Abstract
Objective:
We analyzed the Epstein-Barr nuclear antigen 2 (EBNA2) gene, which contains the most variable region of the viral genome, in persons with multiple sclerosis (MS) and control subjects to verify whether virus genetic variants are involved in disease development.
Methods:
A seminested PCR approach and Sanger sequencing were used to analyze EBNA2 in 53 patients and 38 matched healthy donors (HDs). High-throughput sequencing by Illumina MiSeq was also applied in a subgroup of donors (17 patients and 17 HDs). Patients underwent gadolinium-enhanced MRI and human leucocyte antigen typing.
Results:
MS risk significantly correlated with an excess of 1.2 allele (odds ratio [OR] = 5.13; 95% confidence interval [CI] 1.84–14.32; p = 0.016) and underrepresentation of 1.3B allele (OR = 0.23; 95% CI 0.08–0.51; p = 0.0006). We identified new genetic variants, mostly 1.2 allele- and MS-associated (especially amino acid variation at position 245; OR = 9.4; 95% CI 1.19–78.72; p = 0.0123). In all cases, the consensus sequence from deep sequencing confirmed Sanger sequencing (including the cosegregation of newly identified variants with known EBNA2 alleles) and showed that the extent of genotype intraindividual variability was higher than expected: rare EBNA2 variants were detected in all HDs and patients with MS (range 1–17 and 3–19, respectively). EBNA2 variants did not seem to correlate with human leucocyte antigen typing or clinical/MRI features.
Conclusions:
Our study unveils a strong association between Epstein-Barr virus genomic variants and MS, reinforcing the idea that Epstein-Barr virus contributes to disease development.
Despite converging evidence supporting an etiologic role for Epstein-Barr virus (EBV) in multiple sclerosis (MS), we still do not know through which mechanisms the virus may contribute to disease development.1 The potential pathogenic role of EBV genetic variants in MS may be in keeping with epidemiologic observations, in particular the geographic gradient of MS and the change in MS risk in migrants,2 and with the reported association between virus genetic variants and EBV-related malignancies with different geographic distribution.3–5 Also, the discrepancy between the global diffusion of EBV infection and the low prevalence of MS could be at least in part explained by EBV genomic diversity that differently affects MS development.
Major methodologic difficulties hinder the study of EBV variants through sequencing of the whole viral genome. To date, only 8 complete genome sequences have been described: 5 from patients with EBV-related diseases and 3 from healthy individuals.6 This is mainly attributable to the low frequency of EBV-infected B cells in peripheral blood (2–10 infected cells per million B cells, values that do not differ significantly between healthy individuals and patients with MS).7 To disclose possible MS-related EBV strains, various groups have followed a candidate-gene approach.8–14 Along the same line, we chose to investigate EBV variability in MS by studying Epstein-Barr nuclear antigen 2 (EBNA2). EBNA2 is, in principle, the best candidate for this kind of study because it is the most polymorphic among all EBV genes: 5 major alleles of the EBV type 1 strain, the most frequent strain in the general Caucasian population,15 have been identified based on nucleotide variations within the most variable region of EBNA2, which is also involved in the interaction with host proteins.16 The possibility that genetic variations in this region might have functional consequences prompted us to investigate the association between EBNA2 variants and MS in a population from continental Italy.
METHODS
Subjects and samples.
Blood samples (40 mL) were obtained from 53 patients with MS (50 with a relapsing-remitting and 3 with a progressive form of disease)17 and matched (age, sex, and geographic origin) healthy donors (HDs). At the same time point, patients underwent gadolinium-enhanced MRI.
Peripheral blood mononuclear cells (PBMCs) were obtained by density centrifugation over Ficoll–Hypaque according to standard procedure. PBMCs were stained with anti-human CD19 antibodies (Miltenyi Biotec, Bergisch Gladbach, Germany) and B cells were purified using magnetic bead separation in accordance with the manufacturer's recommended protocol. The final number of CD19+ cells was approximately 4 million, depending on the number of circulating B cells in every subject. Genomic DNA was extracted from PBMCs, CD19+ B cells, EBV-positive (B95-8) and EBV-negative (BJAB) cell lines using a commercial kit (QIAamp DNA mini kit; Qiagen, Venlo, the Netherlands).
Standard protocol approvals, registrations, and patient consents.
The local institutional review board approved the study and all participating subjects gave written informed consent.
Sanger sequencing.
All DNA samples were analyzed by a seminested PCR approach using EBNA2 type-specific primers15 that amplify a region of 500 base pairs (bp) within the coding sequence of the gene. We used Hot Start Taq polymerases (KAPA2G Robust [Kapa Biosystems, Wilmington, MA; Resnova S.r.l., Genzano di Roma, Italy] for the first run, and FastStart [Roche Molecular Biochemicals, Mannheim, Germany] for the second run) for PCR reactions. All PCR products were checked for their length in a gel electrophoresis on 3% agarose gel and assayed by Sanger sequencing analysis.
