine disrupting chemicals (EDCs) and for understanding fundamental biology, including endocrinology.
Endocrine and metabolic disease are among the most common contemporary human afflictions, the prevalence of which has been well defined in large population-based studies (for example, [11,12]). Some of the causes of these are not immediately obvious, but may be related to increasing exposure to EDCs [13]. As a consequence, not only is more testing of potential EDCs needed, but a better understanding of endocrine function and disruption is also required. A solution that is appropriate and in keeping with the three R’s (replacement, refinement and reduction) ethos of animal research [14], is to exploit the use of invertebrate organisms which may offer a simplified model for research, as well as providing a faster, cheaper and more ethically acceptable alternative to mammalian testing, at least during initial chemical screening [15]. As NRs play key roles in endocrine and metabolic functions [16], cross-species comparative studies of the conservation of these genes within invertebrate genomes may identify new model systems for the testing of chemicals with endocrine disrupting potential without using vertebrates. In addition to this, B. glabrata has recently been proposed specifically for developmental toxicity, acute toxicity and mutagenicity testing in order to establish standardised protocols to assess environmental risks [17]. Therefore a better knowledge of normal endocrine function in molluscs will enable us to understand the full impact of EDCs in the environment, where they have been shown to affect both vertebrate and invertebrate species [18].
One class of transcription factor involved in regulating endocrine function in vertebrates are the nuclear receptors (NRs). NRs regulate and coordinate multiple processes by integrating internal and external signals, thereby maintaining homeostasis (reviewed in [16,19]) These proteins exhibit strong similarities in their mode of action due to their conserved molecular structure, which includes a DNA-binding domain (DBD), consisting of two Cys4 zinc fingers, and the ligand-binding domain (LBD), which not only controls signalling by binding small lipophilic molecules, called ligands, but also binds co-activators and co-repressors [20]. Both the LBD and D box domain of the DBD mediate receptor hetero- or homo-dimerization (reviewed in [21]). Biochemical studies and crystal structure of the LBD reveal that ligand binding triggers a conformational change, causing bound co-repressors to be displaced by co-activators, leading to gene expression [22]. NRs are classified into six distinct families by sequence homology using a phylogenetic approach [23]. Based on their conserved nature and their biologically essential roles throughout the Metazoa, NRs are believed to have emerged early in animal evolution, prior to the bilaterian ancestor [24–26].
Molluscs are affected by EDCs and one of the most cited examples is that of the marine pollutant tributyltin (TBT), found in antifouling paint, which is responsible for imposex in at least 195 species of gastropods worldwide (reviewed in [27,28]). TBT has been shown to act through binding to an NR, the retinoid X receptor (RXR) [29,30]. Other environmental pollutants could act through the estrogen receptor (ER) or androgen receptor (AR) and there is also some evidence of a role for androgen and estrogen-like molecules in the reproductive cycle of molluscs [31,32]. ER orthologues have previously been reported in molluscs (eg [33–37]) although their function is currently unconfirmed [38,36,39,37,40], since, for example, the identified ER homologue from the oyster Crassostrea gigas is unresponsive to estrogen [37], The presence of an ER in molluscs and the potential to respond to estrogens is confounded by the absence of aromatase (CYP19) [41], the enzyme required for the conversion of testosterone to estradiol, but it is possible that another enzyme catalyses the aromatase reaction [38]. The presence of ARs in molluscs has been inferred [32,42], but this remains an area of controversy [43] since homologues have not been identified despite investigations specifically searching for the gene [44]. A recent survey of NRs in C. gigas also did not identify an AR [45].
Previously only the RXR has been characterised in B. glabrata [46]. With the recent availability of a draft genome for B. glabrata it is particularly timely to search for the NRs of B. glabrata. The genome of the owl limpet, L. gigantea [47], a marine gastropod, for which 26 NRs have already been identified [25,48,49], was compared with the snail NR repertoire. These species, from different families, represent distinct gastropod lineages. The sequences of identified putative genes were confirmed using transcript data from the L. gigantea genome portal and by amplification of products from B. glabrata mRNA. We assess the conservation, divergence, and uniqueness of gastropod NRs in comparison with the previously characterised receptors of a vertebrate (Homo sapiens), insect (Drosophila melanogaster), nematode worm (Caenorhabditis elegans) (reviewed in [50]) and the parasitic trematode (S. mansoni) (reviewed in [51]).
Methods
Identification of nuclear receptors from B. glabrata and L. gigantea genomes
Human AR (GenBank: EAX05380) and ERα (GenBank: AAI28575) gene sequences were used to search for expressed sequence tags (ESTs) in B. glabrata and L. gigantea at National Centre for Biotechnology Information (NCBI) using TBlastX with default parameters [52]. These ESTs were then used to search for homologues at the preliminary B. glabrata genome (version 4.01: http://129.24.144.93/blast_bg/2index.html) (TBlastX expect limit:1e-04) and assembled L. gigantea transcripts from the filtered gene models in the JGI Genome Portal (http://genome.jgi-psf.org/Lotgi1/Lotgi1.info.html) using BLASTN (expect limit: 1e-99). This was followed by a wider search using NR gene sequences spanning all the groups present in humans and D. melanogaster. A secondary search using the identified L. gigantea and B. glabrata NR genes against both the gastropod genomes using a low expect threshold value identified additional family members, some of which were distinct from human and fly NRs. Query sequences were filtered for low complexity regions. Identified contigs from B. glabrata were downloaded and processed through GENSCAN [53] (http://genes.mit.edu/GENSCAN.html) using default parameters, to identify predicted coding regions, intron-exon boundaries, and peptides. Predicted peptides were assessed using BLASTP (expect limit: 1e-10) against the non-redundant GenBank database to check for the presence of the NR domains (LBD/DBD). The identified nucleotide sequences from both molluscs containing NR domains were analysed for redundancy using clustering analysis with sequence overlap cut-off set at 0.5 and segment coverage cut-off at 0.25 in Seqtools (8.4ver) (http://www.seqtools.dk/).
The identified genes were named based on the phylogenetic analysis and sequence similarity of the full-length sequence to previously characterized human and D. melanogaster NRs according to the recognised nomenclature [23]. Small sequence fragments of putative NRs from B. glabrata too short to be classified were left out of the subsequent analysis. Where there was no vertebrate or invertebrate homologue, subfamily classification was made, with a number representing the subfamily, a capital letter for the group, and a number for the individual gene.
RNA isolation and cDNA synthesis
Total RNA was isolated from whole homogenized adult B. glabrata snails using TRI-reagent (Sigma-Aldrich, St Louis, USA) according to the manufacturer’s protocol and treated with DNAse to eliminate contaminating genomic DNA. Total RNA was extracted from embryonic samples using the RNeasy Fibrous tissue mini kit (Qiagen, Limberg, Netherlands), which included a DNAse treatment as part of the protocol. Quantification and purity of each RNA sample were determined by spectrophotometry (Nanodrop, Thermo Fisher Scientific Ltd. Waltham, USA), and the RNA integrity was visually checked by agarose gel electrophoresis. 4μg of total RNA from adult snails and 1.5μg total RNA from embryonic samples was reverse transcribed in a 20μl reaction using the Superscript III cDNA synthesis kit (Invitrogen, Life Technologies, Carlsbad. USA) with 5 μM of a custom oligo (dTAP) primer (TGACTCGAGTCGACATCGAT20) following the manufacturer’s instructions. Residual RNA was removed by adding 1μl of RNase H (2 U/μl) to the reaction and incubating it at 37°C for 20 min. RT-PCR with 18S primers (18S-F: CGCCCGTCGCTACTATCG and 18S-R: ACGCCAGACCGAGACCAA) [54] verified successful cDNA synthesis.
Polymerase chain reaction
Specific PCR primers (S1 Table) were designed using PRIMER3 (version 0.4.0: http://bioinfo.ut.ee/primer3-0.4.0/), to amplify fragments from DBD and/or LBD for each B. glabrata NR to confirm transcription and sequence. 25μl PCRs contained 2μl cDNA (diluted 1 in 20), 1 X PCR Buffer, 2.5mM MgCl2, 0.2mM dNTPs, 0.5μM forward and reverse primers and 1.25U Taq DNA polymerase (Bioline, London, UK). Reaction conditions were 2 min at 95°C followed by 35 cycles of 30 sec at 95°C, 30 sec at 55–64°C (optimised for each primer pair (S1 Table)) 1 min 30 sec at 72°C, with a final extension of 5 min at 72°C. Amplified products were analysed by gel electrophoresis and products of an appropriate size were gel extracted where necessary and sequenced (Sequencing facility, Wolfson Wellcome Biomedical Laboratory, Natural History Museum, UK). Sequences were deposited in GenBank Accession Nos: JZ390894-JZ390939
Sequence alignments and phylogenetic analysis
The sequenced NR transcripts from B. glabrata were translated using proteomics tools at EXPASY (http://www.expasy.org/). The predicted peptide sequences from both B. glabrata and L. gigantea were analysed using PFAM domain analysis [55] (PF00104 and PF00105) and PANTHER [56], a hidden Markov model-based method (PTHR24082) confirming the NR domains. The NCBI program Simple Modular Architecture Research Tool (SMART, [57]) was used for the identification of DNA-binding domain (DBD) and ligand binding domain (LBD) regions which were then aligned with the DBD and LBD regions of human (H. sapiens), fruit fly (D. melanogaster) nematode (C. elegans) and parasitic trematode (S. mansoni) NRs (Table 1). The NR domains were aligned using default parameters in ClustalX and converted to Nexus format using default parameters in Mesquite v2.75. The DBD and LBD from NRs of the other species were obtained using the conserved domain database (CDD) and reconfirmed using SMART. The NR2A subfamily expansion that contains a large number of the C. elegans NRs [58] was disregarded and only 15 NRs from C. elegans that are broadly conserved among animal phyla were included in this study for comparative analysis.
