Abstract
High mobility group (HMG) proteins concentrate in the nucleus, interacting with chromatin. Amphoterin is an HMG protein (HMGB1) that has been shown to have extranuclear functions and can be secreted from some cell types. Exogenous amphoterin can increase neurite growth, suggesting that the secreted protein may have growth promoting activities in neurons. Consistent with this, we show that depletion of amphoterin mRNA from cultured adult rat DRG neurons attenuates neurite outgrowth, pointing to autocrine or paracrine mechanisms for its growth-promoting effects. The mRNA encoding amphoterin localizes to axonal processes and we showed recently that its 3′-UTR is sufficient for axonal localization of heterologous transcripts (Donnelly et al., 2013). Here, we show that amphoterin mRNA is transported constitutively into axons of adult DRG neurons. A preconditioning nerve injury increases the levels of amphoterin protein in axons without a corresponding increase in amphoterin mRNA in the axons. A 60 nucleotide region of the amphoterin mRNA 3′-UTR is necessary and sufficient for its localization into axons of cultured sensory neurons. Amphoterin mRNA 3′-UTR is also sufficient for axonal localization in distal axons of DRG neurons in vivo. Overexpression of axonally targeted amphoterin mRNA increases axon outgrowth in cultured sensory neurons, but axon growth is not affected when the overexpressed mRNA is restricted to the cell body.
Keywords: axon regeneration, high mobility group protein, HMGB1, mRNA translation, mRNA transport
Introduction
Spatial regulation of protein synthesis contributes to cell polarization and provides means for rapid, autonomous responses of cells to localized stimuli (Gomes et al., 2014; Jung et al., 2014). Neurons are extremely polarized, with cytoplasmic processes in some mammalian species extending to more than 1 m from a cell body that is most often <100 μm in diameter. The mechanisms for spatial control of neuronal translation have been best characterized in dendrites, with trans-synaptic stimuli triggering dendritic protein synthesis (Liu-Yesucevitz et al., 2011). Axonal processes typically extend much longer distances than dendrites, potentially requiring even greater autonomy gained by localized protein synthesis. Translation products of mRNAs localized to distal axons have been shown to facilitate growth, survival, and injury responses of axons (Jung et al., 2012). Studies have shown increasingly complex populations of mRNAs in axons as sensitivity of nonbiased RNA analyses methods has grown. Indeed, recent next-generation sequencing has shown well over a thousand different mRNAs in developing sensory axons (Minis et al., 2014). Despite this increased knowledge of which mRNAs localize into axons, our understanding of how mRNAs are targeted to and locally translated within axons remains limited.
Transport of mRNAs into axons can be regulated by extracellular stimuli (Willis et al., 2007; Andreassi et al., 2010). Axotomy has been shown to alter transport and translation of some axonal mRNAs. Injury conditioning of DRG neurons by axotomy, which increases axon growth after a second injury, increases axonal levels of neuritin (NRN1) and neural membrane protein-35 (NMP35) mRNAs (Merianda et al., 2013a; Merianda et al., 2013b). Axonal mRNA populations similarly shift with axotomy in cultured hippocampal neurons (Taylor et al., 2009). In contrast, we showed previously that amphoterin mRNA is constitutively transported into axons of cultured DRG neurons, not changing after application of extracellular stimuli or after injury (Willis et al., 2007; Yoo et al., 2013).
Amphoterin is a member of the high mobility group (HMG) proteins (it is also known as HMGB1). HMG proteins interact with chromatin in the nucleus (Hock et al., 2007). Amphoterin can be released from dying cells and has been referred to as an “alarmin,” a circulating biomarker for septic shock and ischemia that may contribute to systemic pathologies seen in these conditions (Lotze and Tracey, 2005). Exogenous amphoterin stimulates migration in non-neuronal cells and neurite outgrowth in neurons (Huttunen and Rauvala, 2004). Amphoterin is secreted from some cell types and its mRNA can localize to the cell periphery in glioma cells (Fages et al., 2000). Here, we show that amphoterin mRNA localizes into axons via its proximal 3′-UTR. Translation of the axonal mRNA is upregulated in injury-conditioned neurons and overexpression of the axon-localized, but not cell soma-restricted, mRNA increases neurite outgrowth. This work points to translational regulation of axonally targeted amphoterin mRNA as a means to increase levels of this growth-associated protein locally after axonal injury.