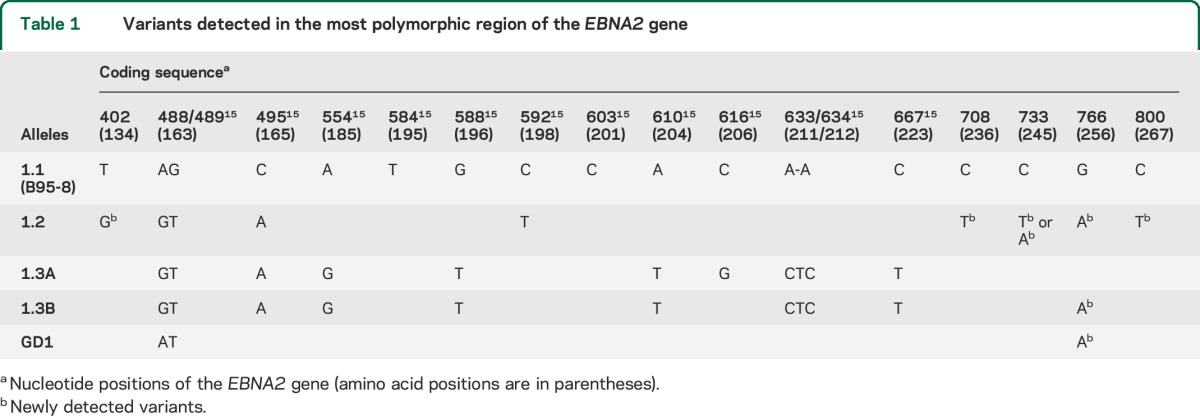
The sequences were aligned with the ClustalW2 multiple sequence alignment program (www.ebi.ac.uk/Tools/msa/clustalw2/) with default settings, and the variants were considered when there was a deviation from the published GD1 (National Center for Biotechnology Information [NCBI] accession number AY961628) and B95.8 (NCBI accession number NC_007605) EBV prototype. Recently, Tierney et al.15 identified allele-specific changes (1.2, 1.3A, 1.3B, and 1.3E) relative to the B95.8 sequence (defined as allele 1.1) by sequencing the coding region corresponding to nucleotides 488–667. In the present work, a larger coding sequence of EBNA2, encompassing nucleotides 402–800, was analyzed (table 1).
Table 1.
Variants detected in the most polymorphic region of the EBNA2 gene
EBNA2 amplicon deep-sequencing and analysis.
We generated amplicon sequences of the EBNA2 gene from 17 patients and 17 HDs (analyzing one sample for each subject) through the Illumina MiSeq platform from template DNA obtained using a multistep PCR and the Illumina Nextera strategy (Illumina, Inc., San Diego, CA).
The identification of EBNA2 variants, using as reference the complete human herpesvirus 4 genome strain B95-8 (NC_007605), was performed using REDItools on quality-filtered sequences.18 The viral allele assignment of both amplicon and Sanger EBNA2 sequences was performed using a methodology implemented in a python script (available on request). Further details on the EBNA2 amplicon deep sequencing and analysis can be found in appendix e-1 on the Neurology® Web site at Neurology.org.
Human leucocyte antigen typing.
Human leukocyte antigen (HLA) typing of Class I HLA-A, HLA-B, and HLA-Cw and Class II DRB1 loci was performed by standard sequence-specific primer PCR,19 using Histo Type DNA well plates (BAG; Formedic Diagnostici, Milan, Italy) according to the manufacturer's instructions. Detection of the alleles recognized by the specific primers was possible after amplification in a GeneAmp PCR 9700 thermocycler (Applied Biosystems, Foster City, CA) and gel electrophoresis on 2% agarose gel. The HLA genotypes together with EBV genotypes are reported in table e-1 for individuals with a complete HLA genotyping at all analyzed HLA loci (33 healthy controls and 38 patients with MS, a random subset of the entire sample).
Statistical analysis.
Fisher exact test and odds ratios (ORs) were calculated using the GraphPad Prism 5 program (GraphPad Software, La Jolla, CA) to compare the proportion of variants in all patients vs HDs. We used a Bayesian Network20 classifier that was configured to use a simple estimator with α = 0.5 and K2 as internal search algorithm. The variables used to train the classifier were the sex, the strain, the 13 position reads (aa134, aa163, aa165, aa185, aa196, aa198, aa204, aaINS 211/212, aa223, aa236, aa245, aa256, aa267), and whether each was a mutation with respect to the strain or not. The final model used to generate the receiver operating characteristic (ROC) utilized in ensemble all these variables and assigned different weights to each one of them. Area under the curve (AUC) significance level (p value) was computed using the method described in the DeLong & DeLong algorithm as difference from the null hypothesis, i.e., AUC = 0.5.
