Summary
While thousands of long noncoding RNAs (lncRNAs) have been identified, few lncRNAs that control neural stem cell (NSC) behavior are known. Here, we identify Pinky (Pnky) as a neural-specific lncRNA that regulates neurogenesis from NSCs in the embryonic and postnatal brain. In postnatal NSCs, Pnky knockdown potentiates neuronal lineage commitment and expands the transit-amplifying cell population, increasing neuron production several-fold. Pnky is evolutionarily conserved and expressed in NSCs of the developing human brain. In the embryonic mouse cortex, Pnky knockdown increases neuronal differentiation and depletes the NSC population. Pnky interacts with the splicing regulator PTBP1, and PTBP1 knockdown also enhances neurogenesis. In NSCs, Pnky and PTBP1 regulate the expression and alternative splicing of a core set of transcripts that relates to the cellular phenotype. These data thus unveil Pnky as a conserved lncRNA that interacts with a key RNA processing factor and regulates neurogenesis from embryonic and postnatal NSC populations.
Introduction
Neural stem cells (NSCs) exist in both the embryonic and postnatal mammalian brain. In the embryonic cortical ventricular zone (VZ) and adult ventricular-subventricular zone (V-SVZ), NSCs are glial cells that can both self-renew and differentiate to yield intermediate progenitors that divide once or more before producing migratory young neurons (Kriegstein and Alvarez-Buylla, 2009). The production of proper numbers of neuronal progenitors from NSCs is a key aspect of brain development, and defects at this stage of the neurogenic lineage may underlie a number of human developmental disorders (Lui et al., 2011).
The mammalian genome encodes many thousands of lncRNAs – transcripts >200 nucleotides long that have no evidence of protein coding potential – and emerging data indicate that lncRNAs can have critical biological functions (Batista and Chang, 2013; Lee, 2012; Mercer and Mattick, 2013; Rinn and Chang, 2012). Although transcription factors, microRNAs, and signaling pathways that control the transition between NSCs and neurogenic progenitors have been studied intensively (Ihrie and Alvarez-Buylla, 2011; Kriegstein and Alvarez-Buylla, 2009; Lui et al., 2011), lncRNAs that regulate this critical, early stage of neurogenesis have not been identified.
Here we describe the unique developmental function of a neural-specific lncRNA we have named Pinky (Pnky) and provide insights into its molecular function. We have previously shown that knockdown of lncRNAs Dlx1as and Six3os in V-SVZ NSCs results in decreased neurogenesis (Ramos et al., 2013). Similarly, loss-of-function studies of different lncRNAs in ESC-derived NSCs (Lin et al., 2014; Ng et al., 2013), zebrafish brain (Ulitsky et al., 2011), and mouse central nervous system (Bond et al., 2009; Rapicavoli et al., 2011; Sauvageau et al., 2013) all demonstrate a loss of neuronal populations. In contrast, postnatal V-SVZ NSCs with Pnky-knockdown (Pnky-KD) generated 3-4 fold more neurons. To understand this phenotype, we used time-lapse microscopy to study lineage commitment and cell proliferation at the single-cell level. We cloned the human PNKY transcript and discovered its expression in the VZ of the embryonic human brain, and we demonstrated a role for Pnky in the development of the embryonic mouse cortex in vivo. Using mass spectrometry, we found that Pnky bound to PTBP1, an RNA-splicing factor that is a potent regulator of neural development (Keppetipola et al., 2012), direct cell reprogramming (Xue et al., 2013), and brain tumor growth (Ferrarese et al., 2014). Further analysis indicated that Pnky and PTBP1 modulate the expression and alternative splicing of an overlapping set of transcripts, and double-knockdown epistasis experiments suggested that Pnky and Ptbp1 function in the same pathway. Overall, our work demonstrates that an evolutionarily conserved lncRNA can regulate neurogenesis from NSCs in both the embryonic and postnatal brain.
Results
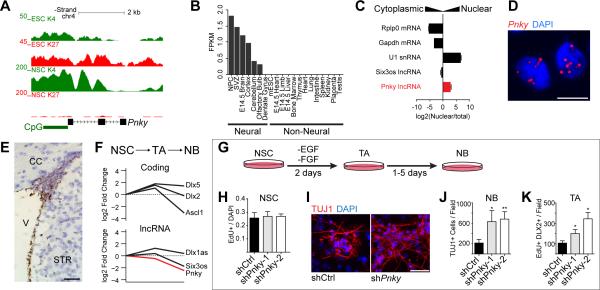
Throughout adult life, V-SVZ NSCs give rise to transit-amplifying (TA) cells, which generate neuroblasts (NB) that migrate to the olfactory bulb where they differentiate into interneurons (Doetsch et al., 1999; Lois and Alvarez-Buylla, 1994; Luskin, 1998; Peretto et al., 1997). Pnky (previously called lnc-pou3f2) is a lncRNA that we initially identified as being expressed in the adult V-SVZ (Ramos et al., 2013). RACE cloning followed by Sanger sequencing demonstrated Pnky to be an 825 nucleotide (nt) polyadenylated RNA encoded from three exons (Figure 1A). Analysis with the Coding Potential Calculator (CPC) (Kong et al., 2007), PhyloCSF (Lin et al., 2011), and the Coding-Potential Assessment Tool (CPAT) (Wang et al., 2013) indicated that the Pnky transcript has no protein-coding potential (Figure S1A). Analysis of available RNA-seq datasets indicated that Pnky is specifically expressed in neural tissues and lineages but is not expressed in the dentate gyrus, which contains another population of adult NSCs (Figure 1B). Furthermore, like many key developmental genes, the promoter of Pnky was “bivalent” with both histone 3 lysine 27-trimethylation (H3K27me3) and histone 3 lysine 4-trimethylation (H3K4me3) in embryonic stem cells (ESCs), coherent with its repressed but “poised” transcriptional state (Figure 1A). In contrast, in ESC-derived NSCs (ESC-NSCs) and V-SVZ NSCs, the Pnky promoter was monovalent with H3K4me3, consistent with its active transcription (Figure 1A and S1B).
