Abstract
Orbital inflammatory diseases include thyroid eye disease (TED), granulomatosis with polyangiitis (GPA), sarcoidosis, and nonspecific orbital inflammation (NSOI). Histopathological diagnosis usually relies on the clinical context and is not always definitive. Gene expression profiling provides diagnostic and therapeutic information in several malignancies, but its role in evaluating nonmalignant disease is relatively untested. We hypothesized that gene expression profiling could provide diagnostic information for NSOI. We collected formalin-fixed, paraffin-embedded orbital biopsies from 10 institutions and 83 subjects including 25 with thyroid eye disease, 25 nonspecific orbital inflammation, 20 healthy controls, 6 with granulomatosis with polyangiitis, and 7 with sarcoidosis. Tissues were divided into discovery and validation sets. Gene expression was quantified using Affymetrix U133 Plus 2.0 microarrays. A random forest statistical algorithm based on data from 39 probe sets identified controls, GPA, or TED with an average accuracy of 76% (p=0.02). Random forest analysis indicated that 52% of tissues from patients with nonspecific inflammation were consistent with a diagnosis of GPA. Molecular diagnosis by gene expression profiling will augment clinical data and histopathology in differentiating forms of orbital inflammatory disease.
Keywords: Orbital inflammation, granulomatosis with polyangiitis, sarcoidosis, thyroid eye disease, gene expression
INTRODUCTION1
Molecular diagnosis using transcriptomics has demonstrated tremendous utility in malignant diseases (Hoadley et al., 2014) such as forms of lymphoma (Bohen et al., 2003; Dave et al., 2006)or ocular melanoma. (van Gils et al., 2008) The added diagnostic value of profiling gene expression in nonmalignant diseases is less certain, but utility has been reported in inflammatory disorders such as myocarditis (Lassner et al., 2014), synovitis (Yeremenko et al., 2013), and esophagitis. (Wen et al., 2013)
Orbital inflammation is an important cause of morbidity that results in pain, diplopia, and sometimes visual loss. Orbital biopsy helps to distinguish malignancy or infection from inflammation. Graves disease is the most common cause of orbital inflammation. It is often referred to as thyroid eye disease (TED). Additional systemic disease associations with orbital disease include sarcoidosis, granulomatosis with polyangiitis (GPA) (formerly Wegener’s granulomatosis), sarcoidosis, Crohn’s disease, IgG4-related disease, and histiocytosis. Many patients suffer from an inflammatory process that is difficult to categorize. These patients are labelled with terms such as nonspecific orbital inflammation (NSOI), orbital pseudotumor, or idiopathic orbital inflammation. Many of these diagnoses are suggested by clinical context and difficult to make based on histopathology alone. For example, the inflammatory infiltrate in TED can be scant such that, when TED affects orbital fat, it is sometimes difficult to distinguish TED from normal orbital adipose tissue. The diagnosis of GPA requires a vasculitis affecting a medium size vessel, but vessels of this size are rarely obtained on an orbital biopsy. Many patients suffer from an inflammatory process that is difficult to categorize. These patients are labelled with terms such as nonspecific orbital inflammation (NSOI), orbital pseudotumor, or idiopathic orbital inflammation.
While orbital inflammation is not rare, many patients do not undergo biopsy which entails some morbidity and expense. In order to understand nonspecific orbital inflammation, we organized an international consortium of orbital surgeons and pathologists. We have recently reported on the transcripts expressed by tissue representing the four most common diagnoses: TED, sarcoidosis, GPA, and NSOI (Rosenbaum et al., manuscript submitted). In this report, we test the hypothesis that a diagnostic algorithm based on a limited number of transcripts could complement observations made by experienced pathologists.
METHODS
Centers, Biopsies, Database
This study was approved by the Institutional Review Board (IRB) at Oregon Health & Science University (IRB00006301) and at each of the other contributing centers. The research adhered to the tenets of the Declaration of Helsinki. Formalin-fixed, paraffin-embedded (FFPE) samples and relevant demographic and clinical data were obtained from 10 institutions. The diagnoses of NSOI, sarcoidosis, GPA, TED, and normal were based on the clinical and histopathological information obtained and submitted by orbital disease specialists and ocular pathologists from their respective institutions. All biopsies were further reviewed by two of the authors (DJW and HEG) as noted below.