ROC curves and the AUC were then computed to quantify the predictive potential of EBNA2 genotyping as marker of disease status.
RESULTS
We obtained blood samples from 91 patients and 56 HDs matched for age, sex, ethnicity, and geographic origin. Although we may consider all patients with MS and most HD subjects as EBV-infected,21 the rate of successful EBNA2 genotyping was lower than 100% and comparable between the 2 groups: 53 of 91 (58%) in patients and 38 of 56 (67%) in HDs. The mean age of patients was 36.7 ± 9.8 years and HDs was 37.5 ± 9.6 years; the male to female ratio was 13 to 40 in patients and 11 to 27 in controls. The clinical characteristics of patients are summarized in table e-2 (most of them were free of disease-modifying therapies [42/53] and were sampled in a stable phase of disease [40/53]). Seven patients and 3 HDs were analyzed at different time points (1–12 months apart); of these, 2 patients and 1 HD were infected with different EBV strains at different time points. By considering each sampling as an independent observation, we analyzed a total of 108 blood samples (66 in patients with MS and 42 in HDs).
We found a significant difference in the distribution of EBV alleles between patients with MS and HDs: the 1.2 allele was predominant in patients, while the 1.3B allele prevailed in controls (table 2). In addition to the variants identified within the EBNA2 region corresponding to the B95.8 coordinates 36,671–36,911 (nucleotides 456–696 of the EBNA2 coding sequence), our analysis unveiled previously undetected variants at nucleotides 402, 708, 733, 766, and 800 (table 1). While the variant at position 766 was present in alleles 1.2, 1.3B, and GD1, without differences between patients and controls, the other 4 variants were associated with the 1.2 alleles and thus significantly correlated to MS status (tables 3 and e-1). The risk of disease development was significantly higher in carriers of the 1.2 alleles (OR = 5.13; 95% confidence interval [CI] 1.84–14.32; p = 0.016) and variants (especially at amino acid 245, OR = 9.4; 95% CI 1.19–78.72; p = 0.0123), while the presence of 1.3B alleles appeared to be protective (OR = 0.23; 95% CI 0.08–0.51; p = 0.0006). To verify the consistency of EBNA2 genotyping data, we implemented a Bayesian Network classifier that was trainable and then testable for its predictive capability on our dataset. The potential of EBV genetic variants to predict MS status was assessed using ROC analysis; the AUC value was 0.715, with a significant difference from null hypothesis (p < 0.001) (figure 1), indicating moderate accuracy of the identified variants in discriminating between patients and HDs.
Table 2.
Frequency of EBNA2 alleles in the peripheral blood of patients with MS and HDs
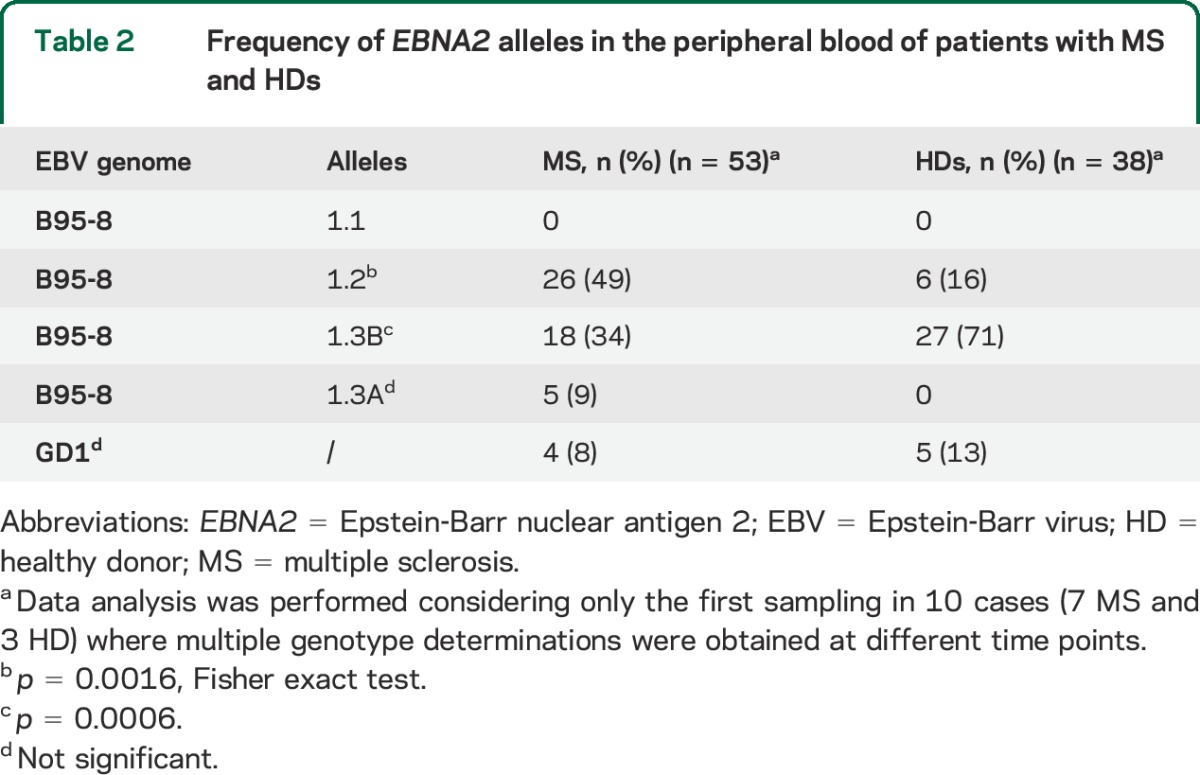
Table 3.