Figure 1. The lncRNA Pinky is expressed in SVZ-NSCs and regulates neuronal differentiation.
A) UCSC genome browser view of the Pnky locus. Also shown are ChIP-seq tracks for H3K4me3 and H3K27me3 in ESCs and V-SVZ NSCs. B) Fragments per kilobase per million mapped reads (FPKM) values for Pnky in indicated tissues. C) Subcellular fractionation followed by RT-qPCR for indicated lncRNAs and mRNAs. Error bars are propagated standard deviation (S.D.) from technical triplicate wells. D) Branched-DNA ISH for Pnky in V-SVZ NSC cultures. Nuclei are counter-stained with 4',6-diamidino-2-phenylindole (DAPI). Scale bar = 10 μm. E) Branched-DNA ISH for Pnky (brown) in adult mouse coronal brain section. Nuclei are counter-stained with hematoxylin. V = ventricle, CC = corpus collosum, STR = striatum. Scale bar = 50 μm. F) Microarray expression analysis from FACS-isolated neural stem cells (NSC), transit-amplifying cells (TA), and neuroblasts (NB). Value in NSCs set to 0. G) Schematic of V-SVZ NSC culture system. H) Quantification of EdU labeling counted from GFP+ cultures of V-SVZ NSCs infected with control or Pnky-KD constructs. I) ICC for TUJ1 (red) after 7d of differentiation in control or Pnky-KD GFP+ cultures. Nuclei are counter-stained with DAPI (blue). Scale bar = 50 μm. J) Quantification of TUJ1+ NBs produced after 7d differentiation. K) Quantification of the number of TA cells after 2d of differentiation. Error bars for H, J, and K are S.D. from triplicate wells, * p < 0.05, **p < 0.01, Student's t-test. See also Figure S1.
Nuclear fractionation of V-SVZ NSC cultures followed by RT-qPCR analysis demonstrated Pnky to be enriched in the nucleus as compared to coding mRNAs, which were enriched in cell lysates containing cytoplasm (Figure 1C). Consistent with the nuclear fractionation studies, in situ hybridization (ISH) for Pnky demonstrated predominantly nuclear localization of the transcript (Figure 1D and S1C).
ISH of adult mouse brain tissue revealed prominent expression of Pnky in the V-SVZ (Figure 1E). To investigate whether Pnky expression is dynamic within the neurogenic lineage, we analyzed gene expression of V-SVZ cells acutely isolated from the brain through fluorescent activated cell sorting (FACS) (Ramos et al., 2013). Briefly, activated neural stem cells (NSCs) express both glial fibrillary acidic protein (GFAP) and the epidermal growth factor receptor (EGFR). Transit-amplifying cells are GFAP- but retain EGFR expression, and neuroblasts express cell surface marker CD24 (Pastrana et al., 2009). Pnky expression was highest in NSCs, and decreased by 5.6-fold (p=0.0003, Student's t-test) in migratory neuroblasts (Figure 1F). Thus, Pnky is normally downregulated during lineage progression in vivo.
To investigate the role of Pnky in neurogenesis, we first used V-SVZ NSC monolayer cultures that recapitulate key features of neuronal differentiation as well as glial cell production (Figure 1G). Pnky was expressed in cultured NSCs in self-renewal conditions. Upon differentiation, Pnky remained expressed in GFAP+ astrocytes, but was down regulated in TUJ1+ neuronal cells (Figure S1D), similar to the pattern of expression seen in vivo. After lentiviral transduction with control (shCtrl-GFP) or Pnky (shPnky-1-GFP and shPnky-2-GFP)knockdown constructs, GFP+ NSCs were isolated through FACS and cultured. Pnky knockdown was efficient in both self-renewing NSCs and after their differentiation (Figure S1E and S1F). Cells in these purified cultures uniformly expressed SOX2, a neural stem cell marker (Figure S1G). Flow cytometric analysis of these infected cells indicated that the majority (66.0%) were also positive for both GFAP and NESTIN (Figure S1H), markers of V-SVZ NSCs. Therefore, the majority of cells transduced with Pnky-KD or control vectors were GFAP+, NESTIN+, SOX2+ V-SVZ NSCs.
Pnky-KD NSCs incorporated the thymidine analog 5-ethynyl-2'-deoxyuridine (EdU) at the same rate as control NSCs, suggesting that proliferation in self-renewal conditions was not affected (Figure 1H and S1I). However, Pnky-KD NSCs generated 3-fold more TUJ1+ neuroblasts after 7 days (7d) of differentiation (Figure 1I and 1J). To further analyze the effect of Pnky-KD in the NSC population, we used a molecular-genetic method of targeting knockdown to V-SVZ NSCs (Doetsch et al., 1999; Park et al., 2014). We derived NSC cultures from G-TVA mice, which express the chicken retroviral receptor TVA from the GFAP promoter. For viral transduction of these cultures, we used shRNA lentiviruses pseudotyped with chicken viral EnvA protein, which restricts infection to the GFAP+ cells (Figure S1J). In G-TVA NSCs, Pnky-KD did not affect NSC proliferation in self-renewal conditions (Figure S1K). However, after differentiation, G-TVA NSC cultures produced a 3.56-fold more neuroblasts as compared to control (Figure S1L). Thus, Pnky-KD targeted to the GFAP+ NSC population in V-SVZ cultures also resulted in increased neuronal production during differentiation.