Biopsies of the orbital adipose tissue from a total of 83 subjects were studied (20 controls with no known orbital disease, 25 with NSOI, 6 with GPA, 7 with sarcoidosis, 25 with TED). The age, gender, diagnoses, and clinical information supporting each diagnosis has been reported elsewhere (Rosenbaum et al., manuscript submitted).
Pathology Review
Two ocular pathologists (D.J.W. and H.E.G.) independently evaluated hematoxylin and eosin stained slides of all samples for histopathological characteristics without reference to the indications for biopsy or other clinical information. After rendering a diagnosis in a masked fashion, the pathologists were informed of the diagnosis from the institution where tissue had been obtained. In some cases, additional stains were requested or additional clinical information was reviewed. A few cases with an ambiguous diagnosis were excluded. In all cases included in this study, the diagnosis reached by Drs. Wilson and Rosenbaum agreed with the contributing center’s diagnosis. Thus, a consensus diagnosis which combined clinical and histopathological information was considered the gold standard to which we compared the diagnoses rendered by either computer algorithm or by an expert pathologist.
Tissue Preparation and Gene Expression Profiling
Prior to gene array analysis, the tissues were roughly evenly divided on the basis of time of receipt at OHSU. This created an initial discovery set and a validation set. RNA extraction and microarray assays were performed in the OHSU Gene Profiling Shared Resource. For each specimen, multiple 10–20 μm sections were collected and total RNA was extracted with the mRNeasy FFPE kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol. RNA was amplified and labeled with SensationPlus FFPE Amplification and 3′ IVT Labeling kits (Affymetrix, Santa Clara, CA) for microarray analysis. For the majority of samples, an input of 50 ng of RNA was used, and a minimum input of 20 ng RNA was used for samples with limiting RNA recoveries. The standard Affymetrix protocol was followed for hybridizing biotin-labeled cDNA targets with a GeneChip Human Genome U133 Plus 2.0 array (Affymetrix, Santa Clara, CA). The Human U133 Plus 2.0 array contains over 54,000 probe sets for 47,000 human transcripts and variants. Following hybridization, arrays were processed and stained according to standard Affymetrix protocols, then scanned on the GeneChip Scanner 3000 7G system (Affymetrix). Affymetrix GeneChip Command Console (AGCC) v. 3.1.1 and Affymetrix Expression Console v. 1.1 software were used for image processing and expression analysis for initial quality control, respectively.
Statistical Analysis
Affymetrix CEL files of each set were preprocessed separately by the Robust Multiarray Analysis. (Irizarry et al., 2003) The posterior logarithm of odds that the gene is differentially expressed (Smyth, 2004) when comparing GPA or TED samples versus uninflamed controls were used to select probe sets for principal coordinate plots (Mardia, 1978) and to classify NSOI samples into non-NSOI disease groups by using random forests, an ensemble learning method for classification. (Breiman, 2001) Prediction accuracies by random forest and by pathologists were compared by confusion matrices. (Kohavi and Provost, 1998) Principal coordinate analyses of the selected probe sets were employed for graphical presentation. Statistical computing was performed with ‘affy’, ‘caret’ ‘MASS’, ‘limma’ and ‘randomForest’ packages in the R project (http://www.r-project.org).
RESULTS
NSOI is heterogeneous in symptoms, histopathology, and response to treatment. The extreme variability of NSOI histopathology is illustrated in Figure 1. Tissue can vary markedly in terms of the degree of inflammation, the extent of uninflamed adipose tissue, the amount of fibrosis, and the presence of granuloma.