Frequency of newly identified EBNA2 variants in patients with MS and HDs
Figure 1. Potential of Epstein-Barr virus genetic variants to predict multiple sclerosis status.
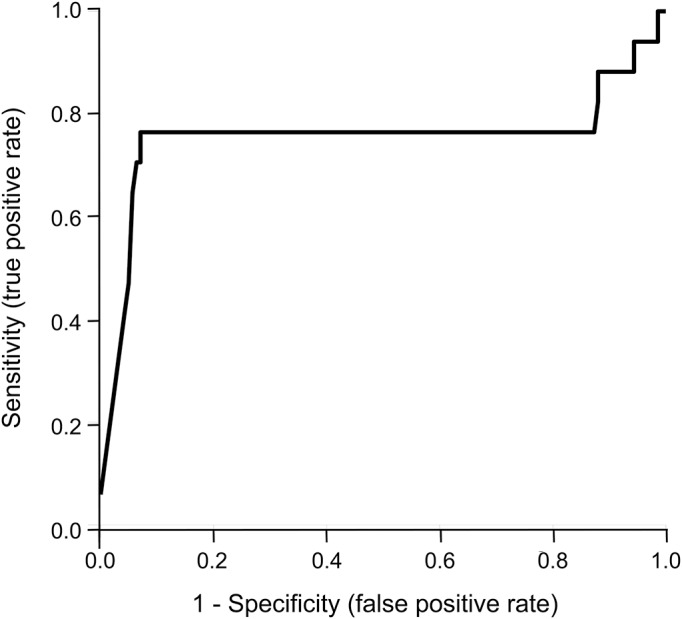
Receiver operating characteristic curve generated by using a Bayesian Network classifier approach that considers sex and Epstein-Barr virus nuclear antigen 2 (EBNA2) genotypes. Area under the curve = 0.715; area under precision-recall curve = 0.633; F measure = 0.640.
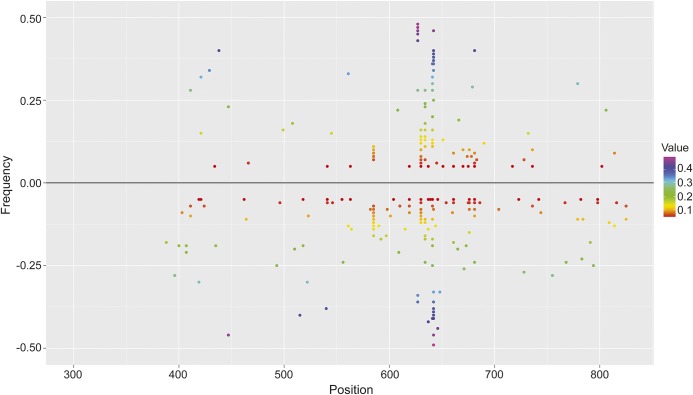
To investigate the occurrence and the extent of intraindividual genotype variability within subjects with MS and HDs, deep sequencing of the EBNA2 region using the Illumina MiSeq platform was performed in a subset of 17 MS and 17 HD samples already characterized by Sanger sequencing. The average fold coverage of the 465-bp-long EBNA2 amplicon was 3,927. To avoid deep-sequencing errors/artifacts, we just considered high-quality variable positions (minimum Phred score ≥25) where the minor allele frequency was ≥5% and fold coverage ≥50. In all cases, the consensus sequence deriving from deep sequencing coincided with the one determined by Sanger sequencing, confirming the cosegregation of newly identified variants with the known EBNA2 alleles. We also found one or more variable positions in all HDs (1–17) and individuals with MS (3–19). For all couples of variants lying on the same read (coverage ≥50), the hypothesis of independent segregation, indicative of random sequencing errors, was rejected through a χ2 test (p < 0.05), thus supporting the coexistence of different genotypes. Overall, a slightly higher prevalence of variant positions was detected in MS with respect to HDs, with an average value per individual of 10.1 and 7.4, respectively. This finding suggests a marginally significant higher occurrence of coexisting alleles, possibly deriving from multiple infections or quasi-speciation, in MS (figure 2) than in HDs (figure 2; t test p < 0.1).15,22 None of the rarer variants identified with the deep-sequencing approach were associated with MS or healthy status.
