Abstract
The aldo-keto reductase (AKR) protein superfamily contains > 190 members that fall into 16 families and are found in all phyla. These enzymes reduce carbonyl substrates such as: sugar aldehydes; keto-steroids, keto-prostaglandins, retinals, quinones, and lipid peroxidation byproducts. Exceptions include the reduction of steroid double bonds catalyzed by AKR1D enzymes (5β-reductases); and the oxidation of proximate carcinogen trans-dihydrodiol polycyclic aromatic hydrocarbons; while the (β-subunits of potassium gated ion channels (AKR6 family) control Kv channel opening. AKRs are usually 37 kDa monomers, have an (α/β)8-barrel motif, display large loops at the back of the barrel which govern substrate specificity, and have a conserved cofactor binding domain. AKRs catalyze an ordered bi bi kinetic mechanism in which NAD(P)H cofactor binds first and leaves last. In enzymes that favor NADPH, the rate of release of NADP+ is governed by a slow isomerization step which places an upper limit on kcat. AKRs retain a conserved catalytic tetrad consisting of Tyr55, Asp50, Lys84, and His117 (AKR1C9 numbering). There is conservation of the catalytic mechanism with short-chain dehydrogenases/reductases (SDRs) even though they show different protein folds. There are 15 human AKRs of these AKR1B1, AKR1C1-1C3, AKR1D1, and AKR1B10 have been implicated in diabetic complications, steroid hormone dependent malignancies, bile acid deficiency and defects in retinoic acid signaling, respectively. Inhibitor programs exist worldwide to target each of these enzymes to treat the aforementioned disorders. Inherited mutations in AKR1C and AKR1D1 enzymes are implicated in defects in the development of male genitalia and bile acid deficiency, respectively, and occur in evolutionary conserved amino acids. The human AKRs have a large number of nsSNPs and splice variants, but in many instances functional genomics is lacking. AKRs and their variants are now poised to be interrogated using modern genomic and informatics approaches to determine their association with human health and disease.
Keywords: hydroxysteroid dehydrogenases, potassium channels, steroid 5β-reductase, quinone reductase, xenobiotics
1. Introduction
The reduction of aldehydes and ketones to primary and secondary alcohols, respectively are formal functionalization reactions and are involved in the phase 1 metabolism of endogenous compounds and xenobiotics bearing these functional groups. These reactions are often catalyzed by proteins that belong to two protein superfamilies, the short-chain dehydrogenases (SDRs) and the aldo-keto reductases (AKRs) [1, 2].
AKRs exist in nearly all phyla, they are mainly monomeric soluble proteins (34–37 kDa), NAD(P)(H) dependent oxidoreductases [3]. While a search of the genome data bases can reveal a large number of in silico sequences that are AKRs, the protein superfamily contains 190 annotated proteins which fall into 16 families [4]. The enzymes have broad substrate specificity and will transform sugar [5] and lipid aldehydes [6, 7], ketosteroids [8], ketoprostaglandins [9, 10] , and chemical carcinogens, e.g. nicotine derived nitrosamines [11] as well as carcinogen metabolites e.g. polycyclic aromatic hydrocarbon trans-dihydrodiols [12, 13] and aflatoxin dialdehyde [14]. Each enzyme is characterized by the same protein fold, a triose-phosphate isomerase TIM barrel or (α/β)8-barrel with the insertion of several additional helices [15, 16] . At the back of the barrel there are three large loops that define substrate specificity. There are currently 119 AKR structures and their complexes in the PDB (as of August 2014). Sequence alignment and structural comparison identifies a common cofactor binding domain which permits pro-R-hydride transfer to the acceptor group, and a conserved catalytic tetrad of Tyr, Lys, His, Asp [3].
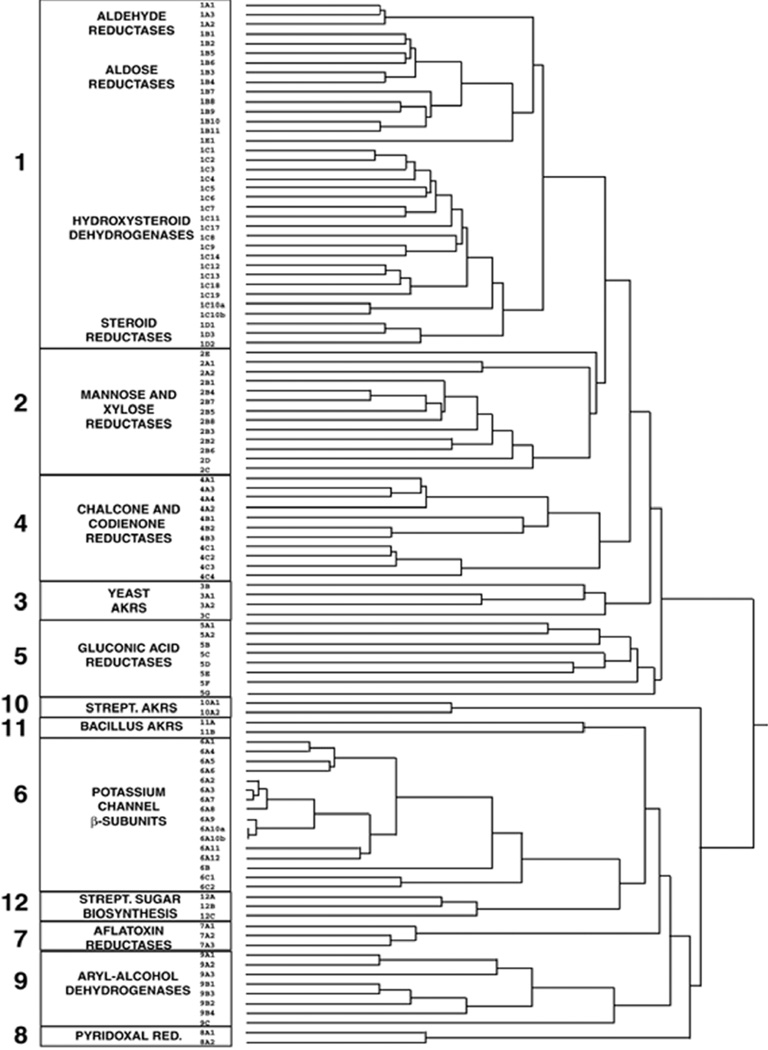
Sequence alignment also allows the identification of the AKR families and sub-families, where related members become grouped based on protein function. In this nomenclature < 40% sequence identity of an AKR identifies the protein as belonging to a new family. Greater than 60% identity between members groups them as members of the same subfamily and a numeral identifies the exact protein member [17]. Thus aldehyde reductase is defined as AKR1A1. The 16 AKR families include: AKR1 (aldehyde reductases, aldose reductases, hydroxysteroid dehydrogenases, and steroid 5β-reductases); AKR2 (manose and xylose reductases); AKR3 (yeast AKRs); AKR4 (chalcone and codienone reductases); AKR5 (gluconic acid reductases); AKR6 (β-subunits of the potassium gated voltage channels); AKR7 (aflatoxin dialdehyde and succinic semialdehyde reductases); AKR8 (pyridoxal reductases); AKR9 (aryl alcohol dehydrogenases); AKR10 (Streptomyces AKRs); AKR11 (Bacillus AKRs); AKR12 (Streptomyces sugar aldehyde reductases); AKR13 (hyperthermophilic bacteria reductases); AKR14 (E. coli reductases), AKR15 (Mycobacterium reductases) and AKR16 (V. cholera reductases), Figure 1 (www.med.upenn.edu/akr). This overview will review common features of the enzymatic properties of the AKRs and on the role of human AKRs in health and disease.
Figure 1.
AKR Superfamily Tree. Dendrogram of AKR families 1–12. The dendrogram was generated in ClustalX using a random number generator seed of 111 and 1000 bootstrap trials [4].. <40% sequence identity defines proteins from different families and >60% sequence identity identifies a sub-family within a family.
2. Enzymological Properties
All AKRs catalyze a sequential ordered bi-bi reaction in which cofactor binds first and leaves last [18, 19]. This has raised issues relating to the identity of the rate-determining step; is the step cofactor binding and release? substrate binding and product release? or the chemical step?