Materials and Methods
Animal surgery.
All animal experiments were approved by Institutional Animal Care and Use Committees of the Alfred I. duPont Hospital for Children or Drexel University. Male Sprague Dawley rats (150–225 g) were anesthetized using isofluorane and then subjected to unilateral sciatic nerve crush at midthigh as described previously (Twiss et al., 2000). Seven days after injury, L4–L5 DRGs and sciatic nerves were collected and processed as outlined below, with DRG and nerve contralateral to crush injury serving as control (i.e., naive).
For lentiviral (LV) transduction of DRGs in vivo, 10 μl of LV at 108 IU/ml (93–190 μg/ml p24) was injected into the L4–L5 nerve roots adjacent to the DRG (Merianda et al., 2013a). Ten days later, animals were subjected to unilateral sciatic nerve crush injury. For analyses of these samples, sciatic nerve from 0.5 cm proximal to 0.5 cm distal to the crush site, injection site in the L4–L5 nerve roots and L4–L5 DRGs were harvested and processed as outlined below.
Neuronal cultures.
DRGs were dissociated using collagenase (500 U/ml; Sigma) and trypsin-EDTA (0.05%; Cellgro) as described previously (Twiss et al., 2000). After a series of washes in DMEM/F12, cells were resuspended in complete medium containing the following: DMEM/F12 with 1 × N1 supplement (Sigma), 10% horse serum (Hyclone), and 10 μm cytosine arabinoside (Sigma). Dissociated DRGs were plated onto poly-l-lysine (0.01%; Sigma) and laminin (6.5 μg/ml; Millipore) substrates. Coated coverslips were used for localization studies. Coated tissue culture plates were used for preparation of RNA from unfractionated cultures. For isolation of axons from cell bodies, cells were cultured onto poly-l-lysine-laminin-coated, porous polyethylene-tetrathalate membrane tissue culture inserts (8 μm pores; Falcon) (Merianda et al., 2013a). Axonal versus cell body compartments were isolated as described previously (Zheng et al., 2001). For replating cultures, DRGs grown on coated tissue culture plates (2 D in vitro) were trypsinized (0.025% trypsin-EDTA; Corning) for 3 min at 37°C. DRGs were then gently triturated using fire-polished Pasteur pipettes in DMEM/F12 with 10% FBS to remove the processes and replated on coated coverslips. These cultures were analyzed the following day.
DNA constructs.
All DNA constructs outlined below were sequence validated before use. For the various UTR-containing and protein overexpression constructs generated, we use 5′ or 3′ followed by all lower-case letters to designate UTRs and all capital letters to designate coding sequences.
Diffusion limited GFPMYR reporter construct with 5′-UTR of CaMKIIα (5′camkII) and 3′ amphoterin (3′-amph) was described previously (Donnelly et al., 2013). This GFPMYR5′camkII/3′-amph construct was used as a template for PCR to generate segments of amphoterin's 3′-UTR with terminal Not1 and Xho1 restriction sites corresponding to nt 738–797, 797–993, and 993–1238 of the rat amphoterin mRNA (GenBank accession no. AF275734).