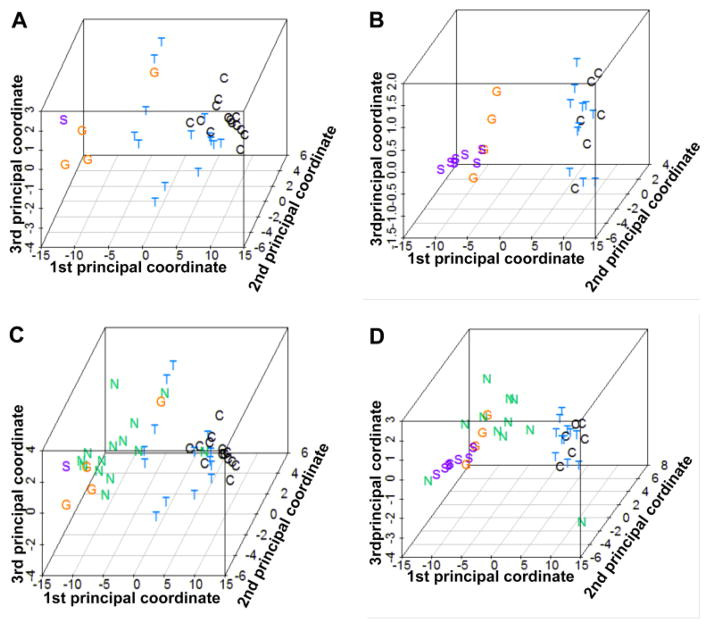
Figure 1.
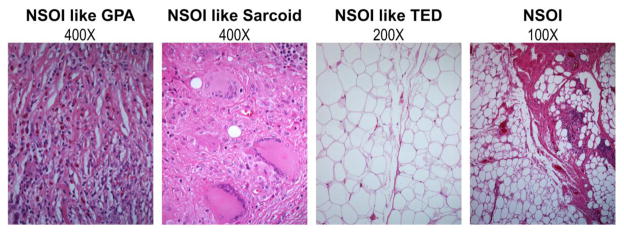
Representative images demonstrate the histologic variability of anterior orbit in subjects diagnosed with NSOI.
Tissues were divided based on the time of collection into a discovery set and a validation set. We previously reported the heterogeneity of gene expression in NSOI tissues and the clustering of the other diseases based on analyses that included all probe sets with significantly different signals for disease versus uninflamed control comparisons in both data sets (Rosenbaum et al., manuscript submitted). In this current study, we distinguish the disease groups based on a minimal number of probe sets. To visualize the complex profiles, we employed a principal coordinate analysis (PCA). In a PCA, the relative similarity of transcriptomes of two samples is indicated by their proximity on a graph.
The previously reported clustering (manuscript submitted) was replicated with a small subset of the data (Figure 2). For each comparison of TED or GPA versus normal, 20 top-ranked probe sets were selected according to the posterior logarithm of odds that the gene is differentially expressed. (Smyth, 2004) One probe set was common to both lists, so the combined list included 39 probe sets. PCA plots based on the 39 selected probe sets for the all of the samples with a consensus diagnosis of control, TED, GPA or sarcoidosis from set 1 or set 2 are shown in Figures 2A and 2B, respectively. The clustering of samples representing normal tissue indicates the relative similarity of the tissue. The samples from subjects with TED are also relatively homogeneous and have the closest proximity to normal samples among the four disease groups. The gene expression profiles from subjects with either sarcoidosis or GPA also tend to cluster. Figures 2C and 2D show the same PCA plots with the addition of the NSOI samples from the respective data sets. The spatial relationships depicted in Figures 2C and 2D are better appreciated in videos (Discovery set PCA video.MP4 and Validation set PCA video.mp4) since the relationships are 3 dimensional. The gene expression profile for patients with NSOI showed more heterogeneity than the other four categories consistent with the hypothesis that NSOI is not a single disease entity. The NSOI sample that clustered with sarcoidosis contained non-caseating granuloma on histopathology. A diagnosis of sarcoidosis was not made because the patient had no pulmonary symptoms and neither a chest x-ray nor a chest CT scan was obtained.
Figure 2.
PCA plots based on 39 probe sets reveal expression profile distances of individual samples within each experimental group. Panels A and B show the relative clustering of data points within the control, GPA, TED, and sarcoidosis groups in the discovery set (A) and validation (B). Panels C (discovery set) and D (validation set) reveal how NSOI samples are distributed with many in close proximity to the GPA samples. Rotatable views of the 3-dimensional plots in panels C and D are in online supplemental files.