2.1. Cofactor Binding
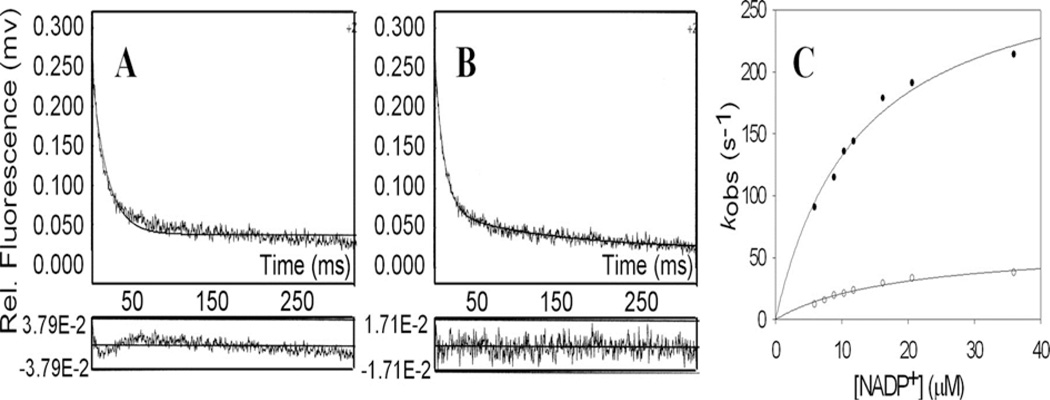
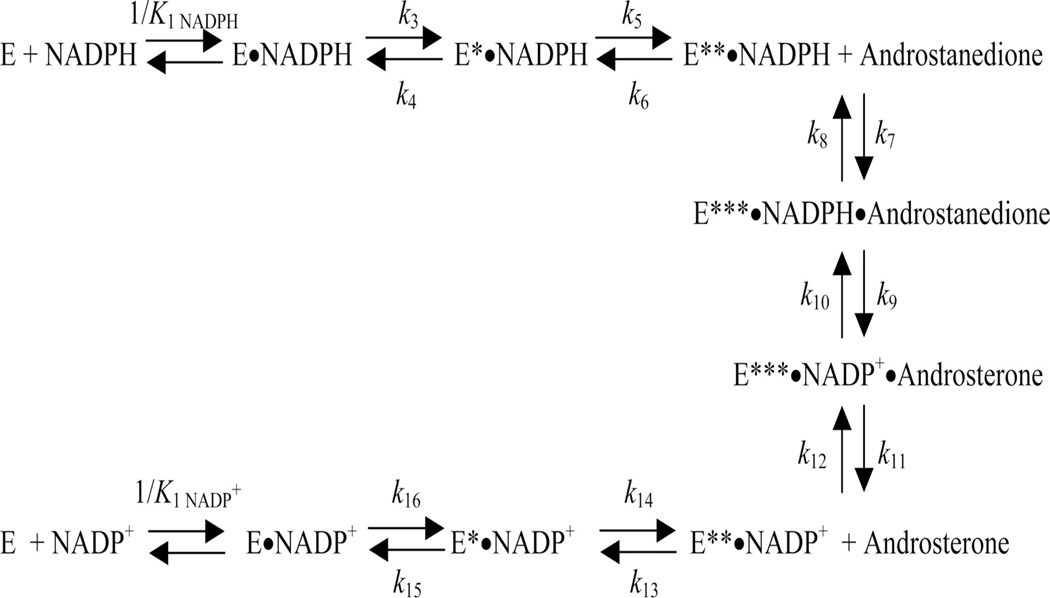
Examination of cofactor binding shows a conserved cofactor binding arrangement [3]. The cofactor is bound in an extended anti-conformation to permit pro-R-hydride transfer. In this conformation the nicotinamide head group is held in the correct orientation by pi-stacking with Tyr16, and by hydrogen bond interaction between the carboxamide side chain with Asn167, Ser166 and Gln190 (AKR1C9 numbering). By contrast the 2’-phosphate of AMP at the tail end is held in place by electrostatic or salt linkages with Arg276, and Arg270. The pyrophosphate bridge may or may not form salt links which can provide a “safety belt” that locks over the cofactor [20, 21]. Examination of cofactor binding can be measured in the stopped flow by measuring the quenching of the intrinsic protein fluorescence as the cofactor binds. Bi-exponential fitting of the fluorescence transient provides the best fit for these data which is interpreted to mean that there is a three step process for cofactor binding, Figure 2. The initial formation of a lose complex is followed by two subsequent isomerization events. A formal kinetic scheme was proposed for AKR1C9 which consisted of 9 enzyme forms and 16 individual microscopic rate constants, whereby the k6 term for NADPH release in the oxidation direction and the k13 term for NADP+ release in the reduction direction are slow steps [18], Figure 3. The first isomerization step is abolished by the R276M mutant in AKR1C9 indicating that cofactor anchoring of the 2’AMP is the event being followed [22]. This electrostatic salt link can be disrupted by high salt and thus cofactor affinity can be affected by salt or buffer concentration [23].
Figure 2.
Cofactor Binding to AKRs Involves Two Sequential Isomerization Steps. Panel A, shows quenching of intrinsic protein (AKR1C9) fluorescence observed with NADP+ fit to a single exponential, Panel B shows quenching of intrinsic protein fluorescence observed with NADP+ fit to a biexponential. AKR1C9 is 0.25 µM and NADP+ is 2.5 µM. The residuals to each fit are shown below the respective panels. Panel C, shows saturation kinetics for both the fast and slow phases. (After, Cooper et al., J. Biol. Chem., 2007 282: 33484; Reproduced with permission from American Society for Biochemistry and Molecular Biology).
Figure 3.
Microscopic Rate Constants in the AKR Kinetic Mechanism. The 9 discrete steps in the kinetic mechanism and the associated 16 rate constants are shown for AKR1C9. (After, Cooper et al., J. Biol. Chem., 2007 282: 33484; Reproduced with permission from American Society for Biochemistry and Molecular Biology).
2. 2. Rate-determination
Multiple-turnover experiments in the stopped-flow for the oxidation of androsterone by NADP+ catalyzed by AKR1C9 at pH 7.0 showed formal burst-phase kinetics followed by a slow product release phase. These experiments showed that kchem = 24.0 s−1, but that the rate of release of NAPH, krNADPH = 0.85 s−1 is identical to kcat. Thus in the oxidation direction rate-determination is driven by cofactor release. By contrast multiple-turnover experiments for the reduction of 5α-androstane-3,17-dione by NADPH catalyzed by AKR1C9 at pH 7.0 gave a kchem = 1.3 s−1 and a rate of release of NADP+, krNADPH = 0.65 s−1 and a kcat = 0.42 s−1. Thus in the reduction direction chemistry and cofactor release both contribute to rate determination [18]. These findings predict that different substrates for pluripotent enzymes will have different rate-determining steps.
2.3. Catalytic-Mechanism
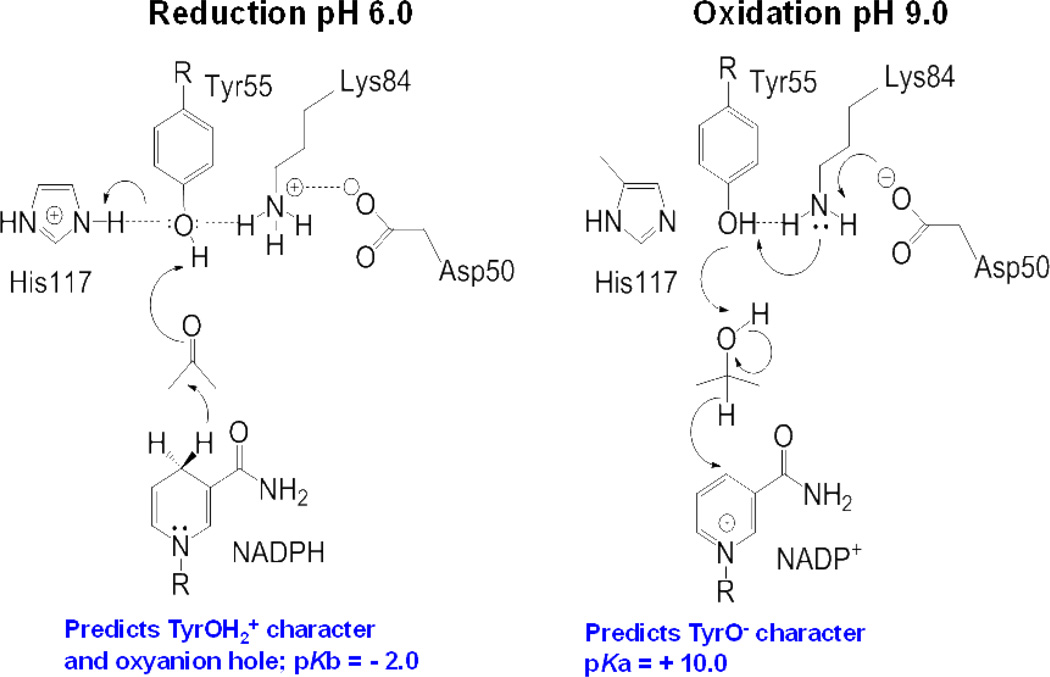
The catalytic mechanism was dissected for AKR1C9 using substrates in which kcat is driven by kchem, site-directed mutagenesis of the catalytic tetrad and kcat/pH rate profiles. This led to the proposal of a “push-pull” mechanism [24], Figure 4. In the reduction direction the catalytic Tyr has TyrOH2+ character. Following hydride transfer to the acceptor carbonyl there is protonation of the carbonyl by the Tyr with a proton relay that involves His117 to bulk water. In the oxidation direction the catalytic Tyr is deprontonated by Lys84 and the phenolate anion then removes a proton from the alcohol with hydride transfer back to the oxidized cofactor. This mechanism requires the enzyme to be diprotic so that microcospic reversibility can occur. The mechanism also demonstrates the importance of the microenvironment since TyrOH2+ has a pKb = −2.0 whereas TyrO− has a pKa = + 10.0. An NMR study is now required to titrate the catalytic Tyr so that its true pKa can be assigned. A similar mechanism was proposed for reduction catalyzed by aldose reductase [25, 26].
Figure 4.
“Push-Pull” Mechanism for Acid-Base Catalysis in AKRs. Left, Reduction of a carbonyl requires Tyr55 to act as a general acid facilitated by His117; Right, Oxidation of an alcohol requires Tyr 55 to act as a general base facilitated by Lys 84 and Asp 50. Amino acid numbering according to AKR1C9.
The protein folds of SDRs and AKRs are unrelated but we noticed that the residues involved in catalysis in the two enzyme superfamilies were similar. In SDRs the catalytic residues are Tyr, Asn, Ser, Lys and in AKRs, the residues are Tyr, Asp, His, Lys. This lead us to overlay the catalytic residues in the x-ray structures of Streptomyces 3α,20β-hydroxysteroid dehydrogenase (a SDR) with those present in AKR1C9 [27]. In conducting this overlay the nicotinamide ring of the cofactor was flipped to account for the difference in stereochemistry of hydride transfer between the two enzyme superfamilies; where SDRs transfer the pro-S-hydrogen. The result of this overlay showed that there was marked conservation of the position of these residues in 3-dimensional space such that the Tyr and Lys residues in both enzymes were superimposed and the Ser and His residues were aligned, Figure 5. We proposed that the catalytic mechanism for both SDRs and AKRs were similar and had been conserved through convergent evolution.
