Abstract
Studies of spontaneous mutations in mice have provided valuable disease models and important insights into the mechanisms of human disease. Ruffled (rul) is a new autosomal recessive mutation causing abnormal hair coat in mice. The rul allele arose spontaneously in the RB156Bnr/EiJ inbred mouse strain. In addition to an abnormal coat texture, we found diffuse epidermal blistering, abnormal electrocardiograms (ECGs), and ventricular fibrosis in mutant animals. Using high-throughput sequencing (HTS) we found a frameshift mutation at 38,288,978 bp of chromosome 13 in the desmoplakin gene (Dsp). The predicted mutant protein is truncated at the c-terminus and missing the majority of the plakin repeat domain. The phenotypes found in Dsprul mice closely model a rare human disorder, Carvajal-Huerta Syndrome. Carvajal-Huerta Syndrome (CHS) is a rare cardiocutaneous disorder that presents in humans with woolly hair, palmoplantar keratoderma and ventricular cardiomyopathy. CHS results from an autosomal recessive mutation on the 3′ end of Desmoplakin (DSP) truncating the full length protein. The Dsprul mouse provides a new model to investigate the pathogenesis of CHS, as well as the underlying basic biology of the adhesion molecules coded by the desmosomal genes.
Keywords: Dsp, hair shaft, gene networks, mouse model, Carvajal-Huerta Syndrome
Introduction
Desmosomes are one of the major cell-cell adhesion complexes, along with adherens, tight and gap junctions (Brooke et al 2012; Galicano et al 1998). The desmosomes consist of three main entities: 1) intercellular desomosomal cadherins (desmocollin and desmoglein), 2) intracellular linker proteins called plakoglobin and plakophillin, and 3) intracellular cytoskeletal linker called desmoplakin (Brooke et al 2012; Galicano et al 1998). The strong cell-cell adhesive bond afforded by the desomosomes is important in maintaining the integrity of epithelia and the resistance to mechanical forces experienced by cardiomyocytes (Vasioukhin et al, 2001). Desmoplakin functions to link the desmosmal complex to intermediate filaments of the cytoskeleton, such as the keratins (Brooke et al 2012; Galicano et al 1998). Previous work has shown that DSP is expressed in all layers of the epidermis (Bierkamp et al, 1999), as well as in the hair follicle outer root sheath, companion layer, and in basal areas of Henle’s and Huxley’s layers (Alibardi and Bernd 2013;Kurzan et al 1989). It is also a critical part of the intercalated discs binding cardiomyocytes together in the heart (Brooke et al 2012; Galicano et al 1998). Human genetic lesions affecting the function of DSP result in a number of disease including dilated cardiomyopathy with wooly hair, keratoderma and tooth agensis (DCWHKTA; OMIM #615821) (Norgett, et al 2006), familial arrhythmogenic right ventricular dysplasia 8 (ARVD8; OMIM #607450) (Rampazzo et al 2002), lethal acantholytic epidermolysis bullosa (OMIM #609638) (Jonkman et al, 2005) and Carvajal-Huerta Syndrome (OMIM #605676) (Carvajal-Huerta, 1998).
We report here a new spontaneous recessive mouse mutation that arose in 1989 in the RB156Bnr/EiJ recombinant inbred strain at The Jackson Laboratory. The phenotype manifests as an abnormal, ruffled coat texture that appears at 10 days of age and is maintained through the life of the animal, hence the allele name, ruffled (rul). In mutant animals, we observed sporadic epidermal blistering, intermittent pelage hair swelling, widespread vibrissa follicular dystrophy, ventricular fibrosis and electrocardiographic abnormalities. Using high-throughput sequencing, we found that rul is a mutation in mouse desmoplakin (Dsp) gene. While targeted alleles of Dsp have been previously reported, rul is the first known spontaneously arising mutation in Dsp. The mutant gene and the phenotype of RB156Bnr/Ei-rul/GrsrJ mice support this strain as a new model of human syndromes that involve integumentary and cardiac defects like Carvajal-Huerta Syndrome.
Materials and Methods
Mice
Phenotypically deviant mice with ruffled coat texture were identified in the RB156Bnr/EiJ inbred strain at The Jackson Laboratory (Bar Harbor, ME). Affected animals were crossed to the C57BL/6EiJ inbred strain to test heritability of the allele. No affected animals were observed in the F1 generation, but affected F2 animals were obtained from subsequent intercrosses between F1 progeny, demonstrating a recessive mode of inheritance for the rul allele. Both male and female rul/rul homozygotes were fertile, and the mutant strain, RB156BNR/Ei/J-rul/J has been maintained since 1989 by crossing homozygotes with phenotypically unaffected heterozygotes. The treatment and use of all mice in this study were compliant with protocols approved by The Jackson Laboratory Animal Care and Use Committee at the Mouse Mutant Resource (MMR) of The Jackson Laboratory (Bar Harbor, ME, USA).
Genetic Linkage Analysis
The rul mutation was genetically mapped in an intercross with CAST/EiJ. CAST/EiJ inbred mice were obtained from The Jackson Laboratory (Bar Harbor, ME, USA). A rul homozygous mouse was mated with a wild type CAST/EiJ mouse. The phenotypically normal F1 progeny from this mating were then intercrossed and affected (rul/rul) mice were genotyped using strain-specific microsatellite markers to distinguish CAST/EiJ alleles from C57BL/6EiJ alleles across all chromosomes.
High-throughput sequencing
Whole exome sequencing was used to identify candidate mutations in the mapped region, as described previously (Fairfield et al. 2011). Briefly, genomic DNA was prepared using Qiagen DNeasy Blood and Tissue kit (Qiagen, Santa Clarita, CA USA) or by phenol/chloroform extraction. The DNA was enriched for coding sequence by hybridization-based capture using SeqCap EZ Mouse Exome SR (Roche NimbleGen). Post-hybridization amplification was completed via Illumina LMPCR protocol as previously described (Fairfield et al. 2011). The resulting enriched libraries were used in cluster formation on an Illumina cBot (Illumina, San Diego, CA). Paired-end sequencing was done using the Illumina HiSeq. The sequencing data were analyzed using tools and workflows provided by GenomeQuest including mapping (HS3), SNP calling, and annotation of variants (GenomeQuest, Boston, MA). Analysis focused on novel variants, which were not positioned in repetitive sequence, and had expected allele ratios (>0.95 for homozygous variants and >0.2 for heterozygous variants), as well as sufficient locus coverage (at least 5X for homozygous variants and 10X for heterozygous variants). Protein coding or splice variants within the mapped interval that were unique when compared to ~100 unrelated exome data sets and inbred strain, variant data generated by the Sanger Mouse Genomes Project (Keane et al. 2011; Mouse Genomes Project) were flagged as candidate mutations. High priority was given to protein coding or splice variants within mapped regions, as well as unique variants that were not found in other exome data sets or in data generated by the Sanger Mouse Genomes Project (Keane et al. 2011; Mouse Genomes Project).