V-SVZ transit-amplifying cells express DLX2 and normally divide several times before giving rise to neuroblasts (Doetsch et al., 2002; Ponti et al., 2013). With Pnky-KD, there were 1.8 to 3.2 fold more EdU+, DLX2+ cells at 2d of differentiation, suggesting that Pnky plays a role at the transit-amplifying stage of neuronal differentiation (Figure 1K and S1M).
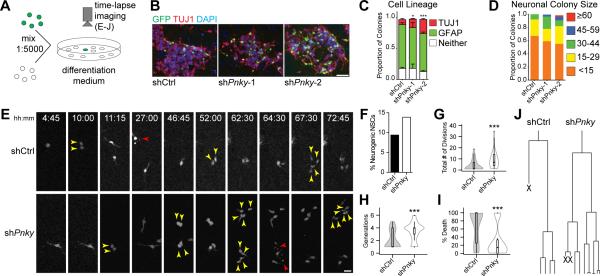
To further investigate how Pnky-KD increases neuronal production, we plated NSCs infected with Pnky-KD or control vector with large numbers of uninfected NSCs (ratio of infected GFP+ NSCs to uninfected NSCs was approximately 1:5000, Fig 2A). After 4d of differentiation, these cultures produced well-isolated clusters of GFP+ cells (Figure 2B). With Pnky knockdown, the proportion of GFP+ cell clusters containing TUJ1+, GFP+ cells was increased (Figure 2C). Furthermore, these clusters also contained greater numbers of TUJ1+, GFP+ neuroblasts (Figure 2D). These data suggested that the increased neurogenesis resulted from both a shift towards neuronal lineage commitment and an increase in cell amplification within the neurogenic lineage.
Figure 2. Pinky knockdown leads to an expansion of neurogenic transit-amplifying progenitors.
A) Schematic of experimental design. B) Representative images of isolated colonies after 4d of differentiation. ICC for TUJ1 (red) and GFP (green). Nuclei are DAPI counterstained (blue). Scale bar = 50 μm. C) Quantification of the fate of isolated GFP+ colonies. Error bars = S.D. of triplicate experiments. D) Quantification of TUJ1+ neuroblasts found in individual neurogenic colonies. E) Representative frames from time-lapse video of control (top) or Pnky-KD (bottom) single cells. Time of differentiation is indicated. Yellow arrows indicate daughter cells resulting from a recent division, red arrows indicate cell death. Scale bar = 25 μm. F) Bar graph representing the percentage of initial tracked progenitors that gave rise to neuroblasts. N= 531 GFP+ shCtrl NSCs and 316 GFP+ shPnky NSCs. G) Violin plots overlaying box-and-whisker plots of total number of divisions undergone by a single initial neurogenic progenitor and all of its daughter cells. N = 44 shCtrl and 33 shPnky progenitors. H) Violin plots overlaying box-and-whisker plots of the number of generations per initial neurogenic progenitor for shCtrl and shPnky. N = 44 shCtrl and 33 shPnky progenitors. I) Violin plots overlaying box-and-whisker plots of % of progeny per single neurogenic progenitor that underwent cell death. N = 44 shCtrl and 33 shPnky progenitors. J) Tree diagram for the frames shown in E and corresponding time-lapse movies. X indicates cell underwent cell death. *p < 0.05 ***p<0.001, Student's t-test. See also Figure S2.
To more directly observe the clonal behavior of individual NSCs with Pnky-KD, we used time-lapse microscopy to image GFP+ cells every 15 min for 3d (Figure 2E). We followed the fate of 316 GFP+ Pnky-KD NSCs and 531 GFP+ control NSCs. After differentiation, we could identify both TUJ1+ neurogenic clones (Figure S2A) and GFAP+ glial clones (Figure S2B and S2C). Clones arising from NSCs with Pnky-KD were 48% more likely to be neurogenic (Figure 2F). Thus, Pnky-KD increased the likelihood that NSCs produced neurogenic progenitors.
Neurogenic clones with Pnky-KD contained 2.74 fold more neuroblasts than control (Figure S2D). This increase in neurogenesis was in part related to an increase in progenitor cell divisions before differentiation into neuroblasts: while control progenitors divided 3.93 (SD=4.68, n=44) times and gave rise to 2.05 (SD=1.42) generations of daughter cells, Pnky-KD resulted in 8.94 (SD=7.64, n=33) divisions and 3.45 (SD=1.41) generations (Figure 2G and 2H). The average cell cycle length was not affected by Pnky-KD (19.4 hours vs. 19.1 hours, p=0.89) (Figure S2E), suggesting that the increased number of divisions related to the maintenance of a proliferative cell state and not an accelerated cell cycle. Time-lapse imaging also enabled direct observation of cell death, and the number of GFP+ neurogenic progeny (neuroblasts) that underwent cell death was reduced 57% by Pnky-KD (Figure 2I). Thus, Pnky-KD promoted neuronal lineage commitment from postnatal V-SVZ NSCs, increased the number of divisions of neurogenic progenitors, and reduced cell death (Figure 2J and S2F).