Figure 5.
Conservation of Catalytic Mechanism in SDRs and AKRs. Superimposition of the catalytic residues from rat liver 3α-HSD (Y55, H117, K84), AKR1C9 in blue with the catalytic residues from Streptomyces 3α,20β-HSD (Y152, S139, K156) an SDR in yellow. The nicotinamide ring of the cofactor in the Streptomyces enzyme was inverted to take into account that the stereochemistry of hydride transfer is pro-S and not pro-R The red sphere indicates the position of a water molecule which assumes the position of the substrate carbonyl. Taken from Bennett et al., Biochemistry 1996, 35: 10702, Reproduced with permission from the American Chemical Society).
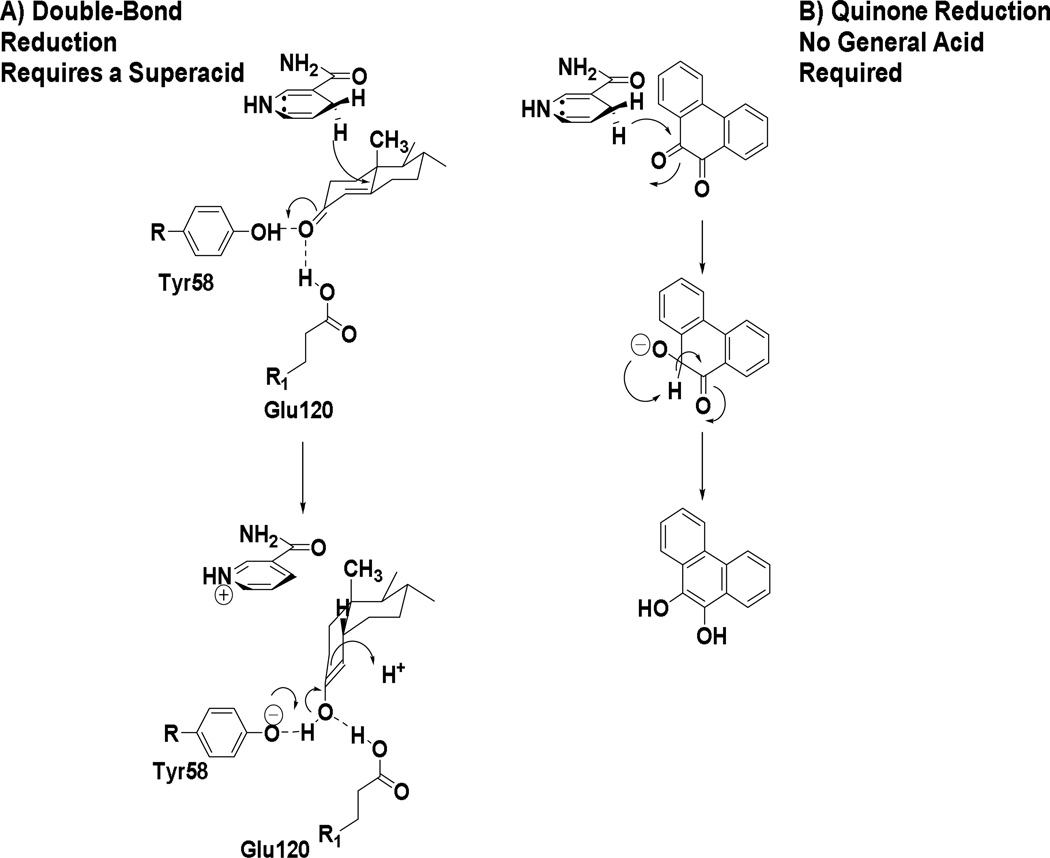
AKRs are remarkably flexible and variations on this catalytic mechanism have evolved to adapt to different substrates. AKR1D family members are not oxidoreductases but catalyze irreversible steroid double-bond reduction by acting as steroid 5β-reductases [28–30]. In this reaction the α,β-unsaturated ketones of steroid hormones are reduced at the C5-position to produce an A/B cis-ring junction so that the 5β-produced products contain a 90° bend in their structure. This reaction is very difficult to perform chemically since reduction of the α,β-unsaturated ketone leads to formation of the allylic alcohol. This reaction is facilitated by AKR1D1 by substitution of the catalytic His with a Glu (Glu120) [31]. This glutamic acid residue assumes an anti-conformation in the AKR1D1 crystal structure which enables the carboxylic acid to remain fully protonated so that the residue can enhance acid catalyzed enolization of the α.β-unsaturated ketone [28, 32], Figure 6A.
Figure 6.
Adaptation of the AKR Catalytic Mechanism for Double Bond and Quinone Reduction. A, steroid double-bond reduction by AKR1D1 requires Glu 120 to act as a super-acid; B, o-quinone reduction by AKR1C9 does not require a general acid and instead intermolecular hydride transfer occurs.
In another variation of this mechanism, a ubiquitous substrate used to assign enzyme activity in AKRs is 9,10-phenanthrenequinone. Remarkably, when the catalytic tyrosine of AKR1C9 was mutated it was found that the Y55F and Y55S mutants retained the kcat of the wild-type enzyme showing that the protonation state of the tyrosine was unimportant for this reaction and that acid-base catalysis was not required [33]. Wild-type enzyme and the active-site mutants Y55F, Y55S, H117A, D50N, K84R, and K84M showed the presence of a new titratable group with a pKb = 8.3–9.9. Thus, the group being titrated is not part of the tetrad. The retention of quinone reductase activity in this AKR in the absence of Tyr55 with kcat versus pH rate profiles and activation energies identical to wild-type enzyme for this reaction suggests that quinone reduction occurs via a mechanism that differs from 3-ketosteroid reduction. In this mechanism, the electron donor (NADPH) and acceptor (o-quinone) are bound in close proximity, which permits hydride transfer without formal protonation of the acceptor carbonyl by Tyr 55. Thus a mechanism was proposed in which following hydride transfer to one carbonyl of the o-quinone there was intermolecular hydride transfer to yield the catechol which air-oxidizes back to the quinone, Figure 6B.
2.4. AKRs Function as Reductases in Vivo
Despite the fact that AKRs can catalyze oxidoreduction in vitro they likely function solely as reductases in vivo. AKRs display nanomolar affinity for NADP(H) with Kd values in the range of 10 – 120 nM but display micromolar affinity for NAD(H) with Kd values between 100–250 µM [23]. Thus the enzymes at first glance are set up to use the prevailing concentrations of NADPH and NAD+ in cells. However, the high affinity for NAD(P)H ensures that there is potent product inhibition of NAD+ dependent oxidation reactions [34]. Further kinetic analysis shows that the Keq and the Haldane equation indicate that the reduction direction is favored [18]. Finally, stable and transiently transfected mammalian cell lines show that when AKRs are forced to use the prevailing cofactor concentration in cells they catalyze reduction reactions only [34–37]. Thus human AKRs have been engineered to act as reductases.
3. Human AKRs and Disease
There are 15 human AKRs see Table 1. These include aldehyde reductase (AKR1A1); aldose reductase and aldose-like reductase proteins (AKR1B1, AKR1B10 and AKR1B15); the hydroxysteroid dehydrogenases (AKR1C1-AKR1C4); steroid 5β-rductase (AKR1D1); 1,5-anhydro-D-fructose reductase (AKR1E2); the β-subunits of the potassium voltage gated channels (AKR6A3, AKR6A5, and AKR6A9) (which form tetramers); and the dimeric aflatoxin aldehyde reductases (AKR7A2 and AKR7A3). These human AKRs are implicated in a variety of disease processes and as such selective enzyme inhibitors have been sought both as chemical probes and as possible therapeutics. In addition natural mutations in human AKRs are also linked with disease, and a number of splice variants are possible.
Table 1.
Human Aldo-Keto Reductases
| Gene | Protein | Chromosomal Localization |
|---|---|---|
| AKR1A1 | Aldehyde Reductase | 1p33-p32 |
| AKR1B1 | Aldose Reductase | 7q35 |
| AKR1B10 | Aldose Reductase | 7q33 |
| AKR1B15 | Aldose Reductase | 7q33 |
| AKR1C1 | 3α(20α)-Hydroxysteroid Dehydrogenase (20α-HSD) | 10p15-10p14 |
| AKR1C2 | Type 3 3α-Hydroxysteroid Dehydrogenase | 10p15-10p14 |
| AKR1C3 | Type 5 17β-Hydroxysteroid Dehydrogenase; Type 2 3α-Hydroxysteroid Dehydrogenase |
10p15-10p14 |
| AKR1C4 | Type 1 3α-Hydroxysteroid Dehydrogenase | 10p15-10p14 |
| AKR1D1 | Steroid 5β-reductase | 7q32-7q33 |
| AKR1E2 | 1,5-Anhydro-D-Fructose Reductase | 10p15.2 |
| AKR6A3 | Potassium voltage gated channel, β-subunit-1 | 3q26.1 |
| AKR6A5 | Potassium voltage gated channel, β-subunit-2 | 1p36.3 |
| AKR6A9 | Potassium voltage gated channel, β-subunit-3 | 17p13.1 |
| AKR7A2 | Aflatoxin Aldehyde Reductase; succinic semialdehyde reductase |
1p35.1-p36.23 |
| AKR7A3 | Aflatoxin Aldehyde Reductase | 1p35.1-p36.23 |
3.1. AKR-Inhibitors
AKR1B1 (aldose reductase) is targeted therapeuticaly due to its ability to convert high circulating concentrations of glucose, observed in diabetics to the hyperosmotic sugar sorbitol (the first step in the polyol pathway) [38], Table 2. The enzyme activity has been linked to the causation of diabetic complications including cataractogenesis [39], retinopathy [40], neuropathy [41] and nephropathy [42] . Specific inhibitors of this enzyme that have been used clinically are sorbinil [43], tolrestat [44], epalrestat [45], ranirestat [46, 47], and fidarestat [48]. The phenotype of AKR1B1 k/o animals protects against the complications of diabetes [49], and suggests that inhibitors would be effective in man. Recent meta-analysis of clinical trials of AKR1B1 inhibitors indicate that they are beneficial in diabetic cardiovascular autonomic neuropathy [50] while others have indicated that only modest improvement in diabetic complications is achieved with epalrestat, ranirestat [46, 47], and fidarestat [51] . AKR1B10 has been implicated in regulating retinoic signaling by converting all-trans-retinaldehyde to retinol [52, 53] and is highly overexpressed in NSCLC [54] and in hepatocarcinogenesis [55] and is considered a tumor marker. Interception of retinoic acid signaling could also prevent the differentiation of cancer stem cells. A sterol based inhibitor oleanolic acid [56] as well as the nonsteroidal anti-inflammatory drug sulindac [57] are lead compounds for inhibitor development for AKR1B10.