Mutation Validation
Candidate mutations were validated by PCR amplification and capillary sequencing of homozygous rul/rul and heterozygous rul/+ genomic DNA samples. Sequencing data were analyzed using Sequencher 4.9 (Gene Codes Corp., Ann Arbor, MI, USA). Primers rul_Dsp1L (AGGAAGACCGGCAGTCAGTA), rul_Dsp1R (GGGAGAGCTTCTGACCAGTG) and rul_Dsp2L (CGACACGACTCCGTGAGTAA), rul_Dsp2L (CTTCTTCGGTGCTGATCCTC) were designed using Primer3 software (Rozen and Skaletsky, 2000). PCR reactions were performed using the following cycling conditions: preheating at 95°C for 3 min; 40 cycles of amplification at 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s. PCR products were purified by The Jackson Laboratory’s Genome Sciences group using Agencout’s AMPure XP magnetic beads (Agencourt, Bioscience Corp., Beverly, MA). Sanger sequencing reations were setup using 5uL of purified PCR product and 1 uL of primer at 5 pmol/ul. Cycle sequencing of DNA samples was performed using ABI BigDye Terminator ready reaction kit v3.1 and run on an ABI 3730xl (Applied Biosystems, Carlsbad, CA). Sequence reads were analyzed and aligned using Sequencher software (Gene Codes Corp., Ann Arbor, MI).
Electrocardiogram (ECG)
Cardiac electrical activity was recorded from conscious mice gently restrained in a Plexiglas cylinder using metal pads to collect the signal. The animals were restrained in a Murine ECG Restrainer (Emka Technologies, Paris, France), and the signal acquisition used a PowerLab System (AD Instruments, Golden CO). The QRS Phenotyping system consists of an ECG platform with conductive metal for foot contact a restrainer tube, with small breathing hole and an adjustable tail gate. There are three different cylinder sizes to accommodate different size animals. The tailgate sizer is used to limit the lateral space within the restrainer (to prevent the mouse from turning around) and is adjusted so that the animal’s front feet and left hind foot are positioned on the appropriate pad. The tail is able to exit the restrainer through a slit in the tail gate. Mice were housed in the testing room overnight for acclimation before testing. After the mouse was gently placed in chamber, the ECG signal was recorded for 5 minutes or until a steady baseline signal was recorded for a 20 to 60 second interval. Some mice take longer to acclimate and settle down, in which case they were left in chamber for longer period. Mice were returned to their home cage and then euthanized for histopathological analysis of heart.
Statistics
All statistics were carried out on ECG data using the statistics program JMP v11 (SAS International, Cary, NC). A Shapiro-Wilk test was used to determine normality of the data with the assumption that normally distributed data will give a p-value >0.05. RR Interval (p=0.4812), QRS (p=0.0803), Heart Rate (p=0.2607), QT Interval (p=0.1739), QTc (p=0.6663), P amplitude (p=0.1060), Q amplitude (p=0.3790), S amplitude (p=0.0615), and T amplitude (p=0.5716) data were found to be normally distributed, while PR Interval (p=0.0163), P duration (p=0.0085), and R amplitude (p=0.0012) were not normally distributed. RR Interval, QRS Interval, Heart rate, QT Interval, QTc, P amplitude, Q amplitude, S amplitude, T amplitude data were analyzed using a Student’s t-test, while PR interval, P duration, and R amplitude data were analyzed by Mann-Whitney tests. P values less than 0.05 were considered significant for all statistical tests.
Histology
To define histological lesions, 12 week old Dsprul/+ and Dsprul/rul animals were heavily sedated by interperitoneal injection of 2% Tribromoethanol (600–800 mg/kg) until no toe pinch reaction was observed. Animals were then perfused with Bouin’s solution by intracardiac perfusion. After perfusion animals were immersed in Bouin’s solution until tissue harvest. Tissues, including brain, spinal cord, thyroid, thymus, heart, lung, liver, diaphragm, kidney, adrenal glands, spleen, intestines, urinary bladder, skin, testis, uterus, ovary, hind legs, tail, and foot were washed overnight, trimmed, processed and embedded routinely in paraffin, sectioned, and stained with hematoxylin and eosin (H&E). Serial sections of neural tissue, brain and spinal cord, were stained with Luxol Fast Blue. Pelage hair samples were manually removed from Dsprul/Dsprul and Dsprul/Dsp+ mice and mounted on glass slides. Histological sections of the heart were obtained after ECG readings were completed. Briefly, 4 and 32 week old animals are euthanized by CO2 asphyxiation and subsequent cervical dislocation. Hearts were harvested and placed in Fekete’s acid-alcoholformalin solution overnight. Hearts were then transferred to 70% ethanol for storage before trimming of tissues, paraffin embedding, and sectioning. Heart sections were stained with Masson’s Trichrome and H&E. All slides are examined by an experienced board certified veterinary anatomic pathologist (JPS).
Western Blotting
Euthanized Dsprul/rul and Dsp+/+ animals were shaved and dorsal skin samples were harvested. Skin was cut into approximately 1 mm × 1 mm pieces, placed in ice-cold extraction buffer (10 mM Imidazole, 100 mM NaCl, 1 mM MgCl2, 5 mM Na2EDTA and 1% Triton X-100) and macerated using a plastic pestle. The lysate was centrifuged at 14,000 g at 4°C for 15 minutes, after which the supernatant was collected and protein concentration was determined using the Bradford assay (Bradford, 1976). Protein samples, 30 μg of soluble extract, were reduced in 6× sample buffer (125 mM Tris-HCl pH 6.8, 2% Sodium dodecylsulfate (SDS), 20% Glycerol, 0.2% Bromophenol Blue and 5% β-Mercaptoethanol (BME)) by boiling for 5 minutes and resolved on a 7.5% SDS-PAGE gel. Subsequently, the gels were electroblotted onto Polyvinylidene fluoride (PVDF) membranes (Invitrogen, Carlsbad, CA) at 115 mA for 9 hrs at room temperature. Non-specific binding of antibodies to the PVDF membrane was blocked by incubation in blocking buffer (5% nonfat dried milk in Tris-buffered saline with 0.1% Tween-20 (TBST)). Next, the blots were incubated with appropriate primary antibody overnight at 4°C or for 2 hours at 37°C diluted in blocking buffer as follows: rabbit anti-mouse Desmoplakin I/II 1:1000 (Santa Cruz Biotechnology, Santa Cruz, CA) and rabbit anti-mouse α-Tubulin 1:5000 (Abcam, Cambridge, MA). This was followed by incubation of the blots for an additional hour in donkey anti-rabbit IgG-HRP conjugate diluted to 1:10,000 (Santa Cruz Biotechnology, Santa Cruz, CA) in blocking buffer. Signal detection was performed using the enhanced chemilluminescence-plus western blotting substrate (ECL+) (Pierce Biotechnology, Rockford, IL) followed by exposure to x-ray film. An internal loading control, α-tubulin, was used to indicate that similar amounts of protein extract were used in all lanes of the Western Blots performed.