Table 1. Sequence identification of all nuclear receptors for species in the study: NRs from fly, human, nematode and trematode (NCBI accession numbers), compared to identified L. gigantea NRs (protein identification numbers: JGI genome portal version 1.1) and B. glabrata NRs (Contig numbers: Preliminary Bg Genomic Data (version 4.01)).
| Group | C. elegans | Accession | S. mansoni | Accession | D. melanogaster | Accession | B. glabrata | Contig | L. gigantea | Protein ID | H. sapiens | Accession |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0A | KNR | CAA31709 | ||||||||||
| KNRL | AAF51627 | |||||||||||
| EGON | CAA34626 | |||||||||||
| 0B | BgDAX | 182 | LgDAX | 153776 | DAX1 | AAH11564 | ||||||
| SHP | AAH30207 | |||||||||||
| 1A | THRa | AAR32912 | BgTHR ‡ | 3.1 | LgTHR | 207623 | THRα | AAH08851 | ||||
| THRb | AAR29359 | THRβ | AAI06931 | |||||||||
| 1B | ODR7 | AAC46497 | RAR-like* | CCD76558 | BgRAR | 398 | LgRAR | 207867 | RARα | AAH08727 | ||
| RARβ | AAH60794 | |||||||||||
| RARγ | AAA63254 | |||||||||||
| 1C | BgPPAR1 | 2052 | LgPPAR1 | 174409 | PPARα | AAB32649 | ||||||
| BgPPAR2 | 1275 | LgPPAR2 | 238472 | PPARβ | AAA36469 | |||||||
| PPARγ | AAH06811 | |||||||||||
| 1D | HR85 | AAO39185 | E75 | AAN11687 | BgE75 | 29 | LgE75 | 136477 | Rev-erb-a | AAH56148 | ||
| BgRev-erb | 160 | LgRev-erb | 168854 | Rev-erb-b | AAH45613 | |||||||
| BgNR1D1 + * | 1939 | |||||||||||
| BgNR1D2 + * | 1958 | |||||||||||
| BgNR1D3 + * | 201 | |||||||||||
| 1E | E78 | AAR30507 | E78 | AAF51692 | BgE78 | 534 | LgE78 | 163301 | ||||
| 1F | HR23 | P41828 | HR3 | AAA28461 | BgHR3 | 73 | LgHR3 | 167096 | RORα | AAH08831 | ||
| BgROR | 74 | LgROR | 155536 | RORβ | AAH93774 | |||||||
| RORγ | AAA64751 | |||||||||||
| 1G | HR14 | AAA96982 | ||||||||||
| 1H | EcR | NP724456 | BgEcR | 1481 | LgEcR | 170342 | LXRα | AAV38218 | ||||
| LXRβ | NP_009052 | |||||||||||
| FXR | AAI30574 | |||||||||||
| 1I | VDR | AAB95155 | ||||||||||
| PXR | AAD05436 | |||||||||||
| CAR | AAY56401 | |||||||||||
| 1J | DAF12 | AAD34462 | HR96α(CAR) * | AAV80235 | HR96 | AAC46928 | BgNR1J1 | 48 | LgNR1J1 | 163618 | ||
| HR8 | AAP31437 | HR96 | AAW88541 | BgNR1J2 | 954 | LgNR1J2 | 63892 | |||||
| HR48 | CAD36502 | BgNR1J3 | 2148 | LgNR1J3 | 163956 | |||||||
| BgNR1J4 | 3233 | |||||||||||
| 1K | HR1 | AAC48174 | ||||||||||
| 2A | HR49 | CAD57702 | HNF4 | CCD80248 | HNF4 | AAF52702 | BgHNF4 | 2702 | LgHNF4 | 73247 | HNF4 | NP_849180 |
| HNF4γ | NP_004124 | |||||||||||
| 2B | RXR1 | CCD59553 | USP | AAF45707 | BgRXR | 3695 | LgRXR | 206562 | RXRα | AAI10999 | ||
| RXR2 | AAD33428 | RXRβ | AAC18599 | |||||||||
| RXRγ | AAA80681 | |||||||||||
| 2C/D | HR41 | NP_500073 | TR | AAV80236 | HR78 | Q24142 | BgTR | 2718 | LgTR* | 231979 | TR2 | AAA36761 |
| TR4 | NP_003289 | |||||||||||
| 2E | FAX | AAD55066 | TLL* | AAW88549 | TLL | AAB71371 | BgTLX* | 3 | LgTLX | 130367 | TLX | AAL05871 |
| HR67 | CAA97428 | PNR* | CCD82728 | PNR | AAF58145 | BgDSF | 42 | LgDSF | 64143 | PNR | AAD28301 | |
| DSF* | AAW88537 | DSF | AAF52303 | BgFAX1 | 148 | LgFAX1 | 168960 | |||||
| FAX1 | AAF54133 | BgPNR ‡ | 3827 | LgNR2E | 232946 | |||||||
| LgPNR | 171552 | |||||||||||
| 2F | UNC55 | CAP16277 | COUP-TFI* | CCD77973 | SVP | NP_524325 | BgCOUP-TFa | 5703 | LgCOUP-TFa | BgCOUP-TFa | COUP-TFα | AAH04154 |
| COUP-TFII* | AAW88536 | COUP-TFβ | AAH42897 | |||||||||
| EAR2 | AAH02669 | |||||||||||
| 3A | BgER | 469 | LgER | 132166 | ERα | AAI28575 | ||||||
| ERβ | AAV31779 | |||||||||||
| 3B | ERR | AAL37554 | BgERR | 751 | LgERR | 168715 | ERRα | AAH63795 | ||||
| ERRβ | AAC99409 | |||||||||||
| ERRγ | EAW93335 | |||||||||||
| 3C | GR | AAH15610 | ||||||||||
| MR | AAA59571 | |||||||||||
| PR | AAA60081 | |||||||||||
| AR | EAX05380 | |||||||||||
| 4A | HR6 | CAA85271 | NR4A5* | AAR28090 | DHR38 | AAF53914 | BgHR38 | 27 | LgHR38 | 94341 | NGFIB | CAG32985 |
| BgNR4a | 2842 | LgNR4a | 182258 | NURR1 | AAH09288 | |||||||
| NOR1 | CAI95138 | |||||||||||
| 5A | HR25 | CAA91028 | FTZ-F1α (SF1)* | CCD77387 | FTZ-F1 | AAN11667 | BgFTZ-F1 | 1318 | LgFTZ-F1 | 196916 | SF1 | AAH32501 |
| LRH1 | AAI18572 | |||||||||||
| 5B | HR39(FTZ-F1) | CCD77680 | DHR39 | AAN11107 | BgHR39 | 3667 | LgHR39 | 94029 | ||||
| 6A | HR91 | CAB60329 | DHR4 | AAX73355 | BgHR4 | 1866 | LgHR4a | 128948 | GCNF | AAB96828 | ||
| LgHR4b | 162354 | |||||||||||
| 2DBDNR | 2DBDα* | AAW88533 | Bg2DBDNR1* | 304 | Lg2DBDNR1* | 168696 | ||||||
| 2DBDβ* | AAW88534 | Bg2DBDNR2* ‡ | 1296 | Lg2DBDNR2* | ||||||||
| 2DBDγ* | AAW88550 | |||||||||||
| Unclassified | BgNRU1 | 221 | ||||||||||
| BgNRU2 | 293 | |||||||||||
| BgNRU3 | 180 | |||||||||||
| BgNRU4* ‡ | 250 |
*NRs not used in phylogenetic tree;
+ NRs are short fragments that could be multiple partial genes or parts of a full transcript;
‡ NRs not found expressed in B. glabrata
Phylogenetic reconstruction was performed using Maximum Likelihood analysis with Bayesian inference and MEGA 6.06. Bayesian inference was conducted with MrBayes version 3.1.2 [59] using the WAG model; the best fitting substitution model determined by both Akaike information criterion and Bayesian information criterion frameworks (ProtTest (v1.4) [60]). Two independent runs of Markov Chain Monte Carlo (MCMC) analysis were performed, with four chains run for 7 million generations with the ‘temperature’ parameter at 0.10, prior probabilities with default values and sampling every 2000 generations. The first 2 million generations were discarded as burn-in because the Log likelihood values were plotted and found to be asymptotic well before the burn-in fraction. Convergence between the independent MCMC runs was examined by the average deviation of the split frequencies and the potential scale-reduction factor (PSRF), which was 1.00. Clades with posterior probabilities >95% were considered well supported. All PSRF values for MrBayes analyses were 1.00. For Maximun Likelihood analysis, the Jones-Taylor-Thornton (JTT) substitution model [61] was used, with a gamma distribution of rates between sites (eight categories, parameter alpha, estimated by the program). Support was evaluated by 1000 bootstrap replicates.
Maximum parsimony was also used to provide additional phylogenetic support for classification, naming, and the phylogenetic relationships observed between B. glabrata, L. gigantea and nuclear receptors from selected species. Maximum parsimony with heuristic searches, branch swapping set to tree-bisection-reconnection, topological constraints not enforced, and 1000 bootstrap replicates were performed using MEGA6.06 [62]. An appropriate out-group to root the sequences was difficult for such a diverse and ancient nuclear receptor family and so the phylogenies were mid-point rooted using Dendroscope (ver3.2.10) [63].
Results and Discussion
Nuclear receptor genes
Thirty nine NR sequences were identified from the genome of B. glabrata and a total of 33 from L. gigantea consisting of 7 newly identified NRs and confirming 26 sequences which had been previously identified from L. gigantea in previous studies [25,48,49] (Table 1). An exhaustive search with mammalian and D. melanogaster NRs and with the identified mollusc NRs found no further NR homologs. Representative genes for each of the major nuclear receptor groups were detected, suggesting the assembled genome for B. glabrata provides a good representation of total gene content with physical genome coverage of approximately 27.5X (Genbank: APKA00000000.1). The numbers found are comparable to the 48 NRs reported in the human genome [64], 49 in mice [65] and 47 in rats [66]. Insects have lower numbers; only 21 NR genes are found in D. melanogaster [67], 22 in Apis mellifera [68], and 21 in Tribolium castaneum [69]. Over 270 NRs have been found in C. elegans [70] but only 21 NRs in the trematode S. mansoni [71]. The unusually large number of NRs in C. elegans is due to extensive proliferation of one gene (HNF4) within the nematode phylum [58]. 43 NRs were recently identified from the Pacific oyster, C. gigas, a marine bivalve, [45], similar numbers to those we identified from the gastropods.
Expression of B. glabrata nuclear receptor mRNAs
Each predicted nuclear receptor was evaluated for expression in B. glabrata adult snails, by amplifying cDNA fragments with receptor-specific primers, to demonstrate that the predicted genes were expressed as transcripts and confirm their sequence. Primer pairs for which fragments were not obtained from adults were also tested on embryos. Expression was confirmed for 34 out of 39 identified receptor genes (Table 1), 31 from cDNA derived from whole adult snails, while 3 nuclear receptors, BgTLX, BgDSF and BgFAX1, were not expressed in adult snails but were identified in 96hrs/120hrs embryos. These receptors belong to NR2E sub family and may be involved in embryonic development as reported in Daphnia pulex [72]. All amplicons corresponded to the predicted sequence length and sequencing confirmed the predicted identity of the gene products (GenBank Accession Nos: JZ390894-JZ390939). For L. gigantea, in silico searches identified sequenced transcripts for 21 out of 33 NR genes (S2 Table). Three B. glabrata putative NRs (BgNR1D1/2/3) for which only partial sequences were obtained from the genome were not included for further phylogenetic analysis, since it was not possible to ascertain if they are multiple partial genes or parts of the same transcript. In addition to this BgTLX, BgNRU4 and LgTR were not analysed further as these were also incomplete sequences. A separate analysis was made of 2DBD genes from B. glabrata and L. gigantea with other previously identified 2DBD sequences from S. mansoni and other species as the 2DBD region could not be aligned with the other NRs.