Amphoterin protein expression plasmids were generated by fusing the 5′-UTR (5′amph) and coding sequence (AMPH) of rat amphoterin cDNA to the N terminus of AcGFP. For this, 200 ng of RNA from rat DRG cultures was processed for RT using iScript cDNA synthesis kit (Bio-Rad) and then amplified by PCR using Pfu DNA polymerase (Stratagene). Primers spanning 5′-UTR plus coding sequence or just coding sequence of amphoterin (nt 1–73 and 90–730, respectively) were used. Purified PCR products were then cloned upstream of AcGFP in pAcGFP1-N3 (Clontech). Varying 3′-UTRs were then cloned downstream of AcGFP. As a control for axonal localization, AcGFP (herein designated GFP) fusion constructs without the localizing 3′-UTRs were also generated. The primers for amplifying the rat 5′Amph + AMPH were engineered to contain 5′ EcoR1 and 3′ BamH1; 3′-UTR segments were engineered to contain 5′ Not1 restriction sites. Constructs generated consisted of AMPH-GFP fusion with 5′-UTR of amphoterin and 3′-UTR of GFP (AMPH-GFP-5′amph/3′gfp) and 5′-UTR of GFP and 3′-UTR of amphoterin (AMPH-GFP-5′gfp/3′-amph). The 3′-UTR of amphoterin in AMPH-GFP-5′gfp/3′-amph and AMPH-GFP-5′/3′-amph was also replaced with nt 738–797 of amphoterin mRNA (AMPH-GFP-5′gfp/3′-amph738–797, AMPH-GFP-5′amph/3′-amph738–797). The amphoterin 738–797 segment was cloned using Nhe1 and Hpa1 restriction sites. Note that the mRNA from these 3′-amph738–797 constructs and those above with shortened 3′-UTR would make use of the SV40 poly-adenylation element present in pAcGFP1-N3, extending the 3′-UTR by at least 128 nt.
For siRNA-resistant AMPH*GFP-5′amph/3′gfp, AMPH*GFP-5′/3′-amph, and AMPH*GFP-5′amph/3′-amph738–797 constructs, four nucleotides in the siRNA-targeted regions (AAGGCTGACAAGGCTCGTTAT) were mutated using QuikChange XL Site-Directed Mutagenesis kit (Stratagene). The following primers were used: sense, 5′ATTTGAAGATATGGCAAAGGC*GACAAGGC*CG*TATGAAAGAGAAATGAAAACC-3′ and antisense, 5′-GGTTTTCATTTCTCTTTCATA*CG*GCCTTGTC*GCCTTTGCCATATCTTCAAAT-3′ (asterisks indicate mutated nucleotides).
For LV constructs, the endogenous polyadenylation signal in the AMPH-GFP-5′amph/3′gfp and AMPH-GFP-5′/3′-amph were mutated to increase efficiency of LV packaging (Vuppalanchi et al., 2010). The QuikChange XL Site-Directed Mutagenesis kit was used for this. AMPH-GFP-5′amph/3′gfp and AMPH-GFP-5′/3′-amph DNAs were subcloned into pENTR shuttle vector (Invitrogen) and recombined into Gateway-compatible derivative of pCDH-CMV-MCS1-Ef1α-copGFP (SBI Systems). LV was then generated as described previously (Blesch, 2004).
Expression of exogenous mRNAs and proteins.
DRGs were resuspended in transfection solution (Rat Neuron Nucleofector Kit; Lonza) along with 5 μg of each plasmid DNA immediately after dissociation. Cells were then transfected using the AMAXA Nucleofector apparatus (Lonza; G-013 program), gently pelleted (100 × g), resuspended in complete medium, and plated as above. For in vitro transduction with LV, serial dilutions of LV preparations were added to dissociated DRGs 4 h after plating. For both transfection and transductions, medium was replaced 24 h later.
siRNA depletion of endogenous amphoterin mRNA.
siRNAs targeting four sequences in rat amphoterin mRNA were initially used to deplete the mRNA (Dharmacon). For “rescue” experiments in which siRNAs and amphoterin expression constructs were cotransfected, the siRNAs from the above pool were individually tested for knock-down efficiency and a single siRNA was subsequently used for the cotransfection experiments. For transfection with siRNAs, dissociated DRGs were cultured for 24 h and then transfected with 300 nm amphoterin siRNA (siAmph) or nontargeting siRNA (siCon) using DharmaFECT3 transfection reagent per manufacturer's protocol (Dharmacon). mRNA and protein depletion were quantitated by RT-qPCR at 72 or 96 h after transfection (for mRNA and protein, respectively).