Results
Dsprul allele causes a sparse, ruffled coat in the RB156Bnr/Ei-rul/GrsrJ inbred strain of mice
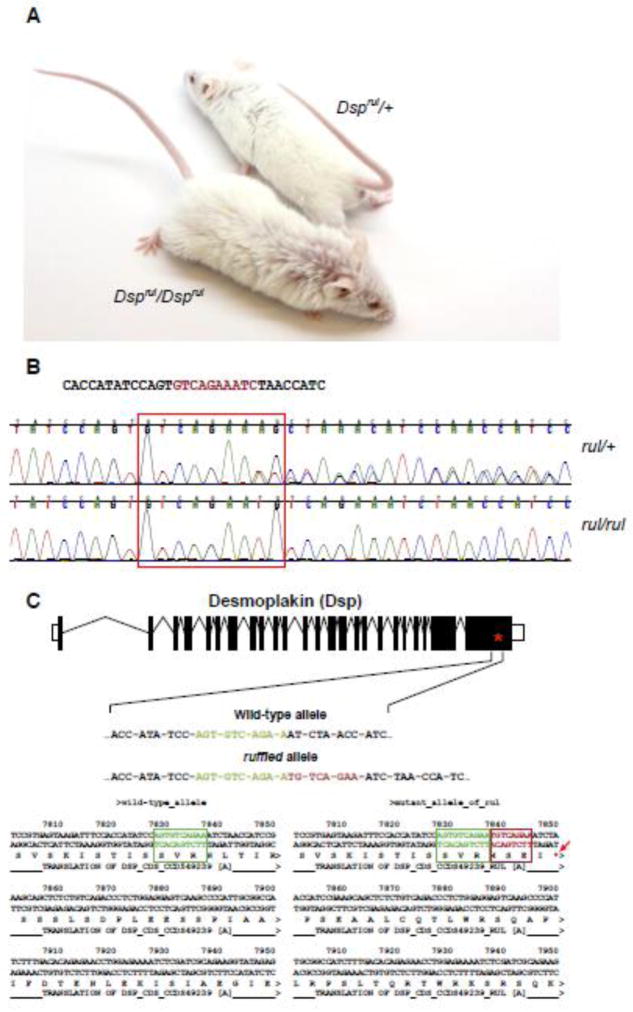
Ruffled (rul) mice exhibit a sparse, disheveled coat, with slight curling of the hair. (Figure 1A). Phenotypically deviant mice can be identified as early as 10 days of age and maintain the ruffled coat throughout their lifetime, while presumptive heterozygote animals are not visibly affected. This is unlike many similar coat texture mutant mice, such as caracul (Krt71Ca) and caracul-like (Cal), which completely lose or have greatly diminished coat texture deviations from the norm as adults. To determine heritability of the mutant allele an RB156Bnr/Ei-rul/GrsrJ mutant (Dsprul/rul) was outcrossed to an inbred C57BL/6JEiJ female. This cross produced normal appearing F1 mice. Subsequent intercrossing of the F1 progeny resulted in affected and unaffected F2 animals in numbers approximating Mendelian expectations for an autosomal recessive inheritance pattern.
Figure 1.
(A) Mice homozygous for the mutant Dsp allele (Dsprul/Dsprul) present with ruffled hair as opposed to heterozygote littermates (Dsprul/+). (B) High throughput sequencing identified a homozygous insertion (highlighted nucleotide sequence) in the Dsp gene in affected mice (red box and arrow) not found in the heterozygous wild-type allele or C57BL/6J-derived sequence. (C) This insertion results in a frame shift of desmoplakin (Dsp) and the introduction of four novel amino acids followed by a premature termination codon.
A complementation test was performed with the rul/rul mice using mutant strains with similar phenotypic features, including lanceolate hair (Dsg4lah), serum/glucocorticoid regulated kinase 3, fuzzy (Sgk3fz), cathepsin L, furless (Ctslfs), GATA binding protein 3, juvenile alopecia (Gata3jal), protease, serine 8 (prostasin), frizzy (Prss8fr), transforming growth factor alpha, waved 1 (Tgfawa1), epidermal growth factor receptor, waved 2 (Egfrwa2), curly whiskers (cw) and wellhaarig (we). In all cases, rul complemented the other mutations indicating that rul is not an allele of any of the test genes.
Genetic linkage analysis was performed using a panel of microsatellite markers spaced across the mouse genome. The ruffled mutation was intercrossed with CAST/EiJ to determine linkage. Affected animals from the RB156Bnr/Ei-rul/GrsrJ x CAST/EiJ cross were mapped using MIT markers whereby the variant allele was localized to a 4.8Mb region on mouse chromosome 13 between D13Mit3 (20,431,342bp) and D13Mit19 (43,767,657bp) (NCBIM38/mm10) which contained 34 protein coding genes.
A Dsp frameshift mutation is likely to be the molecular basis of the mutant ruffled phenotype
Whole exome sequencing of a spleen sample from an affected ruffled animal revealed an insertion allele on Chromosome 13 at position 38,288,978 (GRCm38/mm10) of GTGTCAGAA to GTGTCAGAATGTCAGAA. This single insertion allele nominated for validation in the ruffled mutant correlated with the ruffled coat phenotype when additional affected (rul/rul) and unaffected samples (rul/+) were tested (Figure 1B). The insertion results in a frameshift beginning in the last exon (exon 24) of the desmoplakin gene (Dsp) and results in the introduction of four novel amino acids followed by a premature termination codon (Figure 1C). This nonsense mutation is predicted to result in a truncated DSP. This mutation, which only affects the c-terminus of the protein, is predicted to result in a partially functional protein with spectrin repeat elements and chromosome segregation ATPase domain intact, but partial loss of the highly conserved plectin repeats. Importantly, mice homozygous for a targeted null mutation in Dsp are embryonic lethal (Gallicano et al., 1998), indicating that this frameshift mutation is not likely to result in a completely non-functional DSP protein.