Table 2.
Role of Human AKRs in Health and Disease
| AKR | Reduction Reaction | Associated Disease | Selective Inhibitors (C = used clinically) |
|---|---|---|---|
| AKR1B1 | Glucose → Sorbitol (Polyol Pathway) | Diabetic complications: Cataractogenesis; retinopathy; neuropathy; nephropathy |
Sorbinil (C) Tolrestat (C) Epalrestat (C) Ranirestat (C) Fidarestat (C) |
| AKR1B10 | all-trans-retinaldehyde → retinol | NSCLC Hepatocarcinogenesis |
Oleanolic acid |
| AKR1C1 | Progesterone → 20α-Hydroxypro- gesterone |
Pre-term birth Endometriosis |
Salicylate 3-chloro- phenylsalicylate |
| AKR1C2 | DHT → 3α-androstanediol DHP → Allopregnanolone |
Androgen insufficiency Premenstural syndrome |
Ursodeoxycholate |
| AKR1C3 | Δ4-androstene-3,17-dione→Test osterone 5α-androstane-3,17-dione →5α-DHT Estrone→ 17β-Estradiol PGH2→ PGF2α PGD2→11β-PGF2α |
Advanced prostate cancer Breast Cancer Acute Myeloid Leukemia |
Indomethacin 6-Medroxy- progesterone Acetate (C) |
| AKR1D1 | Δ4-cholesten-3-ones→5β-chole stane-3-ones |
Bile Acid Deficiency | |
| AKR6A | NADPH dependent opening of voltage gated potassium channels |
Aberrant redox regulation of Kev channels and cardiovascular disease |
|
| AKR7A2 | Succinic semialdehyde → γ-hydroxyl- butyrate |
Neuromodulator; succinic semialdehyde dehydrogenase deficiency |
|
| AKR7A3 | Aflatoxin aldehyde → Aflatoxin bis alcohol |
Hepatocarcinogenesis |
AKR1C1 is the predominant 20-ketosteroid reductase in man and metabolizes progesterone to its inactive metabolite 20α-hydroxyprogesterone [8, 37]. Maintenance of circulating progesterone levels is essential to prevent pre-term birth and for blunting the estrogenic effects in endometriosis and endometrial cancer. Salicylate and salicylate derivatives e.g. 3-bromo-5-phenylsalicylate and 3-chloro-5-phenylsalicylate are lead inhibitors for this enzyme [58].
AKR1C2 is a predominant 3-ketosteroid reductase implicated in the metabolism of 5α-dihydrotestosterone to produce the inactive androgen 5α-androstan-3α,17β-diol and in the metabolism of 5β-dihydroprogesterone to produce allopregnanolone, a neuroactive steroid. Inhibitors of AKR1C2 are thus sought to deal with issues of androgen insufficiency. By contrast allopregnanolone is an allosteric effector of the GABAa receptor and increases chloride conductance at the GABA receptor and lack of this neurosteroid is associated with premenstrual syndrome [59]. In this instance an AKR1C2 activator would be beneficial. Thus far no small molecules have been identified as AKR1C activators, but compounds that could facilitate the binding and release of NADP(H) may fulfil this role.
AKR1C3 is also known as type 5 17β-HSD and prostaglandin (PG) F2α synthase [9, 10]. As a 17β-HSD it is involved in the reduction of Δ4-androstene-3,17-dione to testosterone, the reduction of 5α-androstane-3,17-dione to 5α-dihydrotestosterone and the reduction of estrone to 17β-estradiol [8, 35, 36]. Thus inhibitors for the 17β-HSD reaction are sought for advanced prostate cancer and breast cancer where overexpression of the enzyme may provide an escape mechanism for other hormone ablative therapies. As a PGF2α synthase the enzyme also reduces PGH2 to PGF2α and PGD2 to 11β-PGF2α[9]. Production of these ligands for the FP receptor would be pro-proliferative through MAPK signaling [35]. In addition, the latter reaction prevents formation of 15-PGJ2 ligands for PPARγ[60]. Inhibitors available for AKR1C3 include indomethacin [61] and 6-medroxyprogesterone acetate where the latter progestin has been used to target AKR1C3 in the clinic to treat acute myeloid leukemia [62, 63]. A large number of AKR1C3 inhibitor programs have been described and both GTx therapeutics [64] and Astellas Pharma [65] have lead molecules in preclinical and clinical development, respectively.
3. 2. Disease-Based Mutations
A number of diseased based mutations exist in AKRs which demonstrate the importance of these enzymes in human health. AKR1D1 or steroid 5β-reductase catalyzes the metabolism of steroid hormones that contain an α,β-unsaturated ketone in the A-ring. Many of the 5β-dihydrosteroids have biological functions and therefore inhibition of this enzyme is undesirable. AKR1D1 also catalyzes a crucial step in bile-acid biosynthesis in which the planar bile acid precursors are bent at the A/B-ring junction to produce sterols that have emulsifying properties. Natural mutations exist in this enzyme and prevent the conversion of 7α-hydroxycholestenone to bile acid precursors which in turn removes the negative feedback inhibition of 7α-hydroxylase (P4507A1), the rate determining enzyme in the bile acid biosynthetic pathway [66–70]. As a consequence of these mutations bile-acids are not made which are required for the emulsification of fats and the absorption of fat soluble vitamins. 7α-Hydroxycholestenone levels rise due to the blockade in the pathway and the removal of feedback inhibition of 7α-hydroxylase, resulting in the production of excess allo-bile acids which are hepatotoxic. As a result, this deficiency is fatal in the neonate unless infants receive dietary supplementation with bile acids. There are seven point mutations associated with this disease L106F, P133R, P198L, G223E, D241V, R261C and R266Q [66–70]. Mapping of these mutations to the AKR1D1 structure provides no obvious clues as to why the enzymes would lack function, since they are not located in the cofactor binding site, the steroid binding site, or the active site, Figure 7. Sequence alignment however does reveal that these mutated residues occur in highly conserved amino acid residues in the AKR1 family [71, 72].
Figure 7.
Location of Natural Mutations in the AKR1D1 Crystal Structure. (α/β)8-barrel of AKR1D1 (green); NADP+ cofactor (magenta); steroid substrate (blue); position of 7 inherited mutations (black).
Mutations also exist in AKR1C2/AKR1C4 which are associated with the appearance of ambiguous genitalia in new born males [73]. These enzymes play roles in the backdoor pathway into 5α-dihydrotestosterone [74]. In this pathway 5α-reduction occurs at the level of progesterone to produce 5α-dihydroprogesterone which is then converted to allopregnanolone by AKR1C2/AKR1C4. Conversion of this C21 steroid to androsterone is then catalyzed by P450 17α-hydroxylase/17,20-lyase, and androsterone is subsequently converted to 5α-androstane-3α,17β-diol by AKR1C3. The diol is then converted to 5α-DHT by HSD17B6, a SDR [75]. Single and double mutations in AKR1C2 reduce the production of allopregnanolone, e.g. H222Q, or I79V plus either H90G or N300T. However, the effect is further exacerbated by deletion of the AKR1C4 gene altogether [73]. The mutations in AKR1C2 are also in evolutionary conserved amino acids.
AKR6A family members are not considered enzymes but act as β-subunits of the potassium gated voltage channels which are essential for channel opening. They contain the conserved NADPH binding domain and are tetrameric. As a result of the isomerization steps involved in cofactor binding a large number of NADPH-occupancy states can exist in the tetramer which may fine tune channel opening. NADP(H) binding appears to be essential: for the optimal interaction between Kvalpha and beta subunits; for Kvbeta-induced inactivation of Kv currents [76]; and for the differential regulation of Kv currents by oxidized and reduced cofactor [77] .These findings suggest that channel opening may be redox-regulated. Aberrant redox regulation of channel opening due to AKR6A nsSNP variants could contribute to variety of diseases including cardiovascular disease.
AKR7A2 and 7A3 are aflatoxin aldehyde reductases involved in the reduction of aflatoxin dialdehyde to the corresponding mono and bis-alcohols. As such this reaction prevents the aldehydes from forming Schiff’s bases with lysine residues and can reduce the hepatotoxicity of aflatoxin [78]. In addition the AKR7A2 is the major succinic semialdehyde reductase in the CNS and is thus involved in GABA metabolism [79]. The product of this reaction is γ-hydroxybutyrate (GHP) which is a GABA agonist and can also bind to GHP receptors [80]. In succinic semialdehyde dehydrogenase (SSH) deficiency GHP levels will increase leading to enhanced inhibitory effects of GABA [81]. SNP variants which lead to a loss of succinic semialdehyde reductase activity would protect against SSH deficiency.
