Abstract
Context-induced reinstatement of drug seeking is a well established animal model for assessing the neural mechanisms underlying context-induced drug relapse, a major factor in human drug addiction. Neural activity in striatum has previously been shown to contribute to context-induced reinstatement of heroin, cocaine, and alcohol seeking, but not yet for methamphetamine seeking. In this study, we found that context-induced reinstatement of methamphetamine seeking increased expression of the neural activity marker Fos in dorsal but not ventral striatum. Reversible inactivation of neural activity in dorsolateral but not dorsomedial striatum using the GABA agonists muscimol and baclofen decreased context-induced reinstatement. Based on our previous findings that Fos-expressing neurons play a critical role in conditioned drug effects, we assessed whether context-induced reinstatement was associated with molecular alterations selectively induced within context-activated Fos-expressing neurons. We used fluorescence-activated cell sorting to isolate reinstatement-activated Fos-positive neurons from Fos-negative neurons in dorsal striatum and used quantitative PCR to assess gene expression within these two populations of neurons. Context-induced reinstatement was associated with increased expression of the immediate early genes Fos and FosB and the NMDA receptor subunit gene Grin2a in only Fos-positive neurons. RNAscope in situ hybridization confirmed that Grin2a, as well as Grin2b, expression were increased in only Fos-positive neurons from dorsolateral, but not dorsomedial, striatum. Our results demonstrate an important role of dorsolateral striatum in context-induced reinstatement of methamphetamine seeking and that this reinstatement is associated with unique gene alterations in Fos-expressing neurons.
Keywords: extinction, neuroadaptations, neuronal ensembles, renewal, self-administration
Introduction
Environmental contexts associated with drug use can provoke drug craving and relapse even after prolonged abstinence (Wikler, 1973; Robins et al., 1974; O'Brien et al., 1986). We developed a rat model of drug relapse called context-induced reinstatement of drug seeking (Crombag and Shaham, 2002; Crombag et al., 2002, 2008) that is based on the ABA renewal procedure (Bouton and Bolles, 1979; Bouton and Swartzentruber, 1991). In this model, rats are first trained to self-administer a drug in one context (A) and then lever pressing is extinguished in a distinct nondrug context (B). On test day, context-induced reinstatement (renewal) of drug seeking is assessed by returning rats to the training context A in the absence of drug (Crombag and Shaham, 2002). Lever presses are paired with discrete tone-light cues during all phases. This procedure has been used to identify neural mechanisms of context-induced reinstatement of heroin, cocaine, and alcohol seeking (Crombag et al., 2008; Janak and Chaudhri, 2010; Lasseter et al., 2010; Peters et al., 2013; Marchant et al., 2014; McNally, 2014). However, the one study that examined neural mechanisms in context-induced reinstatement of methamphetamine seeking yielded negative results (Widholm et al., 2011).
Studies using the neural activity marker Fos (Morgan and Curran, 1991; Cruz et al., 2014b) indicate that the ventral striatum is activated during context-induced reinstatement of cocaine seeking (Cruz et al., 2014a) while pharmacological inactivation of neural activity in ventral and dorsal striatum attenuates context-induced reinstatement of cocaine (Fuchs et al., 2006, 2008; Cruz et al., 2014a), heroin (Bossert et al., 2007, 2009, 2012), and alcohol (Hamlin et al., 2007; Chaudhri et al., 2008, 2009; Perry and McNally, 2013) seeking. Although noncontingent methamphetamine administration (Merchant et al., 1994; Ohno et al., 1994; Wang et al., 1995) and contingent methamphetamine self-administration (Cadet et al., 2014) have been shown to induce Fos expression in multiple striatal subregions, Fos expression and pharmacological inactivation of these striatal subregions has not been examined for context-induced reinstatement of methamphetamine seeking. We address this question in the first part of our study.
Context-specific activation of Fos-expressing neurons in ventromedial prefrontal cortex and ventral striatum has been shown to play critical roles in context-induced reinstatement of cocaine and heroin seeking (Bossert et al., 2011; Cruz et al., 2014a). Therefore, in the second part of our study, we examined whether context-induced reinstatement of methamphetamine seeking induces unique molecular alterations within Fos-expressing striatal neurons. We used fluorescence-activated cell sorting (FACS; Guez-Barber et al., 2011; Fanous et al., 2013; Liu et al., 2014) to isolate context-activated Fos-positive neurons from the surrounding majority of Fos-negative neurons in dorsal striatum followed by targeted cDNA preamplification and quantitative PCR (qPCR) to assess gene expression alterations. We then used RNAscope in situ hybridization (ISH; Wang et al., 2012) to verify the FACS-based findings and to obtain better anatomical resolution of the molecular alterations within Fos-expressing neurons.
Materials and Methods
Subjects.
We used 130 male Sprague Dawley rats (Charles River) weighing 300–350 g at the time of surgery. Rats were housed individually in the animal facility under a reverse 12 h light/dark cycle (lights off at 8:00 A.M.) with water ad libitum available in their home cages. Food was restricted to 20 g per day of Purina rat chow (given after the daily operant sessions) throughout the experiments. Experiments were performed in accordance to the Guide for the Care and Use of Laboratory Animals (eighth edition, 2011, United States National Research Council, http://grants.nih.gov/grants/olaw/Guide-for-the-Care-and-Use-of-Laboratory-Animals.pdf). From all experiments, we excluded 35 of the 130 rats: 11 rats were excluded either because they did not achieve our pre-established criterion for successful training (<15 infusions/d on the lower methamphetamine training dose of 0.05 mg/kg/infusion) within the allotted time, or the intravenous catheter leaked during the training phase, or they failed to meet extinction criterion (<25 responses per 3 h over 3 d) after 20 extinction sessions; 12 rats were excluded for misplaced cannulae (Experiment 2; rostral to +2.28 mm or caudal to +0.36 mm from bregma); four rats were excluded for loss of head caps; and eight rats were excluded due to health problems.
Intracranial and intravenous surgery.
For all experiments, we anesthetized rats with ketamine and xylazine (80 and 20 mg/kg, i.p.) and implanted catheters into the jugular vein as previously described (Bossert et al., 2004, 2006, 2012; Cruz et al., 2014a). Jugular catheters were guided under the skin over the shoulder blades and attached to a modified 22 gauge cannula that was mounted to the rat's skull with dental cement. A single injection of buprenorphine (0.1 mg/kg, s.c.) was given immediately after surgery to relieve pain and rats were allowed to recover for 7–10 d before starting methamphetamine self-administration training. During the recovery and training phases, catheters were flushed every 24–48 h with gentamicin (Butler Schein; 5 mg/ml) in sterile saline. For Experiment 2, we implanted bilateral guide cannulae and jugular catheters during the same surgical session. Guide cannulae (23 gauge; Plastics One) were implanted bilaterally 1 mm above the target regions using stereotaxic coordinates (Paxinos and Watson, 2005): dorsomedial striatum [anteroposterior (AP) +1.2 mm, mediolateral (ML) ± 2.4 mm (8° angle), and dorsoventral (DV) −4.3 mm] or dorsolateral striatum [AP +1.2 mm, ML ±3.4 mm (2° angle), and DV −4.0 mm] with the nose bar set at −3.3 mm (Bossert et al., 2009). Catheters were then inserted into the jugular vein as described above.
Self-administration training, extinction, and context-induced reinstatement.
Rats were trained and tested in Med Associates self-administration chambers. Each chamber had two levers placed 9 cm above the floor. Presses on the retractable active lever activated the infusion pump while presses on the nonretractable inactive lever had no programmed consequences. Two different contexts A and B were used for the self-administration, extinction, and test environments as previously described (Bossert et al., 2011, 2012; Cruz et al., 2014a). The two contexts contained different auditory (fan turned on or off), visual (red or white house light), tactile (rods of different sizes in floor), and circadian [morning (onset at 8:00–9:00 A.M.) or afternoon (onset at 2:00–3:00 P.M.) sessions] cues. The experiments consisted of three phases: self-administration training (12 d), extinction training (12–20 d), and tests for context-induced reinstatement of methamphetamine-seeking behavior (1 d). The experimental sequence was Context A (training)–Context B (extinction)–Contexts A or B (testing). Although we counterbalanced Contexts A and B in all of our experiments, we refer to the methamphetamine self-administration training context as Context A and the extinction context as Context B to simplify the description of the experiments.
Rats were trained to self-administer methamphetamine 3 h/d for 12 d. (R)-methamphetamine-HCl (National Institute on Drug Abuse Pharmacy) was dissolved in sterile saline and infused at a volume of 100 μl over 3.5 s at a dose of 0.1 (first six sessions) and 0.05 (last six sessions) mg/kg/infusion. During training, methamphetamine infusions were earned on a fixed ratio 1 (FR1) 20 s timeout reinforcement schedule; each drug infusion was paired with a compound tone-light cue (2900 Hz; 20 dB above background, 7.5 W white light) for 2.3 s. During the extinction phase, responses on the previously active lever led to presentation of the tone-light cue but not methamphetamine. Tests for context-induced reinstatement were conducted under extinction conditions and started after a minimum of 12 daily extinction sessions when the rats met extinction criterion of <25 presses per 3 h session on the previously active lever during the last three extinction sessions.
Experiment 1: context-induced reinstatement of methamphetamine seeking and Fos expression.
The test group (A-B-A) underwent methamphetamine self-administration training in Context A, extinction training in Context B, and 90 min reinstatement testing in Context A. The control group (A-B-B) underwent similar training and extinction schedule but were tested in the Extinction Context B for 90 min. Rats in the control and test groups were matched for their methamphetamine intake and number of active lever presses during the training and extinction phases. At the end of the test session, rats were deeply anesthetized with isoflurane and perfused with 100 ml of PBS followed by 400 ml of 4% paraformaldehyde in PBS. The brains were postfixed in paraformaldehyde for 90 min and transferred to 30% sucrose in PBS solution at 4°C for 2–3 d. Brains were frozen in powdered dry ice and kept at −80°C until sectioning. Forty micrometer coronal sections were cut between bregma +2.28 and +0.36 mm (Paxinos and Watson, 2005). Free-floating sections were washed three times in PBS, blocked with 3% normal goat serum (NGS) in PBS containing 0.25% Triton X-100 (PBS-Tx) for 1 h, and incubated 24 h at 4°C with anti-Fos antibody (1:4000 dilution; catalog #sc-52, Santa Cruz Biotechnology) in the same blocking solution. Sections were washed again with PBS and incubated for 2 h in biotinylated goat anti-rabbit secondary antibody (1:600 dilution; Vector Laboratories) in PBS-Tx and 1% NGS. After washing in PBS, sections were incubated for 1 h in avidin-biotin-peroxidase complex (ABC Elite kit, catalog #PK-6100, Vector Laboratories) in PBS containing 0.5% Triton X-100. Finally, sections were washed in PBS and developed in 3,3′-diaminobenzidine for ∼3 min, transferred into PBS, and mounted onto chromalum-gelatin-coated slides. Once dry, the slides were dehydrated through a graded series of alcohol (30, 60, 90, 95, 100, 100% ethanol) and cleared with Citrasolv (Fisher Scientific) before coverslipping with Permount (Sigma-Aldrich).
Bright-field images of immunoreactive (IR) cells in dorsal (medial and lateral) and ventral (accumbens shell and core) striatum were digitally captured using an EXi Aqua camera (QImaging, www.qimaging.com) attached to a Zeiss Axioskop 2 microscope at 100× magnification (Carl Zeiss Microscopy) and iVision software for Macintosh, version 4.0.15 (Biovision). The number of labeled nuclei from two sections (four images) per rat were counted automatically using the iVision software and averaged for a single n value for each brain area of each rat.
Experiment 2: effect of inactivation of dorsolateral and dorsomedial striatum on context-induced reinstatement.
We used the muscimol plus baclofen (muscimol+baclofen) inactivation procedure (McFarland and Kalivas, 2001) to assess a functional role of dorsolateral or dorsomedial striatum in context-induced reinstatement. We used two groups of rats per region (n = 15–19 per group) in a 2 (Drug: Vehicle, Muscimol+baclofen; between-subjects factor) × 2 (Test context: A, B; within-subjects factor) mixed factorial design in which each rat was tested in Contexts A and B. After drug self-administration and extinction training (described above), muscimol+baclofen or vehicle was injected into dorsolateral or dorsomedial striatum 15 min before placing the rats into the test context for 45 min. Muscimol (0.03 nmol/0.5 μl) plus baclofen (0.3 nmol/0.5 μl; Tocris Bioscience) was dissolved in saline vehicle and bilaterally infused (0.5 μl/side) over 1 min using a syringe pump (Harvard Apparatus) and 10 μl Hamilton syringe (Hamilton) connected to a 30 gauge injector (Plastics One). Drug doses were based on previous studies (Koya et al., 2009; Bossert et al., 2012). The injectors extended 1 mm beyond the guide cannula and were left in place for 1 min after the injections. Each rat was tested with either vehicle or muscimol+baclofen infusion using a counterbalanced order of testing: once in Context A (methamphetamine context) and once in Context B (extinction context). The tests were separated by 48 h and rats remained in the animal housing room between tests. At the end of the experiment, the rats were anesthetized; their brains were removed and kept in 10% formaldehyde for 48 h before slicing to check the cannula placement.