Protein analysis
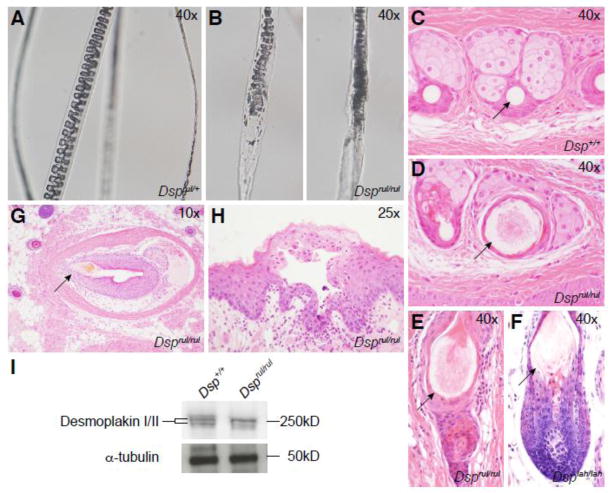
Western blot analysis of dorsal skin samples revealed abnormal protein banding patterns in Dsprul/rul mice as compared to wild type controls (Figure 2I). A doublet at the expected size of 250 kilodaltons (kD), representing both DSP I and DSP II isoforms, was found in wild type samples. Wild type desmoplakin isoforms exhibited nearly identical intensity indicating each DSP isoform is expressed at similar levels. In Dsprul/rul protein extracts, a doublet banding pattern was also observed. However, in mutant samples the intensity of the larger isoform was much higher than that of the smaller isoform indicating differential expression or stability.
Figure 2.
Dsprul homozygous mouse hair shafts displayed abnormal development under both polarized (A) and white (B) light. Abnormally swollen pelage hair fibers were observed in cross sections of the hair follicles from affected mice (D,E) as compared to wild type animals (C), reminiscent of the lesion found in Dsg4lah/lah mice (F). (G) Vibrissae dystrophy was observed throughout the muzzle. (H) Small suprabasal blisters were found in the oral region of deviant mice. (I) Western blot analysis of wild type (lane 1) and Dsprul (lane 2) skin extracts demonstrate a slight difference in molecular weights between wild type and control desmoplakin as well as intensity difference in the bands of the mutant samples.
Histopathology of Dsprul/rul mice revealed both integumental and heart defects
Microscopic examination of hair plucked and mounted from Dsprul/+ (Figure 2A) and Dsprul/rul mice (Figure 2B) revealed uniformly shaped hair shafts with a regular septation or septulation pattern, depending on hair type in the normal mice. By contrast, while many hair shafts were normal in the mutant mice, individual hair shafts exhibited structural deformities with focal loss of septulation patterns. In histological sections of the skin most pelage hair follicles appeared to be relatively normal. In horizontal sections, mutant hair shafts were larger in diameter than in controls with loss of the normal concentric layering (Fig. 2C,D). In vertical sections, focal areas of degenerative change appeared as swellings of the hair shaft with loss of the septulation pattern of the medulla and a pointed tip (Fig. 2E) reminiscent of the lesion seen in Dsg4lah-J mice (Figure 2F) (Kljuic et al 2003; Sundberg et al, 2000). Dsprul/rul mouse also exhibited acanthosis in dorsal skin sections when compared to wild type skin epidermis (data not shown). Examination of the Dsprul/Dsprul skin on the muzzle revealed widespread vibrissae follicular dystrophy (Figure 2G, arrow). Lastly, observations of the oral mucosa of mutant mice uncovered suprabasilar blister and pustule formation manifesting as separations at the epidermal/dermal border in and around the mouth and tongue of affected mice (Figure 2H).
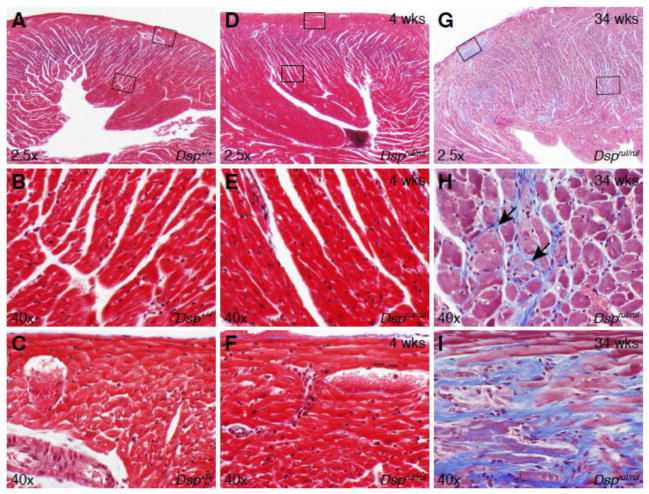
Histological sections of the heart revealed lesions associated with aging of the mutant mice. Initial histological investigations of young (4 weeks) mutant mice did not reveal any abnormal heart findings, but subsequent investigations into older mice (34 weeks) showed cardiomyopathic changes in the heart. Figure 3 compares histological sections of hearts stained with Masson’s Trichrome to easily differentiate between the cardiac muscle (red) and intervening collagen fibers (blue). The wild type (Figure 3A–C) and young mutant mouse (Figure 3D–F) sections are very similar and display no lesions in the left ventricular free wall (Figure 3A,B,D, and E) or near the epicardium (Figure 3A,C,D, and F). Yet, older mutant mouse (34 weeks) heart sections (Figure 3G–I) exhibit extensive areas of fibrosis (Figure 3G and H, arrows) in the left ventricular free wall, as well as a prominent fibrotic lesion near the apex of the heart (Figure 3G and I).
Figure 3.
Dsprul heart sections stained with Masson’s Trichrome. (A) 4 week old wild-type littermate control heart section. High magnification view of (B) left ventricular free wall (lower box in panel A) and (C) epicardium (upper box in panel A). (D) 4 week old homozygous mouse heart section. High magnification view of the (E) left ventricular free wall and (F) epicardium. (G) 34 week old homozygous, left ventricular free wall illustrating extensive fibrosis and a prominent fibrotic lesion. (H) High magnification view of left ventricular free wall fibrosis in aged homozygous mice (middle box in panel G). (I) High magnification view of a fibrotic lesion in the apex of the left ventricle (top, left box in panel G).
Dsprul/rul mice exhibit electrocardiographic defects as compared to wild type littermates
In light of the heart histological findings, electrocardiograms (ECG) were performed on both young (4 weeks old) and old (34 week) mutant and control mice to determine if the mice displayed any cardiac conduction defects as a result of the fibrosis. Table 1 illustrates a comparison between 4 week old Dsprul/+ and Dsprul/rul animals. There are statistically significant (p<0.05) differences in several measured parameters including longer R-R and QRS intervals, smaller R wave and T-wave amplitudes, and a slower heart rate (BPM). The mutant mouse ECG suggests a form of atrial tachycardia. Patients suffering from Carvajal-Huerta Syndrome present with increasing cardiac problems as they age. As such, ECG measurements were taken on 34 week old mice to test whether there was a decrease in cardiac function (Table 2). No changes in cardiac function were identified between affected and unaffected animals. Further work revealed that the RB156Bnr/EiJ background strain has significant cardiac abnormalities as they age including changes in RR interval, heart rate, PR interval, QRS interval, Q amplitude, T amplitude, and RS amplitude (Table 3). Thus, comparisons were done of 34 week old Dsprul/rul ECGs to 4 week old Dsprul/+ ECG to determine if any changes had occurred over time that correspond to the mutant phenotype (Table 4). Comparison of older mutant mice to young unaffected animals revealed significant changes in P amplitude, S amplitude, T amplitude, and RS amplitude. The background strain cardiac issues contribute to the decrease in cardiac function as these mutant animals age making it difficult to isolate the effects of the rul allele on the ECG parameters. ECG data along with the histopathological observations indicate that the mutant allele significantly affects heart structure and function.
