Since Bahl’s statement in 1992[1] that, “bacteria, in general, do not contain glycoproteins, except the obligately halophilic bacteria of the genus Halobacteria[2] and Bacillus stearothermophilus NRS 2004/3a,[3] in which their presence has been demonstrated with some degree of certainty”, a complete chance of perception has taken place. Nowadays, the occurrence of prokaryotic glycoproteins is fully accepted,[4,5] particularly in pathogens.[6,7] Recent work in several laboratories has not only shown a great structural diversity of these glycoconjugates but has also provided key advances in understanding the molecular mechanisms of prokaryotic glycosylation, and the application potential and relevant immunological aspects of selected glycoproteins.[8–13] Among these prokaryotic glycoconjugates are archaeal and bacterial surface layer (S-layer) glycoproteins,[2,14] flagella and pili,[10,15,16] and several intra- and extracellular glycosylated enzymes and haptens.[4]
In the following, the different prokaryotic glycoproteins will be discussed in the order of their discovery. Glycosylated S-layer proteins were the first prokaryotic glycoproteins ever described in detail. Much of the current knowledge has been derived from initial analyses of the N- and O-glycosylated S-layer protein of Halobacterium salinarum by Strominger’s group. That work was then extended by Sumper, Wieland, and co-workers in the 1980s.[2] Another decade later, Eichler and co-workers set out to investigate the molecular mechanisms behind archaeal S-layer protein glycosylation,[17] in particular of the haloarchaeon Haloferax volcanii.[18] Recent studies on the methanoarchaeon Methanococcus voltae revealed the structure of an N-glycan entity decorating several flagellin proteins as well as the S-layer protein.[19] The similar modification of both flagellins and the S-layer glycoprotein points to a common N-glycosylation mechanism in these cells. In Hf. volcanii and Mc. voltae, several agl (archaeal glycosylation) genes have been identified, and their products have been functionally characterized through combined gene-deletion and mass spectrometry approaches.[20] In both organisms, aglB encodes the sole component of the archaeal oligosaccharyltransferase (OTase) that is responsible for the transfer of the completed N-glycan chain to the corresponding acceptor protein. Whereas N-glycosylation of archaeal proteins seems to be more widespread, virtually nothing is known of the archaeal version of O-glycosylation, except the existence of disaccharides linked to threonine residues of the S-layer proteins from Hb. salinarum and Hf. volcanii.[2]
A completely different situation is known from bacterial S-layer glycoproteins. Since the first description of glycosylated bacterial S-layer proteins by Sleytr and Thorne about three decades ago,[14] approximately 15 different, exclusively O-linked, S-layer glycan structures have been fully or at least partially characterized in Gram-positive organisms.[12] Inspection of these glycan structures supports the assumption that the glycans are comparable to lipopolysaccharide (LPS) O-antigens of Gram-negative bacteria.[21] In some S-layer glycoproteins, modifications of the terminal repeating unit at the nonreducing end are present.[3,12] These capping elements are assumed to be involved in determining the chain length of the glycans, comparable to the ABC-transporter-dependent biosynthesis pathway of LPS O-antigens.[21] Very recently, the existence of glycosylated S-layer proteins has been described in a Gram-negative bacterium, namely in the oral pathogen Tannerella forsythia.[22]
In 1999 Guerry’s group, for the first time, described in Campylobacter jejuni 81–176 genes that are part of a general protein glycosylation system, by which up to 40 soluble and membrane proteins are N-glycosylated. Since that discovery, the entire field has moved at an extraordinary pace.[7,23] The genes required for the general N-linked protein glycosylation pathway (pgl) are remarkably conserved and do not seem to have the potential of phase variation. Because pgl gene mutation affects the glycosylation of numerous proteins, there are pleiotropic effects including decreased bacterial attachment and invasion of human epithelial cells, loss of mouse and chick colonization, and reduced protein reactivity to antisera.[24] Of particular interest was the early observation that this N-glycosylation system could be functionally transferred from C. jejuni into Escherichia coli for the production of recombinant glycoproteins in this bacterium.[25] This achievement will open up numerous possibilities for engineering novel glycoconjugates for biotechnological applications.