Two ophthalmic pathologists were asked to render a histological diagnosis in the absence of clinical information. Both pathologists correctly identified the 7 tissues from patients with sarcoidosis on the basis of non-caseating granulomas. The pathologists were not asked to distinguish between NSOI and TED due to the absence of discriminating features. For these cases, the pathologists were credited with the correct diagnosis if the consensus diagnosis based on both histopathology and clinical data was either NSOI or TED.
Pathologist one scored 75/83 tissues because he chose not to evaluate 8 normal tissues (Table 1). This table shows his diagnoses compared to the consensus diagnosis for 38 tissues. He diagnosed “other” for 5 tissues and then used descriptive terms such as “mild inflammation”. These cases are excluded from Table 1. Table 1 also does not include the 25 tissues representing NSOI since by definition, the pathology is nonspecific. Finally, Table 1 excludes the 7 tissues from patients with sarcoidosis as these were correctly identified by both pathologists. For 43 tissues, he selected the NSOI/TED option (Table 1). Of these 43 dual diagnoses, ten subjects had neither of the two options. Pathologist two evaluated 83 tissue samples and indicated the NSOI/TED diagnoses for 70 tissues (Table 1). Twenty times the consensus diagnosis was neither of these.
Table 1.
The expert pathologists could not accurately distinguish between normal control and TED/NSOI tissues and between GPA and TED/NSOI tissues. Results for data sets 1 and 2 are combined, and the erroneous predictions are indicated in bold. The table does not include tissue from patients with sarcoidosis which were diagnosed accurately. It also does not include tissues representing NSOI since by definition the pathology is non-specific.
| Consensus Diagnosis
|
Pathologist 1 Prediction (38 tissues)
|
Pathologist 2 Prediction (51 tissues)
|
||||
|---|---|---|---|---|---|---|
| Control | GPA | TED/NSOI | Control | GPA | TED/NSOI | |
| Control | 6 | 0 | 6 | 2 | 0 | 18 |
| GPA | 0 | 2 | 4 | 0 | 1 | 5 |
| TED | 9 | 0 | 12 | 0 | 0 | 25 |
Pathologist 2 added additional comments to his evaluation of the discovery set of tissues that illustrate limitations of histopathologic evaluation of tissues with lower levels of inflammation. Twelve of 14 normal control tissues were classified as NSOI/TED with 10 having a comment that they may be normal tissues depending on clinical findings. All 14 TED tissues were correctly classified as NSOI/TED, but 8 also had the comment that they could be normal control tissues. Thirteen of 14 NSOI tissues were correctly classified as NSOI/TED with one of the 13 being marked as possibly normal.
We employed the random forest classifier applied to the discovery data set to develop an algorithm to identify either TED or GPA using the same 39 probe sets as used for the PCA plots shown in Figure 2. We then tested this algorithm for its ability to distinguish among the 3 entities (control, TED and GPA) from data set two. We excluded sarcoidosis because this could be accurately diagnosed by histopathology. We excluded NSOI because by definition, its histological features are nonspecific. Results are shown in Table 2. The algorithm performed better than either pathologist (who was not provided with clinical information) as judged by sensitivity and specificity for the identical tissues. The average accuracy of the algorithm was 76% compared to the pathologists who had accuracies of 49% and 58%, respectively. A confusion matrix showed that the algorithm performed significantly better than “no information rate” for which all selections are set to the disease with the highest frequency (p=0.02), while neither pathologist was better than the corresponding no information rate (p>0.24 and p>0.67).
Table 2.