Table 1.
| ECG Measurements | rul/+ (4 wk) (Mean) | STDEV | rul/rul (4 wk)(Mean) | STDEV | p-values |
|---|---|---|---|---|---|
| RR Interval (ms) | 71.6831 | 0.9324 | 82.2624 | 5.6406 | 0.0007 |
| Heart Rate (BPM) | 837.1818 | 10.9558 | 736.7225 | 38.3181 | 0.0001 |
| PR Interval (ms) | 26.3670 | 1.4856 | 30.3536 | 6.6450 | 0.7150 |
| P Duration (ms) | 7.2606 | 1.7714 | 7.9050 | 0.6577 | 0.7150 |
| QRS Interval (ms) | 12.3255 | 0.5390 | 14.2029 | 1.9494 | 0.0243 |
| QT Interval (ms) | 23.1529 | 3.8077 | 26.3332 | 3.1023 | 0.0845 |
| QTc (ms) | 86.4747 | 14.4771 | 92.7726 | 12.5440 | 0.2328 |
| P Amplitude (mV) | 0.1244 | 0.0420 | 0.1372 | 0.0603 | 0.3438 |
| Q Amplitude (mV) | −0.0936 | 0.0396 | −0.0714 | 0.0320 | 0.1709 |
| R Amplitude (mV) | 0.5244 | 0.0521 | 0.5948 | 0.0322 | 0.0285 |
| S Amplitude (mV) | −0.3969 | 0.1571 | −0.3911 | 0.1207 | 0.4735 |
| T Amplitude (mV) | 0.1276 | 0.0206 | 0.1981 | 0.0350 | 0.0012 |
| RS Amplitude (mV) | 0.9194 | 0.0638 | 0.9858 | 0.6989 | 0.2502 |
Table 2.
| ECG Measurements | rul/+ (34 wk) (Mean) | STDEV | rul/rul (34 wk)(Mean) | STDEV | p-values |
|---|---|---|---|---|---|
| RR Interval (ms) | 81.5491 | 1.5094 | 77.6564 | 2.0450 | 0.0543 |
| Heart Rate (BPM) | 736.0320 | 13.8186 | 773.2965 | 20.3491 | 0.0568 |
| PR Interval (ms) | 32.9010 | 4.8816 | 30.9753 | 2.4296 | 0.5637 |
| P Duration (ms) | 8.3810 | 0.2126 | 8.7174 | 0.8733 | 0.5637 |
| QRS Interval (ms) | 16.6060 | 1.7606 | 14.0663 | 1.8921 | 0.1148 |
| QT Interval (ms) | 23.4741 | 2.5729 | 24.9790 | 1.6058 | 0.2332 |
| QTc (ms) | 82.1720 | 8.2113 | 89.7801 | 6.7398 | 0.1672 |
| P Amplitude (mV) | 0.0909 | 0.0419 | 0.0610 | 0.0055 | 0.1379 |
| Q Amplitude (mV) | −0.0041 | 0.0560 | −0.0476 | 0.0225 | 0.1451 |
| R Amplitude (mV) | 0.4090 | 0.2899 | 0.3588 | 0.2238 | 0.5637 |
| S Amplitude (mV) | −0.1923 | 0.0792 | −0.1673 | 0.0377 | 0.3271 |
| T Amplitude (mV) | 0.0520 | 0.0365 | 0.0645 | 0.0175 | 0.3134 |
| RS Amplitude (mV) | 0.6018 | 0.1468 | 0.5262 | 0.1199 | 0.3583 |
Table 3.
| ECG Measurements | rul/+ (4 wk) (Mean) | STDEV | rul/+ (34 wk) (Mean) | STDEV | p-values |
|---|---|---|---|---|---|
| RR Interval (ms) | 71.6831 | 0.9324 | 81.5491 | 1.5094 | 0.0001 |
| Heart Rate (BPM) | 837.1818 | 10.9558 | 736.0320 | 13.8186 | 0.0001 |
| PR Interval (ms) | 26.3670 | 1.4856 | 32.9010 | 4.8816 | 0.0455 |
| P Duration (ms) | 7.2606 | 1.7714 | 8.3810 | 0.2126 | 0.5050 |
| QRS Interval (ms) | 12.3255 | 0.5390 | 16.6060 | 1.7606 | 0.0005 |
| QT Interval (ms) | 23.1529 | 3.8077 | 23.4741 | 2.5729 | 0.4586 |
| QTc (ms) | 86.4747 | 14.4771 | 82.1720 | 8.2113 | 0.6438 |
| P Amplitude (mV) | 0.1244 | 0.0420 | 0.0909 | 0.0419 | 0.1830 |
| Q Amplitude (mV) | −0.0936 | 0.0396 | −0.0041 | 0.0560 | 0.0215 |
| R Amplitude (mV) | 0.5244 | 0.0521 | 0.4090 | 0.2899 | 1.0000 |
| S Amplitude (mV) | −0.3969 | 0.1571 | −0.1923 | 0.0792 | 0.0695 |
| T Amplitude (mV) | 0.1276 | 0.0206 | 0.0520 | 0.0365 | 0.0042 |
| RS Amplitude (mV) | 0.9194 | 0.0638 | 0.6018 | 0.1468 | 0.0437 |
Table 4.