The biosynthesis, assembly and regulation of the flagellar apparatus of Bacteria and Archaea has been the subject of extensive studies over many decades. The characterization of flagellar systems from different bacterial species has revealed subtle yet distinct differences in composition, regulation, and mode of assembly. Glycosylation of the major structural protein, the flagellin, has recently been shown to be an important feature of numerous flagellar systems in both Bacteria and Archaea.[10]
The flagellin of Campylobacter coli VC167 was the first identified flagellar glycoprotein. Early studies indicated considerable heterogeneity in flagellin composition, which was the result of as many as 19 sites of O-linked glycosylation on that protein.[26] The flagellin modifications include pseudaminic acid (Pse5Ac7Ac), a nine-carbon sugar that is structurally similar to sialic acid (Neu5Ac),[27] as well as a second nine-carbon sugar, legionaminic acid and its derivatives.[28] Recent studies have indicated that glycosylation of flagellin is crucial for filament assembly. The flagellin glycans also contribute to the virulence of C. jejuni. The loss of Pse5Am7Ac from the C. jejuni 81–176 flagellin results in reduced adherence and invasion of intestinal epithelial cells and attenuation in ferret diarrheal disease.[10]
Glycosylation of flagellin is extensive throughout the archaeal domain. Detailed studies are scarce, but examples are the flagellins of Hb. salinarum, Mc. voltae, and Mc. maripaludis.[10,29] The first structural characterization of a flagellar glycan was provided by Wieland and co-workers[2] for the flagellin of Hb. salinarum. The flagellins possess the same N-linked glycans as the S-layer proteins. Similar observations were made with the flagellins of the previously mentioned methanococci, thus implying a common N-linked glycosylation pathway for these two major surface protein structures in Archaea.[10,29]
Pilin of pathogenic Neisseria was one of the first examples of an O-glycosylated glycoprotein in a bacterial pathogen. A series of investigations has identified the genes that encode the glycosyltransferases required for the biosynthesis of the pilin glycan.[30–34] Studies in different Neisseria species have proposed that the addition of O-linked glycan to pilin is performed by an OTase that is homologous to the O-antigen ligases that catalyze the addition of O-antigen to LPS.[34,35]
In analogy to the well-accepted general N-glycosylation system of Campylobacter jejuni,[7,24] within one month in the autumn of 2008, three independent research groups submitted a description of their specific model system as an example for a general prokaryotic protein O-glycosylation system. Two of these papers deal with the glycosylation systems of pathogenic Neisseria strains[36,37] and will be discussed later.
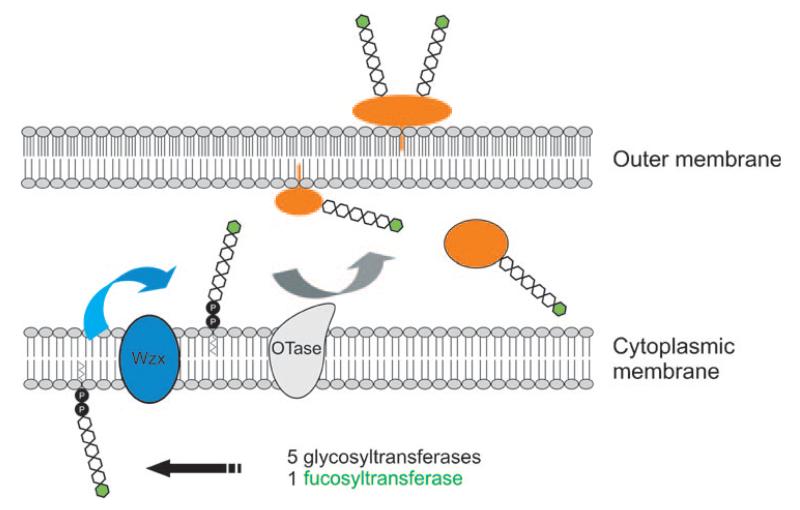
The third one is a seminal paper by Comstock and colleagues who describe a general O-glycosylation system that is important to the physiology of the major human intestinal symbiont Bacteroides fragilis.[38] The Bacteroides are specifically adapted for survival in this ecosystem and collectively comprise one of the most abundant bacterial genera in the human colon. Members of this genus have an unprecedented repertoire of genetic systems devoted to acquiring and metabolizing carbohydrates. This allows the organisms to respond rapidly to food supplies that are shifting and often times scarce. The organisms produce enzymes to harvest l-fucose from host mucosal glycans and harbor a rare bacterial pathway for the incorporation of exogenous fucose into capsular polysaccharides and glycoproteins.