Finally, to rule out that muscimol+baclofen injections into dorsolateral striatum (which decreased context-induced reinstatement) cause nonspecific motor deficits, we trained 18 rats from Experiment 2 to lever press (FR1 20 s timeout reinforcement schedule, 1 h/d) for 45 mg of food pellets (catalog #1811155, TestDiet) for 5 d. We then tested them on days 6 and 8 for food-reinforced responding after injections of vehicle or muscimol+baclofen (counterbalanced) into dorsolateral striatum. We used the same procedure (concentration and volume injected of drugs) as that described above for context-induced reinstatement. At the end of the experiment, we anesthetized the rats, removed their brains and kept them in 10% formaldehyde for 48 h. We sectioned each brain (50 μm sections) using a cryostat (Leica Microsystems), mounted the sections on gelatin-coated slides, and stained them with cresyl violet. We verified cannulae's placement using a light microscope.
Experiment 3: FACS sorting of Fos-labeled neurons after context-induced reinstatement.
We used two groups of rats (n =16–19 per group) in a 2 (Test context: A, B; between-subjects factor) × 2 (Fos expression: Fos-negative, Fos-positive; within-subjects factor) mixed factorial design. Due to the large number of rats in this experiment, we had to perform the experiment in two successive sessions and combine the data. All groups were equally represented in these two sessions. Rats were decapitated after a 90 min test session in Context A or B and dorsal striatum was dissected from 2-mm-thick coronal sections between approximately bregma +2.28 and +0.36 mm (Paxinos and Watson, 2005). Tissue was finely minced with razor blades and placed on ice in a tube containing 1 ml of Hibernate A (catalog #HA-lf, Brain Bits). Afterward, the pieces of tissue were enzymatically digested for 30 min at 4°C with 1 ml of Accutase (catalog #SCR005, Millipore) with end-over-end mixing. Tissue was centrifuged for 2 min at 425 × g and the pellet resuspended in 0.6 ml of ice-cold Hibernate A. Each tissue sample was triturated three times in series using fire-polished glass pipettes with successively smaller diameters (1.3, 0.8, and 0.4 mm). Each trituration step consisted of triturating up and down 10 times followed by 2 min on ice to sediment the debris and undissociated cells. The supernatants containing dissociated cells were collected after each trituration step and combined for a total volume of 1.8 ml. Two aliquots of 800 μl per sample were fixed and permeabilized by adding the same volume of 100% cold ethanol for 15 min with occasional shaking. Cells were centrifuged for 4 min at 1000 × g and resuspended in 1 ml of cold PBS. Then, fixed cells were filtered through 100 and 40 μm cell strainers (Falcon, BD Biosciences). Cell suspension was split into two tubes for each sample. One tube (700 μl) was incubated with the primary and secondary antibodies for sorting cells. The second tube (rest of the cell suspension after filtering ∼250 μl) was used to set fluorescence thresholds for positive NeuN-IR and Fos-IR gates as well as to detect total cell populations with DAPI.
We used the primary antibodies biotinylated mouse anti-NeuN antibody (1:1000 dilution; catalog #MAB377B, Millipore) and rabbit anti-Fos antibody (1:1000 dilution of sc-52; Santa Cruz Biotechnology) for FACS. The cell suspension was rotated end-over-end in primary antibodies for 30 min at 4°C and then washed by adding 0.8 ml of cold PBS with centrifugation for 3 min at 425 × g. The pellet was resuspended in 0.7 ml of cold PBS and incubated at 4°C for 15 min with streptavidin conjugated to R-phycoerythrin (R-PE Streptavidin, 1:1000 dilution; catalog #SA1004-1, Invitrogen, Life Technologies) for neuronal-labeling and the donkey anti-rabbit IgG antibody conjugated to Alexa Fluor 647 (1:300 dilution; catalog #A-31573, Invitrogen, Life Technologies) for Fos labeling. Cells were washed twice consecutively in 0.8 and 1 ml of cold PBS followed by centrifugation for 3 min at 425 × g. The pellet was resuspended in 0.5 ml of cold PBS for FACSAria (BD Biosciences) sorting.
Samples were sorted using a FACS Aria I cell sorter (BD Biosciences). As we previously reported, cells containing neurons can be identified based on their unique forward scattering (FSC) and side scattering (SSC) properties. DAPI staining (labels DNA and nuclei) was used to define the cell and neuron population gates before sorting the main samples. At least 80 and 95% of the events in the cell and neuron gates, respectively, were DAPI-positive events (cells with nuclei). A gating strategy based on FSC width and height was used to select only single cells. We then analyzed and sorted neurons according to phycoerythrin (PE; NeuN-immunopositive) and Alexa Fluor 647 (Fos-immunopositive) fluorescence signal. To set the PE and Alexa Fluor 647 fluorescence thresholds, we used only secondary antibodies for the NeuN gate and all antibodies except the Fos primary antibody for the Fos gate. A maximum of 5000 NeuN-positive and Fos-negative events (Fos-negative neuron population) and all NeuN-positive and Fos-positive events (Fos-positive neuron population) were sorted.
RNA isolation and cDNA synthesis from FACS-isolated neurons.
We used FACS with the multiplex preamplification strategy as described previously (Liu et al., 2014) to analyze single rat brain regions. Briefly, sorted cells were collected directly into 50 μl of the extraction buffer from PicoPure RNA isolation kit (Arcturus Bioscience) and lysed by pipetting up and down 10 times followed by incubation at 42°C for 30 min. The suspension was centrifuged at 1000 × g at 4°C for 2 min and the supernatant was collected for RNA isolation. Column filtration, washing, and elution steps of RNA from the columns are described in section C of the PicoPure RNA isolation protocol. RNA integrity numbers were measured using Agilent RNA 600 Pico kit (Agilent Technologies) and found to be ∼3–4 for the FACS samples. Single-strand cDNAs were synthesized with the Superscript III first strand cDNA synthesis kit according to the manufacturer's protocol (Invitrogen, Life Technologies).
TaqMan PreAmp Master Mix Kit was used for cDNA preamplification (Applied Biosystems, Life Technologies). We used a pooled primer solution of 0.2× concentration of TaqMan ABI primer/probes (100 dilution of stock 20× TaqMan Gene Expression Assay) and 80 nm customer-designed primer sets (Table 1). Each cDNA sample (7.5 μl) was mixed with the pooled primer solution (7.5 μl) and 15 μl of 2× TaqMan PreAmp Master Mix. cDNAs were preamplified in an Applied Biosystems 9700 Thermal Cycler using the following program: 95°C hold for 10 min; then 14 cycles with denaturation at 90°C for 15 s; and annealing and extension at 60°C for 4 min. The preamplification PCR products were immediately diluted five times with molecular biology-grade water (5 Prime) and stored at −20°C or immediately processed for qPCR. Duplex qPCR assays were performed on technical duplicates using a Fam-labeled probe for each target gene (Table 1) and a Vic-labeled probe for the endogenous control gene (Pde10a), along with TaqMan Advanced Fast PCR Master Mix (Life Technologies). Pde10a was used as the control gene because it was relatively abundant, not altered by our methamphetamine self-administration procedure, and not different between Fos-positive and Fos-negative neurons. We had also previously found that this gene was similarly expressed in neurons and other brain cell types (Guez-Barber et al., 2012). Primer Express 3.0 (Applied Biosystems) was used to select primers and probes for preamplification and qPCR that amplified across exon–exon junctions for each target gene. qPCRs were run in a 7500 Fast TaqMan instrument using the following program: 95°C hold for 20 s, then 40 cycles with denaturation at 95°C for 3 s, and annealing and extension at 60°C for 30 s. Calculations of relative expression from Ct data (exponential value) were performed using the formula ΔCt = Ct (target gene) − Ct (Pde10a). For each ΔCt value, we subtracted the average ΔCt from Fos-negative samples from rats tested in Context B (ABB-Fos-negative). Thus, we obtained a ΔΔCt value that was transformed to fold change (linear value) using the formula 2e(−ΔΔCt). This provided the fold change relative to ABB-Fos-negative expression levels for each sorted neuronal population. Uniformity of the preamplification step was verified by comparing cDNA templates from the preamplified and unamplified samples. All ΔΔCt values were within the range of ±1.5 for all target genes between the preamplified and unamplified samples.
Table 1.
Primer/probe table for genes analyzed by qPCR
| Gene | TaqMan probe | Forward primer | Reverse primer |
|---|---|---|---|
| Pde10a | TCCCATCGAGACCGC | CGCTGAACCTCCACAACCA | CGCAGGCAGTCATCATCAAG |
| NeuN | CACTCCAACAGCGTGAC | GGCCCCTGGCAGAAAGTAG | TTCCCCCTGGTCCTTCTGA |
| Fos | Rn00487426_g1* | ||
| Total FosB+ΔFosB | CGAGAAGAAACACTTACC | CCAGAGCCAGGCCTAGAAGA | CTGCGAACCCTTCGTTTTTC |
| ΔFosB | AGAGGAAAAGGCAGAGCT | CGAGAAGAAACAGAAACGGATCA | CCAGGCGTTCCTTCTCTTTTT |
| ΔFosB-2 | CCCTTCCCCGTTGTT | TGGCCGAGTGAAGTTCAAGTC | CGGGCAGGTGAGGACAAA |
| Arc | Rn00571208_g1* | ||
| Homer1 | Rn00581785_m1* | ||
| Homer2 | Rn00584015_m1* | ||
| Drd1 | Rn00432253_m1* | ||
| Drd2 | Rn00561126_m1* | ||
| Grin1 | Rn01436038_m1* | ||
| Grin2a | Rn00561341_m1* | ||
| Gria1 | Rn00709588_m1* | ||
| Gria2 | Rn00568514_m1* | ||
| Cb1r | TGAGAAGGGGTTCC | GTGCCGAGGGAGCTTCTG | GACTCAAGGTGACTGAGAAAGA |
| Trkb1 | CATGAAAGGCCCAGCTT | TGGCGAGACATTCCAAGTTTG | AGAGTCATCGTCGTTGCTGATG |
| Darpp-32 | TCGCCAGAGGAGGAG | TTCCGGGTCTCAGAGCATTC | TGCCCCTCTCCTGAGGTTCT |
| Map2k6 | Rn00586764_m1* | ||
| Mecp2_A2 | ACCAGCTCAAAAAC | GTGACAAGCCACCCTGTATTTG | CCTTGTCTGTCCCAACCTTAGATG |
*TaqMan catalog number for ordering from Applied Biosystems.
Experiment 4: RNAscope ISH assay.
The procedure is based on that described by Wang et al. (2012). Immediately after dissecting 2 mm coronal section containing part of the dorsal striatum for FACS as described before (Exp. 3), the remaining brain with the caudal part of the dorsal striatum was frozen for 20 s in 100 ml −50°C isopentane within 2–3 min of decapitation, wrapped in labeled alumina foil sealed in a zippered plastic bag, stored at −80°C, and cryosectioned for ISH. The rostral face of this remaining brain tissue contained dorsal striatum at approximately bregma +0.24 mm (Paxinos and Watson, 2005) that was immediately contiguous with the tissue used for FACS. Brains were equilibrated in Cryostat (CM 3050S) to −20°C for 2 h and 16 μm coronal sections were cut and thaw-mounted directly onto Super Frost Plus slides (Fisher). These slides were left at −20°C for 10 min and transferred to −80°C until ISH processing.
RNA ISH for Fos, Grin2A, and Grin2B mRNAs was performed manually according to the User Manual for Fresh Frozen Tissue using RNAscope Multiplex Fluorescent Reagent Kit (Advanced Cell Diagnostics). Briefly, the −80°C brain slides were transferred to slide racks and fixed by immersion in 10% neutral buffered formalin (Fisher Scientific) for 20 min at 4°C. Slides were rinsed two times in PBS and dehydrated two times each in 50 and 70% ethanol and twice in 100% ethanol. Slides were transferred to a new 100% ethanol container and kept at −20°C overnight. The slides were dried at room temperature (22°C) for 10 min and a hydrophobic pen (ImmEdger Hydrophobic Barrier Pen, Vector Laboratories) was used to make a physical barrier surrounding the brain sections to contain RNAscope assay solution. We used the HybEZ Hybridization System from Advanced Cell Diagnostics. The protease solution (Pretreatment 4 solution) was incubated with sections at room temperature for 20 min. After washing off the protease solution, 1× target probes for specific RNAs (Fos, Grin2a, and/or Grin2b) were applied to the brain sections and incubated at 40°C for 2 h in the HybEZ oven. Each RNAscope target probe contained a mixture of 20 ZZ oligonucleotide probes that bound to the target RNA. These probes were as follows: Fos-C3 probe (accession number NM_022197.2, target nucleotide region: 473-1497); Grin2a-C1 probe (accession number NM_012573.3; target nucleotide region: 2812-4265); Grin2b-C2 probe (accession number NM_012574.1; target nucleotide region: 3053-3930); Drd1-C1 probe (accession number NM_012546.2; target nucleotide region: 104-1053); and Drd2-C2 probe (accession number NM_012547.1; target nucleotide region: 445-1531).