Phylogenetic analyses
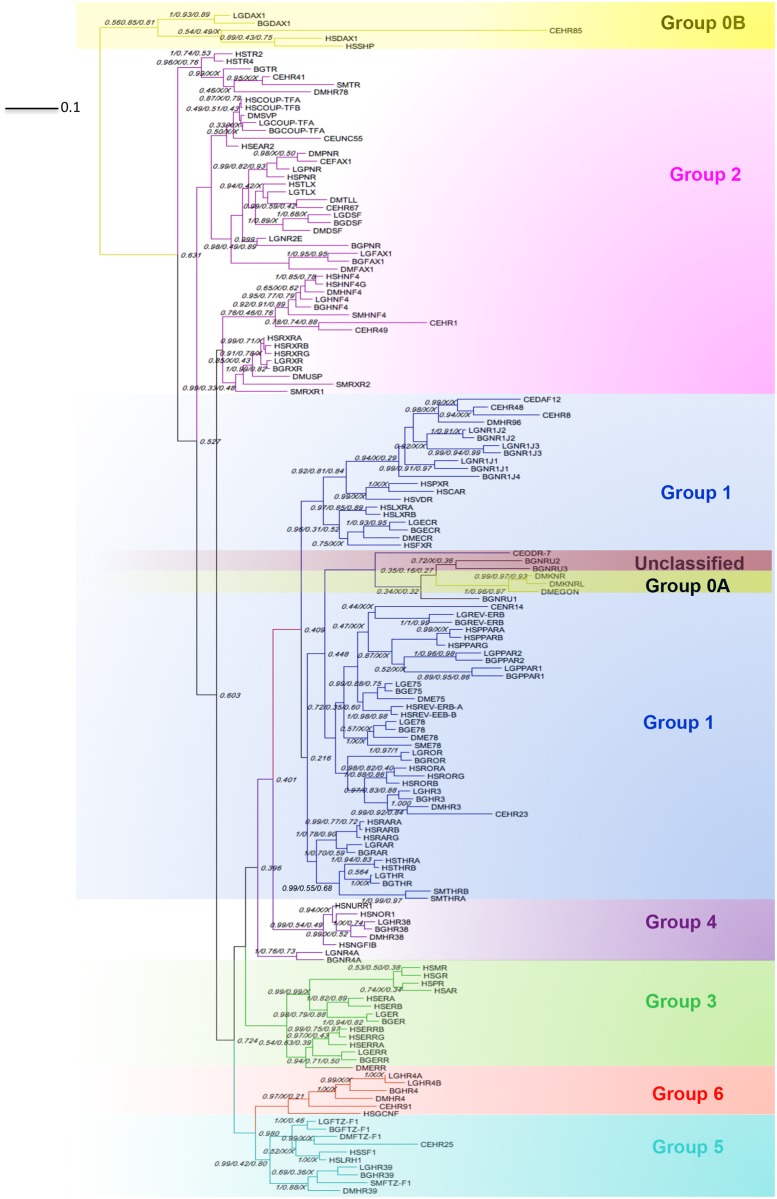
The DBD and LBD regions for the full-length molluscan NRs (32 for B. glabrata and 30 for L. gigantea) were aligned with S. mansoni, D. melanogaster, C. elegans and H. sapiens NR sequences (S1 File). Phylogenetic trees were constructed using 3 different approaches: Bayesian Inference (BI), Maximum Parsimony (MP) and Maximum Likelihood (ML) and the resulting trees assessed for concordance. All trees agree at family and subfamily level (S1 Fig, S2 Fig, S3 Fig) but BI shows the greatest resolution at the base of the phylogram and is shown with nodal support generated from all the tree construction methods (Fig 1). The tree shows the main groupings of the NRs.
Fig 1. Phylogenetic relationships of NRs in molluscs, humans, fly, nematode and trematode.
NRs from six species B. glabrata (Bg), L. gigantea (Lg), H. sapiens (Hs), D. melanogaster (Dm), C. elegans (Ce) and S. mansoni (Sm), were subjected to phylogenetic comparisons using Bayesian inference, maximum parsimony and maximum likelihood methods. The Bayesian tree (midpoint rooted) is shown with posterior probability values from Bayesian inference and bootstrap values from maximum parsimony and maximum likelihood trees. The value of 1 on each node represents 100% posterior probability/bootstrap support; an X indicates an area of disagreement from the Bayesian tree (S1 Fig). Scale bar, 0.1 expected changes/site.
The position of the unclassified B. glabrata NRs (BgNRU) is the same in all trees, close to NR0A subfamily within the NR1 group; however the NR0 subfamily was derived to accommodate nuclear receptors that lack either the DBD or the LBD and these 3 B. glabrata NRs contain both domains (S4 Fig). Therefore these remain as unclassified. The position of members of NR2C/D is also not resolved within Group 2 (Fig 1). Both mollusc genomes each contain one member of this clade that we have designated LgTR/BgTR based on their sequence similarity with HsTR (Table 1). LgTR was not included in the phylogenetic analysis since it was missing the LBD, but BgTR clusters with the other NR2C/D group members Ce_HR41, SmTR and Dm_HR78. The Bayesian tree predicts BgTR as the most distal member of NR2C but the ML and MP trees show both the mollusc sequences to be closely related to DmHR78. However the MP tree placed Ce_HR41 in NR2D, although this NR is usually classified in NR2C. Clearly, the NR2C/D groups seem to be related and not well defined. Within NR1 NR1D was not clearly resolved. In the NR3 subfamily, only the position of the human GR showed non-concordance between the BI tree and the MP tree. All of the trees were in agreement for the NR4/NR5/NR6 subfamilies, with one difference in the position of BgNR4a/LgNR4a and NR5A in the MP tree.
The identified genes have been named in agreement with the unified nomenclature for NRs [23] based on their sequence homology and phylogenetic position. Overall, the identified mollusc NRs encompass almost all the subfamilies, with representatives divided between 21 groups (Table 1) and revealed both similarities and differences in the NR complement between the species used in the study. Seventeen of the 19 subgroups containing human NRs also had representatives in the invertebrate species, including molluscs although in general H. sapiens contained several NRs where snails had only one, most likely the result of gene or lineage-specific duplication events. The presence of these groups among both protostomes and deuterostomes suggests that these receptors originated in a common ancestor of the bilateria [24,25]. The 2 vertebrate groups not represented in molluscs were the Groups 3C and 1I. Three groups that were common to humans and molluscs, but not found in the other invertebrate organisms included here were 0B (DAX), 1C (PPAR) and 3A (ER). The presence of these receptors in molluscs suggests at least the possibility of commonality in shared signalling pathways, elucidating the evolutionary development of the endocrine system. Seven groups were found only in invertebrates; Groups 1G and 1K only in C. elegans; 0A only in D. melanogaster, while groups 1E (E78), 5B (DHR39) 1J and the 2DBD NRs were found in several of the invertebrate species. Representation of these latter groups in L. gigantea and B. glabrata shows that these nuclear receptors pre-date the ecdysozoan/lophotrochozoan split.
Group 3 receptors
The nuclear receptors that bind to steroids, such as androgens, estrogens and progestogens, are responsible for the long-term effects of steroid hormones on reproduction, behaviour, immunity, stress responses and development. The Group 3 steroid receptor family includes the estrogen related receptor (ERR), as well as the estrogen receptor (ER) and group 3C subfamily with the androgen, progesterone, glucocorticoid and mineralocorticoid receptors. Steroid receptors were originally thought to be a vertebrate specific gene family but the identification of the genes with a clear homology to human ERR in trichoplax [73] and an ancestral steroid receptor in amphioxus [74] suggested that these receptors arose early in metazoan evolution and subsequently proliferated in vertebrates through series of gene duplication events [75]. We have identified ER homologues in B. glabrata and L. gigantea (Table 1, Group 3A), which have been previously reported in other molluscan species, including bivalves, C. gigas [37], Mytilus edulis [76] and Chlamys farreri [77] and gastropods, Aplysia californica [38], Lymnaea ollula [78], Marisa cornarietis [33], Thais clavigera [39], Nucella lapillus [79], Bithynia tentaculata [35] and the cephalopod, Octopus vulgaris [36]. The phylogenetic position of the mollusc ER homologue with the vertebrate ER is well supported with a BMCMC posterior probability of 98% and bootstrap values of 0.88 and 0.79 (Fig 1), supporting the suggestion that vertebrate and invertebrate ER diverged from a common ancestor, before the evolution of the deuterostomes [38,75,80]. Structural similarities between ER DBDs of molluscs, annelids, cephalochordates and vertebrates have been documented showing them to bind to and regulate transcription through estrogen response elements [36,40,74,81,82]. The ER LBD in invertebrates may have unique functions, since the mollusc ER homologue, at least in vitro (usually in mammalian reporter cell lines), appears to activate transcription constitutively in the absence of a ligand [36,37,39,40] although in annelids the ER has been shown to activate transcription in the presence of estrogens [82]. We also identified a single estrogen-related receptor (ERR) in both B. glabrata and L. gigantea (Table 1, Group 3B), which clusters with fly and human ERRs. ERRs have been reported previously in molluscs, including M. cornarietis [33], C. gigas (Genbank: EKC20050), Mizuhopecten yessoensis (GenBank: BAN84542.1) and M. edulis [76]. ERRs have no known endogenous ligand, although they are thought to bind to estrogen response elements and may play a role in estrogen signalling and energy metabolism [83].
We identified no convincing homologues in either of the two molluscs for the AR or for any other members of group 3C (Fig 1), which also contains the glucocorticoid receptor (GR), mineralocorticoid receptor (MR) and progesterone receptor (PR). The absence of an AR sequence homologue, both in our systematic exhaustive genomic searches of 2 gastropod species, as well as specific laboratory-based searches in other molluscs [44] using an approach which successfully identified other receptors and another systematic NR survey in the bivalve C. gigas [45] strongly suggests no vertebrate AR homologue exists in gastropod molluscs. The presence of an AR in molluscs has been hotly contested [84], its existence having been inferred from the effects of androgens and anti-androgens in several species of mollusc (eg. [42,85]), rather than by the identification of a homologous sequence. The absence of an AR means that the findings from previous papers requiring its presence to explain their results may require further analysis in the light of this information; although it should also be noted that the effects of steroids, including those reported for androgens in molluscs may not be mediated via NRs. Steroid hormones have also been shown to act via non-genomic mechanisms in vertebrates, using membrane bound receptors from the G protein family [86–88], although homologues for these receptors have also not yet been identified in molluscs.
The absence of a molluscan AR and the constitutive expression of the ER in vitro suggest alternative pathways must exist for spermatogenesis/oogenesis in molluscs and other nuclear receptors have been proposed as initiating these pathways for reproductive processes, some of which exist in vertebrates and which may be particularly important in invertebrates. Although vertebrates have subsequently evolved further processes involving steroid hormones, the presence of orthologous NRs involved in other pathways in molluscs offers the potential opportunity to study these conserved networks in an invertebrate.