Figure 2. EBNA2 position-specific variant frequency observed in sampled healthy donors and individuals with MS.
Upper and lower panels report individual EBNA2 variant frequencies detected in healthy donor and multiple sclerosis (MS) samples, respectively, through the Illumina MiSeq amplicon sequencing (negative values are used for MS variants). Each dot indicates the individual-specific frequency of a variant at a given EBNA2 position, colored according to a heat map. Positions shared among different individuals are displayed by multiple dots in the same position.
We found no correlation between EBV genotypes and clinical characteristics of patients, except for a trend toward lower disease duration in subjects bearing the 1.2 allele. We did not find any correlation between genetic variants of EBV and HLA haplotypes (including those known to be associated with MS)23 of the donors. The HLA frequencies were not significantly different between individuals bearing the 1.2 or the 1.3 EBV alleles, with the exception of a nominally significant higher positivity for A2*0201 (p = 0.025), Cw4*04 (p = 0.04), and DRB1*011 (p = 0.03) alleles among 1.3 EBV–positive individuals, whose nominal p values did not withstand the correction for multiple comparisons (table e-1).
DISCUSSION
In a study that controls for HLA haplotype and clinical/MRI features, we show that the MS status correlates with an excess of EBNA2 1.2 and a defect of 1.3B alleles. Moreover, patients are more likely to harbor newly identified 1.2 allele–related variants, particularly at amino acid position 245. Previous studies on a possible association between EBV genotypes and MS generated mixed results, including the following: no association with EBNA6, EBNA1, and latent membrane protein, or EBNA29–12; “marginally different frequencies” for tegument protein BRRF2 and EBNA113; more frequent EBNA2 type 1 and 2 coinfection in patients than in healthy subjects14; and a broader range of genetic variants in children with MS in the context of an increased rate of EBV reactivation.15 Different patterns of EBV genetic variations in different geographic regions could account for inconsistencies, as suggested by the different geographic distributions of EBV-related malignancies.3–5 By investigating a region of EBNA2 that previous studies in MS cohorts did not investigate, we have detected MS-associated EBV genetic variations that are stronger than those previously reported and seem to be independent of the donors' HLA haplotype.
Apart from the predictive potential of EBNA2 variants, the likelihood that these variants may induce functional consequences, possibly contributing to disease etiology, deserves consideration. In a virus that is not prone to mutations,24 new variants are more likely to have a functional impact. There are only 2 major types of EBV (type 1 and 2), which appear to be identical over the bulk of the genome but show allelic polymorphism in a subset of latent genes: EBNA-LP, EBNA2, EBNA3A, EBNA3B, and EBNA3C.24 In particular, the 2 EBV types share 64% of the nucleotide sequence and 53% of the predicted amino acid sequence of EBNA2, and EBNA2 sequence mutations are known to affect interaction with host proteins.25 We studied the most polymorphic region of EBNA2, where the nucleotide sequence divergence between type 1 and type 2 resides, and found a higher than expected number of new variants, as assessed using a conventional sequencing method and confirmed by next-generation sequencing. The extent of genotype intraindividual variability proved to be high in both patients and controls with a greater occurrence of coexisting alleles in patients, possibly accounted for by the known higher rate of EBV reactivation in MS.26,27 Our study demonstrates the appropriateness of deep-sequencing platforms to investigate patterns of genetic variation of a virus that latently infects peripheral blood cells at a very low frequency. Next-generation sequencing proved to be sensitive to such an extent that when used to study the EBV genotype in immune infiltrates microdissected from postmortem brain samples of a patient with secondary progressive MS (see appendix e-1), it allowed identification of a sequence of the EBNA2 allele 1.1 and related variants (table e-3) that was not detected using a conventional sequencing method. It is plausible that in the near future, deep-sequencing platforms will provide a solution to the controversy on the presence and pathologic significance of a deregulated EBV infection in the MS brain.28