RNA isolation and axonal preparation.
RNA was isolated from DRG cultures using the RNAqueous Micro kit (Ambion). RNA was isolated from DRGs, sciatic nerves, and brain tissues using the RNAqueous kit (Ambion). RNAs were quantified by fluorimetry using RiboGreen (Invitrogen) (Merianda et al., 2013a). For axonal samples, the flow-through from the affinity-based RNA isolation was used to measure the protein content by fluorimetry with NanoOrange (Invitrogen) to normalize for axon content between culture preparations (Willis et al., 2005). All other RNA samples were normalized to RNA content before RT. Forty nanograms of RNA was used for RT (iScript RT kit; Bio-Rad). For axons, ∼40 ng of RNA was used from the samples normalized for protein content. HotstarTaq Mastermix (Qiagen) was used for PCR. RNA isolated from adult rat brain was used as a positive control and RNA processed without the addition of RT served as a negative control.
For standard PCR, 30 cycles were performed consisting of 45 s at 95°C, 45 s at 58°C, and 3 min at 72°C after a 15 min hot start at 95°C. GAPDH primers were used to test for loading. β-actin, γ-actin, and MAP2 mRNAs were used to assess purity of all axonal samples (Willis et al., 2005). For qPCR, 2× SSoFast Evagreen Supermix on a CFX 384 qPCR instrument (Bio-Rad) was used to amplify RT products. ΔCt calculations were done by normalizing to 12S mitochondrial rRNA (Willis et al., 2007). The primers used for rat γ-actin, β-actin, MAP2, and GAPDH mRNAs have been described previously (Willis et al., 2005; Merianda et al., 2013a). Primers for amphoterin mRNA were as follows: sense 5′-GAGTACCGCCCAAAAATCAA-3′ and antisense 5′-TCATCCTCCTCGTCGTCTT-3′.
FISH and immunofluorescence.
Oligonucleotide probes were used to detect endogenous mRNAs as described previously (Vuppalanchi et al., 2010). Antisense oligonucleotide probes to amphoterin mRNA (nt 202–251, 425–474, 525–574) were designed using Oligo6 software (Molecular Biology Insights). These were synthesized with 5′-amino C6 modifier at four thymidines per oligonucleotide and then digoxigenin labeled using digoxigenin succinamide ester (Roche). Digoxigenin-labeled, “scrambled” probes were used as a control. Unless otherwise stated, all steps for FISH and immunofluorescence (IF) were performed at room temperature.
FISH/IF for endogenous amphoterin mRNA in fixed DRG cultures and tissue sections were performed as described previously (Merianda et al., 2013a). Cultured neurons were fixed in 2% paraformaldehyde for 20 min. Tissues were immersion fixed in 2% paraformaldehyde, cryoprotected overnight in buffered 30% sucrose at 4°C, and processed for cryostat sectioning. The following primary antibodies were used: chicken anti-neurofilament (NF) H (1:1000; Millipore), mouse anti-digoxigenin (1:200; Jackson ImmunoResearch), and Cy3-conjugated mouse anti-digoxigenin (1:200; Jackson ImmunoResearch). Secondary antibodies used were as follows: FITC-conjugated donkey anti-chicken (1:200; Jackson ImmunoResearch), AMCA-conjugated anti-chicken (1:200; Jackson ImmunoResearch), and Cy3-conjugated anti-mouse (1:200; Jackson ImmunoResearch). Samples were mounted with Prolong Gold Antifade (Invitrogen).