Sections were then incubated with preamplifier and amplifier probes by applying AMP1 (40°C for 30 min), AMP2 (40°C for 15 min), and AMP3 (40°C for 30 min). Sections were then incubated with the fluorescently labeled probes by selecting a specific combination of colors associated with each channel: green (Alexa 488 nm), orange (Alexa 550 nm), and far-red (Alexa 647 nm). We used AMP4 AltA combination to detect duplex Fos and Grin2a RNAs in far-red and green, respectively; AMP4 AltB to detect duplex Fos and Grin2b RNAs in far-red and green, respectively; and AMP4 AltB to detect triplex Fos, Grin2a, and Grin2b RNAs in far-red, orange, and green respectively. Finally, we incubated the sections for 20 s with DAPI to stain nuclei (blue). Between each step we washed two times with 1× wash buffer supplied with the kit.
The negative control sections received RNase treatment before performing the RNAscope assay. After the fixation and protease digestion, we incubated sections with 5 mg/ml RNase A (Qiagen) for 30 min at 40°C. The slides were washed three times with distilled water and processed with target probe hybridization, followed by the steps described above. Fluorescent images were captured at 200× magnification using a Rolera EM-C2 (QImaging) on a Nikon Eclipse E800 microscope. Images were saved as 16-bit TIFF files, enhanced using Adobe Photoshop CS5 software for analysis. Representative images are shown in Figure 8 after deconvolving single-channel images using Huygens software (v3.7, Scientific Volume Imaging) and then merging the three channels using iVision software. For each Fos-positive cell, we analyzed two of the surrounding Fos-negative cells. For each cell, we counted the total pixels of the fluorescent signal (fluorescent “dots”). We assume that each pixel is supposed to represent a single molecule of mRNA. Selection criterion for Fos-positive cells was to have ≥33 total positive pixels after adjusting automatically the threshold using ImageJ software. Fluorescent dots for Grin2a and Grin2b were counted when their intensity values were greater than 23,387 and 24,000, respectively (threshold was set manually).
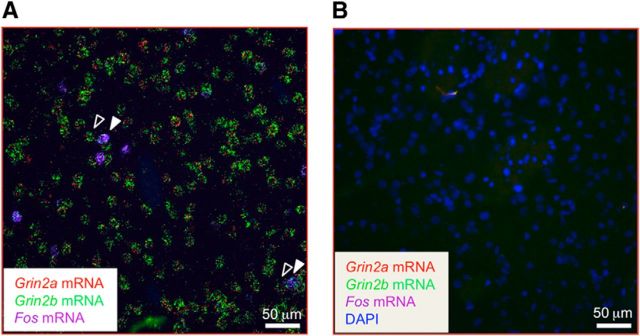
Figure 8.
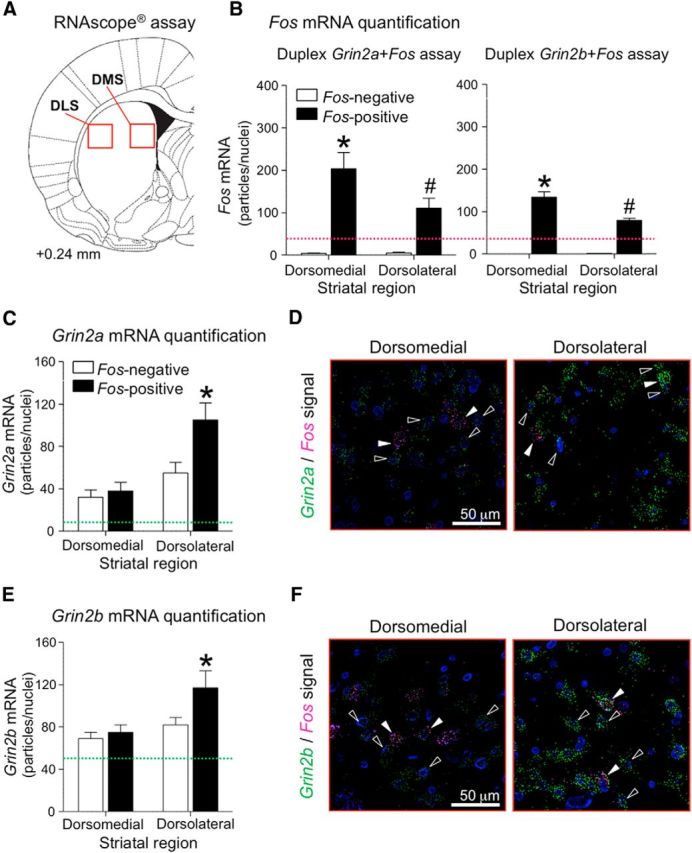
ISH for Fos, Grin2a, and Grin2b mRNAs in dorsal striatum after context-induced reinstatement of methamphetamine seeking. A, Drawing shows approximate brain section level (Paxinos and Watson, 2005) chosen for imaging. Red squares indicate areas analyzed. B, Fos-positive versus Fos-negative neurons were determined by the amount of fluorescent particles (pixels) labeling Fos mRNA. Graphs indicate number of Fos mRNA particles identified within Fos-positive (90–189 total) and Fos-negative (178–378 total) neurons in the dorsomedial or dorsolateral region of the striatum using duplex Grin2a+Fos or Grin2b+Fos probes. Purple dotted line set the threshold (33 particles) for Fos-positive criterion. C, D, Duplex detection of Grin2a (green) and Fos (purple) mRNAs. C, The graph indicates the number of Grin2a mRNA particles identified within Fos-positive (90–107 total) and Fos-negative (178–212 total) neurons (n = 6 rats, 2–3 sections). All selected cells (Fos-positive and Fos-negative) had at least 10 Grin2a mRNA particles (green dotted line). D, Representative images (following deconvolution) of selected cells (Fos-positive, white arrows; Fos-negative, open arrows). E, F, Duplex detection of Grin2b (green) and Fos (purple) mRNAs. E, The graph indicates the number of Grin2b mRNA particles identified within Fos-positive (146–189 total) and Fos-negative (292–378 total) neurons (n = 6 rats, 2–4 sections). All selected cells (Fos-positive and Fos-negative) had ≥53 Grin2b mRNA particles (green dotted line). F, Representative images (following deconvolution) of selected cells (Fos-positive, white arrows; Fos-negative, open arrows). For each Fos-positive cell, two Fos-negative cells were quantified. Nuclei were stained with DAPI (blue).
Statistical analyses.
The behavioral and immunohistochemical data were analyzed by two-way ANOVAs (SPSS version 20, GLM procedure, or GraphPad software). We followed up on significant main or interaction effects (p < 0.05) using Fisher's PLSD post hoc test. A two-way ANOVA (GraphPad Prism 5 software) was performed to analyze FACS-based qPCR and ISH data. For FACS-sorted samples, we used the between-subjects factor of Context (Extinction, Methamphetamine) and the within-subjects factor of neuron type (Fos-negative, Fos-positive). If significant differences were found in ANOVAs, then t tests were used to assess significant differences between Fos-positive and Fos-negative neurons separately for each context. Our criteria for determining significant alterations in gene expression also included a Fos-positive/Fos-negative ratio of >1.5 for gene expression in each context (Table 2). For ISH we used the between-subjects factor of Region (Dorsolateral, Dorsomedial) and the within-subjects factor of Neuron Type (Fos-negative, Fos-positive). If significant differences were found in ANOVAs, then Bonferroni's post hoc test was used to assess significant differences between Fos-positive and Fos-negative neurons separately for each context.
Table 2.
Fos-positive/Fos-negative ratio gene expression in each context
| Gene | Ratio: Fos-positive/Fos-negativea |
Main effect of Fos cell type | Main effect of Context | Interaction | |
|---|---|---|---|---|---|
| Extinction Context B | Methamphetamine Context A | ||||
| Neuronal marker | |||||
| NeuN | 1.1, n = 9 | 1.2, n = 13 | F(1, 20) = 8.4, p < 0.01 | F(1, 20) = 0.1, p = 0.91 | F(1, 20) = 1.3, p = 0.26 |
| IEGs | |||||
| Fosb | 4.7, n = 7 | 2.5, n = 8 | F(1, 13) = 13, p < 0.01 | F(1, 13) = 0.4, p = 0.55 | F(1, 13) = 0.2, p = 0.67 |
| Total FosB+ΔFosBb | 8.1, n = 3 | 2.6, n = 9 | F(1, 10) = 29, p < 0.01 | F(1, 10) = 0.61, p = 0.45 | F(1, 10) = 4.1, p = 0.07 |
| ΔFosB | 1.0, n = 4 | 4.3, n = 10 | F(1,12) = 1.2, p = 0.29 | F(1, 12) = 0.1, p = 0.82 | F(1, 12) = 1.2, p = 0.29 |
| ΔFosB-2 | 1.7, n = 11 | 0.9, n = 13 | F(1, 22) = 0.9, p = 0.36 | F(1, 22) = 1.6, p = 0.21 | F(1, 22) = 1.2, p = 0.29 |
| Arc | 1.6, n = 10 | 1.2, n = 13 | F(1, 21) = 1.7, p = 0.20 | F(1, 21) = 0.1, p = 0.92 | F(1, 21) = 0.4, p = 0.53 |
| Homer1b | 1.6, n = 10 | 1.3, n = 13 | F(1, 21) = 5.6, p < 0.05 | F(1, 21) = 0.7, p = 0.40 | F(1, 21) = 0.8, p = 0.36 |
| Homer2b | 4.3, n = 7 | 2.0, n = 7 | F(1, 12) = 14, p < 0.01 | F(1, 12) = 3.7, p = 0.07 | F(1, 12) = 4.4, p = 0.06 |
| Receptors | |||||
| Drd1 | 1.3, n = 7 | 1.0, n = 13 | F(1, 18) = 0.5, p = 0.48 | F(1, 18) = 1.2, p = 0.22 | F(1, 18) = 0.7, p = 0.41 |
| Drd2 | 1.0, n = 9 | 1.2, n = 13 | F(1, 20) = 1.6, p = 0.22 | F(1, 20) = 3.9, p = 0.06 | F(1, 20) = 1.9, p = 0.18 |
| Grin1 (GluN1) | 1.0, n = 11 | 1.0, n = 13 | F(1, 22) = 0.2, p = 0.63 | F(1, 22) = 0.6, p = 0.44 | F(1, 22) = 1.9, p = 0.19 |
| Grin2a (GluN2a)b | 1.4, n = 12 | 1.6, n = 17 | F(1, 27) = 13, p < 0.01 | F(1, 27) = 1.2, p = 0.28 | F(1, 27) = 0.5, p = 0.47 |
| Gria1 (GluA1) | 1.0, n = 11 | 1.1, n = 13 | F(1, 22) = 1.5, p = 0.23 | F(1, 22) = 0.1, p = 0.94 | F(1, 22) = 0.5, p = 0.51 |
| Gria2 (GluA2) | 1.2, n = 11 | 1.0, n = 13 | F(1, 22) = 6.1, p < 0.05 | F(1, 22) = 0.5, p = 0.49 | F(1, 22) = 0.1, p = 0.79 |
| Cb1rb | 1.7, n = 12 | 1.4, n = 14 | F(1, 24) = 16, p < 0.01 | F(1, 24) = 0.1, p = 0.96 | F(1, 24) = 1.2, p = 0.28 |
| Trkb1 | 1.6, n = 6 | 1.0, n = 6 | F(1, 10) = 3.0, p = 0.11 | F(1, 10) = 0.1, p = 0.95 | F(1, 10) = 1.0, p = 0.33 |
| Other genes | |||||
| Darpp-32 | 1.3, n = 11 | 1.1, n = 13 | F(1, 22) = 1.7, p = 0.20 | F(1, 22) = 0.8, p = 0.37 | F(1, 22) = 0.3, p = 0.59 |
| Mapk2b | 0.8, n = 8 | 1.2, n = 13 | F(1, 19) = 0.1, p = 0.72 | F(1, 19) = 3.3, p = 0.08 | F(1, 19) = 3.9, p = 0.06 |
| Mecp2_A2 | 0.9, n = 11 | 1.0, n = 13 | F(1, 22) = 0.4, p = 0.55 | F(1, 22) = 0.1, p = 0.86 | F(1, 22) = 0.5, p = 0.49 |
aUsing 2e(−ΔΔCt) values from only matched pairs Fos-positive and Fos-negative within the same animal.
bGenes that also passed the criterion of >1.5 for expression in Fos-positive over Fos-negative neurons.