[39] It was shown that B. fragilis produces many glycoproteins that bind the fucose-specific Aleuria aurantia lectin. l-Fucose is an abundant surface molecule of host intestinal epithelial cells, and expression of this surface molecule is coordinated by host and symbiont. The B. fragilis mutant Δgmd-fclΔfkp, defective in the pathways for the biosynthesis of GDP-fucose and consequently unable to incorporate l-fucose into glycoproteins, is also defective in colonizing the mammalian intestine under competitive conditions. Thus, protein fucosylation is central to the physiology of this organism and is necessary for it to competitively colonize the mammalian intestine.[39] The proteins targeted for glycosylation include those predicted to be involved in protein folding, protein–protein interactions, peptide degradation, and surface lipoproteins. To elucidate their localizations, whole bacteria were treated with proteinase K so as to digest surface proteins. As a result, all of the eight selected glycoproteins are secreted from the cytoplasm and targeted either to the periplasm or the inner or outer leaflet of the outer membrane (Figure 1). Glycosylation of periplasmic proteins is unusual. So far, it has only been observed for glycoproteins of Campylobacter jejuni,[7] Desulfovibrio gigas,[40] and Neisseria gonorrhoeae.[37] By site-directed mutagenesis it was further demonstrated that on a selected glycoprotein (BF2494), which is representative of all other identified glycoproteins, the amino acids Thr87, Thr178, and Thr231 are the only glycosylation sites and, therefore, all the glycans of this protein are O-linked.[38] Although O-glycosylation systems in other organisms usually do not require a motif other than Ser, Thr,[8] or Tyr,[4,12] inspection of the protein sequence around the three glycosylation sites of BF2494 revealed that in B. fragilis, a three-residue motif D–S/T–A/I/L/V/M/T for O-glycosylation is present.
Figure 1.
Proposed model for the general O-glycosylation system of Bacteroides fragilis. The glycan chain is synthesized on a lipid carrier on the inner side of the cytoplasmic membrane by the sequential action of five putative glycosyltransferases and a fucosyltransferase not encoded by the lfg region. The glycan is flipped to the periplasmic face of the inner membrane by Wzx (blue). Up to this point, the pathway is common to the synthesis of many O-antigens, capsular polysaccharides and bacterial glycoproteins. The glycosylation of proteins in B. fragilis occurs in the periplasm, by the activity of an undefined OTase (gray), which is likely an integral membrane protein as in other bacteria. This schematic shows the glycosylated proteins (orange) in the three cellular locations that have been experimentally demonstrated. Modified from ref. [38].
Glycosylation of bacterial proteins generally requires the transport of the target proteins from the cytoplasm into the bacterial periplasm.[3,7] This criterion was also tested with B. fragilis; mutation of the signal peptide of BF2494 in B. fragilis showed that the mutant protein is retained in the cytoplasm and that it is not glycosylated.[38] To test whether or not a genomic region of B. fragilis is involved in protein glycosylation, the whole genome was inspected. The region spanning genes BF4298 to BF4306 encodes a putative flippase, five putative glycosyltransferases, and other genes likely to be involved in oligosaccharide synthesis, but no polymerase. This demonstrates that the BF4298–4306 region is involved in protein glycosylation. Furthermore, because this region is required for the synthesis of all the fucosylated glycoproteins of B. fragilis, it appears to be part of a general glycosylation system. Based on these results, the region was named lfg for locus of fragilis glycosylation.