Group 1 and 2 nuclear receptors
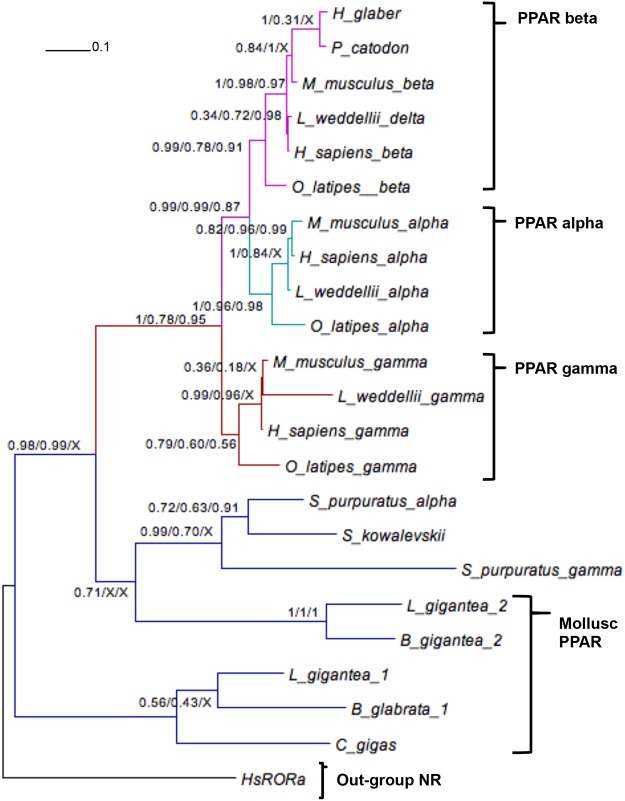
Conservation between other NR-mediated pathways in molluscs and humans, even with the absence of Group 3C NRs, still provides the potential to develop a mollusc model for endocrine physiology. The retinoid X receptor (RXR, Group 2B) has been previously identified in molluscs including B. glabrata, [46], L. gigantea [25] and L. stagnalis [89]. We found that the molluscan RXR sequences grouped with good support (BMCMC posterior probability of 91%) close to the human RXR sequences, with the D. melanogaster ultraspiracle (USP) RXR homologue basal to the clade (Fig 1). BgRXR has been demonstrated to bind and be activated by retinoids, suggesting that retinoid signalling pathways are conserved in the Lophotrochozoa [46]. It was also shown to bind to the response element DR1 either as a homodimer or as a heterodimer with mammalian RARα, LXR, FXR or PPARα [46], which suggests the possibility of conservation of several important signalling pathways in molluscs. The ligand and the co-activator peptide have been previously reported to bind to snail RXR in essentially the same manner as observed in human RXR LBD structures [90], suggesting that the mechanisms of RXR-mediated transcription regulation are very similar in molluscs and humans. The significance of RXR signalling on reproductive physiology in molluscs is strongly supported by the finding that RXR expression is affected during TBT exposure and subsequent development of imposex in N. lapillus [91]; RXR is thought to be the receptor which mediates TBT induction of imposex in this species [29]. It has also been recently demonstrated that, in conjunction with RXR signalling, peroxisome proliferator-activated receptor (PPAR) pathways are activated by organotins [92] and may induce imposex in response to TBT in N. lapillus [93]. Convergence of 9-cis retinoic acid, a natural ligand for RXR, and PPAR signalling pathways through PPAR-RXRα hetero-dimerization is well established in mammals [94]. We identified two PPAR homologues in both L. gigantea and B. glabrata both containing identical sequence to the P-box of human PPAR (CEGCKGFFRRTI) which grouped with the human PPAR genes (Fig 1); however the 3 human genes seem to have diverged after the split with the molluscan paralogues. Examination of PPAR gene relationships in more detail and including some additional species in the phylogenetic analysis, confirms that the 2 molluscan paralogues diverged prior to the split with vertebrates. The genes we have designated PPAR1 lie basal to the clade, while the vertebrate α, β and γ PPAR genes are orthologues of the gene we have designated PPAR2 (Fig 2).
Fig 2. Phylogenetic relationships of peroxisome proliferator-activated receptors (PPAR α, β and γ).
PPAR sequences from different species were subjected to phylogenetic analyses using Bayesian inference, maximum likelihood and maximum parsimony. Accession numbers of the PPARs of the different species in the phylogenetic tree: Heterocephalus glaber XP_004846774; Physeter catodon XP_007109986; Leptonychotes weddellii γ XP_006734113; L. weddellii α XP_006750071; L. weddellii δ XP_006737129; Saccoglossus kowalevskii XP_006819446; Strongylocentrotus purpuratus α XP_781750; S. purpuratus γ XP_784429; C. gigas EKC18691; H. sapiens α AB32649; H. sapiens γ AAH06811; H. sapiens β AAA36469; Mus musculus α NP_035274; M. musculus γ NP_035276; M. musculus β NP_035275; Oryzias latipus α XP_004069934; O. latipus β NP_001265836; O. latipus γ NP_001158348; B. glabrata 1 Contig2052; B. glabrata 2 Contig1275; L. gigantea 1 ProteinID174409; L. gigantea 2 ProteinID238472; H. sapiens ROR AAH0883. Scale bar, 0.1 expected changes/site.
The RXR also binds with the thyroid hormone receptor (THR, Group 1A). Both gastropods contain a THR homologue (Table 1), which has been previously reported in L. gigantea and other invertebrates [49]. Wu et al (2007) confirmed that the S. mansoni THR was able to heterodimerise with the RXR. The molluscan THRs cluster with the human THRα/β (Fig 1). The presence of this NR makes B. glabrata a potential model for thyroid hormone processes. Thyroid hormones (THs) play key roles in growth development and metabolism in vertebrates. In invertebrates exogenous (food) sourced THs have been suggested to be involved in signalling [95].
Ultraspiracle (USP), the fly orthologue of the RXR (reviewed in [96]), has been shown to heterodimerize with the ecdysone receptor (EcR) to control ecdysone signalling in insects driving metamorphosis and moulting [97]. EcR homologues (Group 1H) were identified in both molluscs (Table 1), the first time this has been reported for B. glabrata. The presence of the EcR ligand, ecdysone, has been previously reported in pulmonate snails, in B. glabrata and Lymnaea stagnalis [98], but its origin (endogenous or exogenous) is still contested [99]. Interestingly, ecdysone exposure was found to affect growth and egg production in B. glabrata and it has been suggested that ecdysone secreted from the S. mansoni parasite might provide a mechanism for parasitic castration and gigantism seen in some intermediate hosts when infected [100]. The EcR of L. gigantea has been previously identified and its 3D structure investigated [101] revealing the ligand-binding pocket of the L. gigantea EcR homolog has the potential to bind to ecdysone-related steroids. The observation that EcR is expressed in testis tissue may also indicate a role in molluscan reproductive processes [101].
One retinoic acid receptor (RAR) was identified in each gastropod species. RARs have previously been identified in the gastropod L. stagnalis (GenBank: GU932671), the rock shell, T. clavigera [102], the bivalve, C. gigas [45] and recently in N. lapillus [103]. In vertebrates RAR binds retinoic acid, the biologically active form of vitamin A, which mediates cellular signalling in embryogenic antero-posterior patterning of the central nervous system [104]. However the RAR of N. lapillus did not bind to all-trans retinoic acid or any other retinoid tested nor was it able to activate the transcription of reporter genes in response to stimulation by retinoids [103]. More work is clearly necessary to elucidate the function of RAR in molluscs.
We also identified homologues of Group 2 C/D vertebrate testicular receptor 4 (TR4) in B. glabrata and L. gigantea, (designated as BgTR and LgTR) that in mice have been shown to control spermatogenesis [105] and folliculogenesis [106]. As an orphan nuclear receptor (with no known ligand) the physiological function of TR4 has been difficult to ascertain until the recent development of knock-out mice for this gene (reviewed in [107]). Thus determining the basic biology of vertebrate NRs is dependent on animal experimentation, some of which may be possible in a simplified model system such as a mollusc.
The NR2E nuclear receptors that have been functionally characterized have a common role in nervous system development, for example the tailless (tll) gene of D. melanogaster is involved in embryonic CNS and larval eye development [108] and the mouse Tlx gene has also been found to be a key component of retinal development and vision [109]. In B. glabrata we identified homologues of photoreceptor cell-specific nuclear receptor (PNR), dissatisfaction (DSF), TLX and FAX1, while in L. gigantea, we identified a further NR2E gene (Table 1).
Groups NR1J and NR1I cluster in the tree (Fig 1). No members of the NR1I group, including vitamin D receptor (VDR) or pregnane X receptors (PXR), were identified in B. glabrata and L. gigantea; however NR1J group in protostomes shares similarity with vertebrate NR1I group [110] and there is evidence that both NR1I/NR1J groups share a common ancestor [24]. NR1I receptors are considered as natural sensors and are involved in xenobiotic metabolism in vertebrates [111]; however, other studies have indicated that NR1J members might regulate xenobiotic responses in D. melanogaster and C. elegans [112,113]. Our results show that L. gigantea has three and B. glabrata four NR1J representatives (Table 1). The mollusc NR1J group members possess the well-conserved base contact residues (ESCKAFFR) within the DBD, characteristic of the NR1J sub family [114]. In molluscs the NR1J group receptors may be able to perform the same xenobiotic recognition functions as the closely related NR1I of vertebrates.
Group 5 and 6 nuclear receptors
Germ cell nuclear factor (GCNF) homologues (Group 6) have previously been reported in bilaterians (reviewed in [50]) and we identified them in both gastropod species. This is in contrast to the recently published survey of NRs in the pacific oyster C. gigas [45] which did not identify any NRs from Group 6. Both BgHR4 and LgHR4a/b cluster with good support (posterior probability 1) to the D. melanogaster GCNF homologue, Dm_HR4 (Fig 1). During embryonic stages in vertebrates, GCNF can interfere with retinoic acid signalling affecting the expression of cyp26A1, which is essential for normal hindbrain patterning and early developmental stages [104]. In Group 5 we identified BgFTZ-F1/LgFTZ-F1 and BgHR39/LgHR39, homologues of the D. melanogaster NR5 subfamily members, fushi tarazu factor 1 and hormone receptor 39 respectively, with a conserved stretch of 23 amino acids adjoining the C-terminal end of the zinc finger motif (AVRSDRMRGGRNKFGPMYKRDRA). This sequence is located immediately after the DBD and plays an important role in high affinity interactions of the receptor with DNA [115]. These Group 5 NRs are related to the vertebrate SF-1 which controls reproductive development and regulates the transcription of steroid-modifying cytochrome P450 genes. In Drosophila HR39 is essential for sexual development, required in females both to activate spermathecal secretion and repress male-specific courtship genes, and controls the expression of specific cytochrome P450 genes [116]. The conservation of function between invertebrates and vertebrates in these receptors in Groups NR5 and NR6, suggests the potential for molluscs to be used to model some aspects of mammalian reproductive biology and that further study into the reproductive biology of invertebrates is warranted.