GFP reporter mRNA was detected using cRNA probes. Sense and antisense cRNA probes were generated from linearized pcDNA3-eGFP as template for in vitro transcription with SP6 or T7 RNA polymerases coupled with Digoxigenin-label Nucleotide kit (Roche). The cRNA FISH procedure was performed as described previously (Merianda et al., 2013a). Samples were mounted with Prolong Gold Antifade.
Epifluorescent imaging of FISH/IF for cultured neurons was performed on a Leica DMRXA2 or a Zeiss Axioplan epifluorescent microscope fitted with a Hamamatsu ORCA-ER camera. All images were matched for exposure time, gain, offset, and postprocessing. FISH/IF for tissue sections was imaged by confocal microscopy using a Zeiss LSM700 microscope. Signals were quantitated from raw 16-bit images using ImageJ software to calculate the average pixels per square micrometer in regions of interest over at least three separate experiments for each protein or RNA signal. Neuronal cell body and axons were identified by NF signals and correlation with differential interference contrast (DIC) for cultured neurons (Merianda et al., 2013b).
Immunofluorescence.
Standard immunofluorescence on cultured neurons and tissues was performed as described previously (Merianda et al., 2013a). DRG cultures were fixed in 4% paraformaldehyde for 20 min, rinsed in PBS, and then permeabilized with 0.02% Triton X-100. Tissue were fixed in 4% paraformaldehyde for 2 h, cryoprotected in buffered 30% sucrose at 4°C overnight, and processed for cryostat sectioning. Primary antibodies were as follows: mouse anti-NF (1:400; Sigma), chicken antibodies against recombinant rat amphoterin (0.45 μg/ml; Rouhiainen et al., 2004), and rabbit anti-GFP (1:200; Abcam). Secondary antibodies were as follows: FITC- or Texas red-conjugated donkey anti-chicken, anti-rabbit, or anti-mouse antibodies (1:200; Jackson ImmunoResearch). In some experiments, the SYTOX green Nuclear labeling kit (Life Technologies) was used to identify nuclei. Epifluorescent microscopy was used for cultured cells with a Leica DMRXA2 or Zeiss Axioplan microscope fitted with Hamamatsu ORCA-ER CCD camera. Tissue sections were imaged by confocal microscopy using an LSM700 microscope. Volocity software (PerkinElmer) was used for generating orthogonal projections from XYZ image stacks.
Immunoblotting.
DRG cultures were lysed at 4°C for 20 min in RIPA buffer with protease inhibitor mixture (Sigma). Lysates were cleared by centrifugation at 15,000 × g at 4°C. Cleared lysates were normalized for protein content by Bradford assay, fractionated by SDS/PAGE, and transferred to PVDF membranes (Merianda et al., 2013a). Blots were blocked with 5% nonfat milk and then incubated in chicken anti-amphoterin (0.09 μg/ml; Rouhiainen et al., 2004), rabbit anti-HMGB1 (1:10,000; Abcam), rabbit anti-Erk1/2 (1:1000; Cell Signaling Technology), or rabbit anti-GFP (1:2000; Abcam) antibodies overnight at 4°C. Membranes were rinsed in PBS and then incubated in HRP-conjugated anti-rabbit IgG or anti-chicken IgY (1:5000; Cell Signaling Technology) for 1 h. Signals were detected with ECLplus or ECLadvance reagent (GE Healthcare).
Fluorescence recovery after photobleaching.