Results
Training and extinction for Experiments 1–3
Rats demonstrated reliable methamphetamine self-administration in Experiments 1–3, as indicated by the increase in mean number of infusions (t(1,94) = 22.4) and active lever presses (t(1,94) = 18.1, p < 0.01) when the methamphetamine dose was decreased from 0.1 mg/kg per infusion on training days 1–6 to 0.05 mg/kg per infusion on training days 7–12. Active lever pressing decreased with repeated extinction sessions (p < 0.01; Fig. 1).
Figure 1.
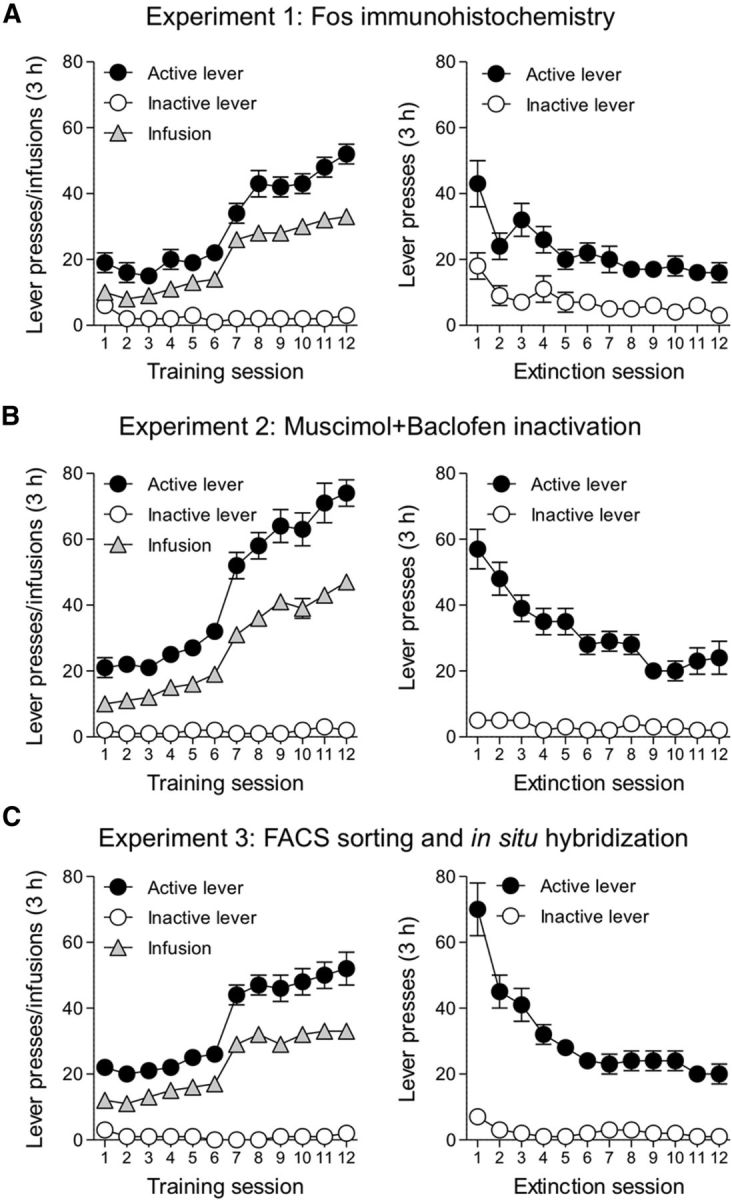
A–C, Methamphetamine self-administration training and extinction of drug-reinforced responding for (A) Experiment 1 (n = 24), (B) Experiment 2 (n = 34), and (C) Experiment 3 (n = 37). Left, Methamphetamine self-administration data during training in Context A: number of infusions, active and inactive lever presses (mean ± SEM) over the 12 d of 3 h daily sessions. Right, Extinction data in Context B: number of nonreinforced active and inactive lever presses (mean ± SEM) over the first 12 d of 3 h daily extinction sessions.
Context-induced reinstatement is associated with increased Fos expression in dorsal but not ventral striatum
In Experiment 1 we determined whether context-induced reinstatement is associated with increased Fos-immunoreactive nuclei (Fos-IR) in dorsal striatum (dorsolateral, dorsomedial) and ventral striatum (accumbens core, shell).
Reinstatement.
During testing, exposure to Methamphetamine Context A, but not Extinction Context B, increased nonreinforced active lever pressing (the operational measure of drug seeking in reinstatement studies; Shaham et al., 2003). Statistical analysis indicated a significant interaction between Context (A, B) and Lever (Active, Inactive; F(1,44) = 21.0, p < 0.01; Fig. 2A).
Figure 2.
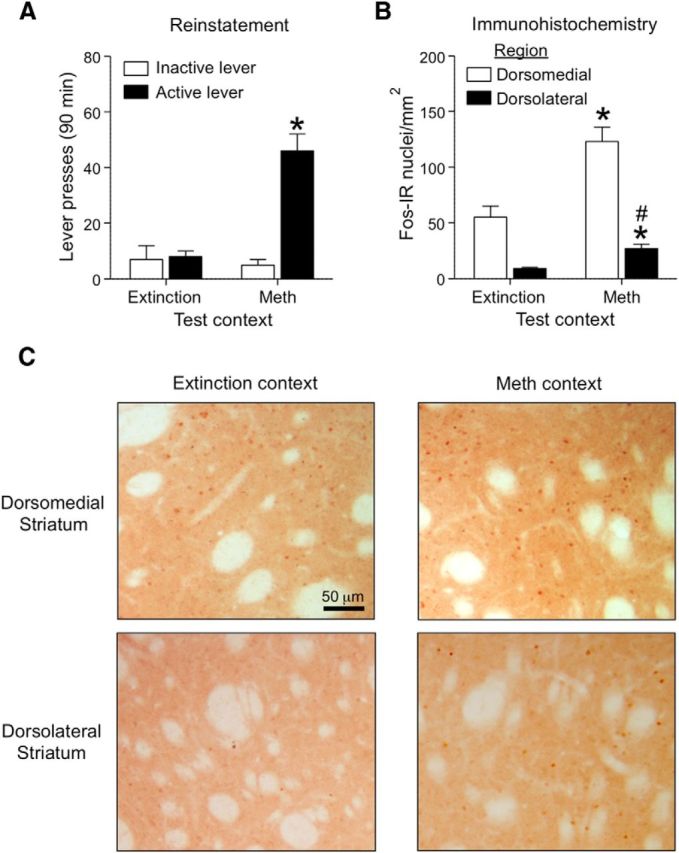
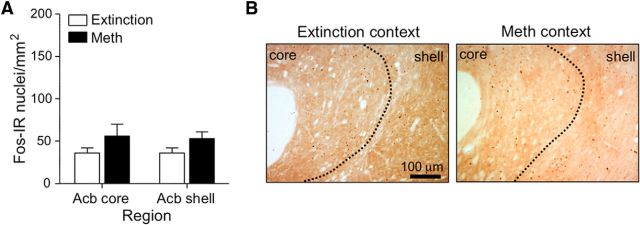
Context-induced reinstatement of methamphetamine seeking is associated with Fos induction in both dorsolateral and dorsomedial striatum. A, Reinstatement test: total number of active and inactive lever presses (mean ± SEM) in rats tested in the Extinction or Methamphetamine Context. Active lever pressing was higher in the Methamphetamine Context than in the Extinction Context (*p < 0.01, n = 10–14 per group). B, Number of Fos-IR nuclei per square millimeter (mean ± SEM) in dorsomedial and dorsolateral striatum. Dorsostriatal Fos-IR was higher in the Methamphetamine Context than in the Extinction Context for both dorsolateral (*p < 0.05) and dorsomedial (*p < 0.01) striatum (n = 10–12 per group). For Methamphetamine Context, the total Fos-IR cells for dorsomedial striatum is significantly higher than the dorsolateral striatum (#p < 0.01). C, Representative images of Fos-IR nuclei in dorsomedial and dorsolateral striatum.
Fos expression.
Exposure to the Methamphetamine Context A also increased the number of Fos-IR nuclei in dorsal (Fig. 2B,C) but not ventral (Fig. 3A,B) striatum. Statistical analysis for dorsal striatum subregions demonstrated a significant interaction between Context (A, B) and Subregions (Dorsomedial, Dorsolateral; F(1,40) = 8.14, p < 0.01), main effect of Context (F(1,40) = 24.1, p < 0.01), and main effect of Subregions (F(1,40) = 65.6, p < 0.01). As can be seen in Figure 2, Fos-IR levels for both contexts (A and B) were higher in dorsomedial than in dorsolateral striatum (main effect of Subregion: F(1,40) = 65.6, p < 0.01). However fold induction (Context A/Context B) of Fos expression was greater in dorsolateral striatum (3.0-fold) than in dorsomedial striatum (2.2-fold). Context-induced reinstatement was not associated with increased Fos-IR in the accumbens core or shell (p > 0.05).
Figure 3.
Context-induced reinstatement of methamphetamine seeking has no effect on Fos-IR in nucleus accumbens. A, Number of Fos-IR nuclei per square millimeter (mean ± SEM) in accumbens core or shell. B, Fos-IR nuclei in accumbens core and accumbens shell captured at 100× magnification. Fos-IR was not different from Extinction Context for the ventral striatum areas quantified (n = 10–12 per group).
Inactivation of dorsolateral but not dorsomedial striatum decreased context-induced reinstatement
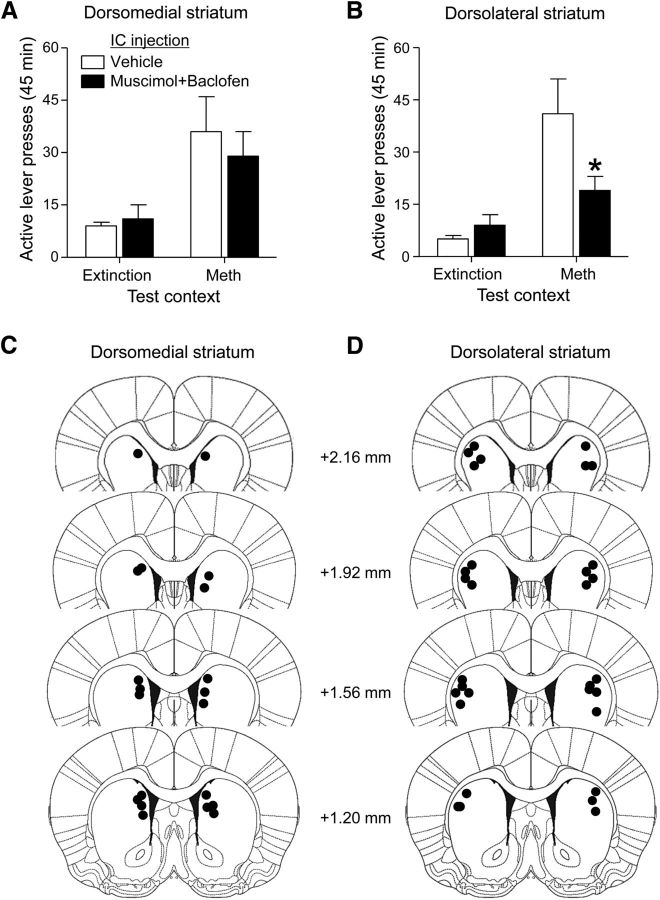
In Experiment 1, we found that context-induced reinstatement is associated with increased Fos expression in dorsolateral and dorsomedial striatum but not in ventral striatum. In Experiment. 2, we determined the functional role of the dorsal striatum subregions in context-induced reinstatement by reversibly inactivating these regions with muscimol+baclofen 15 min before reinstatement tests in Context A or B after extinction. We found that muscimol+baclofen injections into dorsolateral but not dorsomedial striatum decreased context-induced reinstatement (Fig. 4A,B). Statistical analysis of the dorsolateral striatum data indicated a significant interaction between Context (A, B), Lever (Active, Inactive), and Drug (Vehicle, Muscimol+baclofen; F(1,34) = 6.3, p < 0.05). Statistical analysis of the dorsomedial striatum data indicated a significant interaction between Context and Lever (F(1,56) = 11.52, p < 0.01) but no main effect of Drug or an interaction between Drug and the other two factors (p > 0.05). Post hoc analysis indicated that muscimol+baclofen injections into dorsolateral but not dorsomedial striatum decreased active lever pressing in the Methamphetamine Context A (p < 0.01), but not in Extinction Context B. The different experimental manipulations had no effect on inactive lever presses, which were very low (<4 average presses per session). Placements of the injector tips within the dorsolateral and dorsomedial striatum are shown in Figure 4C,D.
Figure 4.
Local inactivation of dorsolateral but not dorsomedial striatum with muscimol+baclofen decreased context-induced reinstatement of methamphetamine seeking. A, B, Bilateral injections of muscimol+baclofen into (B) dorsolateral striatum, but not (A) dorsomedial striatum, decreased active lever pressing (mean ± SEM) during reinstatement tests in the Methamphetamine Context but not in the Extinction Context (*different from the Vehicle Group, *p < 0.01, n = 7–12 per group). C, D, Dots indicate approximate placements (millimeters from bregma) of injector tips into the (C) dorsomedial or (D) dorsolateral striatum (Paxinos and Watson, 2005).