Viral genotype variants are crucial for the outcome of host–virus interactions. The polymorphisms we found in the EBNA2 gene may have functional consequences as supported by recent data showing that a single amino acid in EBNA2 determines superior B lymphoblastoid cell line growth maintenance.29 The allelic variants we described may affect the host–virus interplay at different levels: (1) the same region is involved in interactions with cellular proteins such as Nur7730 and SMARCB131 that have been associated with MS susceptibility32 and antiviral responses33; (2) the gene region we investigated is important for successful strategies of EBV immune evasion34,35; (3) last, but not least, the EBNA2 messenger RNA, where we found MS-related variants, belongs to the targetome of human micro-RNAs that may serve to keep in check EBV infection.36
MS may resemble other immune-mediated diseases in which infectious agents are thought to have a key role. For example, enterovirus strains, through persistent infection of pancreatic beta cells, may trigger type 1 diabetes mellitus in susceptible hosts.37 The murine norovirus strain CR6 that establishes persistent infection in mice, but not the nonpersistent strain CW3, contributes to a phenotype of Crohn disease in mice bearing the risk allele of the autophagy gene ATG16L1.38 Finally, distinct hepatitis C virus genotypes are known to affect the clinical outcome (clearance/persistence) and the response to therapy through differential induction of interferon-inducible genes and signaling pathways.39 The present work shows that the reported associations between viral variants and other immune-mediated diseases may also be true for MS, through variants with a potentially functional impact on the host. These results may be complementary to our recent studies showing that a portion of the genetic predisposition to MS may be attributable to variants in genes that interact with EBV.40 Specifically, EBNA2 binding motifs were found to be significantly enriched in genomic intervals associated with MS (Ricigliano et al., unpublished), suggesting that a complex interplay between both host and viral genetic variants may contribute to disease development. Besides EBNA2, it cannot be excluded that additional regions of the complex EBV genome may be relevant as well.
Supplementary Material
ACKNOWLEDGMENT
The authors thank Dr. Francesca Aloisi and Dr. Nicola Vanacore (Istituto Superiore di Sanità, Rome) for helpful discussion and suggestions. The authors thank all patients and controls for their participation in this study.
GLOSSARY
- AUC
area under the curve
- CI
confidence interval
- EBNA
Epstein-Barr nuclear antigen
- EBV
Epstein-Barr virus
- HD
healthy donor
- HLA
human leucocyte antigen
- MS
multiple sclerosis
- NCBI
National Center for Biotechnology Information
- OR
odds ratio
- PBMC
peripheral blood mononuclear cell
- ROC
receiver operating characteristic
Footnotes
Supplemental data at Neurology.org
AUTHOR CONTRIBUTIONS
R.M., C.M., G.R., G.P., M.S., F.R.G., S.D. conceived and designed the experiments. R.M., C.P., C.M., A.A., C.A., B.S., B.R. performed the experiments. R.M., C.M., R.U., E.P., A.M.D. analyzed the data. R.M., C.P., V.A., D.F.A., M.C.B., A.F., F.B., V.A.G.R., S.R., D.C. selected patients and collected samples. R.M., G.R., G.P., M.S. wrote the manuscript. R.M., G.P., L.B., G.R., M.S. provided data interpretation. L.B., S.D., F.R.G. provided critical reading of the manuscript.
STUDY FUNDING
This study was supported by grants from Fondazione Italiana Sclerosi Multipla (2011/R/31 to M.S., 2011/R/14 to S.D., and 2013/R/12 to L.B.), from Ministero della Salute (Progetto Strategico 2007 to G.R. and M.S., Progetto Strategico grant 107 to M.S. and L.B.), Fondazione Cariplo (grant 2010-0728 to S.D.), and Ministero dell'Istruzione dell'Università e della Ricerca (PRIN2008 to S.D. and M.S.).
DISCLOSURE
R. Mechelli holds a patent application concerning EBV genotypes in MS. C. Manzari reports no disclosures relevant to the manuscript. C. Policano holds a patent application concerning EBV genotypes in MS. A. Annese and E. Picardi report no disclosures relevant to the manuscript. R. Umeton holds a patent application concerning EBV genotypes in MS. A. Fornasiero, A. D'Erchia, M. Buscarinu, C. Agliardi, V. Annibali, B. Serafini, B. Rosicarelli, S. Romano, D. Angelini, V. Ricigliano, F. Buttari, and L. Battistini report no disclosures relevant to the manuscript. D. Centonze acted as an advisory board member of Merck Serono, Teva, Bayer Schering, Biogen Idec, and Novartis, and received funding for traveling and honoraria for speaking or consultation fees from Merck Serono, Teva, Novartis, Bayer Schering, Sanofi-Aventis, and Biogen Idec. He is the principal investigator in clinical trials for Novartis, Merck Serono, Teva, Bayer Schering, Sanofi-Aventis, Biogen Idec, and Roche. F. Guerini, S. D'Alfonso, and G. Pesole report no disclosures relevant to the manuscript. M. Salvetti holds a patent application concerning EBV genotypes in MS. M. Salvetti receives research support and has received fees as speaker from Sanofi-Aventis, Biogen, Bayer Schering, and Merck Serono. G. Ristori holds a patent application concerning EBV genotypes in MS. Go to Neurology.org for full disclosures.