| rul/+ (4 wk) (Mean) | STDEV | rul/rul (34 wk)(Mean) | STDEV | p-values | |
|---|---|---|---|---|---|
| RR Interval (ms) | 71.6831 | 0.9324 | 77.6564 | 2.0450 | 0.1165 |
| Heart Rate (BPM) | 837.1818 | 10.9558 | 773.2965 | 20.3491 | 0.0923 |
| PR Interval (ms) | 26.3670 | 1.4856 | 30.9753 | 2.4296 | 0.6547 |
| P Duration (ms) | 7.2606 | 1.7714 | 8.7174 | 0.8733 | 0.1797 |
| QRS Interval (ms) | 12.3255 | 0.5390 | 14.0663 | 1.8921 | 0.4630 |
| QT Interval (ms) | 23.1529 | 3.8077 | 24.9790 | 1.6058 | 0.2587 |
| QTc (ms) | 86.4747 | 14.4771 | 89.7801 | 6.7398 | 0.3606 |
| P Amplitude (mV) | 0.1244 | 0.0420 | 0.0610 | 0.0055 | 0.0395 |
| Q Amplitude (mV) | −0.0936 | 0.0396 | −0.0476 | 0.0225 | 0.1535 |
| R Amplitude (mV) | 0.5244 | 0.0521 | 0.3588 | 0.2238 | 0.1797 |
| S Amplitude (mV) | −0.3969 | 0.1571 | −0.1673 | 0.0377 | 0.0115 |
| T Amplitude (mV) | 0.1276 | 0.0206 | 0.0645 | 0.0175 | 0.0005 |
| RS Amplitude (mV) | 0.9194 | 0.0638 | 0.5262 | 0.1199 | 0.0116 |
Molecular pathway analysis
The clinical association of “woolly hair” and juvenile cardiomyopathies is regarded as so coherent that dermatological presentation of woolly hair in human patients is a strong indication that cardiac investigations should be made to exclude the possibility of presence of the cardiocutaneous syndromes, Naxos and Carvajal-Huerta syndromes (Williams et al, 2011). The association between woolly hair and cardiac abnormalities extends to other syndromes such as Noonan syndrome and cardiofaciocutaneous (CFC) syndrome, and sporadic disease (Chien et al, 2006; Thergaonkar and Bhat 2013). It is now established that significant phenotypic overlap between diseases often indicates underlying lesions either in the same gene or in genes acting epistatically within the same physiological pathway or network. The strong associations in the cardiocutaneous syndromes suggest that the underlying pathways affected are common at least in part, to both cardiac and skin development and physiology. Naxos disease (Protonotarios and Tsatsopoulou 2005) and Carvajal-Huerta Syndrome (Carvajal-Huerta, 1998) both present with woolly hair, cardiomyopathy and palmoplantar keratoderma. Cardiopathology in Naxos disease generally presents as an arrhythmogenic right ventricular dysplasia (ARVD) while in Carvajal syndrome left ventricular failure often predominates.
Most mutations in ARVD patients are found in plakophilin-2 (PKP2) followed by the desmoglein-2 (DSG2), desmoplakin (DSP), and desmocollin-2 (DSC2) (Simpson et al 2009) all genes involved in the formation and function of the desmosome. More recently mutations in the DSG2 gene have been associated with left ventricular (LV) cardiac phenotypes. Autosomal recessive forms of ARVD often manifest as part of Naxos or Carvajal-Huerta Syndromes. Naxos disease involving predominantly the right ventricle is caused by homozygous junction plakoglobin (JUP) mutations, ARVD is associated with palmoplantar keratoderma and woolly hair, whereas Carvajal-Huerta Syndromes is associated with homozygous DSP mutations.
Using pathway analysis, combined with phenotypic similarity scores we surveyed existing human and mouse data to establish whether further genes may be implicated in the common network underlying Naxos and Carvajal-Huerta Syndromes, of which Dsp is a key member.
The Ingenuity Pathways Analysis© software (Invitrogen, Carlsbad, CA) was used to determine the relationship of Dsp to other genes known to cause similar clinical features to ruffled mice including Prss8, Krt14, Gata3, Ctsl, Egfr, and Tgfa (Figure S1; blue circles). As expected Dsp also displayed a close relationship with other desmosomal genes, such as Dsg4, Dsc1, Dsc2, Dsc3 (Figure S1; green circles). Results confirmed a close association with networks associated with dermatologic diseases and conditions (1.7E-8) confirming the importance of Dsp in hair and skin development and function (Figure S1). Additional searching for protein-protein and regulatory interactions in the STRING database (Franceschini et al, 2013) (Figure S2) further showed direct interactions between DSP, PKP1, 2 and 4, together with JUP, and DSG2. Evidence for direct association between DSP and PKP3 was provided through the BioGraph database (Bonne et al, 2003; Liekens et al, 2011).
The Pkp3 gene knockout (Sklyarova et al, 2008), shows a closely related phenotype to that of its interacting partner JUP (Bierkamp et al, 1996), with abnormal hair and skin in Pkp3 mice, and skin lesions in the Jup mice, though these died in utero from the severe effects on cardiogenesis. Pkp3 is not thought to be expressed in the heart and so no effect has been reported there. Pkp4 has not yet been knocked out in mice. In humans, a single proband with a PKP4 mutation and ARVC was not reported to have dermatological abnormalities, strongly suggesting that the shared cardiocutaneous phenotypes in Naxos and Carvajal-Huerta Syndromes is critically dependent on the Pkp2/Dsp/Jup axis and that these are the key network components for hair morphogenesis.
Phenotypic semantic similarity analysis using phenotypic definitions of Naxos and Carvajal-Huerta syndromes provided in the databases was carried out using the Aber-OWL dermophenotype resource (http://jagannath.pdn.cam.ac.uk/aber-owl/dermophenotypes/) Phenomenet (Hoehndorf et al, 2011) and Phenodigm (Smedley et al, 2013) which returned mouse mutants with related phenotypes including, from Aber–OWL Grhl1. The Grhl1 knockout (Wilanowski et al, 2008) shows marked similarities to the skin and hair phenotypes seen in Dsprul, with palmoplantar keratoderma and reduced levels of Dsg1 expression consistent with its binding to the Dsg 1 promoter. DSG1 interacts directly with PKP2, (Chen et al, 2002) DSP and JUP via PKP2 and DSC (Smith et al, 1998). Together this network data suggests that Grhl1 may be an additional functional component of the desmosomal pathway required for normal skin and hair morphogenesis.
Other components implicated in the network include catenin beta 1 (Ctnnb1) (Schmidt-Ullrich et al, 2001), both via direct protein-protein interaction (PPI) and also phenotypically identified via Phenomenet, integrin beta 4, (b1 implicated by PPI in STRING), and the EGF receptor.
Conclusions
A large number of molecular pathways come together to coordinate the development of the hair and skin, yet the mechanisms regulating these pathways are nearly unknown. We demonstrate here that a spontaneous, frame shift mutation in the Dsp gene results in an abnormal, ruffled coat. Histologically, the ruffled mutation results in irregular pelage hair shaft development, intermittent mucosal blistering, and widespread follicular dystrophy of the vibrissae. Of the 5 mutations genetically engineered in the Dsp gene, three are ES cell lines (no live mice) and the other two (Dsptm1Efu and Dsptm2Efu) are embryonic lethal due to the loss of tissue integrity in cardiac muscle and epidermal tissues (Brooke et al 2012; Galicano et al 1998; http://www.informatics.jax.org/allele/summary?markerId=MGI:109611). As such, the Dsprul allele is the only mutant line that produces viable mice that can be studied. Due to the phenotypic differences in the genetically engineered null alleles and the spontaneous Dsprul allele reported here, the Dsprul is therefore not likely to result in a completely non-functional DSP, but is predicted to affect binding of DSP to intracellular structural filaments, thus inhibiting cell adhesion.