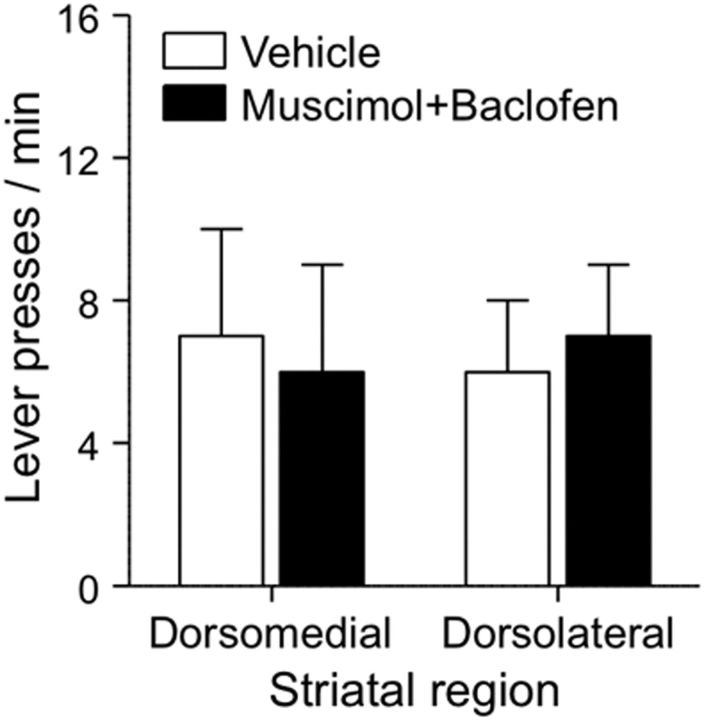
Finally, to rule out the possibility that the effect of muscimol+baclofen injections into dorsolateral striatum were due to motor deficits, we trained 18 rats that previously participated in Experiment 2 (11 rats with cannulae in dorsolateral striatum and seven rats with cannulae in dorsomedial striatum) to lever press for food pellets. After stable responding was observed, we determined the effect of muscimol+baclofen or vehicle injections into dorsomedial or dorsolateral striatum on high-rate operant responding for food. These injections had no effect of food-reinforced responding [interaction between Drug (Vehicle, Muscimol+baclofen) and Subregions (Dorsomedial, Dorsolateral), F(1,32) = 0.04, p > 0.1; effect of Drug, F(1,32) < 0.01, p > 0.1; effect of Subregions, F(1,32) < 0.01, p > 0.1], indicating that muscimol+baclofen effects on context-induced reinstatement were not due to motor deficits (Fig. 5).
Figure 5.
Local inactivation of dorsolateral or dorsomedial striatum with muscimol+baclofen had no effect on high-rate food reinforced responding. Rats were tested after 6 d of food self-administration training (n = 7–11 per group).
Together, the results of Experiment 2 indicate that neuronal activity in dorsolateral but not dorsomedial region plays an important role in context-induced reinstatement.
FACS sorting of context-activated Fos-expressing neurons
In Experiment 3, we used FACS to isolate Fos-positive and Fos-negative neurons from dorsal striatum after context-induced reinstatement of methamphetamine seeking and assessed differences of gene expression in these two neuron populations. Exposure to Methamphetamine Context A, but not Extinction Context B, increased lever pressing on test day (Fig. 6A). Statistical analysis indicated a significant interaction between the Context and Lever (F(1,70) = 21.7, p < 0.01).
Figure 6.
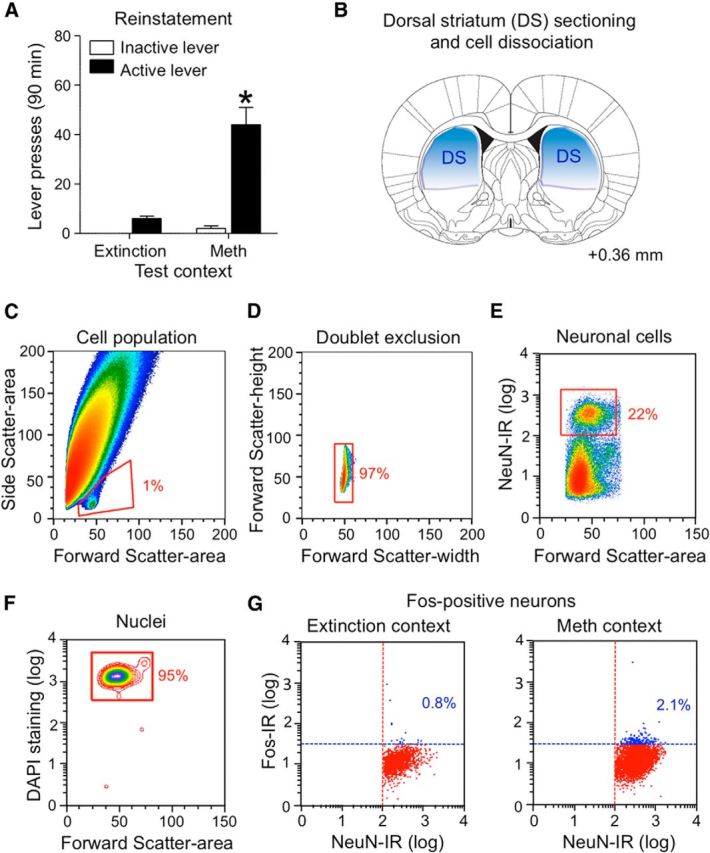
FACS of dorsostriatal neurons activated during reinstatement of methamphetamine seeking. A, Reinstatement test: number of active and inactive lever presses (mean ± SEM). Active lever pressing was higher in the Methamphetamine Context than in the Extinction Context (*p < 0.01, n = 17–20 per group). B, A representative drawing of the caudal face (millimeters from bregma) of a slice used for dissecting dorsal striatum (DS, blue; Paxinos and Watson, 2005). C–G, Representative density plots (C, D, F) and scattergrams (E, G) used to gate Fos-expressing neurons for FACS isolation. C, Cells (red gate): linear plot of forward scatter area (x-axis, cell size) and side scatter area (y-axis, granularity). D, Single cells (red gate): linear plot of forward scatter-height (y-axis) and forward scatter-width (x-axis, width size of the events). E, Neurons (red gate): logarithmic plot of fluorescence for NeuN-positive neurons (R-phycoerythrin on y-axis) indicates neurons in the upper cluster of events (red gate) and non-neurons in the lower cluster. F, Nuclei (neuronal cell gate): logarithmic plot of DAPI fluorescence (y-axis) indicates 95% of nuclei were found within the neuronal population. G, Fos-positive and Fos-negative neurons: logarithmic plots of neurons that were double-labeled for NeuN (R-phycoerythrin on x-axis) and Fos (Alexa 647 on y-axis) immunofluorescence. Selected Fos-positive neurons (upper right quadrants) after exposure to the Extinction Context (mean: 0.8 ± 0.1% of all neurons, n = 16) or Methamphetamine Context (2.1 ± 0.2% of all neurons, n = 19). Limited 5000 Fos-negative neurons (lower right quadrant) and all Fos-positive neurons were collected into microtubes for RNA isolation and real-time qPCR analysis.
We then used our modified FACS procedure (Liu et al., 2014) to isolate Fos-positive and Fos-negative neurons from the dorsal striatum of each rat. Dorsal striatum was dissected from 2-mm-thick sections (Fig. 6B) immediately after the reinstatement test and cells were dissociated and immunolabeled for Fos and the general neuronal marker NeuN. Threshold parameters were set before sorting by using small amounts of dissociated cell samples obtained from rats that were exposed to the Extinction Context or the Methamphetamine Context. Every particle (cell or debris) that passed through the flow cytometer was an “event” indicated by a dot in scattergrams that represent the different light characteristics of these events. The light scattergram (Fig. 6C, cell population) is shown as a density plot of events where FSC indicates particle size and SSC indicates granularity of the particle.
The cell population plot indicates a large heterogeneous population with different sizes and granularity along with a small relatively homogeneous population (∼1% of all events). We selected or “gated” the smaller population of events as the “cell population” based on previously determined FSC and SSC characteristics (Guez-Barber et al., 2011; Liu et al., 2014). From the “cell population” gate, we gated the single-cell population based on the height and width of the FSC signal to exclude incompletely dissociated cells (Fig. 6D, doublet exclusion); events with higher FSC width contained cell doublets.
From the “single cell” gate, we gated neurons that were immunolabeled with the NeuN antibody fluorescently labeled with R-phycoerythrin (Fig. 6E, neuronal cells). Neurons formed a cluster of events with high levels of NeuN labeling and comprised 22% of events from the previous single-cell gate. In a separate experiment, we found that 95% of all neurons were labeled with the nuclear DNA marker DAPI (Fig. 6F, nuclei). From the “neuronal cell” gate, we gated Fos-positive and Fos-negative neurons based on their degree of immunolabeling with the Fos antibody fluorescently labeled with Alexa 647 (Fig. 6G, Fos-positive neurons). Neurons with higher levels of Fos labeling were considered Fos-positive neurons (blue dots) while the remaining neurons were considered Fos-negative (red dots). Dotted lines indicate the thresholds used to separate the positive from the negative population for NeuN (red line) and Fos (blue line). Off-line analysis indicates that the Methamphetamine Context A induced more than twice as many Fos-positive neurons (2.1%) than the Extinction Context B (0.8%; t(1,33) = 5.2, p < 0.01). The number of neurons obtained from dorsal striatum were not different between rats tested in the Methamphetamine Context (15,100 ± 2433, n = 19) and Extinction Context (15,426 ± 3140, n = 16).
qPCR analyses of gene expression in Fos-positive and Fos-negative neurons
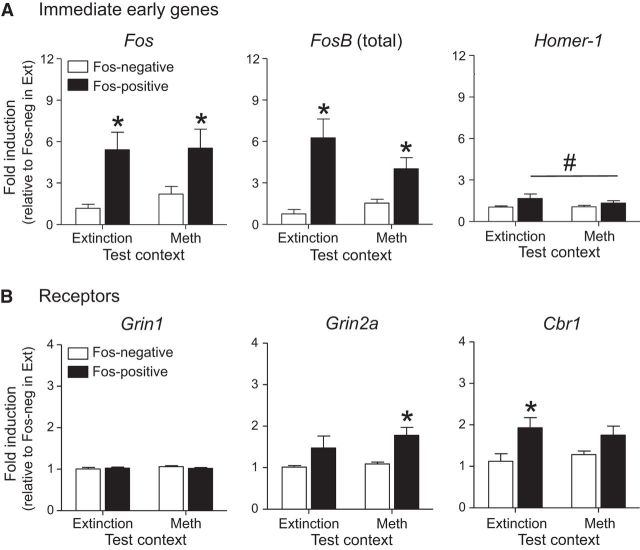
The mean number of FACS-isolated neurons collected for RNA analysis was 149 Fos-positive and 4587 Fos-negative neurons from each Methamphetamine Context A rat and 85 Fos-positive and 4831 Fos-negative neurons from each Extinction Context B rat. Gene expression was assessed using qPCR and the data are expressed as fold change over expression levels in Fos-negative neurons from Extinction Context rat brains (Fig. 7). Detailed statistics for each gene are described in Table 2. Post hoc analyses of Methamphetamine Context A samples alone indicated that expression levels of the immediate-early genes (IEGs) Fos and FosB (total) and the glutamate receptor subunit gene (Grin2a) were higher in the Fos-positive neurons than in the Fos-negative neurons. Expression levels were not different for genes encoding the dopamine receptors D1dr and D2dr, or for the AMPA receptor subunits Gria1 and Gria2. Thus Grin2a, which encodes the GluN2A subunit of the NMDA receptor, was the only receptor gene upregulated by the Methamphetamine Context A in Fos-positive neurons (F(1,27) = 13.0, p < 0.01). Analysis of Extinction Context B samples alone indicated that expression levels of Homer-1 (F(1,21) = 5.6, p < 0.05), Homer-2 (F(1,12) = 14.0, p < 0.01), and the cannabinoid receptor gene (Cbr1; F(1,24) = 16.0, p < 0.01) were higher in the Fos-positive neurons than in the Fos-negative neurons. Expression levels for all other genes were not different between groups.
Figure 7.
Gene expression in Fos-positive and Fos-negative neurons that were FACS-isolated from dorsal striatum after context-induced reinstatement of methamphetamine. A, IEGs (Fos, FosB (total), Homer-1). B, Genes encoding NMDA receptor subunits (Grin1, Grin2a) and cannabinoid receptor (Cbr1). Exposure to the Extinction or Methamphetamine (Meth) Contexts increased gene expression in Fos-positive neurons relative to the Fos-negative neurons (*p < 0.05; #, main effect for Fos cell type; Extinction Context, n = 3–11; Methamphetamine Context, n = 7–17). See Table 2 summary for all genes analyzed.