An algorithm based on GPA versus control and TED versus control gene expression differences in set 1 did better at distinguishing control, GPA, and TED samples in set 2 than predictions based on histopathology alone (Table 1). Tissues from patients with NSOI or sarcoidosis were not included as explained in the legend for Table 1.
| Consensus Diagnosis
|
Algorithm Prediction (21 Tissues)
|
||
|---|---|---|---|
| Control | GPA | TED | |
| Control | 6 | 0 | 5 |
| GPA | 0 | 4 | 0 |
| TED | 0 | 0 | 6 |
Finally we applied the algorithm to the 25 tissues from patients with NSOI. The algorithm which was designed to distinguish among GPA, TED, and normal, assigned a diagnosis of GPA to 13 tissues, a diagnosis of TED to 10, and a diagnosis of normal to 3. We interpret this as support of the PCA plots indicating that a subset of patients with NSOI has a gene expression profile very similar to GPA. The algorithm, however, was not designed to identify sarcoidosis and classified the sarcoid-like NOSI sample as GPA. Further, the algorithm cannot assign a diagnosis of “none of the above”; i.e., the algorithm cannot recognize the possibility that a subset of patients with NSOI might have neither GPA nor TED. Of the 13 tissues assigned a diagnosis of GPA by the algorithm, only 2 were classified by a pathologist as GPA and another was noted by a pathologist to be possibly GPA. Two of the TED tissues were also noted by a pathologist to be possibly GPA.
DISCUSSION
While molecular diagnosis has demonstrated marked success in subdividing malignant diseases, it has had fewer applications in inflammatory disease. Orbital inflammatory disease is ideal to test the potential of molecular diagnosis. First, there is a broad differential diagnosis. Second, the various diagnoses often have therapeutic implications. Standard therapy, for example, for GPA differs from standard therapy for sarcoidosis. Third, evaluation by histopathology alone does not always allow a precise diagnosis, as is clearly shown in this study. In the absence of clinical data, the expert pathologists in this study were not able to accurately differentiate normal tissue from TED, or vice versa. In addition, there were not sufficient histopathological clues to establish a diagnosis of GPA, in the absence of the clinical information. The gene expression algorithm was able to differentiate these diagnoses to a greater degree of specificity, suggesting the information available in gene expression profiling adds to the diagnostic process independent from the visual clues evident on histopathological examination. Finally, the heterogeneous cases classified together as NSOI can potentially be divided into meaningful subsets based on the combination of clinical data, histopathological evaluation, and gene expression
A molecular approach to the diagnosis of GPA would have great value. Diagnosing GPA when it involves the lung and kidney and a positive anti-neutrophil cytoplasmic antibody (ANCA) test is usually not a great challenge. But many patients with GPA have a limited form of the disease involving tissues such as sinus, sclera, orbit, nasal mucosa, or subglottis. The ANCA is often negative in limited disease. (Reid, 1979) Furthermore a definitive diagnosis from biopsy of a tissue like sinus or orbit is very low, in part because medium size vessels are not typically biopsied. A molecular signature for GPA could have implications for biopsy of several different tissues. NSOI patients should be studied prospectively with gene expression profiling to see if a group of patients with a GPA phenotype can be identified. These patients might benefit from a more specific treatment regimen.
Our study provides quantitative data indicating that even experts may struggle to make a single diagnosis from an orbital biopsy in the absence of clinical information. In actual practice, of course, pathologists do not render a diagnosis in the absence of clinical information just as a clinician does not rely solely on laboratory tests without observations from a medical history and physical examination. In this study, the pathologists were deprived of clinical information to reduce their accuracy artificially in order to create a comparator for the algorithm. An algorithm should be tested prospectively to confirm if it can add to information gathered by histopathology. Our result suggests that in practice, wherein diagnoses are not made in the absence of clinical data, an algorithm based on expression levels of a small number of genes could augment diagnosis and choice of therapy. Because our study included a small number of patients with known GPA, further refinement of the algorithm that we employed is required. Our random forest analysis supports a conclusion which we reported elsewhere (Rosenbaum, JT, in review): many patients with NSOI may have a limited form of GPA. Our observations strongly support the concept that molecular diagnosis can contribute greatly to the understanding of inflammatory diseases.
Supplementary Material
Highlights.
Orbital inflammation has a broad differential diagnosis.
An international team quantified specific RNA transcripts in orbital biopsies.
This information enhanced the diagnostic accuracy of biopsies.