REFERENCES
- 1.Lünemann JD. Epstein-Barr virus in multiple sclerosis: a continuing conundrum. Neurology 2012;78:11–12. [DOI] [PubMed] [Google Scholar]
- 2.Ascherio A, Munger KL. Environmental risk factors for multiple sclerosis: part I: the role of infection. Ann Neurol 2007:61:288–299. [DOI] [PubMed] [Google Scholar]
- 3.Chang CM, Yu KJ, Mbulaiteye SM, Hildesheim A, Bhatia K. The extent of genetic diversity of Epstein-Barr virus and its geographic and disease patterns: a need for reappraisal. Virus Res 2009;143:209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen JN, Ding YG, Feng ZY, et al. Association of distinctive Epstein-Barr virus variants with gastric carcinoma in Guangzhou, southern China. J Med Virol 2010;82:658–667. [DOI] [PubMed] [Google Scholar]
- 5.Sawada A, Croom-Carter D, Kondo O, et al. Epstein-Barr virus latent gene sequences as geographical markers of viral origin: unique EBNA3 gene signatures identify Japanese viruses as distinct members of the Asian virus family. J Gen Virol 2011;92:1032–1043. [DOI] [PubMed] [Google Scholar]
- 6.Santpere G, Darre F, Blanco S, et al. Genome-wide analysis of wild-type Epstein-Barr virus genomes derived from healthy individuals of the 1000 Genomes Project. Genome Biol Evol 2014;6:846–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Babcock GJ, Decker LL, Volk M, Thorley-Lawson DA. EBV persistence in memory B cells in vivo. Immunity 1998;9:395–404. [DOI] [PubMed] [Google Scholar]
- 8.Munch M, Hvas J, Christensen T, Møller-Larsen A, Haahr S. A single subtype of Epstein-Barr virus in members of multiple sclerosis clusters. Acta Neurol Scand 1998;98:395–399. [DOI] [PubMed] [Google Scholar]
- 9.Lindsey JW, Patel S, Zou J. Epstein-Barr virus genotypes in multiple sclerosis. Acta Neurol Scand 2008;117:141–144. [DOI] [PubMed] [Google Scholar]
- 10.Simon KC, Yang X, Munger KL, Ascherio A. EBNA1 and LMP1 variants in multiple sclerosis cases and controls. Acta Neurol Scand 2011;124:53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lay ML, Lucas RM, Toi C, et al. Epstein-Barr virus genotypes and strains in central nervous system demyelinating disease and Epstein-Barr virus-related illnesses in Australia. Intervirology 2012;55:372–379. [DOI] [PubMed] [Google Scholar]
- 12.Brennan RM, Burrows JM, Bell MJ, et al. Strains of Epstein-Barr virus infecting multiple sclerosis patients. Mult Scler 2010;16:643–651. [DOI] [PubMed] [Google Scholar]
- 13.Santón A, Cristóbal E, Aparicio M, et al. High frequency of co-infection by Epstein-Barr virus types 1 and 2 in patients with multiple sclerosis. Mult Scler 2011;17:1295–1300. [DOI] [PubMed] [Google Scholar]
- 14.Yea C, Tellier R, Chong P, et al. Epstein-Barr virus in oral shedding of children with multiple sclerosis. Neurology 2013;81:1392–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tierney RJ, Edwards RH, Sitki-Green D, et al. Multiple Epstein-Barr virus strains in patients with infectious mononucleosis: comparison of ex vivo samples with in vitro isolates by use of heteroduplex tracking assays. J Infect Dis 2006;193:287–297. [DOI] [PubMed] [Google Scholar]
- 16.Ling PD, Hayward SD. Contribution of conserved amino acids in mediating the interaction between EBNA2 and CBF1/RBPJk. J Virol 1995;69:1944–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Polman CH, Reingold SC, Banwell B, et al. Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann Neurol 2011;69:292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Picardi E, Pesole G. REDItools: high-throughput RNA editing detection made easy. Bioinformatics 2013;29:1813–1814. [DOI] [PubMed] [Google Scholar]
- 19.Olerup O, Zetterquist H. HLA-DR typing by PCR amplification with sequence-specific primers (PCR-SSP) in 2 hours: an alternative to serological DR typing in clinical practice including donor-recipient matching in cadaveric transplantation. Tissue Antigens 1992;39:225–235. [DOI] [PubMed] [Google Scholar]
- 20.Friedman N, Geiger D, Goldszmidt M. Bayesian Network classifiers. Machine Learning 1997;29:131. [Google Scholar]
- 21.Cohen JI. Epstein-Barr virus infection. N Engl J Med 2000;343:481–492. [DOI] [PubMed] [Google Scholar]