Carvajal-Huerta Syndrome (CHS) is a rare cardiocutaneous disorder that presents with woolly hair, palmoplantar keratoderma and left ventricular cardiomyopathy (Carvajal-Huerta, 1998). CHS results from an autosomal recessive mutation on the 3′ end of the human Desmoplakin gene (DSP) resulting in the truncation of the full length protein. The mutated DSP protein is expressed, but missing the c-terminal portion theoretically resulting in disruption of the intermediate filament binding domain. This truncation would result in weaker associations between cells at the desmosomal complex and explains the origin of many of the phenotypes observed in human patients diagnosed with CHS. The Dsprul allelic mutation of desmoplakin in the laboratory mouse closely resembles human CHS clinically, histologically, and the type and location of the mutation in the homologous gene, i.e. truncation of the c-terminal plakin domain. As the mice are viable, this is represents a valuable new model for cardiocutaneous syndromes and CHS in particular.
The relationship between phenotype and genotype for these desmosomal-associated protein mutations is complex. In humans, C terminal mutations in DSP have been associated with a full cardiocutaneous phenotype, for example the 7901delG reported by Norgett et al., (Norgett et al, 2006) and the frameshift mutation detected by Williams et al. (Williams et al, 2001), yet Xu et al (Xu et al, 2010) report four DSP lesions, two in the C terminal portion of the gene, without reported skin phenotypes, as does Yang et al. (Yang et al, 2006). We must conclude that the nature of the mutation is critical in the pathophysiology, and possibly also in embryonic viability as some may be incompatible with survival to term, as seen with the mouse Dsp knockouts. The fact that many ARVC patients are now being found who are heterozygous for multiple abnormal genes in the pathway (Xu et al, 2010) suggests that penetrance or expressivity might be very much lesion-dependent. This may explain why the cardiovascular phenotypes described here in the rul mouse may not completely recapitulate those in humans with related lesions, and it would be interesting to examine the effects of compound mutations with additional lesions, for example in the Pkp2 gene, mirroring the situation reported in some human patients with ARVC.
This new recessive mouse mutation causes abnormal ruffled looking hair that can be clinically identified in affected mice at about 10 days of age. In light of this mutation, we hypothesize that Dsp works in conjunction with a large complex of genes that affects development of the hair shaft as well as epidermal integrity. Initial network analysis indicates an important role for Dsp in hair development; however, further work is needed to establish the underlying mechanisms influencing these phenotypic differences.
Supplementary Material
Network analysis of the relationship of Dsp to other genes known to cause similar clinical features to ruffled mice (Dsg4, Dsc1, Dsc2, Dsc3, Ctsl, Prss8, Gata3, Tgfa, Egfr) was conducted using Ingenuity Pathways Analysis© software (Invitrogen). Results confirmed a close association with networks associated with dermatologic diseases and conditions (1.7E-8). Dsp is circled in red, genes coding for desomosmal proteins are circles in green, genes with mutant alleles producing phenocopies to Dsprul are circled in blue.
Protein-protein and regulatory interactions in the STRING database further showed direct interactions between DSP, PKP1, 2 and 4, together with JUP, and DSG2.
Highlights.
Spontaneous coat texture mutant discovered in a colony of RB156Bnr/EiJ inbred mice.
The causative mutation was found to be a frameshift in the desmoplakin (Dsp) gene.
Mutant mice exhibit phenotypes similar to Carvajal-Huerta Syndrome.
Epidermal blistering, abnormal electrocardiograms (ECGs), and ventricular fibrosis.
Acknowledgments
The authors thank J. Hammer for assistance with the graphics.
This work was supported by the National Institutes of Health (AR053639, AR056635, AR063781, and OD010972), Council for Nail Disorders, Cicatricial Alopecia Research Foundation and mentorship grants from the North American Hair Research Society. Shared scientific services were supported in part by a Basic Cancer Center Core Grant from the National Cancer Institute (CA34196).
Abbreviations
- CHS
Carvajal-Huerta Syndrome
- rul
ruffled
- Dsp/DSP
Desmoplakin gene/protein
- tm
targeted mutation
- HTS
High Throughput Sequencing
- kD
kilodalton
Footnotes
Conflict of Interest: The authors of this manuscript have no conflicts to disclose.
Author contributions are as follows: CHP, CSP, HF, IG, BSH, JPS performed the research, CHP, CSP, LGR, DEB, SSD, JPS designed the study, CHP, CSP, LGR, DEB, SSD, BTL, JPS analyzed the data and wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alibardi L, Bernd N. Immunolocalization of junctional proteins in human hairs indicates that the membrane complex stabilizes the inner root sheath while desmosomes contact the companion layer through specific keratins. Acta Histochemica. 2013;115:519–526. doi: 10.1016/j.acthis.2012.11.011. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Bierkamp C, Schwarz H, Huber O, et al. Desomosomal localization of β-catenin in the skin of plakoglobin null-mutant mice. Development. 1999;126:371–381. doi: 10.1242/dev.126.2.371. [DOI] [PubMed] [Google Scholar]
- Bonné S, Gilbert B, Hatzfeld M, et al. Defining desmosomal plakophilin-3 interactions. The Journal of Cell Biology. 2003;161:403–416. doi: 10.1083/jcb.200303036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooke MA, Nitoiu D, Kelsell DP. Cell-cell connectivity: desmosomes and disease. J Pathol. 2012 Jan;226(2):158–71. doi: 10.1002/path.3027. Epub 2011 Nov 14. [DOI] [PubMed] [Google Scholar]
- Carvajal-Huerta L. Epidermolytic palmoplantar keratoderma with woolly hair and dilated cardiomyopathy. J Am Acad Dermatol. 1998 Sep;39(3):418–21. doi: 10.1016/s0190-9622(98)70317-2. [DOI] [PubMed] [Google Scholar]
- Chen X, Bonne S, Hatzfeld M, et al. Protein binding and functional characterization of plakophilin 2. Evidence for its diverse roles in desmosomes and beta -catenin signaling. The Journal of biological chemistry. 2002;277:10512–10522. doi: 10.1074/jbc.M108765200. [DOI] [PubMed] [Google Scholar]
- Chien AJ, Valentine MC, Sybert VP. Hereditary woolly hair and keratosis pilaris. Journal of the American Academy of Dermatology. 2006;54:S35–S39. doi: 10.1016/j.jaad.2005.01.092. [DOI] [PubMed] [Google Scholar]