Acknowledgments
RNA extraction and microarray assays were performed in the OHSU Gene Profiling Shared Resource. We are grateful to Kristina Vartanian for her excellent technical support for the microarray work.
Financial support was from NIH Grants EY020249, EY010572 and RR024140, Research to Prevent Blindness, the William and Mary Bauman Foundation, the Mas Family Foundation, and the Stan and Madelle Rosenfeld Family Trust.
APPENDIX I
Members of the Orbital Disease Consortium
Columbia University, New York, NY, USA: Michael Kazim, Payal Patel
Emory University, Atlanta, GA, USA: Hans E. Grossniklaus
King Khaled Eye Specialist Hospital, Riyadh, Saudi Arabia: Deepak P. Edward, Hind Alkatan, Hailah al Hussain
Medical College of Wisconsin, Milwaukee, WI USA: Gerald J Harris
Ohio State University, Columbus, OH, USA: Jill A. Foster
Ohio University, Columbus, OH, USA: Craig N. Czyz
Oregon Health & Science University, Portland OR, USA: Dongseok Choi, Roger A. Dailey, Christina A. Harrington, John D. Ng, Stephen R. Planck*, James T. Rosenbaum*, Cailin Sibley, Patrick Stauffer, Eric A. Steele, David J. Wilson
Royal Adelaide Hospital, Adelaide, SA, Australia: Dinesh Selva
University of British Columbia, Vancouver, BC, Canada: Valerie White, Peter Dolman
University of California, San Diego, CA, USA: Bobby Korn, Don Kikkawa
University of Miami, FL, USA: David Tse, Chris Alabiad, Sander Dubovy, Prashant Parekh
Wake Forest University, Winston-Salem, NC, USA: Patrick Yeatts
*also Legacy Health System, Portland, OR, USA
Footnotes
Abbreviations: GPA - granulomatosis with polyangiitis; NSOI – nonspecific orbital inflammation; TED – thyroid eye disease
Potential Conflict of Interest: James Rosenbaum has in the past consulted for Genentech and was a co-investigator on a studied funded by Genentech to evaluate the use of rituximab for orbital inflammatory diseases. The other authors report no potential competing interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bohen SP, et al. Variation in gene expression patterns in follicular lymphoma and the response to rituximab. Proc Natl Acad Sci USA. 2003;100:1926–1930. doi: 10.1073/pnas.0437875100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L. Random Forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- Dave SS, et al. Molecular diagnosis of Burkitt’s lymphoma. N Engl J Med. 2006;354:2431–42. doi: 10.1056/NEJMoa055759. [DOI] [PubMed] [Google Scholar]
- Hoadley KA, et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158:929–44. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, et al. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohavi R, Provost R. Glossary of Terms. Machine Learning. 1998;30:271–274. [Google Scholar]
- Lassner D, et al. Improved diagnosis of idiopathic giant cell myocarditis and cardiac sarcoidosis by myocardial gene expression profiling. Eur Heart J. 2014;35:2186–2195. doi: 10.1093/eurheartj/ehu101. [DOI] [PubMed] [Google Scholar]
- Mardia KV. Some properties of classical multi-dimensional scaling. Commun Stat Theory Methods. 1978;A7:1233–1241. [Google Scholar]
- Reid L. Bronchopulmonary dysplasia--pathology. J Pediatr. 1979;95:836–41. doi: 10.1016/s0022-3476(79)80446-1. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article 3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- van Gils W, et al. Expression profiling in uveal melanoma provides a strong marker for survival and reveals two regions on chromosome 3p related to prognosis. Invest Ophthalmol Vis Sci. 2008;49:4254–4262. doi: 10.1167/iovs.08-2033. [DOI] [PubMed] [Google Scholar]
- Wen T, et al. Molecular diagnosis of eosinophilic esophagitis by gene expression profiling. Gastroenterology. 2013;145:1289–99. doi: 10.1053/j.gastro.2013.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeremenko N, et al. Disease-specific and inflammation-independent stromal alterations in spondylarthritis synovitis. Arthritis Rheum. 2013;65:174–85. doi: 10.1002/art.37704. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.