RNAscope ISH analyses
The results from Experiment 2 indicated that the dorsolateral but not dorsomedial striatum is critical for context-induced reinstatement. Due to detection limitations of the current FACS procedure, we analyzed gene expression in the entire dorsal striatum rather than in the separate dorsolateral and dorsomedial subregions. Therefore, we used the recently developed RNAscope ISH procedure (Wang et al., 2012) to determine whether context-induced reinstatement is associated with a subregion-specific increases of Grin2a expression in Fos-positive dorsal striatum neurons. This also gave us the opportunity to assess subregion-specific expression levels of Grin2b, the gene encoding the GluN2B subunit of the NMDA receptor, in Fos-positive neurons.
We determined increased expression of Fos, Grin2a, and Grin2b in Fos-positive and Fos-negative neurons in dorsolateral and dorsomedial striatum from rats in the Methamphetamine Context A test group. We used coronal sections cut from 12 brains that were kept frozen from Experiment 3 and double-labeled them for Fos and Grin2a mRNAs or Fos and Grin2b mRNAs (Fig. 8A). Fos-positive neurons were labeled with Grin2a or Grin2b and a minimum of 33 Fos-labeled pixels (Fig. 8B) while Fos-negative neurons were selected from cells below 33 Fos-labeled pixels and ≥10 for Grin2a (Fig. 8C) or 53 labeled pixels for Grin2b (Fig. 8E). Initial analysis indicated that independent of the cell type (Fos-positive, Fos-negative), Grin2a and Grin2b levels were higher in the dorsolateral than in the dorsomedial striatum [Grin2a (F(1,10) = 9.3, p < 0.05) and Grin2b (F(1,10) = 4.9, p = 0.051)].
Higher levels of Grin2a mRNA were found in Fos-positive neurons in the dorsolateral but not dorsomedial striatum (Fig. 8C); representative images are shown in Figure 8D. Statistical analysis indicated a significant interaction (F(1,10) = 25.0, p < 0.01) between the within-subjects factors of Subregion (Dorsolateral, Dorsomedial) and Neuron type (Fos-positive, Fos-negative). Post hoc analysis indicated Grin2a levels were higher in Fos-positive neurons than in Fos-negative neurons in the dorsolateral striatum (F(1,10) = 40.0, p < 0.01), but not different in the dorsomedial striatum.
Higher levels of Grin2b mRNA were also found in Fos-positive neurons in the dorsolateral but not dorsomedial striatum (Fig. 8E); representative images are shown in Figure 8F. Statistical analysis indicated a significant interaction (F(1,10) = 5.7, p < 0.05) between the within-subjects factors of Subregion (Dorsolateral, Dorsomedial) and Neuron type (Fos-positive, Fos-negative). Post hoc analysis indicated Grin2b levels were higher in Fos-positive neurons than in Fos-negative neurons in dorsolateral striatum (F(1,10) = 12.0, p < 0.01), but not different in dorsomedial striatum.
In a separate control experiment, sections were labeled with all three probes: Fos, Grin2a, and Grin2b (Fig. 9). The pixels for Grin2a and Grin2b labeling did not overlap, which confirmed specificity of these two probes. No labeling was observed when these sections were incubated with RNase before hybridization with the specific probes in the RNAscope assay.
Figure 9.
A, B, Triplex detection of Grin2a (red), Grin2b (green), and Fos (purple) mRNAs in dorsolateral striatum in the (A) absence or (B) presence of 5 mg/ml RNase treatment. A, Triple labeling for Grin2a, Grin2b, and Fos in the absence of RNase treatment. Fos-positive (white arrows) and Fos-negative (open arrows) cells contained positive signals for both Grin2a and Grin2b mRNAs. The fluorescence channel for DAPI in this image was removed to more clearly observe the particles for each mRNA. B, Triple labeling following prior treatment with 5 mg/ml RNase. Only blue DAPI-labeled DNA was observed. Positive signals were not observed for Grin2a, Grin2b, or Fos.
Finally, we determined the phenotype of the reinstatement-activated Fos-positive neurons by assessing coexpression of Fos mRNA with Drd1 and Drd2 mRNA. We used coronal sections cut from nine brains that were kept frozen from Experiment 3 and triple-labeled them for Fos, Drd1, and Drd2 mRNAs (Fig. 10A). The majority (95–97%) of the Fos-positive cells coexpressed either Drd1 or Drd2 mRNA with similar percentages for Drd1 versus Drd2 mRNA in the dorsolateral striatum (Fig. 10B). A significantly higher percentage of Fos-positive neurons coexpressed Drd1 than Drd2 in the dorsomedial striatum (F(1,32) = 17.4, p < 0.01). Only 3–5% of Fos-positive cells did not coexpress Drd1 or Drd2 mRNA. Representative images are shown in Figure 10C.
Figure 10.
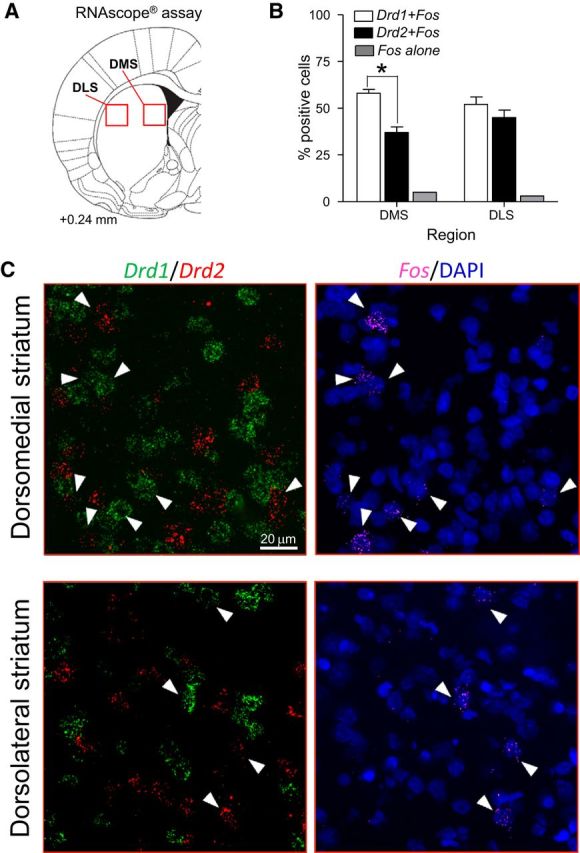
Triplex detection of Drd1 (green), Drd2 (red), and Fos (purple) mRNAs in dorsal striatum after context-induced reinstatement of methamphetamine seeking. A, Drawing shows approximate brain section level (Paxinos and Watson, 2005) chosen for imaging. Red squares indicate areas analyzed. B, Graphs indicate the percentage of Fos-positive neurons (n = 9 rats, 1 section each) that coexpressed Drd1 (Drd1+Fos) or Drd2 (Drd2+Fos) mRNA in the dorsomedial or dorsolateral region of the striatum. A small number of Fos-positive neurons did not coexpress either Drd1 or Drd2 mRNA (Fos alone). C, Representative images of selected cells (Fos-positive, white arrows). Images on the left show the merged channels for Drd1 (green) and Drd2 (red) signals. Images on the right show the merged channels for Fos (purple) and DAPI (blue) signals from the same brain sections and fields than the right side. The fluorescent signals for Fos and Drd2 signals had similar colors; although sufficiently different for quantitation, it is easier to see Fos and DAPI signals separately from those for Drd1 and Drd2 signals.
Discussion
Activation of dorsolateral striatal neurons is important for context-induced reinstatement of methamphetamine seeking. Context-induced reinstatement of methamphetamine seeking induced Fos expression in the dorsolateral and dorsomedial striatum, but not in accumbens core or shell. Reversible inactivation of the dorsolateral, but not dorsomedial, striatum with muscimol+baclofen decreased context-induced reinstatement of drug seeking. Thus only neurons in the dorsolateral striatum were found to be both activated and necessary for this reinstatement. FACS with qPCR analysis indicated that context-induced reinstatement of methamphetamine seeking also induced unique gene alterations within the activated Fos-positive neurons but not in Fos-negative neurons. These alterations included genes involved in transcription regulation (Fos, FosB; Nestler, 2001) and synaptic plasticity associated with glutamate transmission (Homer-1, Homer-2; Kalivas et al., 2004; Lominac et al., 2005; Cozzoli et al., 2009; Ary et al., 2013), as well as Grin2a, the gene encoding the GluN2A subunit of the NMDA receptor (Buller et al., 1994). RNAscope ISH (Wang et al., 2012) indicated that expression levels of the genes Grin2a and Grin2b, which encode the GluN2A and GluN2B NMDA receptor subunits, were selectively increased in Fos-positive neurons in the dorsolateral, but not dorsomedial, striatum. Together, our results indicate that neuronal activity in the dorsolateral striatum contributes to context-induced reinstatement of methamphetamine seeking and that the context-activated Fos-expressing neurons in this striatal subregion undergo unique neuroadaptations.
Role of dorsal and ventral striatum in context-induced reinstatement of drug seeking
Our results are consistent with those of previous studies on the role of dorsolateral striatum on context-induced and cue-induced reinstatement of drug seeking (Bossert et al., 2013; Marchant et al., 2014). Specifically, reversible inactivation (tetrodotoxin or muscimol+baclofen) of dorsolateral striatum decreased cue-induced reinstatement of cocaine and heroin seeking and context-induced reinstatement of cocaine seeking (Fuchs et al., 2006; Rogers et al., 2008). Additionally, blockade of D1-family receptors with SCH23390 in dorsolateral but not dorsomedial striatum decreased context-induced reinstatement of heroin seeking (Bossert et al., 2009). Thus, while several studies indicate that different brain areas mediate relapse induced by discrete cues and contexts associated with opiate versus psychostimulant drugs (Badiani et al., 2011; Badiani, 2013; Bossert et al., 2013; Peters et al., 2013), the dorsolateral striatum is consistently a critical site for context-induced and cue-induced drug seeking across drug classes (Vanderschuren and Everitt, 2005; Vanderschuren et al., 2005).
In a recent study, we found that context-induced reinstatement of cocaine seeking was associated with increased Fos expression in accumbens shell and core and that selective inhibition of the activated Fos-expressing neurons in accumbens shell (but not core) using the Daun02 procedure (Cruz et al., 2013) decreased this reinstatement (Cruz et al., 2014a). Based on these and related findings on the effect of cocaine cues on accumbens Fos expression (Ciccocioppo et al., 2001; Kufahl et al., 2009), it was surprising to find that context-induced reinstatement was not associated with increased Fos expression in either accumbens core or shell. However, our current finding is consistent with previous reversible inactivation studies that indicate functional dissociation between brain areas involved in reinstatement of methamphetamine versus cocaine seeking (Bossert et al., 2013). For example, reversible inactivation of ventral medial prefrontal cortex causes reinstatement of cocaine but not methamphetamine seeking after extinction (Peters et al., 2008; Rocha and Kalivas, 2010). Reversible inactivation of the same brain area decreases cue-induced methamphetamine but not cocaine seeking after extinction (McLaughlin and See, 2003; Rocha and Kalivas, 2010). Finally, reversible activation of the ventral medial prefrontal cortex decreases the time-dependent increases in cue-induced cocaine but not methamphetamine seeking (incubation of drug craving; Koya et al., 2009; Li et al., 2014).
The use of FACS to identify unique molecular changes in context-activated and cue-activated neurons
In previous studies, we used the Daun02 inactivation method (Koya et al., 2009) to demonstrate a causal role of behaviorally activated Fos-expressing neurons in both context-induced and cue-induced drug seeking (Bossert et al., 2011; Fanous et al., 2012; Cruz et al., 2014a). Based on these findings and the extensive literature on the role of drug-induced neuroadaptations in drug reward and relapse (Nestler, 2001; Kalivas, 2009; Van den Oever et al., 2010; Wolf and Ferrario, 2010), we proposed that unique alterations of transcription and synaptic plasticity in these Fos-expressing neurons are likely to play a unique role in drug addiction (Cruz et al., 2014b). Unlike the majority of addiction studies, we could not use brain homogenates to assess these alterations, because specific molecular alterations within the small population of Fos-expressing neurons would be diluted or masked within the whole homogenate (Cruz et al., 2013; Liu et al., 2014).
Thus we used FACS to isolate Fos-expressing neurons for molecular analysis. The use of FACS with adult brain tissue was first used to purify striatal neurons expressing GFP that were constitutively driven by the promoters for D1 and D2 receptors in mice (Lobo et al., 2006; Lobo, 2009). We developed an antibody-based FACS method to purify activated Fos-expressing neurons from adult wild-type rats (Guez-Barber et al., 2012) and used this protocol to assess molecular alterations in Fos-expressing neurons that were activated during the expression of cocaine sensitization and cue-induced heroin seeking (Guez-Barber et al., 2011; Fanous et al., 2013). However, this protocol required pooled brain regions (entire striatum or PFC) and pooled samples (≤10 rats per sample) that made it difficult to implement in studies using complex behavioral models of drug addiction and relapse (Cruz et al., 2013; Liu et al., 2014). Based on these considerations, we added targeted preamplification of cDNA to enhance sensitivity of our qPCR assay for gene expression from small numbers of FACS-isolated neurons in single rat dorsal striatum (Liu et al., 2014).