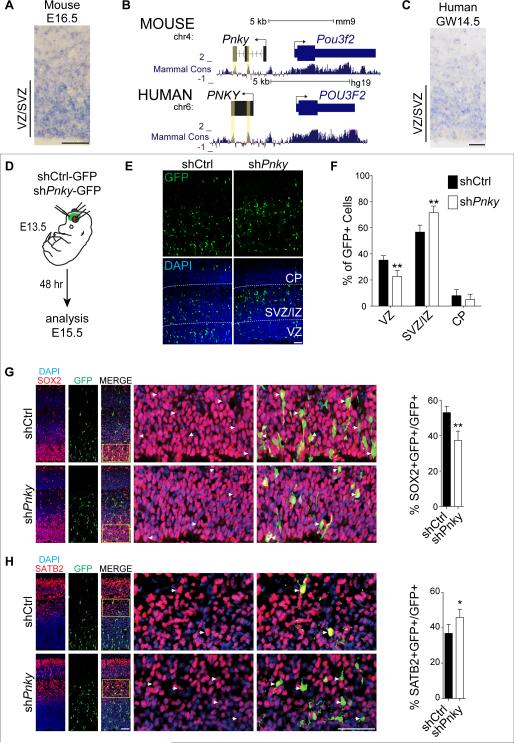
To investigate whether Pnky also regulates neurogenesis from other NSC populations, we next examined the embryonic brain. Pnky transcripts were detected in embryonic mouse brain tissue by RNA-seq (Figure 1B), and ISH revealed its expression in the cortical VZ – where embryonic NSCs reside – at embryonic day 14.5 (E14.5) and E16.5 (Figure 3A, S3A, and S3B). Pnky has two regions of high conservation among vertebrates (Figure 3B and S3C), and using strand-specific RNA-seq of gestational week 16 (GW16) human cortical samples, we detected a non-coding transcript divergent to POU3F2 that included the conserved sequences. RACE cloning identified human PINKY (PNKY) as a polyadenylated 1592 nt transcript containing the two conserved elements and expressed from a conserved promoter region (Figure 3B). As in the developing mouse brain, ISH of GW14.5 human cortex demonstrated PNKY expression in the VZ, with decreased levels in the subventricular zone and intermediate zone (SVZ/IZ), where young neurons begin to differentiate (Figure 3C and S3D).
Figure 3. Pinky is expressed in the developing mouse and human cortex and regulates the differentiation of mouse cortical progenitors in vivo.
A) ISH for Pnky in embryonic day 16.5 (E16.5) mouse brain. B) Genome browser track showing two regions of high conservation, determined by PhyloP score (yellow boxes). Human PNKY genomic region is shown below with conservation indicated. For clarity, the mouse – strand is shown left to right. C) ISH for PNKY in gestational week 14.5 (GW14.5) human brain. D) Schematic of in utero.electroporation of mouse embryonic brain. E) Cortical sections at E15.5, 2d after electroporation with shCtrl (left) or shPnky (right). IHC for GFP (top) and with DAPI counterstain (bottom). VZ = ventricular zone, SVZ/IZ = subventricular zone/intermediate zone, CP = cortical plate. F) Quantification of GFP+ cell distribution in indicated zones as a percentage of total GFP+ cells. G) Left: Cortical sections at E15.5, 2d after electroporation with shCtrl or shPnky. IHC for GFP (green) and SOX2 (red), with DAPI nuclear counterstain (blue). Yellow box indicates region enlarged in the adjacent panels. Arrowheads indicate co-labeled cells. Right: quantification of SOX2+, GFP+ cells as a percentage of total GFP+ cells. H) Left: Cortical sections immunostained for GFP (green) and SATB2 (red), with DAPI nuclear counterstain (blue). Yellow box indicates region enlarged in the adjacent panels. Right: Quantification of SATB2+, GFP+ cells as a percentage of total GFP+ cells. All error bars are S.D., n=4 brains of each condition from 3 separate surgeries. *p<0.05, **p<0.01. All scale bars = 50 μm. See also Figure S3.
To investigate the role of Pnky in the mouse embryonic VZ, we electroporated Pnky-KD (shPnky-2-GFP) or control (shCtrl-GFP) construct into VZ cells at E13.5 (Figure 3D). Two days later, we analyzed GFP+ cells in the VZ, SVZ/IZ, and cortical plate (CP) (Figure 3E). With Pnky knockdown, there was a 35% decrease in the proportion of GFP+ cells in the VZ and a 26% increase in GFP+ cells in the SVZ/IZ (Figure 3F). We did not detect apoptotic GFP+ cells as assessed by cleaved caspase-3 staining (not shown). At this time point in cortical development, SOX2+ NSCs in the VZ produce proliferative TBR2+ progenitor cells in the SVZ/IZ that differentiate into SATB2+ young neurons. With Pnky knockdown, the proportion of SOX2+, GFP+ cells was reduced (Figure 3G). While Pnky knockdown did not affect the proportion of TBR2+, GFP+ TA cells or their proliferation (Figure S3E-J), the proportion of SATB2+, GFP+ young neurons was increased (Figure 3H). These data indicate that Pnky regulates the production of young neurons from embryonic brain NSCs in vivo (Figure S3K).
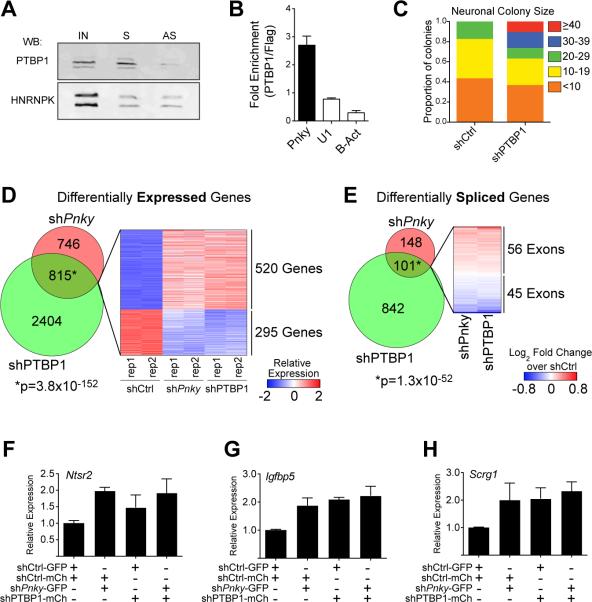
Many lncRNAs regulate gene expression through interactions with specific protein partners. To identify Pnky-interacting proteins, we incubated biotinylated Pnky (S) or antisense Pnky (AS) control RNA with V-SVZ NSC nuclear extract and used mass spectrometry to identify proteins that bound to the transcripts (Figure S4A and S4B). Polypyrimidine tract-binding protein 1 (PTBP1) was identified as a binding partner of Pnky, and this interaction was confirmed by Western blot analysis (Figure 4A). While both Pnky and control RNA nonspecifically bound other RNA binding proteins including HNRNPK and ELAVL1 (Figure 4A and S4C), only Pnky RNA enriched for PTBP1, suggesting that the interaction between Pnky and PTBP1 is specific (Figure 4A). Furthermore, RNA immunoprecipitation with PTBP1 antibodies enriched for Pnky transcript but not U1 or beta-actin mRNA (Figure 4B).