[38] Interestingly, the lfg region is adjacent to metG, which encodes the essential enzyme methionyl-tRNA synthetase. The transcriptional linkage of the protein translation machinery (metG) with the protein glycosylation machinery (lfg) suggests a high level of importance for protein glycosylation in B. fragilis. The analysis of five other intestinal Bacteroides species demonstrated that they all have regions similar to lfg, with a flippase gene downstream from metG and several glycosyltransferase genes. This finding, coupled with the observation that other Bacteroides species produce fucosylated glycoproteins, was a strong hint for the presence of a conserved general O-glycosylation system in B. fragilis and in other intestinal Bacteroides species. Alignment of the sequence of BF2494 with the sequences from the other five orthologues confirmed that they perfectly match with the glycosylation motif D–S/T–A/I/L/V/M/T. These data suggest that the intestinal Bacteroides species possess similar general O-glycosylation systems, one of the few general glycosylation systems known in bacteria.[7,36–38]
The mechanism of O-glycosylation in B. fragilis appears to have some similarities to protein glycosylation in other bacteria (Figure 1). It appears to be similar to pilin O-glycosylation in Neisseria meningitidis and N. gonorrhoeae, and in some strains of Pseudomonas aeruginosa,[41] and to a recently described S-layer O-glycosylation system of Geobacillus stearothermophilus,[3] but different from O-glycosylation in eukaryotes.[7,38,42]
As mentioned before, similar to the proposed general O-glycosylation of B. fragilis, in pathogenic Neisseria meningitidis, an O-glycosylation pathway modifies both a single abundant protein, pilin, the subunit protein that forms pili, and an outer membrane glycoprotein, the nitrite reductase AniA.[36] Since pilin is under intense immune selection and is the archetypal example for antigenic variation, the decoration of the surface-exposed, flexible, C-terminal domain of AniA with the same phase-variable O-linked glycan modification might also be an immune evasion strategy. Immune selection acting on the surface proteins of this host-adapted pathogen might have been the driving force for the evolution of this proposed general O-glycosylation pathway.[36]
In the related Gram-negative bacterium Neisseria gonorrhoeae, a general O-linked glycosylation system was proposed that targets structurally and functionally diverse groups of membrane-associated proteins.[37] The analyzed glycoproteins are implicated in activities that vary from protein folding, disulfide bond formation, and solute uptake to both aerobic and anaerobic respiration. As in eukaryotes, the broad scope of this system is dictated by the relaxed specificity of the OTase as well as the bulk properties and context of the protein-targeting signal rather than by a strict amino acid consensus sequence. Together, these findings reveal previously unrecognized commonalities connecting O-linked protein glycosylation in distantly related life forms.[37] They also extend the ways in which the N. gonorrhoeae O-linked system emulates the C. jejuni N-linked systems to include the targeting of multiple periplasmic substrates.[7,42]
As in the C. jejuni system, these findings raise obvious questions as to what biologic significance global protein glycosylation might have and what forces and processes have shaped glycoproteome content.[37] The great importance and potential for exploitation in this area of prokaryotic protein glycosylation are certainly very clear.
Acknowledgements
I thank Christina Schäffer for her input the during preparation of this contribution, my colleagues in the NanoGlycobiology group for sharing ideas, and Uwe B. Sleytr for his interest. Work in the laboratory is supported by the Austrian Science Fund and by the Federal Ministry of Science and Education.