- Fairfield H, Gilbert GJ, Barter M, et al. Mutation discovery in mice by whole exome sequencing. Genome Biol. 2011;12(9):R86. doi: 10.1186/gb-2011-12-9-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleckman P, Jaeger K, Silva KA, et al. Comparative anatomy of mouse and human nail unit. Anat Rec (Hoboken) 2013;296(3):521–32. doi: 10.1002/ar.22660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini A, Szklarczyk D, Frankild S, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic acids research. 2013;41:D808–815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallicano GI, Kouklis P, Bauer C, et al. Desmoplakin is required early in development for assembly of desmosomes and cytoskeletal linkage. J Cell Biol. 1998;143:2009–2022. doi: 10.1083/jcb.143.7.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoehndorf R, Schofield PN, Gkoutos GV. PhenomeNET: a whole-phenome approach to disease gene discovery. Nucleic acids research. 2011;39:e119. doi: 10.1093/nar/gkr538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkman MF, Pasmooij AMG, Pasmans SGMA, et al. Loss of desmoplakin tail causes lethal acantholytic epidermolysis bullosa. Am J Hum Genet. 2005;77:653–660. doi: 10.1086/496901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane TM, Goodstadt L, Danecek P, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477(7364):289–94. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kljuic A, Bazzi H, Sundberg JP, et al. Desmoglein 4 in hair follicle differentiation and epidermal adhesion: evidence from inherited hypotrichosis and acquired pemphigus vulgaris. Cell. 2003;113:249–260. doi: 10.1016/s0092-8674(03)00273-3. [DOI] [PubMed] [Google Scholar]
- Kurzan H, Moll I, Moll R, et al. Compositionally different desmosomes in the various compartments of the human hair follicle. Differentiation. 1989;63:295–304. doi: 10.1046/j.1432-0436.1998.6350295.x. [DOI] [PubMed] [Google Scholar]
- Liekens AM, De Knijf J, Daelemans W, et al. BioGraph: unsupervised biomedical knowledge discovery via automated hypothesis generation. Genome biology. 2011;12:R57. doi: 10.1186/gb-2011-12-6-r57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norgett EE, Lucke TW, Bowers B, et al. Early death from cardiomyopathy in a family with autosomal dominant striate palmoplantar keratoderma and wooly hair associated with a novel insertion mutation in desmoplakin (Letter) J Invest Derm. 2006;126:1651–1654. doi: 10.1038/sj.jid.5700291. [DOI] [PubMed] [Google Scholar]
- Protonotarios N and Tsatsopoulou A. Naxos Disease. Indian Pacing and Electrophysiology Journal. 2005;5:76–80. [PMC free article] [PubMed] [Google Scholar]
- Rampazzo A, Nava A, Malacrida S, et al. Mutation in human desmoplakin domain binding to plakoglobin causes a dominant form of arrhythmogenic right ventricular cardiomyopathy. Am J Hum Genet. 2002;71:1200–1206. doi: 10.1086/344208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Ullrich R, Aebischer T, Hulsken J, et al. Requirement of NF-kappaB/Rel for the development of hair follicles and other epidermal appendices. Development. 2001;128:3843–3853. doi: 10.1242/dev.128.19.3843. [DOI] [PubMed] [Google Scholar]
- Silva KA, Sundberg JP. Necropsy methods. In: Hedrich HJ, editor. The Laboratory Mouse. London: Academic Press; 2012. pp. 779–806. [Google Scholar]
- Simpson MA, Mansour S, Ahnood D, et al. Homozygous Mutation of Desmocollin-2 in Arrhythmogenic Right Ventricular Cardiomyopathy with Mild Palmoplantar Keratoderma and Woolly Hair. Cardiology. 2009;113:28–34. doi: 10.1159/000165696. [DOI] [PubMed] [Google Scholar]
- Sklyarova T, Bonne S, D’Hooge P, et al. Plakophilin-3-deficient mice develop hair coat abnormalities and are prone to cutaneous inflammation. The Journal of investigative dermatology. 2008;128:1375–1385. doi: 10.1038/sj.jid.5701189. [DOI] [PubMed] [Google Scholar]
- Smedley D, Oellrich A, Kohler S, et al. PhenoDigm: analyzing curated annotations to associate animal models with human diseases. Database: the journal of biological databases and curation. 2013;2013:bat025. doi: 10.1093/database/bat025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EA, Fuchs E. Defining the interactions between intermediate filaments and desmosomes. J Cell Biol. 1998;141:1229–1241. doi: 10.1083/jcb.141.5.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg JP, Boggess D, Bascom C, et al. Lanceolate hair-J (lahJ): a mouse model for human hair disorders. Exp. Dermatol. 2000;9:206–218. doi: 10.1034/j.1600-0625.2000.009003206.x. [DOI] [PubMed] [Google Scholar]
- Thergaonkar RW, Bhat V. Cardiofaciocutaneous syndrome. Medical Journal, Armed Forces India. 2013;69:175–177. doi: 10.1016/j.mjafi.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasioukhin V, Bowers E, Bauer C, et al. Desmoplakin is essential in epidermal sheet formation. Nat Cell Biol. 2001;3:1076–1085. doi: 10.1038/ncb1201-1076. [DOI] [PubMed] [Google Scholar]
- Wilanowski T, Caddy J, Ting SB, et al. Perturbed desmosomal cadherin expression in grainy head-like 1-null mice. The EMBO Journal. 2008;27:886–897. doi: 10.1038/emboj.2008.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T, Machann W, Kühler L, et al. Novel desmoplakin mutation: juvenile biventricular cardiomyopathy with left ventricular non-compaction and acantholytic palmoplantar keratoderma. Clinical Research in Cardiology. 2011;100:1087–1093. doi: 10.1007/s00392-011-0345-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Yang Z, Vatta M, et al. Compound and digenic heterozygosity contributes to arrhythmogenic right ventricular cardiomyopathy. J Am Coll Cardiol. 2010;55:587–597. doi: 10.1016/j.jacc.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Bowles NE, Scherer SE, et al. Desmosomal dysfunction due to mutations in desmoplakin causes arrhythmogenic right ventricular dysplasia/cardiomyopathy. Circulation research. 2006;99:646–655. doi: 10.1161/01.RES.0000241482.19382.c6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Network analysis of the relationship of Dsp to other genes known to cause similar clinical features to ruffled mice (Dsg4, Dsc1, Dsc2, Dsc3, Ctsl, Prss8, Gata3, Tgfa, Egfr) was conducted using Ingenuity Pathways Analysis© software (Invitrogen). Results confirmed a close association with networks associated with dermatologic diseases and conditions (1.7E-8). Dsp is circled in red, genes coding for desomosmal proteins are circles in green, genes with mutant alleles producing phenocopies to Dsprul are circled in blue.
Protein-protein and regulatory interactions in the STRING database further showed direct interactions between DSP, PKP1, 2 and 4, together with JUP, and DSG2.