Figure 4. Pinky interacts with PTBP1 and regulates transcript expression and differential splicing.
A) Immunoblot for PTBP1 or HNRNPK following RNA-pulldown with biotin-labeled sense (S) or anti-sense (AS) Pnky RNA incubated with V-SVZ NSC nuclear extract. IN = input V-SVZ nuclear extract. B) RT-qPCR detection for indicated RNA recovered by PTBP1-specific antibody normalized to control FLAG antibody. Error bars are propagated S.D. from technical triplicates. C) Quantification of number of TUJ1+ neuroblasts found in individual neurogenic colonies with Ptbp1-knockdown or control. D) Venn Diagram demonstrating the overlap between genes differentially expressed (FDR < 0.05) upon Ptbp1 or Pnky knockdown. Heatmap representation of the differential expression of the overlapping gene set in shCtrl, shPnky, and shPtbp1 biological duplicate cultures. E) Venn Diagram demonstrating the overlap between exons showing differential usage (FDR < 0.01) upon Ptbp1 or Pnky knockdown. Heatmap representation of the differential usage of the overlapping gene set in shPnky and shPtbp1 cultures compared to control. F-H) RT-qPCR detection of expression of indicated genes normalized to Gapdh. Expression in each condition is shown relative to control (shCtrl-GFP, shCtrl-mCherry). Error bars are 95% confidence intervals from 3 separate cultures. See also Figure S4 and Tables S1-S3.
PTBP1 is expressed in NSCs and functions as a repressor of neuronal differentiation. In the embryonic brain, loss of Ptbp1 results in precocious neuronal differentiation (Shibasaki et al., 2013), and in fibroblasts, Ptbp1 knockdown leads to direct neuronal trans-differentiation (Xue et al., 2013). PTBP1 was detected in the nucleus of V-SVZ NSCs (Figure S4D), and Ptbp1 knockdown resulted in larger neuronal colonies (Figure 4C), similar to the phenotype observed with Pnky-KD (Figure 2D).
In NSCs, PTBP1 negatively regulates the expression of Ptpb2, which is required for the generation of neuronal precursors (Boutz et al., 2007; Licatalosi et al., 2012). Using a PTBP2-specific antibody (Polydorides et al., 2000), we found the Pnky transcript did not specifically bind PTBP2 (Figure S4E). During neurogenesis, Ptbp1 is normally down-regulated, and Ptbp2 is increased (Keppetipola et al., 2012). Therefore we next characterized the relationship between Ptbp1, Ptbp2, and Pnky expression. Upon Ptbp1 knockdown in V-SVZ NSCs, Ptbp2 transcripts were upregulated, as expected (Figure S4F). Ptbp1 knockdown did not decrease Pnky expression (Figure S4F). Pnky-KD had no effect on Ptbp1 expression and caused a small reduction (25%) in Ptbp2 (Figure S4G). Thus, the increase in neuronal differentiation observed with Pnky-KD does not appear to relate to changes in Ptbp1 or Ptbp2 expression.
PTBP1 regulates mRNA transcript levels and pre-mRNA splicing during neuronal differentiation (Keppetipola et al., 2012; Yap et al., 2012; Zheng et al., 2012). To determine whether Pnky and PTBP1 regulate a common set of transcripts, we performed RNA-seq of V-SVZ NSCs with either Pnky knockdown or Ptbp1 knockdown. The overlap of differentially expressed genes was highly significant (Figure 4D and Table S1, 815 genes, P = 3.8×10−152) and enriched for Gene Ontology terms (Huang et al., 2009) related to cell-cell adhesion, synpatogenesis and neurogenesis (Table S2). Furthermore, using DEX-seq (Anders et al., 2012) to analyze differential exon usage, we found that Pnky knockdown and Ptbp1 knockdown also resulted in a common set of splice variants (Figure 4E and Table S3 101 exons, P = 1.3×10−52). Thus, these data demonstrate that Pnky and its associated protein PTBP1 regulate an overlapping set of transcripts that underlies their role in regulating neurogenesis from V-SVZ NSCs.
To further assess whether Pnky and Ptbp1 function in the same pathway, we performed an epistasis experiment. The lack of synergistic or additive effects upon combined loss of an lncRNA and coding gene pair suggests that they function in the same molecular pathway (Dimitrova et al., 2014). Therefore, we used FACS to isolate NSCs with double knockdown of Pnky and Ptbp1 as well as NSCs with each single knockdown (Figure S4H-I). We then analyzed the expression of several genes (Ntsr2, Igfbp5, Scrg1, and Ppp1r3c) that had been commonly upregulated by both Pnky-KD and PTBP1 knockdown in RNA-seq experiments (Figure 4D). As expected, Ntsr2, Igfbp5, Scrg1, and Ppp1r3c all had increased expression in the cells with single-knockdown as compared to control (Figure 4F-H and S4J). Importantly, the combined knockdown of Pnky and Ptbp1 did not further enhance this gene expression (Figure 4F-H and S4J). These data indicate a genetic interaction between Pnky and Ptbp1 and suggest that they function in the same pathway to regulate gene expression in NSCs.