Fluorescent recovery after photobleaching (FRAP) was used to monitor localized translation of diffusion-limited GFPMYR mRNA as described previously (Yudin et al., 2008). Briefly, transfected DRG cultures were analyzed 48–72 h after transfection for intraaxonal GFP fluorescence. A Leica TCS/SP2 confocal microscope fitted with an environmental chamber was used for scanning and photobleaching; cultures were maintained at 37°C, 5% CO2 over the duration of the imaging sequences. Terminal axons were imaged every 30 s for 2 min with a 488 nm laser (7% power) to establish a baseline fluorescence intensity (“prebleach”). An ROI consisting of terminal axon (≥400 μm from the cell body) was then exposed to 75% power 488 nm laser over 40 frames at 1.6 s intervals for photobleaching. Recovery was monitored by 7% power 488 nm laser excitation 60 s for a 20 min duration (“postbleach”). GFP emission was collected with a band filter of 498–530 nm for each sequence. To test for translation dependence, 150 μg/ml cycloheximide (Sigma) was added to the DRG culture 30 min before the FRAP sequence. ImageJ software was used to calculate the average pixels per square micrometer in the ROIs of the raw confocal images. Signals were normalized between cells and transfections by defining prebleach intensity of 100% and t = 0 min postbleach intensity as 0%.
Analyses of neurite growth.
Growth of DRG neurites, which have been shown previously to have axonal and not dendritic characteristics (Smith and Skene, 1997; Zheng et al., 2001; Vuppalanchi et al., 2010), was tested in cultures transfected with siRNAs and/or amphoterin expression constructs at 72 h after transfection. For siRNAs, transfected neurons were identified by siGLO detecting reagent added during transfection. For AMPH-GFP and GFP expression constructs, transfected neurons were identified by GFP fluorescence. Overall axonal length was assessed from digital images using ImageJ (Donnelly et al., 2011). At least three separate culture preparations and transfections were analyzed for each condition.
Statistical analyses.
The GraphPad Prism 4 software package was used for all statistical analyses. Student's t test was used to compare two means of independent groups in the axonal growth assays and fluorescent intensity comparisons from FISH/IF images. Student's t test was used to compare between two groups for the RT-qPCR studies. For FRAP data, a one-way ANOVA with Bonferroni post hoc test was used to test the significance of multiple independent groups (Merianda et al., 2013a).
Results
Amphoterin protein increases in axons of injury-conditioned neurons
We showed previously that axonal levels of amphoterin mRNA are not changed after a 4 h exposure to neurotrophins, myelin-associated glycoprotein, or semphaphorin-3A (Willis et al., 2007). Although we had not seen changes in axonal amphoterin mRNA after crush injury in DRGs and sciatic nerve (Yoo et al., 2013), increased amphoterin levels were reported after spinal cord injury (Kawabata et al., 2010; Chen et al., 2011; Fang et al., 2014). Therefore, we investigated whether intraaxonal levels of amphoterin protein might change with axonal injury. For this, we used L4–L5 DRG neurons cultured from animals that had been conditioned 7 d previously by unilateral sciatic nerve crush to determine whether injury conditioning alters axonal amphoterin mRNA levels. Dissociated DRGs were cultured on a porous membrane insert for isolation of axonal versus cell body compartments (Zheng et al., 2001). Using protein lysates isolated from axonal versus cell body compartments for immunoblotting, amphoterin protein appeared to shift from being cell body predominant in the naive neurons to being axon predominant in the injury-conditioned neurons (Fig. 1A). Probing blots of cell body and axonal protein lysates for Erk 1/2, which is not synthesized in axons at appreciable level in adult DRG axons (Willis et al., 2007), showed equivalent protein loading for the naive and injury-conditioned protein lysates. Exposure matched immunofluorescent images of cultured neurons showed a similar trend, with higher cell body signals in naive neurons and higher axonal signals in injury-conditioned neurons (Fig. 1B,C). Amphoterin protein also showed a shift in levels in vivo after injury. Similar to the DRG cultures, L4–L5 DRGs from naive animals had higher amphoterin protein signals than those from 7 d injured animals (Fig. 1D, top, E). Sciatic nerve sections taken proximal to the crush site showed the opposite result, with amphoterin protein much higher in axons of the 7 d injured than in the naive sciatic nerve (Fig. 1D, bottom, E).
Figure 1.