Using FACS+qPCR analysis, we found that exposure to both the Methamphetamine Context and the Extinction Context induced IEGs in only the Fos-expressing dorsal striatal neurons. This finding replicates our previous findings that IEGs, such as Fos and FosB, are induced only in Fos-expressing neurons and not in the less activated Fos-negative majority of neurons in striatum and medial prefrontal cortex after exposure to drugs or drug cues (Guez-Barber et al., 2011; Fanous et al., 2013). Although the functional significance of inducing these IEGs is unknown, Fos and FosB encode transcription factor proteins that can induce subsequent waves of gene expression selectively with Fos-expressing neurons (Cruz et al., 2014b).
FACS+qPCR analysis and RNAscope ISH together indicated that context-induced reinstatement of methamphetamine seeking increased mRNA levels of Grin2a and Grin2b, but not Grin1, in Fos-positive neurons in the dorsolateral, but not dorsomedial, striatum. On the other hand, the Gria1 and Gria2 genes, which encode the AMPA receptor subunits GluA1 and GluA2, were not different between Fos-positive and Fos-negative neurons. The Grin2a, Grin2b, and Grin1 genes encode GluN2A, GluN2B, and GluN1 subunits of the NMDA receptor, which are expressed at moderate to high levels in striatum (Buller et al., 1994; Standaert et al., 1999). Although the functional significance of inducing these genes is unknown, increased levels of Grin2a and Grin2b may be related to synaptic alterations that were selectively induced in Fos-expressing neurons in the accumbens and medial prefrontal cortex (Cifani et al., 2012), or to GluN2B-containing silent synapses induced in all neurons in these brain areas following cocaine locomotor sensitization (Huang et al., 2009; Brown et al., 2011) or cocaine self-administration (Lee et al., 2013; Ma et al., 2014).
Exposure to the Extinction Context, but not the Methamphetamine Context, induced Homer-1, Homer-2, and Cbr1 gene expression in Fos-expressing neurons. The functional significance of unique molecular alterations in Fos-expressing neurons in the Drug versus Extinction Contexts is unknown but is in agreement that different mechanisms control reward learning versus extinction learning (Peters et al., 2009).
Another finding from qPCR assays was that the mRNA encoding D1 and D2 receptors did not differ between the Fos-positive and Fos-negative neurons. The RNAscope data confirmed that Fos mRNA was similarly induced in D1 and D2 neurons, most likely in medium spiny neurons, which make up ∼95% of neurons in the dorsal striatum (Tepper et al., 2007). These results extend our previous studies showing that the context-activated Fos-expressing neurons are not confined to a particular cell type in the striatum (Cruz et al., 2014a) or medial prefrontal cortex (Bossert et al., 2011).
Concluding remarks
We demonstrated that the dorsolateral striatum is a critical brain substrate for context-induced reinstatement of methamphetamine seeking, which together with previous results indicate that neuronal activity in this brain region is a common substrate for context-induced reinstatement of drug seeking for both psychostimulant and opiate drugs. Additionally, our FACS+qPCR and RNAscope ISH data indicate that context-induced reinstatement of methamphetamine seeking is associated with unique molecular alterations of NMDA-type glutamate receptor subunit genes in context-activated Fos-expressing neurons in dorsal striatum. It is currently not possible to manipulate gene expression selectively within these Fos-expressing neurons. Thus the functional significance of inducing unique molecular alterations within Fos-expressing neurons activated by exposure to the Extinction or Methamphetamine Contexts is currently unknown. Future experiments are necessary to determine whether selective regulation of these genes in Fos-expressing neurons plays a functional role in context-induced reinstatement of drug seeking.
Footnotes
This work was supported by the National Institute on Drug Abuse Intramural Research Program, National Institutes of Health. F.J.R. was supported by an appointment to the National Institute on Drug Abuse Research Participation Program sponsored by the National Institutes of Health and administered by the Oak Ridge Institute for Science and Education, and received additional financial support from Becas-Chile Scholarship managed by the Comisión Nacional de Investigación Científica y Tecnológica and the Universidad de los Andes, Santiago, Chile. The Johns Hopkins FACS Core facility was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institute of Health under Award Number P30AR053503.
The authors declare no competing financial interests.
References
- Ary AW, Lominac KD, Wroten MG, Williams AR, Campbell RR, Ben-Shahar O, von Jonquieres G, Klugmann M, Szumlinski KK. Imbalances in prefrontal cortex CC-Homer1 versus CC-Homer2 expression promote cocaine preference. J Neurosci. 2013;33:8101–8113. doi: 10.1523/JNEUROSCI.1727-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badiani A. Substance-specific environmental influences on drug use and drug preference in animals and humans. Curr Opin Neurobiol. 2013;23:588–596. doi: 10.1016/j.conb.2013.03.010. [DOI] [PubMed] [Google Scholar]
- Badiani A, Belin D, Epstein D, Calu D, Shaham Y. Opiate versus psychostimulant addiction: the differences do matter. Nat Rev Neurosci. 2011;12:685–700. doi: 10.1038/nrn3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossert JM, Liu SY, Lu L, Shaham Y. A role of ventral tegmental area glutamate in contextual cue-induced relapse to heroin seeking. J Neurosci. 2004;24:10726–10730. doi: 10.1523/JNEUROSCI.3207-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossert JM, Gray SM, Lu L, Shaham Y. Activation of group II metabotropic glutamate receptors in the nucleus accumbens shell attenuates context-induced relapse to heroin seeking. Neuropsychopharmacology. 2006;31:2197–2209. doi: 10.1038/sj.npp.1300977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossert JM, Poles GC, Wihbey KA, Koya E, Shaham Y. Differential effects of blockade of dopamine D1-family receptors in nucleus accumbens core or shell on reinstatement of heroin seeking induced by contextual and discrete cues. J Neurosci. 2007;27:12655–12663. doi: 10.1523/JNEUROSCI.3926-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossert JM, Wihbey KA, Pickens CL, Nair SG, Shaham Y. Role of dopamine D(1)-family receptors in dorsolateral striatum in context-induced reinstatement of heroin seeking in rats. Psychopharmacology (Berl) 2009;206:51–60. doi: 10.1007/s00213-009-1580-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossert JM, Stern AL, Theberge FR, Cifani C, Koya E, Hope BT, Shaham Y. Ventral medial prefrontal cortex neuronal ensembles mediate context-induced relapse to heroin. Nat Neurosci. 2011;14:420–422. doi: 10.1038/nn.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossert JM, Stern AL, Theberge FR, Marchant NJ, Wang HL, Morales M, Shaham Y. Role of projections from ventral medial prefrontal cortex to nucleus accumbens shell in context-induced reinstatement of heroin seeking. J Neurosci. 2012;32:4982–4991. doi: 10.1523/JNEUROSCI.0005-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossert JM, Marchant NJ, Calu DJ, Shaham Y. The reinstatement model of drug relapse: recent neurobiological findings, emerging research topics, and translational research. Psychopharmacology (Berl) 2013;229:453–476. doi: 10.1007/s00213-013-3120-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouton ME, Bolles RC. Role of conditioned contextual stimuli in reinstatement of extinguished fear. J Exp Psychol Anim Behav Process. 1979;5:368–378. doi: 10.1037/0097-7403.5.4.368. [DOI] [PubMed] [Google Scholar]
- Bouton ME, Swartzentruber D. Sources of relapse after extinction in Pavlovian and instrumental learning. Clin Psychol Rev. 1991;11:123–140. [Google Scholar]
- Brown TE, Lee BR, Mu P, Ferguson D, Dietz D, Ohnishi YN, Lin Y, Suska A, Ishikawa M, Huang YH, Shen H, Kalivas PW, Sorg BA, Zukin RS, Nestler EJ, Dong Y, Schlüter OM. A silent synapse-based mechanism for cocaine-induced locomotor sensitization. J Neurosci. 2011;31:8163–8174. doi: 10.1523/JNEUROSCI.0016-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buller AL, Larson HC, Schneider BE, Beaton JA, Morrisett RA, Monaghan DT. The molecular basis of NMDA receptor subtypes: native receptor diversity is predicted by subunit composition. J Neurosci. 1994;14:5471–5484. doi: 10.1523/JNEUROSCI.14-09-05471.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadet JL, Brannock C, Jayanthi S, Krasnova IN. Transcriptional and epigenetic substrates of methamphetamine addiction and withdrawal: evidence from a long-access self-administration model in the rat. Mol Neurobiol. 2014 doi: 10.1007/s12035-014-8776-8. Advance online publication. Retrieved Feb. 27, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhri N, Sahuque LL, Cone JJ, Janak PH. Reinstated ethanol-seeking in rats is modulated by environmental context and requires the nucleus accumbens core. Eur J Neurosci. 2008;28:2288–2298. doi: 10.1111/j.1460-9568.2008.06517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhri N, Sahuque LL, Janak PH. Ethanol seeking triggered by environmental context is attenuated by blocking dopamine D1 receptors in the nucleus accumbens core and shell in rats. Psychopharmacology (Berl) 2009;207:303–314. doi: 10.1007/s00213-009-1657-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccocioppo R, Sanna PP, Weiss F. Cocaine-predictive stimulus induces drug-seeking behavior and neural activation in limbic brain regions after multiple months of abstinence: reversal by D(1) antagonists. Proc Natl Acad Sci U S A. 2001;98:1976–1981. doi: 10.1073/pnas.98.4.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifani C, Koya E, Navarre BM, Calu DJ, Baumann MH, Marchant NJ, Liu QR, Khuc T, Pickel J, Lupica CR, Shaham Y, Hope BT. Medial prefrontal cortex neuronal activation and synaptic alterations after stress-induced reinstatement of palatable food seeking: a study using c-fos-GFP transgenic female rats. J Neurosci. 2012;32:8480–8490. doi: 10.1523/JNEUROSCI.5895-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozzoli DK, Goulding SP, Zhang PW, Xiao B, Hu JH, Ary AW, Obara I, Rahn A, Abou-Ziab H, Tyrrel B, Marini C, Yoneyama N, Metten P, Snelling C, Dehoff MH, Crabbe JC, Finn DA, Klugmann M, Worley PF, Szumlinski KK. Binge drinking upregulates accumbens mGluR5-Homer2-PI3K signaling: functional implications for alcoholism. J Neurosci. 2009;29:8655–8668. doi: 10.1523/JNEUROSCI.5900-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crombag HS, Shaham Y. Renewal of drug seeking by contextual cues after prolonged extinction in rats. Behav Neurosci. 2002;116:169–173. doi: 10.1037/0735-7044.116.1.169. [DOI] [PubMed] [Google Scholar]
- Crombag HS, Grimm JW, Shaham Y. Effect of dopamine receptor antagonists on renewal of cocaine seeking by reexposure to drug-associated contextual cues. Neuropsychopharmacology. 2002;27:1006–1015. doi: 10.1016/S0893-133X(02)00356-1. [DOI] [PubMed] [Google Scholar]