Discussion
Taken together, our data indicate that Pnky is a conserved, neural-specific, nuclear lncRNA that interacts with PTBP1 and regulates the production of neurons from NSCs. While other lncRNAs have been found to function in central nervous system development – with lncRNA loss-of-function resulting in decreased neurogenesis (Bond et al., 2009; Chalei et al., 2014; Lin et al., 2014; Ng et al., 2013; Rapicavoli et al., 2011; Sauvageau et al., 2013; Ulitsky et al., 2011) – Pnky knockdown increased the production of neurons, suggesting a distinct developmental role for this lncRNA in controlling neurogenesis from NSCs. Our data are consistent with a model in which Pnky serves to “restrain” neuronal commitment, regulating the production of young neurons from NSCs in the developing embryonic cortex as well as those in the postnatal V-SVZ.
In the developing cortex, we found that Pnky-KD resulted in an increase in young neurons and decreased the NSC population in the VZ. This did not appear to involve a change in the proliferation of the transit amplifying cells of this embryonic lineage. In V-SVZ NSCs from the postnatal brain, Pnky-KD also promoted neuronal differentiation, and we additionally observed greater numbers of divisions of the neurogenic progenitor cells. Thus, the phenotypes of Pnky-KD in these two very different NSC populations are largely similar in that neuronal differentiation is increased. Interestingly, Pnky does not appear to regulate all NSC populations, as we did not detect Pnky transcripts in the dentate gyrus, which harbors another population adult NSCs.
Our studies indicated that Pnky interacts with PTBP1, which is a key regulator of neural development (Shibasaki et al., 2013). PTBP1 is also a powerful mediator of cell reprogramming (Xue et al., 2013), and its depletion from fibroblasts can lead to direct trans-differentiation into neurons. Moreover, PTBP1 is a potent driver of brain tumor growth and invasiveness (Ferrarese et al., 2014). During neuronal differentiation, PTBP1 is normally downregulated, and there is an increase in PTBP2. We did not find evidence for a specific interaction between Pnky and PTPB2 or other nuclear RNA-binding proteins such as HNRNPK and ELAVL1. These data suggest that Pnky and PTPB1 – a splicing factor with critical functions in both normal development and brain tumors – are part of a specific ribonucleoprotein complex in NSCs.
Given this physical interaction between Pnky and PTBP1, we worked towards understanding whether this lncRNA and protein partner function together. First, we found that both Pnky and PTPB1 knockdown in V-SVZ NSCs promoted neurogenesis. Furthermore, Pnky-KD promoted neuronal differentiation in the developing cortex, and mice with Ptbp1 loss-of-function also exhibit precocious cortical neurogenesis. Thus, Pnky-KD and loss of PTBP1 produce similar phenotypic results. Second, we found that Pnky-KD and PTBP1-knockdown produced gene expression changes as well as splicing changes that were highly similar, indicating that Pnky and PTPB1 regulate a common set of transcripts related to neuronal differentiation. Third, our epistasis experiment indicated that Pnky and PTBP1 double knockdown does not produce changes greater than single knockdowns. Thus, the physical interaction between Pnky and PTBP1 along with the evidence for their genetic interaction is consistent with a model in which they function together in a common molecular pathway. Whether the physical interaction between Pnky and PTPB1 is required for their regulation of neuronal differentiation remains to be tested directly. Future work may reveal an interplay between lncRNAs and PTBP1 in different biological contexts, including cancer and direct cell reprogramming.
Experimental Procedures
V-SVZ NSC cultures
Postnatal day 5-7 (P5-7) C57/B6 mouse brains were used to derive V-SVZ cultures as in (Park et al., 2014). Cells were split 1:2 to passage 5 or 6 before switching from self-renewal to differentiation medium as described (Park et al., 2014).
In situ hybridization
Branched DNA in situ was performed on adult tissue with the RNAScope 2.0 high definition BROWN kit (ACD). In situs on embryonic tissue were performed as described (Wallace and Raff, 1999) with DIG-labeled RNA probes.
Time-lapse imaging
Cultures were established by trypsinization and subsequent mixing of infected cultures (shCtrl or shPnky) with wildtype cells at a ratio of 1:200 to give ~15 GFP+ cells/high power field. Cultures were switched to differentiation medium and imaged on a Leica SP5 inverted confocal microscope fitted with a Life Imaging Services microscope temperature control system. 8 optical sections were taken every 15 minutes for 3 days. Cell fate was determined by morphology, and representative fields were analyzed by ICC for Tuj1 and GFAP.
Human Fetal Tissue
Fetal cortical tissue was collected from elective pregnancy termination specimens at San Francisco General Hospital, usually within 2h of the procedure. Research protocols were approved by the UCSF Committee on Human Research.
In utero electroporation
In utero electroporation was performed on E13.5 embryos from timed-pregnant wildtype Swiss-Webster mice (Simonsen labs) as described (Saito, 2006). Constructs used were PsicoR-shLuciferase (shCtrl), and PsicoR-shPnky-2 (shPnky). Embryos were harvested 48 hours later. 1-3 non-adjacent coronal sections per brain were imaged for quantification. Four animals from three separate surgeries were quantified for each experiment.