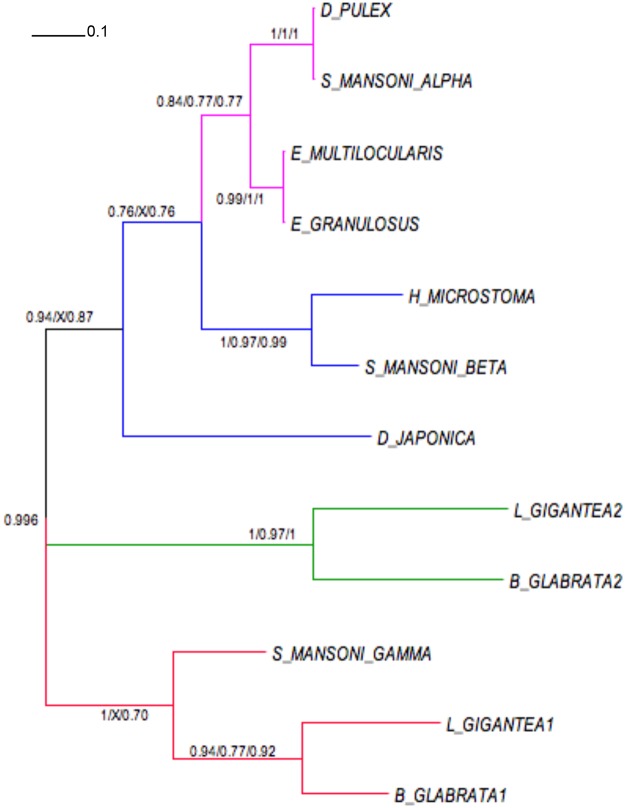
2DBD nuclear receptors
We identified two 2DBD-NRs in B. glabrata as well as the two 2DBD-NRs previously reported from L. gigantea [48]. We compared these to the 3 previously identified from S. mansoni [71] as well as 2DBD NRs from other organisms (Fig 3). The gastropod 2DBD1 clusters with Sm2DBDγ while the position of the second gastropod 2DBD was not determined within the clade. Both the identified Bg 2DBD-NRs possess the two tandem DNA binding domains, and a well-conserved P-box sequence (CEACKK) in the first DBD and a P-box (CEGCK) in the second DBD. The P-box of the second DBD is similar to DBD of NR1 family but the P-box of the first DBD is unique to this NR and may determine novel targets [48].
Fig 3. Phylogenetic relationships of 2DBD nuclear receptors (alpha, beta and gamma).
2DBD NR sequences from different species were subjected to phylogenetic analyses using Bayesian inference, maximum likelihood and maximum parsimony. Accession numbers of the 2DBD NRs of the different species in the phylogenetic tree: D. pulex FE382753; Hymenolepis microstoma CDS31978; Echinococcus multilocularis CDI99377; Echinococcus granulosus CDS24085; S. mansoni α AAW88533; S. mansoni β AAW88534; S. mansoni γ AAW88550; Dugesia japonica BP186725; L. gigantea 2DBD1 ProteinID168696; L. gigantea 2DBD2 ProteinID173632; B. glabrata 2DBD1 Contig304; B. glabrata 2DBD2 Contig1296. Scale bar, 0.1 expected changes/site.
NRs in parasite and intermediate host
The discovery of differences and similarities in the nuclear receptor repertoire of snail host and S. mansoni may open up avenues to further characterize host-parasite interactions and potentially to interfere with schistosome development within the host. Parasites are known to interfere with the snail reproductive system, since one effect of trematode infection in snails is parasitic castration (reviewed in [117]). The intimate host-parasite and intermediate host-parasite relationships, makes a comparison of nuclear receptors in all three organisms significant. It has been suggested, at least for the definitive host, that the ability of the parasite to exploit the hosts’ hormonal microenvironment may be critical to allow it to establish, grow and reproduce [118]. This may also be significant in enabling the parasite to inhabit its intermediate host and may depend upon shared nuclear receptors. All 3 organisms have one or more THR, RAR, HNF4, RXR, TR, COUP-TF and similar NRs in Groups 2E, 4A and 5A. Only snails and schistosomes have the E78, HR39, HR96 and 2DBD NRs, while snails and humans only have DAX1, PPAR, Rev-erb, ROR, ER, ERR and group 6A NRs. NRs which are specific to the snails (in this three-way comparison) are the EcR and the group of unclassified NRs, designated as BgNRU1, BgNRU2, BgNRU3, and BgNRU4 respectively and placed in a separate group (Table 1). These unclassified receptors may have originated from a specific duplication event in a B. glabrata precursor, or alternatively, they could be remnants of ancient NR subfamilies, whose representatives have been secondarily lost in the other represented species. With regard to the EcR, given that snails have a EcR homologue and schistosomes do not, the suggestion that β-ecdysterone acts as an attractant in host location by miracidia and affects the rate of growth and maturation in snails [100] is potentially interesting, although the ability of snails to make ecdysone is not proven [99]. Both organisms have homologues of the D. melanogaster NRs E78 and HR39 which are active in ecdysone signalling [116,119].
In addition to discovering the intricacies of the molecular interactions between host and parasite with a view to disrupting the development of the schistosome, the other key strategy for schistosomiasis control is to reduce the numbers and/or distribution of the intermediate host snails. Currently molluscicides, such as the commercially available niclosamide-based Bayluscide, are effectively employed in schistosomiasis control programs; however the problems of resistance and toxicity to other organisms means that the search for alternative, more selective, compounds is on-going. The capacity of nuclear receptors to bind small ligands, including exogenous substances such as natural products and synthetic chemicals, makes them potential targets for molluscicides [120].
Biomphalaria glabrata as a model organism
There is a long tradition in biology of examining biochemical processes in simplified models to elucidate mechanisms in more complex organisms. We have demonstrated that there are some NR groups with a single representative in the mollusc species’ examined instead of the multiple genes from humans. For example in the NR1 subfamily, both molluscs have single homologues of several significant NRs, such as the retinoic acid receptor (RAR), THR and ROR. For these groups, B. glabrata could provide a simple model system, not only to study the development and diversification of endocrine systems, but also to investigate and test gene function and response to external stimuli. The identification of many vertebrate-like NRs in B. glabrata could make it a suitable model candidate to investigate functional relationships of individual receptors. The ultimate choice of model organisms lies not only with the biology of the organism, but also on its tractability for study and manipulation. Aside from the genome, the full range of genomic tools available for B. glabrata, such as RNAi [8,121], BAC library [5] and microarrays [6,122,123] as well as the potential to use classical approaches of enhancer and suppressor genetics and transgenics to explore regulatory networks, make B. glabrata a good candidate. However, the advent of next generation sequencing makes transcriptome study from non-model organisms a real possibility and therefore other mollusc species may ultimately make better model systems. In particular, this approach could be used to identify inputs from signal transduction pathways, potential hormone metabolic genes, co-activators, co-repressors and other unknown factors that may impinge upon receptor activity. Based on the absence of Group3C NRs, it can be concluded that B. glabrata is an inappropriate model for mammalian steroid hormone function mediated via NRs, as the genes for several major steroid hormone receptors are not present. However since steroid hormones have also been shown to act via non-genomic mechanisms in vertebrates, using membrane bound receptors (mPR and GPR30) [87] there is still potential to examine alternative pathways in these organisms.
Conclusions
We identified 39 nuclear receptor genes in B. glabrata and 33 in L. gigantea representing all seven principal vertebrate nuclear receptor groups. Molluscan endocrinology is poorly understood, so further study to determine the functionality of these identified NRs promises new insights, especially concerning the many unanswered questions regarding the effects of steroids and other EDCs on molluscs. The importance of the snail as an intermediate host for schistosomes, also justifies further investigation into the function of such genes. The absence of vertebrate NRs such as VDR, CAR and PXR, as well as steroid hormone NRs, AR/PR/GR/MR for which we found no orthologues in the two mollusc species examined in this study, indicates that several significant signalling pathways are absent in gastropods. Nevertheless, we have identified an array of NRs common to both vertebrates/molluscs and molluscs/flies. These results add weight to Thummel’s speculation about the convergent regulation of NRs in vertebrates and invertebrates [124]. Elucidation of NR targets in molluscs may unlock their potential as new model organisms allowing the discovery of new pathways leading to similar phenotypes found in vertebrates or, indeed, similar pathways that produce a different phenotype; both of which could potentially form simple test assays, for determining gene function or drug/chemical testing. The range of phenotypes, and their underlying genetic mechanisms, available for study in different species may enable the identification of alternative pathways mediated by NRs that might also be exploited [125]. The potential of many invertebrate species for endocrine study is yet to be explored, but the underlying fundamental similarities and differences between molluscs and vertebrates may be the solution to determining not only the endocrine mechanisms of molluscs but also the full intricacies of our own.
Supporting Information
The NRs from six different species were subjected to phylogenetic comparisons using Bayesian inference. The Bayesian tree (midpoint rooted) with posterior probability values is shown. Notations Bg, Lg, Hs, Dm, Ce and Sm in association with receptor names denote sequences from B. glabrata, L. gigantea, H. sapiens, D. melanogaster, C. elegans and S. mansoni respectively.
(PDF)
The nuclear receptors from six different species were subjected to phylogenetic comparison using maximum likelihood method using Jones-Taylor-Thornton (JTT) substitution model. Node labels indicate bootstrap values. Notations Bg, Lg, Hs, Dm, Sm and Ce in association with the receptor name denote sequences from B. glabrata, L. gigantea, H. sapiens, D. melanogaster, S, mansoni and C. elegans, respectively.
(PDF)
The nuclear receptors from six different species were subjected to phylogenetic comparison using maximum parsimony. Node labels indicate bootstrap values. Notations Bg, Lg, Hs, Dm, Sm and Ce in association with the receptor name denote sequences from B. glabrata, L. gigantea, H. sapiens, D. melanogaster, S. mansoni and C. elegans, respectively.
(PDF)
(PDF)
Notations Bg, Lg, Hs, Dm, Ce and Sm in association with receptor names denote sequences from B. glabrata, L. gigantea, H. sapiens, D. melanogaster, C. elegans, and S. mansoni respectively.
(TXT)
Forward and reverse primer sequences with their product size (as submitted to GenBank), their corresponding GenBank accession numbers and their optimised annealing temperatures.
(DOCX)
Detailed summary of all the top BLAST expressed sequence tag (EST) hits for L. gigantea NRs, including the EST description, GenBank accession number and E-value.
(PDF)
Acknowledgments
We thank the B. glabrata Genome Initiative steering committee for their approval to publish this analysis from the unannotated B. glabrata genome (version 4.01).
Data Availability
All sequence data is available either in the supporting information (predicted protein sequence) or available from GenBank (sequenced fragments: Accession numbers: JZ390894-JZ390939).