References
- [1].Bahl OP. In: Glycoconjugates–Composition, Structure, and Function. Allen HJ, Kisailus EC, editors. Marcel Dekker; New York: 1992. pp. 1–12. [Google Scholar]
- [2].Sumper M, Wieland FT. In: Glycoproteins. Montreuil J, Vliegenthart JFG, Schachter H, editors. Elsevier; Amsterdam: 1995. pp. 455–473. [Google Scholar]
- [3].Steiner K, Novotny R, Werz DB, Zarschler K, Seeberger PH, Hofinger A, Kosma P, Schäffer C, Messner P. J. Biol. Chem. 2008;283:21120–21133. doi: 10.1074/jbc.M801833200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Messner P, Schäffer C. In: Progress in the Chemistry of Organic Natural Products. Herz W, Falk H, Kirby GW, editors. Vol. 85. Springer; Wien: 2003. pp. 51–124. [DOI] [PubMed] [Google Scholar]
- [5].Messner P. J. Bacteriol. 2004;186:2517–2519. doi: 10.1128/JB.186.9.2517-2519.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Benz I, Schmidt MA. Mol. Microbiol. 2002;45:267–276. doi: 10.1046/j.1365-2958.2002.03030.x. [DOI] [PubMed] [Google Scholar]
- [7].Szymanski CM, Wren BW. Nat. Rev. Microbiol. 2005;3:225–237. doi: 10.1038/nrmicro1100. [DOI] [PubMed] [Google Scholar]
- [8].Eichler J, Adams MWW. Microbiol. Mol. Biol. Rev. 2005;69:393–425. doi: 10.1128/MMBR.69.3.393-425.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Feldman MF, Wacker M, Hernandez M, Hitchen PG, Marolda CL, Kowarik M, Morris HR, Dell A, Valvano MA, Aebi M. Proc. Natl. Acad. Sci. USA. 2005;102:3016–3021. doi: 10.1073/pnas.0500044102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Logan SM. Microbiology. 2006;152:1249–1262. doi: 10.1099/mic.0.28735-0. [DOI] [PubMed] [Google Scholar]
- [11].Fletcher CM, Coyne MJ, Bentley DL, Villa OF, Comstock LE. Proc. Natl. Acad. Sci. USA. 2007;104:2413–2418. doi: 10.1073/pnas.0608797104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Messner P, Steiner K, Zarschler K, Schäffer C. Carbohydr. Res. 2008;343:1934–1951. doi: 10.1016/j.carres.2007.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Yurist-Doutsch S, Chaban B, VanDyke DJ, Jarrell KF. J. Eichler, Mol. Microbiol. 2008;68:1079–1084. doi: 10.1111/j.1365-2958.2008.06224.x. [DOI] [PubMed] [Google Scholar]
- [14].Sleytr UB, Thorne KJI. J. Bacteriol. 1976;126:377–383. doi: 10.1128/jb.126.1.377-383.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Castric P. Microbiology. 1995;141:1247–1254. doi: 10.1099/13500872-141-5-1247. [DOI] [PubMed] [Google Scholar]
- [16].Stimson E, Virji M, Makepeace K, Dell A, Morris HR, Payne G, Saunders JR, Jennings MP, Barker S, Panico M, Blench I, Moxon ER. Mol. Microbiol. 1995;17:1201–1214. doi: 10.1111/j.1365-2958.1995.mmi_17061201.x. [DOI] [PubMed] [Google Scholar]
- [17].Abu-Qarn M, Eichler J. Mol. Microbiol. 2006;61:511–525. doi: 10.1111/j.1365-2958.2006.05252.x. [DOI] [PubMed] [Google Scholar]
- [18].Abu-Qarn M, Giordano F, Battaglia A, Trauner A, Hitchen PG, Morris HR, Dell A, Eichler J. J. Bacteriol. 2008;190:3140–3146. doi: 10.1128/JB.00056-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Voisin S, Houliston RS, Kelly J, Brisson J-R, Watson D, Bardy SL, Jarrell KF, Logan SM. J. Biol. Chem. 2005;280:16586–16593. doi: 10.1074/jbc.M500329200. [DOI] [PubMed] [Google Scholar]
- [20].Chaban B, Voisin S, Kelly J, Logan SM, Jarrell KF. Mol. Microbiol. 2006;61:259–268. doi: 10.1111/j.1365-2958.2006.05226.x. [DOI] [PubMed] [Google Scholar]
- [21].Raetz CRH, Whitfield C. Annu. Rev. Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lee S-W, Sabet M, Um H-S, Yang J, Kim HC, Zhu W. Gene. 2006;371:102–111. doi: 10.1016/j.gene.2005.11.027. [DOI] [PubMed] [Google Scholar]