RNA-pulldown and Mass Spectrometry
Biotinylated RNA pulldown was performed as in (Hacisuleyman et al., 2014). Selected SDS PAGE-separated bands were excised and in-gel digested with trypsin as in (Jiménez et al., 2001). LC-MS analyses of tryptic peptides utilized LTQ Orbitrap Velos mass spectrometer (Thermo Scientific) equipped with a NanoLC Ultra System (Eksigent), as described (Roan et al., 2014).
RNA-seq analysis
Cluster generation and high-throughput sequencing were performed on a HiSeq 2500 (Illumina), using the paired-end 100 bp protocol. Reads were aligned to the mouse genome mm9 using Tophat v2.0.10 (Trapnell et al., 2009). Differential expression was assessed using Cuffdiff v2.1.1 (Trapnell et al., 2010). Alternative splicing was analyzed using DEXSeq v1.8.0 (Anders et al., 2012), using an FDR threshold of 0.01. Data are deposited in NCBI GEO under accession number GSE65542.
Supplementary Material
Acknowledgements
We thank Robert Darnell for the PTBP2 antibody, David Weinberg for helpful discussions, and Ewa Witkowski, Steven Hall, and The Sandler-Moore Mass Spectrometry Core Facility at UCSF for help with mass spectrometry. This project was supported by NIH DP2-OD006505-01 and VA 1I01 BX000252-04 to D.A.L, NIH R01 NS35710 to A.R.K., NIH 1F31NS080501-01A1 to A.D.R., National Science Foundation Graduate Research Fellowship Grant No. 1144247 to R.E.A., MSTP training grant 2T32GM007618-34 to S.J.L., SFSU CIRM Bridges TB1-01194 fellowship to S.H., HHMI Medical Research Fellowship to H.Z., and facilities and resources provided by the San Francisco Veterans Affairs Medical Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
ADR and REA designed and performed experiments, interpreted data, and wrote the manuscript. SJL performed RNA-seq analysis. TJN performed in utero electroporations and human tissue staining. CG assisted with time-lapse imaging. SJH processed and interpreted electroporation experiments. RDS developed biochemical assays. HZ performed histological work. DAL supervised research and helped write the manuscript. All authors edited the manuscript.
We declare no competing financial interests.
References
- Anders S, Reyes A, Huber W. Detecting differential usage of exons from RNA-seq data. Genome Res. 2012;22:2008–2017. doi: 10.1101/gr.133744.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Chang HY. Long Noncoding RNAs: Cellular Address Codes in Development and Disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond AM, Vangompel MJW, Sametsky EA, Clark MF, Savage JC, Disterhoft JF, Kohtz JD. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat Neurosci. 2009;12:1020–1027. doi: 10.1038/nn.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutz PL, Stoilov P, Li Q, Lin C-H, Chawla G, Ostrow K, Shiue L, Ares M, Black DL. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalei V, Sansom SN, Kong L, Lee S, Montiel JF, Vance KW, Ponting CP. The long non-coding RNA Dali is an epigenetic regulator of neural differentiation. Elife. 2014;3:e04530. doi: 10.7554/eLife.04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova N, Zamudio JR, Jong RM, Soukup D, Resnick R, Sarma K, Ward AJ, Raj A, Lee JT, Sharp PA, et al. LincRNA-p21 Activates p21 In cis to Promote Polycomb Target Gene Expression and to Enforce the G1/S Checkpoint. Mol Cell. 2014;54:777–790. doi: 10.1016/j.molcel.2014.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetsch F, Caillé I, Lim DA, García-Verdugo JM, Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97:703–716. doi: 10.1016/s0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- Doetsch F, Petreanu L, Caille I, Garcia-Verdugo JM, Alvarez-Buylla A. EGF converts transit-amplifying neurogenic precursors in the adult brain into multipotent stem cells. Neuron. 2002;36:1021–1034. doi: 10.1016/s0896-6273(02)01133-9. [DOI] [PubMed] [Google Scholar]
- Ferrarese R, Harsh GR, Yadav AK, Bug E, Maticzka D, Reichardt W, Dombrowski SM, Miller TE, Masilamani AP, Dai F, et al. Lineage-specific splicing of a brain-enriched alternative exon promotes glioblastoma progression. J. Clin. Invest. 2014;124:2861–2876. doi: 10.1172/JCI68836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacisuleyman E, Goff LA, Trapnell C, Williams A, Henao-Mejia J, Sun L, McClanahan P, Hendrickson DG, Sauvageau M, Kelley DR, et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nature Structural &Amp; Molecular Biology. 2014;21:198–206. doi: 10.1038/nsmb.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Ihrie RA, Alvarez-Buylla A. Lake-front property: a unique germinal niche by the lateral ventricles of the adult brain. Neuron. 2011;70:674–686. doi: 10.1016/j.neuron.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez CR, Huang L, Qiu Y, Burlingame AL. In-Gel Digestion of Proteins for MALDI-MS Fingerprint Mapping. John Wiley & Sons, Inc.; Hoboken, NJ, USA: 2001. [DOI] [PubMed] [Google Scholar]