- Crombag HS, Bossert JM, Koya E, Shaham Y. Review. Context-induced relapse to drug seeking: a review. Philos Trans R Soc Lond B Biol Sci. 2008;363:3233–3243. doi: 10.1098/rstb.2008.0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz FC, Koya E, Guez-Barber DH, Bossert JM, Lupica CR, Shaham Y, Hope BT. New technologies for examining the role of neuronal ensembles in drug addiction and fear. Nat Rev Neurosci. 2013;14:743–754. doi: 10.1038/nrn3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz FC, Babin KR, Leao RM, Goldart EM, Bossert JM, Shaham Y, Hope BT. Role of nucleus accumbens shell neuronal ensembles in context-induced reinstatement of cocaine-seeking. J Neurosci. 2014a;34:7437–7446. doi: 10.1523/JNEUROSCI.0238-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz FC, Rubio FJ, Hope BT. Using c-fos to study neuronal ensembles in corticostriatal circuitry of addiction. Brain Res pii:S0006-8993(14)01552-2. 2014b doi: 10.1016/j.brainres.2014.11.005. Advance online publication. Retrieved Feb. 27, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanous S, Goldart EM, Theberge FR, Bossert JM, Shaham Y, Hope BT. Role of orbitofrontal cortex neuronal ensembles in the expression of incubation of heroin craving. J Neurosci. 2012;32:11600–11609. doi: 10.1523/JNEUROSCI.1914-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanous S, Guez-Barber DH, Goldart EM, Schrama R, Theberge FR, Shaham Y, Hope BT. Unique gene alterations are induced in FACS-purified Fos-positive neurons activated during cue-induced relapse to heroin seeking. J Neurochem. 2013;124:100–108. doi: 10.1111/jnc.12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs RA, Branham RK, See RE. Different neural substrates mediate cocaine seeking after abstinence versus extinction training: a critical role for the dorsolateral caudate-putamen. J Neurosci. 2006;26:3584–3588. doi: 10.1523/JNEUROSCI.5146-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs RA, Ramirez DR, Bell GH. Nucleus accumbens shell and core involvement in drug context-induced reinstatement of cocaine seeking in rats. Psychopharmacology (Berl) 2008;200:545–556. doi: 10.1007/s00213-008-1234-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guez-Barber D, Fanous S, Golden SA, Schrama R, Koya E, Stern AL, Bossert JM, Harvey BK, Picciotto MR, Hope BT. FACS identifies unique cocaine-induced gene regulation in selectively activated adult striatal neurons. J Neurosci. 2011;31:4251–4259. doi: 10.1523/JNEUROSCI.6195-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guez-Barber D, Fanous S, Harvey BK, Zhang Y, Lehrmann E, Becker KG, Picciotto MR, Hope BT. FACS purification of immunolabeled cell types from adult rat brain. J Neurosci Methods. 2012;203:10–18. doi: 10.1016/j.jneumeth.2011.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamlin AS, Newby J, McNally GP. The neural correlates and role of D1 dopamine receptors in renewal of extinguished alcohol-seeking. Neuroscience. 2007;146:525–536. doi: 10.1016/j.neuroscience.2007.01.063. [DOI] [PubMed] [Google Scholar]
- Huang YH, Lin Y, Mu P, Lee BR, Brown TE, Wayman G, Marie H, Liu W, Yan Z, Sorg BA, Schlüter OM, Zukin RS, Dong Y. In vivo cocaine experience generates silent synapses. Neuron. 2009;63:40–47. doi: 10.1016/j.neuron.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janak PH, Chaudhri N. The potent effect of environmental context on relapse to alcohol-seeking after extinction. Open Addict J. 2010;3:76–87. doi: 10.2174/1874941001003020076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalivas PW. The glutamate homeostasis hypothesis of addiction. Nat Rev Neurosci. 2009;10:561–572. doi: 10.1038/nrn2515. [DOI] [PubMed] [Google Scholar]
- Kalivas PW, Szumlinski KK, Worley P. Homer2 gene deletion in mice produces a phenotype similar to chronic cocaine treated rats. Neurotox Res. 2004;6:385–387. doi: 10.1007/BF03033313. [DOI] [PubMed] [Google Scholar]
- Koya E, Golden SA, Harvey BK, Guez-Barber DH, Berkow A, Simmons DE, Bossert JM, Nair SG, Uejima JL, Marin MT, Mitchell TB, Farquhar D, Ghosh SC, Mattson BJ, Hope BT. Targeted disruption of cocaine-activated nucleus accumbens neurons prevents context-specific sensitization. Nat Neurosci. 2009;12:1069–1073. doi: 10.1038/nn.2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufahl PR, Zavala AR, Singh A, Thiel KJ, Dickey ED, Joyce JN, Neisewander JL. c-Fos expression associated with reinstatement of cocaine-seeking behavior by response-contingent conditioned cues. Synapse. 2009;63:823–835. doi: 10.1002/syn.20666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasseter HC, Xie X, Ramirez DR, Fuchs RA. Prefrontal cortical regulation of drug seeking in animal models of drug relapse. Curr Top Behav Neurosci. 2010;3:101–117. doi: 10.1007/7854_2009_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BR, Ma YY, Huang YH, Wang X, Otaka M, Ishikawa M, Neumann PA, Graziane NM, Brown TE, Suska A, Guo C, Lobo MK, Sesack SR, Wolf ME, Nestler EJ, Shaham Y, Schlüter OM, Dong Y. Maturation of silent synapses in amygdala-accumbens projection contributes to incubation of cocaine craving. Nat Neurosci. 2013;16:1644–1651. doi: 10.1038/nn.3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zeric T, Kambhampati S, Bossert JM, Shaham Y. The central amygdala nucleus is critical for incubation of methamphetamine craving. Neuropsychopharmacology. 2014 doi: 10.1038/npp.2014.320. Advance online publication. Retrieved Feb. 28, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu QR, Rubio FJ, Bossert JM, Marchant NJ, Fanous S, Hou X, Shaham Y, Hope BT. Detection of molecular alterations in methamphetamine-activated Fos-expressing neurons from a single rat dorsal striatum using fluorescence-activated cell sorting (FACS) J Neurochem. 2014;128:173–185. doi: 10.1111/jnc.12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo MK. Molecular profiling of striatonigral and striatopallidal medium spiny neurons past, present, and future. Int Rev Neurobiol. 2009;89:1–35. doi: 10.1016/S0074-7742(09)89001-6. [DOI] [PubMed] [Google Scholar]
- Lobo MK, Karsten SL, Gray M, Geschwind DH, Yang XW. FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nat Neurosci. 2006;9:443–452. doi: 10.1038/nn1654. [DOI] [PubMed] [Google Scholar]
- Lominac KD, Oleson EB, Pava M, Klugmann M, Schwarz MK, Seeburg PH, During MJ, Worley PF, Kalivas PW, Szumlinski KK. Distinct roles for different Homer1 isoforms in behaviors and associated prefrontal cortex function. J Neurosci. 2005;25:11586–11594. doi: 10.1523/JNEUROSCI.3764-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma YY, Lee BR, Wang X, Guo C, Liu L, Cui R, Lan Y, Balcita-Pedicino JJ, Wolf ME, Sesack SR, Shaham Y, Schlüter OM, Huang YH, Dong Y. Bidirectional modulation of incubation of cocaine craving by silent synapse-based remodeling of prefrontal cortex to accumbens projections. Neuron. 2014;83:1453–1467. doi: 10.1016/j.neuron.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant NJ, Kaganovsky K, Shaham Y, Bossert JM. Role of corticostriatal circuits in context-induced reinstatement of drug seeking. Brain Res pii:S0006-8993(14)01191-3. 2014 doi: 10.1016/j.brainres.2014.09.004. Advance online publication. Retrieved Feb. 28, 2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarland K, Kalivas PW. The circuitry mediating cocaine-induced reinstatement of drug-seeking behavior. J Neurosci. 2001;21:8655–8663. doi: 10.1523/JNEUROSCI.21-21-08655.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin J, See RE. Selective inactivation of the dorsomedial prefrontal cortex and the basolateral amygdala attenuates conditioned-cued reinstatement of extinguished cocaine-seeking behavior in rats. Psychopharmacology (Berl) 2003;168:57–65. doi: 10.1007/s00213-002-1196-x. [DOI] [PubMed] [Google Scholar]
- McNally GP. Extinction of drug seeking: neural circuits and approaches to augmentation. Neuropharmacology. 2014;76:528–532. doi: 10.1016/j.neuropharm.2013.06.007. [DOI] [PubMed] [Google Scholar]
- Merchant KM, Hanson GR, Dorsa DM. Induction of neurotensin and c-fos mRNA in distinct subregions of rat neostriatum after acute methamphetamine: comparison with acute haloperidol effects. J Pharmacol Exp Ther. 1994;269:806–812. [PubMed] [Google Scholar]
- Morgan JI, Curran T. Stimulus-transcription coupling in the nervous system: involvement of the inducible proto-oncogenes fos and jun. Annu Rev Neurosci. 1991;14:421–451. doi: 10.1146/annurev.ne.14.030191.002225. [DOI] [PubMed] [Google Scholar]
- Nestler EJ. Molecular basis of long-term plasticity underlying addiction. Nat Rev Neurosci. 2001;2:119–128. doi: 10.1038/35053570. [DOI] [PubMed] [Google Scholar]
- O'Brien CP, Ehrman RN, Ternes JW. Classical conditioning in human opioid dependence. In: Goldberg S, Stolerman I, editors. Behavioral analysis of drug dependence. Orlando, FL: Academic; 1986. pp. 329–356. [Google Scholar]
- Ohno M, Yoshida H, Watanabe S. NMDA receptor-mediated expression of Fos protein in the rat striatum following methamphetamine administration: relation to behavioral sensitization. Brain Res. 1994;665:135–140. doi: 10.1016/0006-8993(94)91163-0. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Watson C. The rat brain in stereotaxic coordinates. fifth edition. Amsterdam: Elsevier Academic; 2005. [Google Scholar]
- Perry CJ, McNally GP. mu-Opioid receptors in the nucleus accumbens shell mediate context-induced reinstatement (renewal) but not primed reinstatement of extinguished alcohol seeking. Behav Neurosci. 2013;127:535–543. doi: 10.1037/a0032981. [DOI] [PubMed] [Google Scholar]
- Peters J, LaLumiere RT, Kalivas PW. Infralimbic prefrontal cortex is responsible for inhibiting cocaine seeking in extinguished rats. J Neurosci. 2008;28:6046–6053. doi: 10.1523/JNEUROSCI.1045-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J, Kalivas PW, Quirk GJ. Extinction circuits for fear and addiction overlap in prefrontal cortex. Learn Mem. 2009;16:279–288. doi: 10.1101/lm.1041309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J, Pattij T, De Vries TJ. Targeting cocaine versus heroin memories: divergent roles within ventromedial prefrontal cortex. Trends Pharmacol Sci. 2013;34:689–695. doi: 10.1016/j.tips.2013.10.004. [DOI] [PubMed] [Google Scholar]
- Robins LN, Davis DH, Goodwin DW. Drug use by U.S. Army enlisted men in Vietnam: a follow-up on their return home. Am J Epidemiol. 1974;99:235–249. doi: 10.1093/oxfordjournals.aje.a121608. [DOI] [PubMed] [Google Scholar]
- Rocha A, Kalivas PW. Role of the prefrontal cortex and nucleus accumbens in reinstating methamphetamine seeking. Eur J Neurosci. 2010;31:903–909. doi: 10.1111/j.1460-9568.2010.07134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers JL, Ghee S, See RE. The neural circuitry underlying reinstatement of heroin-seeking behavior in an animal model of relapse. Neuroscience. 2008;151:579–588. doi: 10.1016/j.neuroscience.2007.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaham Y, Shalev U, Lu L, De Wit H, Stewart J. The reinstatement model of drug relapse: history, methodology and major findings. Psychopharmacology (Berl) 2003;168:3–20. doi: 10.1007/s00213-002-1224-x. [DOI] [PubMed] [Google Scholar]
- Standaert DG, Friberg IK, Landwehrmeyer GB, Young AB, Penney JB., Jr Expression of NMDA glutamate receptor subunit mRNAs in neurochemically identified projection and interneurons in the striatum of the rat. Brain Res Mol Brain Res. 1999;64:11–23. doi: 10.1016/S0169-328X(98)00293-9. [DOI] [PubMed] [Google Scholar]
- Tepper JM, Abercrombie ED, Bolam JP. Basal ganglia macrocircuits. Prog Brain Res. 2007;160:3–7. doi: 10.1016/S0079-6123(06)60001-0. [DOI] [PubMed] [Google Scholar]
- Van den Oever MC, Spijker S, Smit AB, De Vries TJ. Prefrontal cortex plasticity mechanisms in drug seeking and relapse. Neurosci Biobehav Rev. 2010;35:276–284. doi: 10.1016/j.neubiorev.2009.11.016. [DOI] [PubMed] [Google Scholar]
- Vanderschuren LJ, Everitt BJ. Behavioral and neural mechanisms of compulsive drug seeking. Eur J Pharmacol. 2005;526:77–88. doi: 10.1016/j.ejphar.2005.09.037. [DOI] [PubMed] [Google Scholar]
- Vanderschuren LJ, Di Ciano P, Everitt BJ. Involvement of the dorsal striatum in cue-controlled cocaine seeking. J Neurosci. 2005;25:8665–8670. doi: 10.1523/JNEUROSCI.0925-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, Wu X, Vo HT, Ma XJ, Luo Y. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2012;14:22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JQ, Smith AJ, McGinty JF. A single injection of amphetamine or methamphetamine induces dynamic alterations in c-fos, zif/268 and preprodynorphin messenger RNA expression in rat forebrain. Neuroscience. 1995;68:83–95. doi: 10.1016/0306-4522(95)00100-W. [DOI] [PubMed] [Google Scholar]
- Widholm JJ, Gass JT, Cleva RM, Olive MF. The mGluR5 positive allosteric modulator CDPPB does not alter extinction or contextual reinstatement of methamphetamine-seeking behavior in rats. J Addict Res Ther. 2011 doi: 10.4172/2155-6105.S1-004. S1:pii:004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikler A. Dynamics of drug dependence. Implications of a conditioning theory for research and treatment. Arch Gen Psychiatry. 1973;28:611–616. doi: 10.1001/archpsyc.1973.01750350005001. [DOI] [PubMed] [Google Scholar]
- Wolf ME, Ferrario CR. AMPA receptor plasticity in the nucleus accumbens after repeated exposure to cocaine. Neurosci Biobehav Rev. 2010;35:185–211. doi: 10.1016/j.neubiorev.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]