3. 3. Single-Nucleotide Polymorphisms
An examination of the NCBI data base leads to the identification of a large number of nsSNPs for the human AKRs. To systematically name these SNP variants we recommend a nomenclature that has been used in other gene superfamilies where AKR1C1 (would be the individual member); AKR1C1*1 (would be the wild-type allele) which can be followed by a letter to indicate that a sub-allele AKR1C1*1A exists but that the resultant single nucleotide substitution does not change the protein sequence. AKR1C1*2 (would be a mutant allele due to a (nsSNP) and the sub-allelle “A” would again indicate a single nucleotide substitution that does not change the sequence of the mutant protein. The numbering and lettering of the nsSNP’s and the sub-alleles, respectively is in chronological order of discovery.
A subset of the nSNPs were found to be 100% conserved in the AKR1C and 1D subfamilies, Table 3. Bioinformatic tools such as SIFT or PolyPhen-2 which have a prediction accuracy of 76% indicated that these amino acid substitutions would be harmful or deleterious to protein function [82] . To test this prediction we have conducted site-directed mutagenesis on AKR1C2 to introduce the relevant nsSNPs into the protein and have found that the resultant enzymes to be devoid of enzyme activity (Jin, Chen & Penning, unpublished observations). With the onset of personalized genomics, rules will be required to predict the function of nsSNPs since there will be too many to analyze by functional genomic approaches. Based on the natural mutations in AKRs, bioinformatics and functional analysis we predict that nsSNPs in evolutionary conserved amino acids within a protein superfamily are deleterious to function.
Table 3.
nsSNPs in Evolutionarily Conserved Amino Acids in AKR1C and AKR1D Subfamilies
| AKR | SNP | MAF* | Position in Structure |
AKR | SNP | MAF* | Position in Structure |
|---|---|---|---|---|---|---|---|
| AKR1C2 | T23I | nd | β1-α1 loop | AKR1C4 | R76T | nd | α2-β3 loop |
| P119T | nd | Loop A | G135E | 0.114 | Tight turn Loop A |
||
| K185E | nd | α6-β6 loop | E192A | 0.001 | β6 | ||
| R258C | nd | α7 | Q262R | nd | α7 | ||
| AKR1C3 | L85F | nd | β3 | R263H | nd | α7 | |
| P180S | 0.014 | Tight turn α5-β6 loop |
AKR1D1 | V77L | nd | α2-β3 loop | |
| K183M | 0.029 | α5-β6 loop | R261H | nd | A7 | ||
| Q199R | nd | β6-α6 loop | |||||
| K247R | nd | Helix-1 | |||||
| R258C | 0.03 | α7 | |||||
| M293I | nd | Helix-2 |
MAF = minor allelic frequency
ND = not determined
3. 4. Splice Variants
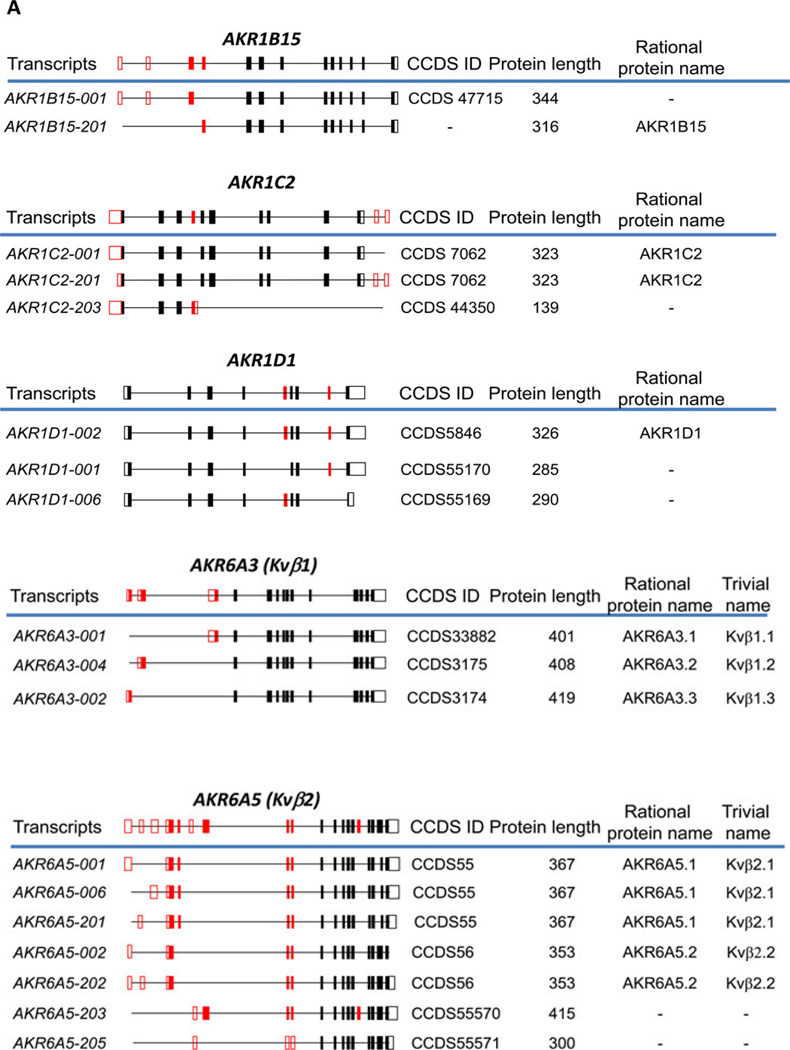
With the introduction of next generation sequencing techniques, RNAseq experiments have detected splice variants of the human AKR transcripts, see Figure 8. To accommodate these findings we have expanded the AKR protein nomenclature. We recommend using the Ensembl data base transcript number which has a unique consensus coding sequence (CCDS number). When the native protein is expressed it is referred to as the regular AKR name followed by period and numeral “1” e.g. AKR6A3.1; subsequent protein isoforms are numbered in the sequence of discovery e.g. AKR6A3.2 and AKR6A.3.3 [83].
Figure 8.
Human AKR Splice Variants. Gene structure is shown above the line; coding exons are solid boxes; transcripts identified in Ensembl are below the line, left; CCDS, consensus coding sequence; rational protein and trivial names are also given (taken from ref. 83).
For AKR1B15, a long and short protein form exist, AKR1B15.2 and AKR1B15.1 (which are encoded by transcripts AKR1B15-001 and AKR1B15-201) the longer form contains a leader sequence, that may dictate subcellular localization. However, evidence that these transcripts are translated into these protein isoforms has yet to be obtained. AKR1C2 gives rise to three splice variants. Two transcripts AKR1C2-001 and AKR1C2–201 give rise to the same full length mature protein of 323 amino acids, however, AKR1C2–203 transcript gives rise to a severely truncated protein of 139 amino acids which is predicted to be inactive due to the loss of functional domains. AKR1D1 gives rise to three splice variants but only one transcript, AKR1D1-002 gives rise to the mature 326 amino-acid protein. The other two transcripts would give rise to proteins of 285 amino acids in length (AKR1D1-001) and 290 amino acids in length (AKR1D1-006), respectively; the former lacks an important part of the steroid binding site and the latter lacks a portion of the C-terminus, and both would be predicted to be inactive. It is important to emphasize that none of the proteins for these AKR1D1 transcripts have yet been detected in cells.
In contrast to the splice variants described for the AKR1 family, three splice variants of AKR6A3 (AKR6A3-001, AKR6A3–004, and AKR6A3-002) have been detected as expressed proteins of 401 amino acids in length AKR6A3.1; 408 amino acids in length AKR6A3.2; and 419 amino acids in length AKR6A.3; in addition seven splice variants have been detected for AKR6A5 which give rise to two different proteins AKR6A5.1 and AKR6A5.2 of 367 amino acids and 353 amino acids in length, respectively. How these isoforms influence potassium channel will be important to determine.
4. Summary
AKRs are pluripotent enzymes that play a role in phase 1 metabolism of endogenous substrates and xenobiotics. Rate-determination is substrate dependent and may be driven by cofactor release, the chemical step or by a combination of these events. The catalytic Tyr and Glu acid residues conduct chemistry in violation of their pKa values at physiological pH. Determination of their pKa values in the protein microenvironment by NMR will be an important undertaking to further elucidate the AKR catalytic mechanisms. Isoform specific inhibitors of the human enzymes are required to target specific diseases with the understanding that off-target effects may occur when these compounds bind to related AKRs; a lack of appreciation for the similarity in AKR structure-function could lead to the failure of therapeutic leads. Naturally occurring inherited mutations in the AKR1C and AKR1D1 genes demonstrate that they cause serious defects in steroid metabolism leading to disease and suggests that inherited mutations in other AKRs may account for other disease phenotypes. Informatics tools will be required to predict whether nsSNPs in AKRs are deleterious to function as we enter into the era of personalized genomics. Importantly some AKRs work sequentially within the same metabolic pathway so that the combined effect of individual nsSNPs may be magnified. Each human AKR gene can give rise to a number of splice variants, where truncations may affect protein function and extensions may determine subcellular localization.
Supplementary Material
Highlights Points.