Funding Statement
This work was funded by the National Centre for the Replacement, Refinement and Reduction of Animals in Research, Grant Ref:G0900802 to CSJ, LRN, SJ & EJR [www.nc3rs.org.uk]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Chitsulo L, Loverde R, Engels D, Barakat R, Colley D, Cioli D, et al. Schistosomiasis. Nat Rev Microbiol. 2004;2: 12–13. [DOI] [PubMed] [Google Scholar]
- 2. Lockyer AE, Spinks J, Noble LR, Rollinson D, Jones CS. Identification of genes involved in interactions between Biomphalaria glabrata and Schistosoma mansoni by suppression subtractive hybridization. Mol Biochem Parasitol. 2007;151: 18–27. 10.1016/j.molbiopara.2006.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lockyer AE, Spinks JN, Walker AJ, Kane RA, Noble LR, Rollinson D, et al. Biomphalaria glabrata transcriptome: Identification of cell-signalling, transcriptional control and immune-related genes from open reading frame expressed sequence tags (ORESTES). Dev Comp Immunol. 2007;31: 763–782. 10.1016/j.dci.2006.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mitta G, Galinier R, Tisseyre P, Allienne JF, Girerd-Chambaz Y, Guillou F, et al. Gene discovery and expression analysis of immune-relevant genes from Biomphalaria glabrata hemocytes. Dev Comp Immunol. 2005;29: 393–407. [DOI] [PubMed] [Google Scholar]
- 5. Adema CM, Luo MZ, Hanelt B, Hertel LA, Marshall JJ, Zhang SM, et al. A bacterial artificial chromosome library for Biomphalaria glabrata, intermediate snail host of Schistosoma mansoni . Mem Inst Oswaldo Cruz. 2006;101: 167–177. [DOI] [PubMed] [Google Scholar]
- 6. Lockyer AE, Emery AM, Kane RA, Walker AJ, Mayer CD, Mitta G, et al. Early differential gene expression in haemocytes from resistant and susceptible Biomphalaria glabrata strains in response to Schistosoma mansoni . PLoS One. 2012;7: e511020 10.1371/journal.pone.0051102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Adema CM, Hanington PC, Lun C-M, Rosenberg GH, Aragon AD, Stout BA, et al. Differential transcriptomic responses of Biomphalaria glabrata (Gastropoda, Mollusca) to bacteria and metazoan parasites, Schistosoma mansoni and Echinostoma paraensei (Digenea, Platyhelminthes). Mol Immunol. 2010;47: 849–860. 10.1016/j.molimm.2009.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jiang YG, Loker ES, Zhang SM. In vivo and in vitro knockdown of FREP2 gene expression in the snail Biomphalaria glabrata using RNA interference. Dev Comp Immunol. 2006;30: 855–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hanington PC, Forys MA, Dragoo JW, Zhang S-M, Adema CM, Loker ES. Role for a somatically diversified lectin in resistance of an invertebrate to parasite infection. Proc Natl Acad Sci USA. 2010;107: 21087–21092. 10.1073/pnas.1011242107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Raghavan N, Knight M. The snail (Biomphalaria glabrata) genome project. Trends Parasitol. 2006;22: 148–151. [DOI] [PubMed] [Google Scholar]
- 11. Ford ES, Li C, Zhao G, Pearson WS, Mokdad AH. Prevalence of the metabolic syndrome among US adolescents using the definition from the International Diabetes Federation. Diabetes Care. 2008;31: 587–589. [DOI] [PubMed] [Google Scholar]
- 12. Golden SH, Robinson KA, Saldanha I, Anton B, Ladenson PW. Prevalence and incidence of endocrine and metabolic disorders in the United States: a comprehensive review. J Clin Endocrinol Metab. 2009;94: 1853–1878. 10.1210/jc.2008-2291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schug TT, Janesick A, Blumberg B, Heindel JJ. Endocrine disrupting chemicals and disease susceptibility. J Steroid Biochem Mol Biol. 2011;127: 204–215. 10.1016/j.jsbmb.2011.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schiffelers M-JWA, Blaauboer BJ, Bakker WE, Beken S, Hendriksen CFM, Koeter HBWM, et al. Regulatory acceptance and use of 3R models for pharmaceuticals and chemicals: Expert opinions on the state of affairs and the way forward. Regul Toxicol Pharmacol. 2014;69: 41–48. 10.1016/j.yrtph.2014.02.007 [DOI] [PubMed] [Google Scholar]
- 15. Matthiessen P. An assessment of endocrine disruption in mollusks and the potential for developing internationally standardized mollusk life cycle test guidelines. Integr Environ Assess Manag. 2008;4: 274–284. 10.1897/IEAM_2008-003.1 [DOI] [PubMed] [Google Scholar]
- 16. Sladek FM. What are nuclear receptor ligands? Mol Cell Endocrinol. 2011;334: 3–13. 10.1016/j.mce.2010.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tallarico L de F, Borrely SI, Hamada N, Grazeffe VS, Ohlweiler FP, Okazaki K, et al. Developmental toxicity, acute toxicity and mutagenicity testing in freshwater snails Biomphalaria glabrata (Mollusca: Gastropoda) exposed to chromium and water samples. Ecotoxicol Environ Saf. 2014;110: 208–215. 10.1016/j.ecoenv.2014.09.005 [DOI] [PubMed] [Google Scholar]
- 18. Tyler C, Jobling S, Sumpter J. Endocrine disruption in wildlife: A critical review of the evidence. Crit Rev Toxicol. 1998;28: 319–361. 10.1080/10408449891344236 [DOI] [PubMed] [Google Scholar]
- 19. Pawlak M, Lefebvre P, Staels B. General molecular biology and architecture of nuclear receptors. Curr Top Med Chem. 2012;12: 486–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14: 121–141. [PubMed] [Google Scholar]
- 21. Aranda A, Pascual A. Nuclear hormone receptors and gene expression. Physiol Rev. 2001;81: 1269–1304. [DOI] [PubMed] [Google Scholar]
- 22. Wurtz J-M, Bourguet W, Renaud J-P, Vivat V, Chambon P, Moras D, et al. A canonical structure for the ligand-binding domain of nuclear receptors. Nat Struct Mol Biol. 1996;3: 87–94. [DOI] [PubMed] [Google Scholar]
- 23. Auwerx J, Baulieu E, Beato M, Becker-Andre M, Burbach PH, Camerino G, et al. A unified nomenclature system for the nuclear receptor superfamily. Cell. 1999;97: 161–163. [DOI] [PubMed] [Google Scholar]
- 24. Bertrand S, Brunet FG, Escriva H, Parmentier G, Laudet V, Robinson-Rechavi M. Evolutionary genomics of nuclear receptors: from twenty-five ancestral genes to derived endocrine systems. Mol Biol Evol. 2004;21: 1923–1937. [DOI] [PubMed] [Google Scholar]
- 25. Bridgham JT, Eick GN, Larroux C, Deshpande K, Harms MJ, Gauthier ME, et al. Protein evolution by molecular tinkering: diversification of the nuclear receptor superfamily from a ligand-dependent ancestor. PLoS Biol. 2010;8: e1000497 10.1371/journal.pbio.1000497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Markov GV, Laudet V. Origin and evolution of the ligand-binding ability of nuclear receptors. Mol Cell Endocrinol. 2011;334: 21–30. 10.1016/j.mce.2010.10.017 [DOI] [PubMed] [Google Scholar]
- 27. Matthiessen P, Gibbs P. Critical appraisal of the evidence for tributyltin-mediated endocrine disruption in mollusks. Environ Toxicol Chem. 1998;17: 37–43. [Google Scholar]
- 28. Oehlmann J, Schulte-Oehlmann U. Endocrine disruption in invertebrates. Pure Appl Chem. 2003;75: 2207–2218. [Google Scholar]
- 29. Castro LFC, Lima D, Machado A, Melo C, Hiromori Y, Nishikawa J, et al. Imposex induction is mediated through the Retinoid X Receptor signalling pathway in the neogastropod Nucella lapillus . Aquat Toxicol. 2007;85: 57–66. [DOI] [PubMed] [Google Scholar]
- 30. Sternberg RM, Gooding MP, Hotchkiss AK, LeBlanc GA. Environmental-endocrine control of reproductive maturation in gastropods: implications for the mechanism of tributyltin-induced imposex in prosobranchs. Ecotoxicology. 2010;19: 4–23. 10.1007/s10646-009-0397-z [DOI] [PubMed] [Google Scholar]
- 31. Lafont R, Mathieu M. Steroids in aquatic invertebrates. Ecotoxicology. 2007;16: 109–130. 10.1007/s10646-006-0113-1 [DOI] [PubMed] [Google Scholar]
- 32. Koehler H-R, Kloas W, Schirling M, Lutz I, Reye AL, Langen J-S, et al. Sex steroid receptor evolution and signalling in aquatic invertebrates. Ecotoxicology. 2007;16: 131–143. 10.1007/s10646-006-0111-3 [DOI] [PubMed] [Google Scholar]
- 33. Bannister R, Beresford N, May D, Routledge EJ, Jobling S, Rand-Weaver M. Novel estrogen receptor-related transcripts in Marisa cornuarietis; a freshwater snail with reported sensitivity to estrogenic chemicals. Environ Sci Technol. 2007;41 10.1021/es062565m [DOI] [PubMed] [Google Scholar]
- 34. Stange D, Sieratowicz A, Horres R, Oehlmann J. Freshwater mudsnail (Potamopyrgus antipodarum) estrogen receptor: Identification and expression analysis under exposure to (xeno-) hormones. Ecotoxicol Environ Saf. 2012;75: 94–101. 10.1016/j.ecoenv.2011.09.003 [DOI] [PubMed] [Google Scholar]
- 35. Hultin CL, Hallgren P, Persson A, Hansson MC. Identification of an estrogen receptor gene in the natural freshwater snail Bithynia tentaculata . Gene. 2014;540: 26–31. 10.1016/j.gene.2014.02.039 [DOI] [PubMed] [Google Scholar]
- 36. Keay J, Bridgham JT, Thornton JW. The Octopus vulgaris estrogen receptor is a constitutive transcriptional activator: Evolutionary and functional implications. Endocrinology. 2006;147: 3861–3869. 10.1210/en.2006-0363 [DOI] [PubMed] [Google Scholar]
- 37. Matsumoto T, Nakamura AM, Mori K, Akiyama I, Hirose H, Takahashi Y. Oyster estrogen receptor: cDNA cloning and immunolocalization. Gen Comp Endocrinol. 2007;151: 195–201. 10.1016/j.ygcen.2007.01.016 [DOI] [PubMed] [Google Scholar]
- 38. Thornton JW, Need E, Crews D. Resurrecting the ancestral steroid receptor: Ancient origin of estrogen signaling. Science. 2003;301: 1714–1717. 10.1126/science.1086185 [DOI] [PubMed] [Google Scholar]
- 39. Kajiwara M, Kuraku S, Kurokawa T, Kato K, Toda S, Hirose H, et al. Tissue preferential expression of estrogen receptor gene in the marine snail, Thais clavigera . Gen Comp Endocrinol. 2006;148: 315–326. 10.1016/j.ygcen.2006.03.016 [DOI] [PubMed] [Google Scholar]
- 40. Bannister R, Beresford N, Granger DW, Pounds NA, Rand-Weaver M, White R, et al. No substantial changes in estrogen receptor and estrogen-related receptor orthologue gene transcription in Marisa cornuarietis exposed to estrogenic chemicals. Aquat Toxicol. 2013;140: 19–26. 10.1016/j.aquatox.2013.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Markov GV, Tavares R, Dauphin-Villemant C, Demeneix BA, Baker ME, Laudet V. Independent elaboration of steroid hormone signaling pathways in metazoans. Proc Natl Acad Sci USA. 2009;106: 11913–11918. 10.1073/pnas.0812138106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Omran NE-SE-S. Testosterone, gonadotropins and androgen receptor during spermatogenesis of Biomphalaria alexandrina snails (Pulmonata: Basommatophora). Reprod Biol. 2012;12: 301–308. 10.1016/j.repbio.2012.10.002 [DOI] [PubMed] [Google Scholar]