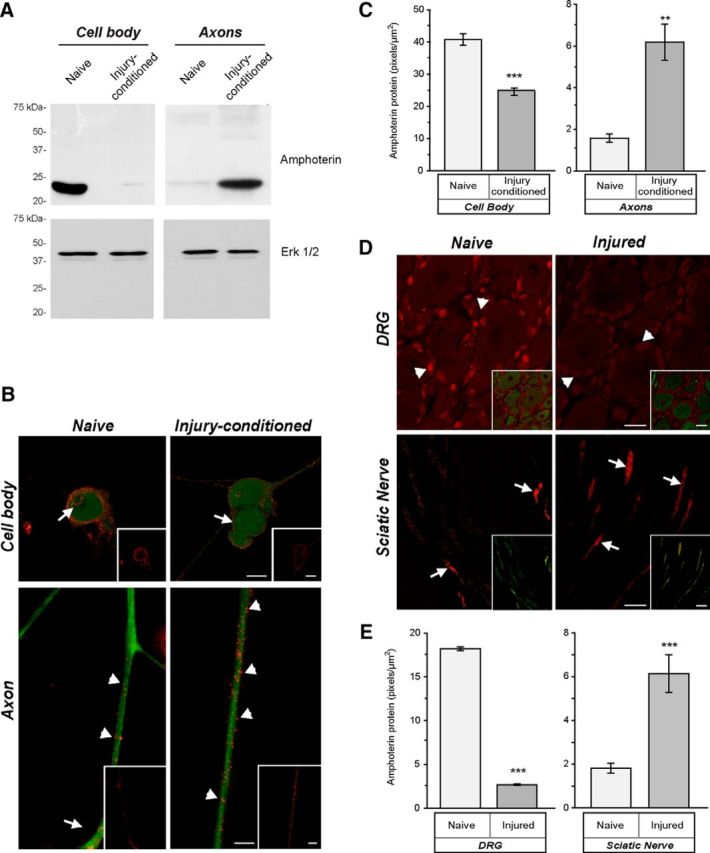
Amphoterin protein is enriched in axons of injury-conditioned neurons. A, Axonal versus cell body compartments were used for isolation of protein from DRG cultures. By immunoblotting, amphoterin protein is shown to be cell body predominant in naive DRG cultures and axon predominant in injury-conditioned DRG cultures. Replicate blots probed with anti-Erk 1/2 shows approximately equal loading between the naive and injury-conditioned lysates. B, C, Representative, exposure-matched epifluorescent images of naive and injury-conditioned DRG cultures stained for amphoterin (red) and NF (green) protein are shown in B. Consistent with the immunoblotting in A, amphoterin protein is higher in cell bodies of naive compared with injury-conditioned neurons. There is a clear increase in amphoterin protein in the axons of the injury-conditioned neurons and the signal appears concentrated along the periphery of the axon (arrowheads). Cell body signals are noted along the cell periphery (arrows) and in the nucleus of the naive neurons, whereas the injury-conditioned neurons show predominantly cell periphery signals (arrows). Quantification immunoreactivity from exposure-matched image sets in C shows increased levels of amphoterin protein in axons of injury-conditioned versus naive DRG cultures, whereas cell bodies show a decrease in amphoterin protein in injury-conditioned versus naive DRG cultures (n ≥ 25 for cell body and n ≥ 30 for axons from at least 3 separate experiments; **p ≤ 0.01, ***p ≤ 0.001 by Student's t test) Scale bars, 10 μm. D, Representative, exposure-matched confocal images of L4 DRGs and sciatic nerves of naive and 7 d postsciatic nerve crush animals with the amphoterin protein in red and the NF protein in green. Similar to the cultured neurons in B, there is a clear increase in axonal amphoterin protein (arrows) in the injured compared with naive nerve. Amphoterin protein signals in the neuronal perikaryon also decrease in the injured compared with naive DRGs. Satellite cells in the DRG show a strong, apparently nuclear signal for amphoterin that similarly declines with injury (arrowheads). E, Quantification of amphoterin protein signals from the tissue sections comparing axonal levels in naive versus injured nerve and neuronal cell body levels in naive versus injured DRGs (n ≥ 25 for cell body and n ≥ 30 for axons from at least 3 separate experiments; **p ≤ 0.01, ***p ≤ 0.001 by Student's t test). Scale bars: DRG panels, 25 μm; sciatic nerve panels, 10 μm.