Aldo-keto reductases (AKRs) are a major superfamily of NAD(P)H-dependent oxidoreductases
They use a conserved catalytic mechanism and rate is often governed by cofactor release
There are 15 human AKRs implicated in health and disease
Inhibitor programs exist world-wide to develop therapeutics and chemical probes for AKRs
Natural mutations in human AKRs are responsible for disease phenotypes
Abbreviations
- AKR
aldo-keto reductase
- PG
prostaglandin
- SDR
short-chain dehydrogenase reductase
- nsSNP
non-synonymous single nucleotide polymorphism
- SSH
succinic semialdehyde dehydrogenase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jin Y, Penning TM. Aldo-keto reductases and bioactivation/detoxication, Annual Review of Pharmacology & Toxicology. Annual Reviews, Palo Alto CA2007. 2007;47:263–292. doi: 10.1146/annurev.pharmtox.47.120505.105337. [DOI] [PubMed] [Google Scholar]
- 2.Oppermann U. Annual Reviews of Pharmacology & Toxicology. Vol. 47. Palo Alto, CA2007: Annual Reviews; 2007. Carbonyl reductases: The complex relationship of mammalian carbonyl and quinone-reducing enzymes and their role in physiology; pp. 293–322. [DOI] [PubMed] [Google Scholar]
- 3.Jez JM, Bennett MJ, Schelgel BP, Leiws M, Penning TM. Comparative anatomy of the aldo-keto reductase superfamily. Biochem. J. 1997;326:625–636. doi: 10.1042/bj3260625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hyndman D, Bauman DR, Heredia VV, Penning TM. The aldo-keto reductase superfamily homepage. Chem. Biol. Inter. 2003;143–144:621–631. doi: 10.1016/s0009-2797(02)00193-x. [DOI] [PubMed] [Google Scholar]
- 5.Bohren KM, Bullock B, Wermuth B, Gabbay KH. The aldo-keto reductase superfamily. cDNAs and deduced amino acid sequences of human aldehyde and aldose reductases. J. Biol. Chem. 1989;264:9547–9551. [PubMed] [Google Scholar]
- 6.Burczynski ME, Sridhar GR, Palackal NT, Penning TM. The reactive oxygen species--and Michael acceptor-inducible human aldo-keto reductase AKR1C1 reduces the α,β-unsaturated aldehyde 4-hydroxy-2-nonenal to 1,4-dihydroxy-2-nonene. J. Biol. Chem. 2001;276:2890–2897. doi: 10.1074/jbc.M006655200. [DOI] [PubMed] [Google Scholar]
- 7.Srivastava S, Chandra A, Bhatnagar A, Srivastava SK, Ansari NH. Lipid peroxidation product, 4-hydroxynonenal and its conjugate with GSH are excellent substrates of bovine lens aldose reductase. Biochem Biophys Res Commun. 1995;217:741–746. doi: 10.1006/bbrc.1995.2835. [DOI] [PubMed] [Google Scholar]
- 8.Penning TM, Burczynski ME, Jez JM, Hung C-F, Lin H-K, Ma H, Moore M, Palackal N, Ratnam K. Human 3α-hydroxysteroid dehydrogenase isoforms (AKR1C1-AKR1C4) of the aldo keto reductase superfamily: functional plasticity and tissue distribution reveals roles in the inactivation and formation of male and female sex hormones. Biochem. J. 2000;351:67–77. doi: 10.1042/0264-6021:3510067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Komoto J, Yamada T, Watanabe K, Takusagawa F. Crystal structure of human prostaglandin F synthase (AKR1C3) Biochemistry. 2004;43:2188–2198. doi: 10.1021/bi036046x. [DOI] [PubMed] [Google Scholar]
- 10.Suzuki-Yamamoto T, Nishizawa M, Fukui M, Okuda-Ashitaka E, Nakajima T, Ito S, Watanabe K. cDNA cloning, expression and characterization of human prostaglandin F synthase. FEBS Lett. 1999;462:335–340. doi: 10.1016/s0014-5793(99)01551-3. [DOI] [PubMed] [Google Scholar]
- 11.Atalla A, Breyer-Pfaff U, Maser E. Purification and characterization of oxidoreductases-catalyzing carbonyl reduction of the tobacco-specific nitrosamine 4-methylnitrosamino-1-(3-pyridyl)-1-butanone (NNK) in human liver cytosol. Xenobiotica. 2000;30:755–769. doi: 10.1080/00498250050119826. [DOI] [PubMed] [Google Scholar]
- 12.Burczynski ME, Harvey RG, Penning TM. Expression and characterization of four recombinant human dihydrodiol dehydrogenase isoforms: oxidation of trans-7, 8-dihydroxy-7,8-dihydrobenzo[a]pyrene to the activated o-quinone metabolite benzo[a]pyrene-7,8-dione. Biochemistry. 1998;37:6781–6790. doi: 10.1021/bi972725u. [DOI] [PubMed] [Google Scholar]
- 13.Palackal NT, Lee SH, Harvey RG, Blair IA, Penning TM. Activation of polycyclic aromatic hydrocarbon trans-dihydrodiol proximate carcinogens by human aldo-keto reductase (AKR1C) enzymes and their functional overexpression in human lung carcinoma (A549) cells. J Biol Chem. 2002;277:24799–24808. doi: 10.1074/jbc.M112424200. [DOI] [PubMed] [Google Scholar]
- 14.Kozma E, Brown E, Ellis EM, Lapthorn AJ. The crystal structure of rat liver AKR7A1. A dimeric member of the aldo-keto reductase superfamily. Chem. Biol. Inter. 2003;143–144:289–297. doi: 10.1074/jbc.M110808200. [DOI] [PubMed] [Google Scholar]
- 15.Hoog SS, Pawlowski JE, Alzari PM, Penning TM, Lewis M. Three-dimensional structure of rat liver 3α-hydroxysteroid/dihydrodiol dehydrogenase: a member of the aldo-keto reductase superfamily. Proc. Natl. Acad. Sci. U S A. 1994;91:2517–2521. doi: 10.1073/pnas.91.7.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilson DK, Bohren KM, Gabbay KH, Quiocho FA. An unlikely sugar substrate site in the 1.65 A structure of the human aldose reductase holoenzyme implicated in diabetic complications. Science. 1992;257:81–84. doi: 10.1126/science.1621098. [DOI] [PubMed] [Google Scholar]
- 17.Jez JM, Flynn TG, Penning TM. A new nomenclature for the aldo-keto reductase superfamily. Biochem Pharmacol. 1997;54:639–647. doi: 10.1016/s0006-2952(97)84253-0. [DOI] [PubMed] [Google Scholar]
- 18.Cooper WC, Jin Y, Penning TM. Elucidation of a complete kinetic mechanisms for a mammalian hydroxysteroid dehydrogenase (HSD) and identification of all enzyme forms on the reaction coordinate: The example of rat liver 3α-HSD (AKR1C9) J. Biol. Chem. 2007;282:33484–33493. doi: 10.1074/jbc.M703414200. [DOI] [PubMed] [Google Scholar]
- 19.Grimshaw CE, Bohren KM, Lai CJ, Gabbay KH. Human aldose reductase: rate constants for a mechanism including interconversion of ternary complexes by recombinant wild-type enzyme. Biochemistry. 1995;34:14356–14365. doi: 10.1021/bi00044a012. [DOI] [PubMed] [Google Scholar]
- 20.Borhani DW, Harter TM, Mark PJ. The crystal structure of the aldose reductase·NADPH binary complex. J. Biol. Chem. 1992;267:24841–24847. doi: 10.2210/pdb1abn/pdb. [DOI] [PubMed] [Google Scholar]
- 21.El-Kabbani O, Judge K, Ginell SL, Myles DAA, DeLucas LJ, Geoffrey FT. Structure of porcine aldehyde reductase holoenzyme. Nature Strutural Biology. 1995;2:687–692. doi: 10.1038/nsb0895-687. [DOI] [PubMed] [Google Scholar]
- 22.Ratnam K, Ma H, Penning TM. The arginine 276 anchor for NADP(H) dictates fluorescence kinetic transients in 3α-hydroxysteroid dehydrogenase, a representative aldo-keto reductase. Biochemistry. 1999;38:7856–7864. doi: 10.1021/bi982838t. [DOI] [PubMed] [Google Scholar]
- 23.Ma H, Ratnam K, Penning TM. Mutation of nicotinamide pocket residues in rat liver 3α-hydroxysteroid dehydrogenase reveals different modes of cofactor binding. Biochemistry. 2000;39:102–109. doi: 10.1021/bi991659o. [DOI] [PubMed] [Google Scholar]
- 24.Schlegel BP, Jez JM, Penning TM. Mutagenesis of 3α-hydroxysteroid dehydrogenase reveals a “push-pull” mechanism for proton transfer in aldo-keto reductases. Biochemistry. 1998;37:3538–3548. doi: 10.1021/bi9723055. [DOI] [PubMed] [Google Scholar]
- 25.Bohren KM, Grimshaw CE, Lai CJ, Harrison DH, Ringe D, Petsko GA, Gabbay KH. Tyrosine-48 is the proton donor and histidine-110 directs substrate stereochemical selectivity in the reduction reaction of human aldose reductase: enzyme kinetics and crystal structure of the Y48H mutant enzyme. Biochemistry. 1994:2021–2032. doi: 10.1021/bi00174a007. [DOI] [PubMed] [Google Scholar]
- 26.Grimshaw CE, Bohren KM, Lai CJ, Gabbay KH. Human aldose reductase: pK of tyrosine 48 reveals the preferred ionization state for catalysis and inhibition. Biochemistry. 1995:14374–14384. doi: 10.1021/bi00044a014. [DOI] [PubMed] [Google Scholar]
- 27.Bennett MJ, Schlegel BP, Jez JM, Penning TM, Lewis M. Structure of 3α- hydroxysteroid/dihydrodiol dehydrogenase complexed with NADP+ . Biochemistry. 1996;35:10702–10711. doi: 10.1021/bi9604688. [DOI] [PubMed] [Google Scholar]