- 22.Gutierrez MI, Splangler G, Kingma D, et al. Epstein-Barr virus in nasal lymphomas contains multiple ongoing mutations in the EBNA-1 gene. Blood 1998;92:600–606. [PubMed] [Google Scholar]
- 23.International Multiple Sclerosis Genetics Consortium, Wellcome Trust Case Control Consortium 2, Sawcer S, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 2011;476:214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sample J, Young L, Martin B, et al. Epstein-Barr virus types 1 and 2 differ in their EBNA-3A, EBNA-3B, and EBNA-3C genes. J Virol 1990;64:4084–4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klein G, Klein E, Kashuba E. Interaction of Epstein-Barr virus (EBV) with human B-lymphocytes. Biochem Biophys Res Commun 2010;396:67–73. [DOI] [PubMed] [Google Scholar]
- 26.Angelini DF, Serafini B, Piras E, et al. Increased CD8+ T cell response to Epstein-Barr virus lytic antigens in the active phase of multiple sclerosis. PLoS Pathog 2013;9:e1003220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mechelli R, Anderson J, Vittori D, et al. Epstein-Barr virus nuclear antigen-1 B-cell epitopes in multiple sclerosis twins. Mult Scler 2011;17:1290–1294. [DOI] [PubMed] [Google Scholar]
- 28.Lassmann H, Niedobitek G, Aloisi F, Middeldorp JM; NeuroproMiSe EBV Working Group. Epstein-Barr virus in the multiple sclerosis brain: a controversial issue—report on a focused workshop held in the Centre for Brain Research of the Medical University of Vienna, Austria. Brain 2011;134:2772–2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tzellos S, Correia PB, Karstegl CE, et al. A single amino acid in EBNA-2 determines superior B lymphoblastoid cell line growth maintenance by Epstein-Barr virus type 1 EBNA-2. J Virol 2014;88:8743–8753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee JM, Lee KH, Weidner M, Osborne BA, Hayward SD. Epstein-Barr virus EBNA2 blocks Nur77-mediated apoptosis. Proc Natl Acad Sci USA 2002;99:11878–11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwiatkowski B, Chen SY, Schubach WH. CKII site in Epstein-Barr virus nuclear protein 2 controls binding to hSNF5/Ini1 and is important for growth transformation. J Virol 2004;78:6067–6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Achiron A, Feldman A, Gurevich M. Characterization of the multiple sclerosis traits: nuclear receptors (NR) impaired apoptosis pathway and the role of 1-alpha 25-dihydroxyvitamin D(3). J Neurol Sci 2011;311:9–14. [DOI] [PubMed] [Google Scholar]
- 33.Cui K, Tailor P, Liu H, et al. The chromatin-remodeling BAF complex mediates cellular antiviral activities by promoter priming. Mol Cell Biol 2004;24:4476–4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lucchesi W, Brady G, Dittrich-Breiholz O, et al. Differential gene regulation by Epstein-Barr virus type 1 and type 2 EBNA2. J Virol 2008;82:7456–7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee JM, Lee KH, Farrell CJ, et al. EBNA2 is required for protection of latently Epstein-Barr virus-infected B cells against specific apoptotic stimuli. J Virol 2004;78:12694–12697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skalsky RL, Corcoran DL, Gottwein E, et al. The viral and cellular microRNA targetome in lymphoblastoid cell lines. PLoS Pathog 2012;8:e1002484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hober D, Sauter P. Pathogenesis of type 1 diabetes mellitus: interplay between enterovirus and host. Nat Rev Endocrinol 2010;6:279–289. [DOI] [PubMed] [Google Scholar]
- 38.Cadwell K, Patel KK, Maloney NS, et al. Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine. Cell 2010;141:1135–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Donlin MJ, Cannon NA, Aurora R, et al. Contribution of genome-wide HCV genetic differences to outcome of interferon-based therapy in Caucasian American and African American patients. PLoS One 2010;5:e9032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mechelli R, Umeton R, Policano C, et al. A “candidate-interactome” aggregate analysis of genome-wide association data in multiple sclerosis. PLoS One 2013;8:e63300. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.