- 43. Scott AP. Do mollusks use vertebrate sex steroids as reproductive hormones? Part I: Critical appraisal of the evidence for the presence, biosynthesis and uptake of steroids. Steroids. 2012;77: 1450–1468. 10.1016/j.steroids.2012.08.009 [DOI] [PubMed] [Google Scholar]
- 44. Sternberg RM, Hotchkiss AK, LeBlanc GA. The contribution of steroidal androgens and estrogens to reproductive maturation of the eastern mud snail Ilyanassa obsoleta . Gen Comp Endocrinol. 2008;156: 15–26. 10.1016/j.ygcen.2007.12.002 [DOI] [PubMed] [Google Scholar]
- 45. Vogeler S, Galloway TS, Lyons BP, Bean TP. The nuclear receptor gene family in the Pacific oyster, Crassostrea gigas, contains a novel subfamily group. BMC Genomics. 2014;15: 369 10.1186/1471-2164-15-369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bouton D, Escriva H, De Mendonca RL, Glineur C, Bertin B, Noel C, et al. A conserved retinoid X receptor (RXR) from the mollusk Biomphalaria glabrata transactivates transcription in the presence of retinoids. J Mol Endocrinol. 2005;34 10.1677/jme.1.01766 [DOI] [PubMed] [Google Scholar]
- 47. Simakov O, Marletaz F, Cho S-J, Edsinger-Gonzales E, Havlak P, Hellsten U, et al. Insights into bilaterian evolution from three spiralian genomes. Nature. 2013;493: 526–531. 10.1038/nature11696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wu W, Niles EG, Hirai H, LoVerde PT. Evolution of a novel subfamily of nuclear receptors with members that each contain two DNA binding domains. BMC Evol Biol. 2007;7: 27 10.1186/1471-2148-7-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wu W, Niles EG, Loverde PT. Thyroid hormone receptor orthologues from invertebrate species with emphasis on Schistosoma mansoni . BMC Evol Biol. 2007;7: 150 10.1186/1471-2148-7-150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Enmark E, Gustafsson J-Å. Comparing nuclear receptors in worms, flies and humans. Trends Pharmacol Sci. 2001;22: 611–615. [DOI] [PubMed] [Google Scholar]
- 51. Wu W, LoVerde PT. Nuclear hormone receptors in parasitic helminths. Mol Cell Endocrinol. 2011;334: 56–66. 10.1016/j.mce.2010.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic Local Alignment Search Tool. J Mol Biol. 1990;215: 403–410. [DOI] [PubMed] [Google Scholar]
- 53. Burge CB, Karlin S. Finding the genes in genomic DNA. Curr Opin Struct Biol. 1998;8: 346–354. [DOI] [PubMed] [Google Scholar]
- 54. Hertel LA, Adema CM, Loker ES. Differential expression of FREP genes in two strains of Biomphalaria glabrata following exposure to the digenetic trematodes Schistosoma mansoni and Echinostoma paraensei . Dev Comp Immunol. 2005;29: 295–303. [DOI] [PubMed] [Google Scholar]
- 55. Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, et al. The Pfam protein families database. Nucleic Acids Res. 2004;32: D138–D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13: 2129–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 2006;34: 242–244. 10.1093/nar/gkj079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Robinson-Rechavi M, Maina CV, Gissendanner CR, Laudet V, Sluder A. Explosive lineage-specific expansion of the orphan nuclear receptor HNF4 in nematodes. J Mol Evol. 2005;60: 577–586. [DOI] [PubMed] [Google Scholar]
- 59. Ronquist F, Huelsenbeck J. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- 60. Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21: 2104–2105. 10.1093/bioinformatics/bti263 [DOI] [PubMed] [Google Scholar]
- 61. Jones D, Taylor W, Thornton J. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8: 275–282. [DOI] [PubMed] [Google Scholar]
- 62. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 2013;30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Huson DH, Richter DC, Rausch C, Dezulian T, Franz M, Rupp R. Dendroscope: An interactive viewer for large phylogenetic trees. BMC Bioinformatics. 2007;8: 460 10.1186/1471-2105-8-460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Robinson-Rechavi M, Carpentier AS, Duffraisse M, Laudet V. How many nuclear hormone receptors are there in the human genome? Trends Genet. 2001;17: 554–556. 10.1016/S0168-9525(01)02417-9 [DOI] [PubMed] [Google Scholar]
- 65. Robinson-Rechavi M, Garcia HE, Laudet V. The nuclear receptor superfamily. J Cell Sci. 2003;116: 585–586. [DOI] [PubMed] [Google Scholar]
- 66. Zhang ZD, Burch PE, Cooney AJ, Lanz RB, Pereira FA, Wu JQ, et al. Genomic analysis of the nuclear receptor family: New insights into structure, regulation, and evolution from the rat genome. Genome Res. 2004;14 10.1101/gr.2160004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, et al. The genome sequence of Drosophila melanogaster . Science. 2000;287: 2185–2195. [DOI] [PubMed] [Google Scholar]
- 68. Velarde RA, Robinson GE, Fahrbach SE. Nuclear receptors of the honey bee: annotation and expression in the adult brain. Insect Mol Biol. 2006;15: 583–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bonneton F, Chaumot A, Laudet V. Annotation of Tribolium nuclear receptors reveals an increase in evolutionary rate of a network controlling the ecdysone cascade. Insect Biochem Mol Biol. 2008;38: 416–429. 10.1016/j.ibmb.2007.10.006 [DOI] [PubMed] [Google Scholar]
- 70. Sluder AE, Maina CV. Nuclear receptors in nematodes: themes and variations. Trends Genet. 2001;17: 206–213. [DOI] [PubMed] [Google Scholar]
- 71. Wu W, Niles EG, El-Sayed N, Berriman M, LoVerde PT. Schistosoma mansoni (Platyhelminthes, Trematoda) nuclear receptors: Sixteen new members and a novel subfamily. Gene. 2006;366: 303–315. 10.1016/j.gene.2005.09.013 [DOI] [PubMed] [Google Scholar]
- 72. Thomson SA, Baldwin WS, Wang YH, Kwon G, LeBlanc GA. Annotation, phylogenetics, and expression of the nuclear receptors in Daphnia pulex . BMC Genomics. 2009;10: 500 10.1186/1471-2164-10-500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Baker ME. Trichoplax, the simplest known animal, contains an estrogen-related receptor but no estrogen receptor: Implications for estrogen receptor evolution. Biochem Biophys Res Commun. 2008;375: 623–627. 10.1016/j.bbrc.2008.08.047 [DOI] [PubMed] [Google Scholar]
- 74. Katsu Y, Kubokawa K, Urushitani H, Iguchi T. Estrogen-dependent transactivation of amphioxus steroid hormone receptor via both estrogen and androgen response elements. Endocrinology. 2010;151: 639–648. 10.1210/en.2009-0766 [DOI] [PubMed] [Google Scholar]
- 75. Baker ME. Co-evolution of steroidogenic and steroid-inactivating enzymes and adrenal and sex steroid receptors. Mol Cell Endocrinol. 2004;215: 55–62. [DOI] [PubMed] [Google Scholar]
- 76. Puinean AM, Labadie P, Hill EM, Osada M, Kishida M, Nakao R, et al. Laboratory exposure to 17 beta-estradiol fails to induce vitellogenin and estrogen receptor gene expression in the marine invertebrate Mytilus edulis . Aquat Toxicol. 2006;79: 376–383. 10.1016/j.aquatox.2006.07.006 [DOI] [PubMed] [Google Scholar]
- 77. Zhang H, Pan L, Zhang L. Molecular cloning and characterization of estrogen receptor gene in the scallop Chlamys farreri: Expression profiles in response to endocrine disrupting chemicals. Comp Biochem Physiol C-Toxicol Pharmacol. 2012;156: 51–57. 10.1016/j.cbpc.2012.03.007 [DOI] [PubMed] [Google Scholar]
- 78. Kumkate S, Onmek N, Boonburapong B, Singhakaew S, Leardkamolkarn V. Estrogen-related fecundity reduction of Lymnaea ollula following Fasciola gigantica infection. J Parasitol. 2009;95: 1391–1396. 10.1645/GE-2080.1 [DOI] [PubMed] [Google Scholar]
- 79. Castro LFC, Melo C, Guillot R, Mendes I, Queiros S, Lima D, et al. The estrogen receptor of the gastropod Nucella lapillus: Modulation following exposure to an estrogenic effluent? Aquat Toxicol. 2007;84: 465–468. 10.1016/j.aquatox.2007.07.008 [DOI] [PubMed] [Google Scholar]
- 80. Eick GN, Thornton JW. Evolution of steroid receptors from an estrogen-sensitive ancestral receptor. Mol Cell Endocrinol. 2011;334: 31–38. 10.1016/j.mce.2010.09.003 [DOI] [PubMed] [Google Scholar]
- 81. Bridgham JT, Brown JE, Rodríguez-Marí A, Catchen JM, Thornton JW. Evolution of a new function by degenerative mutation in cephalochordate steroid receptors. PLoS Genet. 2008;4: e1000191 10.1371/journal.pgen.1000191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Keay J, Thornton JW. Hormone-activated estrogen receptors in annelid invertebrates: Implications for evolution and endocrine disruption. Endocrinology. 2009;150: 1731–1738. 10.1210/en.2008-1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Bardet PL, Laudet V, Vanacker JM. Studying non-mammalian models? Not a fool’s ERRand! Trends Endocrinol Metab. 2006;17: 166–171. 10.1016/j.tem.2006.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Scott AP. Do mollusks use vertebrate sex steroids as reproductive hormones? Part II. Critical review of the evidence that steroids have biological effects. Steroids. 2013;78: 268–281. 10.1016/j.steroids.2012.11.006 [DOI] [PubMed] [Google Scholar]
- 85. Tillmann M, Schulte-Oehlmann U, Duft M, Markert B, Oehlmann J. Effects of endocrine disruptors on prosobranch snails (Mollusca: Gastropoda) in the laboratory. Part III: Cyproterone acetate and vinclozolin as antiandrogens. Ecotoxicology. 2001;10: 373–388. 10.1023/A:1012279231373 [DOI] [PubMed] [Google Scholar]
- 86. Thomas P, Dong J. Binding and activation of the seven-transmembrane estrogen receptor GPR30 by environmental estrogens: a potential novel mechanism of endocrine disruption. J Steroid Biochem Mol Biol. 2006;102: 175–179. [DOI] [PubMed] [Google Scholar]
- 87. Levin ER. Rapid signaling by steroid receptors. Am J Physiol Regul Integr Comp Physiol. 2008;295: R1425–R1430. 10.1152/ajpregu.90605.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Simoncini T, Genazzani AR. Non-genomic actions of sex steroid hormones. Eur J Endocrinol. 2003;148: 281–292. [DOI] [PubMed] [Google Scholar]
- 89. Carter CJ, Farrar N, Carlone RL, Spencer GE. Developmental expression of a molluscan RXR and evidence for its novel, nongenomic role in growth cone guidance. Dev Biol. 2010;343: 124–137. 10.1016/j.ydbio.2010.03.023 [DOI] [PubMed] [Google Scholar]
- 90. De Groot A, De Rosny E, Juillan-Binard C, Ferrer JL, Laudet V, Pierce RJ, et al. Crystal structure of a novel tetrameric complex of agonist-bound ligand-binding domain of Biomphalaria glabrata retinoid X receptor. J Mol Biol. 2005;354: 841–853. [DOI] [PubMed] [Google Scholar]