- 28.L Di Costanzo, Drury JE, Penning TM, Christianson DW. Crystal structure of human liver Δ4-3-ketosteroid 5β-reductase (AKR1D1) and implications for substrate binding and catalysis. J Biol Chem. 2008;283:16830–16839. doi: 10.1074/jbc.M801778200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kondo K, Kai M, Setoguchi Y, Eggertsen G, Sjoborn P, Setoguchi T, Okuda K, Bjorkhem I. Cloning and expression of cDNA of human Δ4-3-oxosteroid-5β-reductase and substrate specificity of the expressed enzyme. Eur J Biochem. 1994;219:357–363. doi: 10.1111/j.1432-1033.1994.tb19947.x. [DOI] [PubMed] [Google Scholar]
- 30.Okuda A, Okuda K. Purification and characterization of Δ4-3-ketosteroid 5β-reductase. J. Biol. Chem. 1984;259:7519–7524. [PubMed] [Google Scholar]
- 31.Jez JM, Penning TM. Engineering steroid 5β-reductase activity into rat liver 3α- hydroxysteroid dehydrogenase. Biochemistry. 1998;37:9695–9703. doi: 10.1021/bi980294p. [DOI] [PubMed] [Google Scholar]
- 32.Di Costanzo L, Drury JE, Christianson DW, Penning TM. Structure and catalytic mechanism of steroid 5β-reductase (AKR1D1) Mol. Cell. Endocrinol. 2009;301:191–198. doi: 10.1016/j.mce.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schlegel BP, Ratnam K, Penning TM. Retention of NADPH-linked quinone reductase activity in an aldo-keto reductase following mutation of the catalytic tyrosine. Biochemistry. 1998;37:11003–11011. doi: 10.1021/bi980475r. [DOI] [PubMed] [Google Scholar]
- 34.Rizner T, Lin H-K, Peehl DM, Steckelbroeck S, Bauman DR, Penning TM. Human type 3 3α-hydroxysteroid dehydrogenase (AKR1C2) and androgen metabolism in prostate cells. Endocrinology. 2003;144:2922–2932. doi: 10.1210/en.2002-0032. [DOI] [PubMed] [Google Scholar]
- 35.Byrns MC, Duan L, Lee S-H, Blair IA, Penning TM. Aldo-keto reductase 1C3 expression in MCF-7 cells reveals roles in steroid hormone and prostaglandin metabolism that may explain its overexpression in breast cancer. J. Steroid Biochem. Mol. Biol. 2010;118:177–187. doi: 10.1016/j.jsbmb.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Byrns MC, Mindnich R, Duan L, Penning TM. Overexpression of aldo-keto reductase 1C3 (AKR1C3) in LNCaP cells diverts androgen metabolism towards testosterone resulting in resistance to the 5α-reductase inhibitor finasteride. J. Steroid Biochem. Mol. Biol. 2012;130:7–15. doi: 10.1016/j.jsbmb.2011.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rizner TL, Smuc T, Rupreht R, Sinkovec J, Penning TM. AKR1C1 and AKR1C3 may determine progesterone and estrogen ratios in endometrial cancer. Mol. Cell & Endocrinol. 2006;248:126–135. doi: 10.1016/j.mce.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 38.Gabby KH. Hyperglycemia, polyol metabolism, and complications of diabetes mellitus. Ann Rev. Med. 1975;26:521–536. doi: 10.1146/annurev.me.26.020175.002513. [DOI] [PubMed] [Google Scholar]
- 39.Murata M, Ohta N, Sakurai S, Alam S, Tsai J, Kador PF, Sato S. The role of aldose reductase in sugar cataract formation: aldose reductase plays a key role in lens epithelial cell death (apoptosis) Chemico-Biol. Inter. 2001;130–132:617–625. doi: 10.1016/s0009-2797(00)00289-1. [DOI] [PubMed] [Google Scholar]
- 40.Chang KC, Ponder J, Labarbera DV, Petrash JM. Aldose reductase inhibition prevents endotoxin-induced inflammatory responses in retinal microglia. Invest Ophthalmol Vis Sci. 2014;55:2853–2861. doi: 10.1167/iovs.13-13487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Judzewitsch RG, Jaspan JB, Polonsky KS, Weinberg CR, Halter JB, Halar E, Pfeifer MA, Vukadinovic C, Bernstein L, Schneider M, Liang KY, Gabbay KH, Rubenstein AH, Porte D., Jr Aldose reductase inhibition improves nerve conduction velocity in diabetic patients. N. Engl. J. Med. 1983;308:119–125. doi: 10.1056/NEJM198301203080302. [DOI] [PubMed] [Google Scholar]
- 42.Liu H, Luo Y, Zhang T, Zhang Y, Wu Q, Yuan L, Chung SS, Oates PJ, Yang JY. Genetic deficiency of aldose reductase counteracts the development of diabetic nephropathy in C57BL/6 mice. Diabetologia. 2011;54:1242–1251. doi: 10.1007/s00125-011-2045-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beyer-Mears A, Cruz E, Varagiannis E. Reversal of stage-I sugar cataract by Sorbinil, an aldose reductase inhibitor. Pharmacology. 1985;31:88–96. doi: 10.1159/000138103. [DOI] [PubMed] [Google Scholar]
- 44.Sestanj K, Bellini F, Fung S, Abraham N, Treasurywala A, Humber L, Simard-Duquesne N, Dvornik D. N-[5-(trifluoromethyl)-6-methoxy-1-naphthalenyl]thioxomethyl]- N-methylglycine (Tolrestat), a potent, orally active aldose reductase inhibitor. J Med Chem. 1984;27:255–256. doi: 10.1021/jm00369a003. [DOI] [PubMed] [Google Scholar]
- 45.Hotta N, Kawamori R, Fukuda M, Shigeta Y. Aldose Reductase Inhibitor-Diabetes Complications Trial Study Group, Long-term clinical effects of epalrestat, an aldose reductase inhibitor, on progression of diabetic neuropathy and other microvascular complications: multivariate epidemiological analysis based on patient background factors and severity of diabetic neuropathy. Diabet. Med. 2012;29:1529–1533. doi: 10.1111/j.1464-5491.2012.03684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matsumoto T, Ono Y, Kuromiya A, Toyosawa K, Ueda Y, Bril V. Long-term treatment with ranirestat (AS-3201), a potent aldose reductase inhibitor, suppresses diabetic neuropathy and cataract formation in rats. J Pharmacol Sci. 2008;107:340–348. doi: 10.1254/jphs.08071fp. [DOI] [PubMed] [Google Scholar]
- 47.Negoro T, Murata M, Ueda S, Fujitani B, Ono Y, Kuromiya A, Komiya M, Suzuki K, Matsumoto J. Novel, highly potent aldose reductase inhibitors: (R)-(–)-2-(4-bromo-2-fluorobenzyl)-1,2,3,4- tetrahydropyrrolo[1,2-a]pyrazine −4-spiro-3’-pyrrolidine-1,2’,3,5’-tetrone (AS-3201) and its congeners. J. Med. Chem. 1998;41:4118–4129. doi: 10.1021/jm9802968. [DOI] [PubMed] [Google Scholar]
- 48.Hotta N, Toyota T, Matsuoka K, Shigeta Y, Kikkawa R, Kaneko T, Takahashi A, Sugimura K, Koike Y, Ishii J, Sakamoto N. SNK-860 Diabetic Neuropathy Study Group, Clinical efficacy of fidarestat, a novel aldose reductase inhibitor, for diabetic peripheral neuropathy: a 52-week multicenter placebo-controlled double-blind parallel group study. Diabetes Care. 2001;24:1776–1782. doi: 10.2337/diacare.24.10.1776. [DOI] [PubMed] [Google Scholar]
- 49.Chung SS, amd Chung SK. Genetic analysis of aldose reductase in diabetic complications. Curr Med. Chem. 2003;10:1375–1387. doi: 10.2174/0929867033457322. [DOI] [PubMed] [Google Scholar]
- 50.Hu X, Li S, Yang G, Liu H, Boden G, Li L. Efficacy and safety of aldose reductase inhibitor for the treatment of diabetic cardiovascular autonomic neuropathy: systematic review and meta-analysis. PLoS One. 2014:e87096. doi: 10.1371/journal.pone.0087096. eCollection 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schemmel KE, Padiyara RS, D’Souza JJ. Aldose reductase inhibitors in the treatment of diabetic peripheral neuropathy: a review. J Diabetes Complications. 2010:354–360. doi: 10.1016/j.jdiacomp.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 52.Gallego O, Belyaeva OV, Porté S, Ruiz FX, Stetsenko AV, Shabrova EV, Kostereva NV, Farrés J, Parés X, Kedishvili NY. Comparative functional analysis of human medium-chain dehydrogenases, short-chain dehydrogenases/reductases and aldo-keto reductases with retinoids. Biochem J. 2006;399:101–109. doi: 10.1042/BJ20051988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gallego O, Ruiz FX, Ardèvol A, Domínguez M, Alvarez R, de Lera AR, Rovira C, Farrés J, Fita I, Parés X. Structural basis for the high all-trans-retinaldehyde reductase activity of the tumor marker AKR1B10. Proc Natl Acad Sci U S A. 2007;104:20764–20769. doi: 10.1073/pnas.0705659105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fukumoto S-I, Yamauchi N, Moriguchi H, Hippo Y, Watanbe A, Shibahara J, Taniguichi H, Ishikawa S, Ito H, Yamamato S, Iwanari H, Horonaka M, Ishikawa H, Niki T, Sohara Y, Kodama T, Mishimura M, Fukayama M, Doska-Akita H, Auratani H. Overexpression of the aldo-keto reductase family protein AKR1B10 is highly correlated with smokers non-small cell lung carcinoma. Clin. Cancer Res. 2005;11:1776–1785. doi: 10.1158/1078-0432.CCR-04-1238. [DOI] [PubMed] [Google Scholar]
- 55.Liu Z, Yan R, Al-Salman A, Shen Y, Bu Y, Ma J, Luo DX, Huang C, Jiang Y, Wilber A, Mo YY, Huang MC, Zhao Y, Cao D. Epidermal growth factor induces tumour marker AKR1B10 expression through activator protein-1 signalling in hepatocellular carcinoma cells. Biochem. J. 2012;442:273–282. doi: 10.1042/BJ20111322. [DOI] [PubMed] [Google Scholar]