- Keppetipola N, Sharma S, Li Q, Black DL. Neuronal regulation of pre-mRNA splicing by polypyrimidine tract binding proteins, PTBP1 and PTBP2. Crit. Rev. Biochem. Mol. Biol. 2012;47:360–378. doi: 10.3109/10409238.2012.691456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Zhang Y, Ye Z-Q, Liu X-Q, Zhao S-Q, Wei L, Gao G. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35:W345–W349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein A, Alvarez-Buylla A. The Glial Nature of Embryonic and Adult Neural Stem Cells. Annu. Rev. Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Yano M, Fak JJ, Mele A, Grabinski SE, Zhang C, Darnell RB. Ptbp2 represses adult-specific splicing to regulate the generation of neuronal precursors in the embryonic brain. Genes & Development. 2012;26:1626–1642. doi: 10.1101/gad.191338.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MF, Jungreis I, Kellis M. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics. 2011;27:i275–i282. doi: 10.1093/bioinformatics/btr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin N, Chang K-Y, Li Z, Gates K, Rana ZA, Dang J, Zhang D, Han T, Yang C-S, Cunningham TJ, et al. An Evolutionarily Conserved Long Noncoding RNA TUNA Controls Pluripotency and Neural Lineage Commitment. Mol Cell. 2014 doi: 10.1016/j.molcel.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois C, Alvarez-Buylla A. Long-distance neuronal migration in the adult mammalian brain. Science. 1994;264:1145–1148. doi: 10.1126/science.8178174. [DOI] [PubMed] [Google Scholar]
- Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luskin MB. Neuroblasts of the postnatal mammalian forebrain: their phenotype and fate. J. Neurobiol. 1998;36:221–233. [PubMed] [Google Scholar]
- Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nature Structural &Amp; Molecular Biology. 2013;20:300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- Ng S-Y, Bogu GK, Soh BS, Stanton LW. The Long Noncoding RNA RMST Interacts with SOX2 to Regulate Neurogenesis. Molecular Cell. 2013;51:349–359. doi: 10.1016/j.molcel.2013.07.017. [DOI] [PubMed] [Google Scholar]
- Park DH, Hong SJ, Salinas RD, Liu SJ, Sun SW, Sgualdino J, Testa G, Matzuk MM, Iwamori N, Lim DA. Activation of neuronal gene expression by the JMJD3 demethylase is required for postnatal and adult brain neurogenesis. CellReports. 2014;8:1290–1299. doi: 10.1016/j.celrep.2014.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastrana E, Cheng L-C, Doetsch F. Simultaneous prospective purification of adult subventricular zone neural stem cells and their progeny. Proceedings of the National Academy of Sciences. 2009;106:6387–6392. doi: 10.1073/pnas.0810407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peretto P, Merighi A, Fasolo A, Bonfanti L. Glial tubes in the rostral migratory stream of the adult rat. Brain Res. Bull. 1997;42:9–21. doi: 10.1016/s0361-9230(96)00116-5. [DOI] [PubMed] [Google Scholar]
- Polydorides AD, Okano HJ, Yang YY, Stefani G, Darnell RB. A brain-enriched polypyrimidine tract-binding protein antagonizes the ability of Nova to regulate neuron-specific alternative splicing. Proc. Natl. Acad. Sci. U.S.a. 2000;97:6350–6355. doi: 10.1073/pnas.110128397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponti G, Obernier K, Alvarez-Buylla A. Lineage progression from stem cells to new neurons in the adult brain ventricular-subventricular zone. Cell Cycle. 2013;12:1649–1650. doi: 10.4161/cc.24984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos AD, Diaz A, Nellore A, Delgado RN, Park K-Y, Gonzales-Roybal G, Oldham MC, Song JS, Lim DA. Integration of genome-wide approaches identifies lncRNAs of adult neural stem cells and their progeny in vivo. Cell Stem Cell. 2013;12:616–628. doi: 10.1016/j.stem.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapicavoli NA, Poth EM, Zhu H, Blackshaw S. The long noncoding RNA Six3OS acts in trans to regulate retinal development by modulating Six3 activity. Neural Development. 2011;6:32. doi: 10.1186/1749-8104-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roan NR, Chu S, Liu H, Neidleman J, Witkowska HE, Greene WC. Interaction of Fibronectin With Semen Amyloids Synergistically Enhances HIV Infection. J. Infect. Dis. 2014 doi: 10.1093/infdis/jiu220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T. In vivo electroporation in the embryonic mouse central nervous system. Nat Protoc. 2006;1:1552–1558. doi: 10.1038/nprot.2006.276. [DOI] [PubMed] [Google Scholar]
- Sauvageau M, Goff LA, Lodato S, Bonev B, Groff AF, Gerhardinger C, Sanchez-Gomez DB, Hacisuleyman E, Li E, Spence M, et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife. 2013;2:e01749. doi: 10.7554/eLife.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibasaki T, Tokunaga A, Sakamoto R, Sagara H, Noguchi S, Sasaoka T, Yoshida N. PTB deficiency causes the loss of adherens junctions in the dorsal telencephalon and leads to lethal hydrocephalus. Cerebral Cortex. 2013;23:1824–1835. doi: 10.1093/cercor/bhs161. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnology. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace VA, Raff MC. A role for Sonic hedgehog in axon-to-astrocyte signalling in the rodent optic nerve. Development. 1999;126:2901–2909. doi: 10.1242/dev.126.13.2901. [DOI] [PubMed] [Google Scholar]
- Wang L, Park HJ, Dasari S, Wang S, Kocher J-P, Li W. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013;41:e74–e74. doi: 10.1093/nar/gkt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Ouyang K, Huang J, Zhou Y, Ouyang H, Li H, Wang G, Wu Q, Wei C, Bi Y, et al. Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell. 2013;152:82–96. doi: 10.1016/j.cell.2012.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap K, Lim ZQ, Khandelia P, Friedman B, Makeyev EV. Coordinated regulation of neuronal mRNA steady-state levels through developmentally controlled intron retention. Genes & Development. 2012;26:1209–1223. doi: 10.1101/gad.188037.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Gray EE, Chawla G, Porse BT, O'Dell TJ, Black DL. PSD-95 is post-transcriptionally repressed during early neural development by PTBP1 and PTBP2. Nat Neurosci. 2012;15:381–8–S1. doi: 10.1038/nn.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.