- 91. Lima D, Reis-Henriques MA, Silva R, Santos AI, Castro LFC, Santos MM. Tributyltin-induced imposex in marine gastropods involves tissue-specific modulation of the retinoid X receptor. Aquat Toxicol. 2011;101: 221–227. 10.1016/j.aquatox.2010.09.022 [DOI] [PubMed] [Google Scholar]
- 92. Le Maire A, Grimaldi M, Roecklin D, Dagnino S, Vivat-Hannah V, Balaguer P, et al. Activation of RXR—PPAR heterodimers by organotin environmental endocrine disruptors. EMBO Rep. 2009;10: 367–373. 10.1038/embor.2009.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Pascoal S, Carvalho G, Vasieva O, Hughes R, Cossins A, Fang Y, et al. Transcriptomics and in vivo tests reveal novel mechanisms underlying endocrine disruption in an ecological sentinel, Nucella lapillus . Mol Ecol. 2012;22: 1589–1608. 10.1111/mec.12137 [DOI] [PubMed] [Google Scholar]
- 94. Kliewer S, Umesono K, Noonan D, Heyman R, Evans R. Convergence of 9-cis retinoic acid and peroxisome proliferator signaling pathways through heterodimer formation of their receptors. Nature. 1992;358: 771–774. 10.1038/358771a0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Heyland A, Moroz LL. Cross-kingdom hormonal signaling: an insight from thyroid hormone functions in marine larvae. J Exp Biol. 2005;208: 4355–4361. 10.1242/jeb.01877 [DOI] [PubMed] [Google Scholar]
- 96. King-Jones K, Thummel C. Nuclear receptors—A perspective from Drosophila . Nat Rev Genet. 2005;6: 311–323. 10.1038/nrg1581 [DOI] [PubMed] [Google Scholar]
- 97. Li T, Bender M. A conditional rescue system reveals essential functions for the ecdysone receptor (EcR) gene during molting and metamorphosis in Drosophila . Development. 2000;127: 2897–2905. [DOI] [PubMed] [Google Scholar]
- 98. Whitehead DL, Sellheyer K. The identification of ecdysterone (20-hydroxyecdysone) in 3 species of mollusks (Gastropoda, Pulmonata). Experientia. 1982;38 10.1007/BF01959769 [DOI] [Google Scholar]
- 99. Garcia M, Griffond B, Lafont R. What are the origins of ecdysteroids in gastropods? Gen Comp Endocrinol. 1995;97: 76–85. [DOI] [PubMed] [Google Scholar]
- 100. Shiff C, Dossaji S. Ecdysteroids as regulators of host and parasite interactions—a study. Trop Med Parasitol. 1991;42: 11–16. [PubMed] [Google Scholar]
- 101. Laguerre M, Veenstra JA. Ecdysone receptor homologs from mollusks, leeches and a polychaete worm. FEBS Lett. 2010;584: 4458–4462. 10.1016/j.febslet.2010.10.004 [DOI] [PubMed] [Google Scholar]
- 102. Urushitani H, Katsu Y, Ohta Y, Shiraishi H, Iguchi T, Horiguchi T. Cloning and characterization of the retinoic acid receptor-like protein in the rock shell, Thais clavigera . Aquat Toxicol. 2013;142: 403–413. 10.1016/j.aquatox.2013.09.008 [DOI] [PubMed] [Google Scholar]
- 103. Gutierrez-Mazariegos J, Kumar Nadendla E, Lima D, Pierzchalski K, Jones JW, Kane M, et al. A mollusk Retinoic Acid Receptor (RAR) ortholog sheds light on the evolution of ligand binding. Endocrinology. 2014; en.2014–1181. 10.1210/en.2014-1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Chung AC, Cooney AJ. The varied roles of nuclear receptors during vertebrate embryonic development. Nucl Recept Signal. 2003;1 10.1621/nrs.01007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Mu X, Lee Y-F, Liu N-C, Chen Y-T, Kim E, Shyr C-R, et al. Targeted inactivation of testicular nuclear orphan receptor 4 delays and disrupts late meiotic prophase and subsequent meiotic divisions of spermatogenesis. Mol Cell Biol. 2004;24: 5887–5899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Chen L-M, Wang R-S, Lee Y-F, Liu N-C, Chang Y-J, Wu C-C, et al. Subfertility with defective folliculogenesis in female mice lacking testicular orphan nuclear receptor 4. Mol Endocrinol. 2008;22: 858–867. 10.1210/me.2007-0181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Ding X, Yu S, Chen B, Lin S, Chang C, Li G. Recent advances in the study of testicular nuclear receptor 4. J Zhejiang Univ-SCI B. 2013;14: 171–177. 10.1631/jzus.B1200357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Daniel A, Dumstrei K, Lengyel JA, Hartenstein V. The control of cell fate in the embryonic visual system by atonal, tailless and EGFR signaling. Development. 1999;126: 2945–2954. [DOI] [PubMed] [Google Scholar]
- 109. Yu RT, Chiang MY, Tanabe T, Kobayashi M, Yasuda K, Evans RM, et al. The orphan nuclear receptor Tlx regulates Pax2 and is essential for vision. Proc Natl Acad Sci USA. 2000;97: 2621–2625. 10.1073/pnas.050566897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Snow MI, Larsen PL. Structure and expression of daf-12: a nuclear hormone receptor with three isoforms that are involved in development and aging in Caenorhabditis elegans . BBA-Gene Struct Expr. 2000;1494: 104–116. [DOI] [PubMed] [Google Scholar]
- 111. Benoit G, Malewicz M, Perlmann T. Digging deep into the pockets of orphan nuclear receptors: insights from structural studies. Trends Cell Biol. 2004;14: 369–376. 10.1016/j.tcb.2004.05.007 [DOI] [PubMed] [Google Scholar]
- 112. King-Jones K, Horner MA, Lam G, Thummel CS. The DHR96 nuclear receptor regulates xenobiotic responses in Drosophila . Cell Metab. 2006;4: 37–48. [DOI] [PubMed] [Google Scholar]
- 113. Lindblom TH, Dodd AK. Xenobiotic detoxification in the nematode Caenorhabditis elegans . J Exp Zool Part A Comp Exp Biol. 2006;305: 720–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Antebi A, Yeh W, Tait D, Hedgecock E, Riddle D. daf-12 encodes a nuclear receptor that regulates the dauer diapause and developmental age in C. elegans . Genes Dev. 2000;14: 1512–1527. [PMC free article] [PubMed] [Google Scholar]
- 115. Ueda H, Sun GC, Murata T, Hirose S. A novel DNA-binding motif abuts the zinc finger domain of insect nuclear hormone receptor ftz-f1 and mouse embryonal long terminal repeat-binding protein. Mol Cell Biol. 1992;12: 5667–5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Allen AK, Spradling AC. The SF1-related nuclear hormone receptor HR39 regulates Drosophila female reproductive tract development and function. Development. 2008;135: 311–321. 10.1242/dev.015156 [DOI] [PubMed] [Google Scholar]
- 117. Lockyer AE, Jones CS, Noble LR, Rollinson D. Trematodes and snails: an intimate association. Can J Zool. 2004;82: 251–269. [Google Scholar]
- 118. Escobedo G, Roberts CW, Carrero JC, Morales-Montor J. Parasite regulation by host hormones: an old mechanism of host exploitation? Trends Parasitol. 2005;21: 588–593. 10.1016/j.pt.2005.09.013 [DOI] [PubMed] [Google Scholar]
- 119. Stone B, Thummel C. The Drosophila 78c early-late puff contains E78, an ecdysone-inducible gene that encodes a novel member of the nuclear hormone-receptor superfamily. Cell. 1993;75: 307–320. 10.1016/0092-8674(93)80072-M [DOI] [PubMed] [Google Scholar]
- 120. Rapado LN, Freitas GC, Polpo A, Rojas-Cardozo M, Rincon JV, Scotti MT, et al. A benzoic acid derivative and flavokawains from Piper species as schistosomiasis vector controls. Molecules. 2014;19: 5205–5218. 10.3390/molecules19045205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Knight M, Miller A, Liu Y, Scaria P, Woodle M, Ittiprasert W. Polyethyleneimine (PEI) mediated siRNA gene silencing in the Schistosoma mansoni snail host, Biomphalaria glabrata . PLoS Neglect Trop Dis. 2011;5: e1212 10.1371/journal.pntd.0001212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Lockyer AE, Spinks J, Kane RA, Hoffmann KF, Fitzpatrick JM, Rollinson D, et al. Biomphalaria glabrata transcriptome: cDNA microarray profiling identifies resistant- and susceptible-specific gene expression in haemocytes from snail strains exposed to Schistosoma mansoni . BMC Genomics. 2008;9: 634 10.1186/1471-2164-9-634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Hanington PC, Lun C-M, Adema CM, Loker ES. Time series analysis of the transcriptional responses of Biomphalaria glabrata throughout the course of intramolluscan development of Schistosoma mansoni and Echinostoma paraensei . Int J Parasitol. 2010;40: 819–831. 10.1016/j.ijpara.2009.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Thummel C. Flies on steroids—Drosophila metamorphosis and the mechanisms of steroid hormone action. Trends Genet. 1996;12: 306–310. 10.1016/0168-9525(96)10032-9 [DOI] [PubMed] [Google Scholar]
- 125. Greenspan RJ. No critter left behind: An invertebrate renaissance. Curr Biol. 2005;15: R671–R672. 10.1016/j.cub.2005.08.018 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The NRs from six different species were subjected to phylogenetic comparisons using Bayesian inference. The Bayesian tree (midpoint rooted) with posterior probability values is shown. Notations Bg, Lg, Hs, Dm, Ce and Sm in association with receptor names denote sequences from B. glabrata, L. gigantea, H. sapiens, D. melanogaster, C. elegans and S. mansoni respectively.
(PDF)
The nuclear receptors from six different species were subjected to phylogenetic comparison using maximum likelihood method using Jones-Taylor-Thornton (JTT) substitution model. Node labels indicate bootstrap values. Notations Bg, Lg, Hs, Dm, Sm and Ce in association with the receptor name denote sequences from B. glabrata, L. gigantea, H. sapiens, D. melanogaster, S, mansoni and C. elegans, respectively.
(PDF)
The nuclear receptors from six different species were subjected to phylogenetic comparison using maximum parsimony. Node labels indicate bootstrap values. Notations Bg, Lg, Hs, Dm, Sm and Ce in association with the receptor name denote sequences from B. glabrata, L. gigantea, H. sapiens, D. melanogaster, S. mansoni and C. elegans, respectively.
(PDF)
(PDF)
Notations Bg, Lg, Hs, Dm, Ce and Sm in association with receptor names denote sequences from B. glabrata, L. gigantea, H. sapiens, D. melanogaster, C. elegans, and S. mansoni respectively.
(TXT)
Forward and reverse primer sequences with their product size (as submitted to GenBank), their corresponding GenBank accession numbers and their optimised annealing temperatures.
(DOCX)
Detailed summary of all the top BLAST expressed sequence tag (EST) hits for L. gigantea NRs, including the EST description, GenBank accession number and E-value.
(PDF)
Data Availability Statement
All sequence data is available either in the supporting information (predicted protein sequence) or available from GenBank (sequenced fragments: Accession numbers: JZ390894-JZ390939).