- 56.Takemura M, Endo S, Matsunaga T, Soda M, Zhao HT, El-Kabbani O, Tajima K, Iinuma M, Hara A. Selective inhibition of the tumor marker aldo-keto reductase family member 1B10 by oleanolic acid. J Nat Prod. 2011;74:1201–1206. doi: 10.1021/np200118q. [DOI] [PubMed] [Google Scholar]
- 57.Li H, Yang AL, Chung YT, Zhang W, Liao J, Yang GY. . Sulindac inhibits pancreatic carcinogenesis in LSL-KrasG12D–LSL-Trp53R172H–Pdx-1-Cre mice via suppressing aldo-keto reductase family 1B10 (AKR1B10). Carcinogenesis. 2013;34:2090–2098. doi: 10.1093/carcin/bgt170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.El-Kabbani O, Scammells PJ, Day T, Dhagat U, Endo S, Matsunaga T, Soda M, Hara A. Structure-based optimization and biological evaluation of human 20α-hydroxysteroid dehydrogenase (AKR1C1) salicylic acid-based inhibitors. Eur. J. Med. Chem. 2010:5309–5317. doi: 10.1016/j.ejmech.2010.08.052. [DOI] [PubMed] [Google Scholar]
- 59.Bicíková M, Dibbelt L, Hill M, Hampl R, Stárka L. Allopregnanolone in women with premenstrual syndrome. Horm. Metab. Res. 1998;30:227–230. doi: 10.1055/s-2007-978871. [DOI] [PubMed] [Google Scholar]
- 60.Desmond JC, Mountford JC, Drayson MT, Walker EA, Hewison M, Ride JP, Luong QT, Hayden RE, Vanin EF, Bunce CM. The aldo-keto reductase AKR1C3 is a novel suppressor of cell differentiation that provides a plausible target for the non-cyclooxygenase-dependent antineoplastic actions of nonsteroidal anti-inflammatory drugs. Cancer Res. 2003;63:505–512. [PubMed] [Google Scholar]
- 61.Byrns MC, Steckelbroeck S, Penning TM. An indomethacin analogue N-(4-chlorobenzoyl)melatonin is a selective inhibitor of aldo-keto reductase 1C3 (type 2 3α-HSD, type 5 17β-HSD and prostaglandin F synthase), a potential target for the treatment of hormone dependent and hormone independent malignancies. Biochem. Pharmacol. 2008;75:484–493. doi: 10.1016/j.bcp.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.D.N. Khanim F, Veliça P, Hayden R, Ride J, Pararasa C, Chong MG, Gunther U, Veerapen N, Winn P, Farmer R, Trivier E, Rigoreau L, Drayson M, Bunce C. Selective AKR1C3 inhibitors do not recapitulate the anti-leukaemic activities of the pan-AKR1C inhibitor medroxyprogesterone acetate. Br J Cancer. 2014;110:1506–1516. doi: 10.1038/bjc.2014.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Khanim FL, Hayden RE, Birtwistle J, Lodi A, Tiziani S, Davies NJ, Ride JP, Viant MR, Gunther UL, Mountford JC, Schrewe H, Green RM, Murray JA, Drayson MT, Bunce CM. Combined bezafibrate and medroxyprogesterone acetate: potential novel therapy for acute myeloid leukaemia. PLoS One. 2009 Dec 7;4(12):e8147. doi: 10.1371/journal.pone.0008147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yepuru M, Wu Z, Kulkarni A, Yin F, Barrett CM, Kim J, Steiner MS, Miller DD, Dalton JT, Narayanan R. Steroidogenic enzyme AKR1C3 is a novel androgen receptor-selective coactivator that promotes prostate cancer growth. Clin Cancer Res. 2013;19:5613–5625. doi: 10.1158/1078-0432.CCR-13-1151. [DOI] [PubMed] [Google Scholar]
- 65.Kikuchi A, Furutani T, Azami H, Watanabe K, Niimi T, Kamiyama Y, Kuromitsu S, Baskin-Bey E, Heeringa M, Ouatas T, Enjo K. In vitro and in vivo characterisation of ASP9521: a novel, selective, orally bioavailable inhibitor of 17β-hydroxysteroid dehydrogenase type 5 (17βHSD5; AKR1C3) Invest New Drugs. 2014 Jul;:1. doi: 10.1007/s10637-014-0130-5. [Epub ahead of print], (2014) [DOI] [PubMed] [Google Scholar]
- 66.Gonzales E, Crestel D, Baussan C, Dabadie A, Gerhardt M-F, Jacquemin E. SRD5B1 (AKR1D1) gene analysis in delta-4-3-ketosteroid 5β-reductase deficiency: evidence for primary genetic defect. J. Hepatology. 2004;40:715–719. doi: 10.1016/j.jhep.2003.12.024. [DOI] [PubMed] [Google Scholar]
- 67.Kimura A, Nittono H, Takei H, Kurosawa T. Abnormally low ratio of cholic acid to chenodeoxycholic acid due to a deficiency of 3-oxo-delta-4-steroid 5β-reductase. Pediatrics International. 2000;42:594. doi: 10.1046/j.1442-200x.2000.01284.x. [DOI] [PubMed] [Google Scholar]
- 68.Lemonde HA, Custard EJ, Bouquet J, Duran M, Overmars H, Scambler PJ, Clayton PT. Mutations in SRD5B1 (AKR1D1), the gene encoding delta-4-3-oxosteroid 5β-reductase, in hepatitis and liver failure in infancy. Gut. 2003:1494–1499. doi: 10.1136/gut.52.10.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shneider BL, Setchell KD, Whitington PF, Neilson KA, Suchy FJ. Δ4-3-oxosteroid 5β-reductase deficiency causing neonatal liver failure and hemochromatosis. J. Pediatrics. 1994;124:234–238. doi: 10.1016/s0022-3476(94)70310-8. [DOI] [PubMed] [Google Scholar]
- 70.Sumazaki R, Nakamura N, Shoda J, Kurosawa T, Tohma M. Gene analysis in delta-4-3- oxosteroid 5β-reductase deficiency. The Lancet. 1997;349:329. doi: 10.1016/s0140-6736(05)62828-0. [DOI] [PubMed] [Google Scholar]
- 71.Drury JE, Mindnich R, Penning TM. Characterization of disease-related 5β-reductase (AKR1D1) mutations reveals their potential to cause bile acid deficiency. J. Biol. Chem. 2010;285:24529–24537. doi: 10.1074/jbc.M110.127779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mindnich R, Drury JE, Penning TM. The effect of disease associated point mutations on 5β-reductase (AKR1D1) enzyme function. Chem. Biol. Interact. 2011;191:250–254. doi: 10.1016/j.cbi.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Flück CE, Meyer-Böni M, Pandey AV, Kempná P, Miller WL, Schoenle EJ, Biason- Lauber A. Why boys will be boys: two pathways of fetal testicular androgen biosynthesis are needed for male sexual differentiation. Am J Hum Genet. 2011;89:201–218. doi: 10.1016/j.ajhg.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Auchus RJ. The backdoor pathway to dihydrotestosterone, Trends Endocrinol. Metab. 2004;15:432–438. doi: 10.1016/j.tem.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 75.Bauman DR, Steckelbroeck S, Williams MV, Peehl DM, Penning TM. Identification of the major oxidative 3a–hydroxysteroid dehydrogenase in human prostate that converts 5α-andostane- 3α,17β-diol to 5α-dihydrotestosterone. A potential therapeutic target for androgen dependent disease. Mol. Endocrinol. 2006;20:444–458. doi: 10.1210/me.2005-0287. [DOI] [PubMed] [Google Scholar]
- 76.Tipparaju SM, Liu SQ, Barski OA, Bhatnagar A. NADPH binding to β-subunit regulates inactivation of voltage-gated K(+) channels. Biochem Biophys Res Commun. 2007;359:269–276. doi: 10.1016/j.bbrc.2007.05.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tipparaju SM, Li XP, Kilfoil PJ, Xue B, Uversky VN, Bhatnagar A, Barski OA. Interactions between the C-terminus of Kv1.5 and Kvβ regulate pyridine nucleotide-dependent changes in channel gating. Pflugers Arch. 2012;463:799–918. doi: 10.1007/s00424-012-1093-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bodreddigari S, Jones LK, Egner PA, Groopman JD, Sutter CH, Roebuck BD, Guengerich FP, Kensler TW, Sutter TR. Protection against aflatoxin B1-induced cytotoxicity by expression of the cloned aflatoxin B1-aldehyde reductases rat AKR7A1 and human AKR7A3. Chem. Res. Toxicol. 2008;21:1134–1142. doi: 10.1021/tx7004458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lyon RC, Johnston SM, Watson DG, McGarvie G, Ellis EM. Synthesis and catabolism of gamma-hydroxybutyrate in SH-SY5Y human neuroblastoma cells: role of the aldo-keto reductase AKR7A2. J. Biol. Chem. 2007;282:25986–25992. doi: 10.1074/jbc.M702465200. [DOI] [PubMed] [Google Scholar]
- 80.Snead OCI. Evidence for a G Protein-Coupled γ-Hydroxybutyric Acid Receptor. J. Neurochem. 2000;75:1986–1996. doi: 10.1046/j.1471-4159.2000.0751986.x. [DOI] [PubMed] [Google Scholar]
- 81.Kim KJ, Pearl PL, Jensen K, Snead OC, Malaspina P, Jakobs C, Gibson KM. Succinic semialdehyde dehydrogenase: biochemical-molecular-clinical disease mechanisms, redox regulation, and functional significance. Antioxd Redox Signal. 2011;15:691–718. doi: 10.1089/ars.2010.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kumar P, Heinkoff S, Ng PC. Predicting the effects of coding non-synnonymous varaints on protein function using the SIFT alogrithm. Nature Protocols. 2009;4:1073–1082. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 83.Barski OA, Mindnich R, Penning TM. Alternative splicing in the aldo-keto reductase superfamily: implications for protein nomenclature. Chem Biol Interact. 2013;202:153–158. doi: 10.1016/j.cbi.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.