- [23].Guerry P, Szymanski CM. Trends Microbiol. 2008;16:428–435. doi: 10.1016/j.tim.2008.07.002. [DOI] [PubMed] [Google Scholar]
- [24].Nothaft H, Amber S, Aebi M, Szymanski CM. In: Campylobacter. 3rd ed Nachamkin I, Szymanski CM, Blaser MJ, editors. ASM Press; Washington, DC: 2008. pp. 447–469. [Google Scholar]
- [25].Wacker M, Linton D, Hitchen PG, Nita-Lazar M, Haslam SM, North SJ, Panico M, Morris HR, Dell A, Wren BW, Aebi M. Science. 2002;298:1790–1793. doi: 10.1126/science.298.5599.1790. [DOI] [PubMed] [Google Scholar]
- [26].Logan SM, Kelly JF, Thibault P, Ewing CP, Guerry P. Mol. Microbiol. 2002;46:587–597. doi: 10.1046/j.1365-2958.2002.03185.x. [DOI] [PubMed] [Google Scholar]
- [27].Thibault P, Logan SM, Kelly JF, Brisson J-R, Ewing CP, Trust TJ, Guerry P. J. Biol. Chem. 2001;276:34862–34870. doi: 10.1074/jbc.M104529200. [DOI] [PubMed] [Google Scholar]
- [28].McNally DJ, Aubry AJ, Hui JPM, Khieu NH, Whitfield D, Ewing CP, Guerry P, Brisson J-R, Logan SM, Soo EC. J. Biol. Chem. 2007;282:14463–14475. doi: 10.1074/jbc.M611027200. [DOI] [PubMed] [Google Scholar]
- [29].VanDyke DJ, Wu J, Logan SM, Kelly JF, Mizuno S, Aizawa S-I, Jarrell KF. Mol. Microbiol. 2009;72:633–644. doi: 10.1111/j.1365-2958.2009.06671.x. [DOI] [PubMed] [Google Scholar]
- [30].Stimson E, Virji M, Makepeace K, Dell A, Morris HR, Payne G, Saunders JR, Jennings MP, Barker S, Panico M, Blench I, Moxon ER. Mol. Microbiol. 1995;17:1201–1214. doi: 10.1111/j.1365-2958.1995.mmi_17061201.x. [DOI] [PubMed] [Google Scholar]
- [31].Virji M. Gene. 1997;192:141–147. doi: 10.1016/s0378-1119(97)00082-6. [DOI] [PubMed] [Google Scholar]
- [32].Marceau M, Forest K, Beretti J-L, Tainer J, Nassif X. Mol. Microbiol. 1998;27:705–715. doi: 10.1046/j.1365-2958.1998.00706.x. [DOI] [PubMed] [Google Scholar]
- [33].Banerjee A, Ghosh SK. Mol. Cell. Biochem. 2003;253:179–190. doi: 10.1023/a:1026058311857. [DOI] [PubMed] [Google Scholar]
- [34].Power PM, Seib KL, Jennings MP. Biochem. Biophys. Res. Commun. 2006;347:904–908. doi: 10.1016/j.bbrc.2006.06.182. [DOI] [PubMed] [Google Scholar]
- [35].Aas FE, Vik Å, Vedde J, Koomey M, Egge-Jacobsen W. Mol. Microbiol. 2007;65:607–624. doi: 10.1111/j.1365-2958.2007.05806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ku SC, Schulz BL, Power PM, Jennings MP. Biochem. Biophys. Res. Commun. 2009;378:84–89. doi: 10.1016/j.bbrc.2008.11.025. [DOI] [PubMed] [Google Scholar]
- [37].Vik Å, Aas FE, Anonsen JH, Bilsborough S, Schneider A, Egge-Jacobsen W, Koomey M. Proc. Natl. Acad. Sci. USA. 2009;106:4447–4452. doi: 10.1073/pnas.0809504106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Fletcher CM, Coyne MJ, Villa OF, Chatzidaki-Livanis M, Comstock LE. Cell. 2009;137:321–331. doi: 10.1016/j.cell.2009.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Coyne MJ, Reinap B, Lee MM, Comstock LE. Science. 2005;307:1778–1781. doi: 10.1126/science.1106469. [DOI] [PubMed] [Google Scholar]
- [40].Santos-Silva T, Dias JM, Dolla A, Durand MC, Goncalves LL, Lampreia J, Moura I, Romao MJ. J. Mol. Biol. 2007;370:659–673. doi: 10.1016/j.jmb.2007.04.055. [DOI] [PubMed] [Google Scholar]
- [41].Faridmoayer A, Fentabil MA, Mills DC, Klassen JS, Feldman MF. J. Bacteriol. 2007;189:8088–8098. doi: 10.1128/JB.01318-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Helenius A, Aebi M. Annu. Rev. Biochem. 2004;73:1019–1049. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]