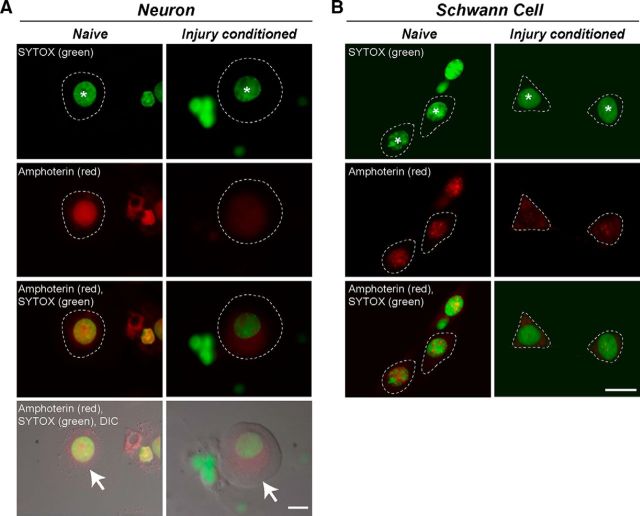
As an HMG protein, amphoterin is regarded as a nuclear protein. The DRG cultures and sections analyzed in Figure 1 showed varying degrees of nuclear amphoterin signal in the perikaryon of the DRG neurons and satellite cells. Therefore, we used confocal microscopy in DRG cultures colabeled with a nuclear stain to get a better assessment of amphoterin's distribution in the DRG cell body. Confocal sections through the center of the neuronal nucleus in DRG cultures showed a strong intranuclear amphoterin signal in the naive cultures, but this appeared to decrease in cultures from 7 d injury-conditioned animals (Fig. 2A). Merged images and overlay with DIC showed that cytoplasmic amphoterin immunoreactivity increased in the DRG neurons from injury-conditioned compared with naive animals (Fig. 2A, bottom two rows). Schwann cells in these cultures showed a similar decrease in nuclear staining, but not to the same extent as seen in the neurons (Fig. 2B). These data suggest that the amphoterin protein seen in the axons after injury conditioning could represent a separate cohort from the amphoterin population in the neuronal cell body.
Figure 2.
Nuclear amphoterin protein shifts to cytoplasm in injury-conditioned DRGs. Confocal microscopy was used to gain a better assessment of amphoterin protein localization in DRG neurons and glial cells. A, Representative single optical planes through center of neuronal perikaryon and nucleus of naive (left column) and injury-conditioned (right column) DRG cultures that were immunostained for amphoterin protein and counterstained with SYTOX to highlight the nucleus. The optically sectioned nucleus is marked with an asterisk in the first row. The cytoplasm of the neurons is outlined with a dashed line in the fluorescent images and marked with arrows in the images merged with DIC (fourth row). Note that amphoterin signal is strongly nuclear in the naive DRG neuron but shifts to more cytoplasmic in the injury-conditioned DRG neuron. B, Representative single optical planes taken through the center of the Schwann cells in these DRG cultures (left, naïve; right, injury-conditioned). The optically sectioned Schwann cell nuclei are marked with asterisks in the first row. The cytoplasm of the Schwann cell is outlined with a dashed line in the images. The Schwann cell amphoterin signal is strongly nuclear in the naive DRG neuron, but also shifts to more cytoplasmic in the Schwann cells after injury-conditioning. Scale bars, 10 μm.
Amphoterin mRNA is constitutively transported into DRG axons
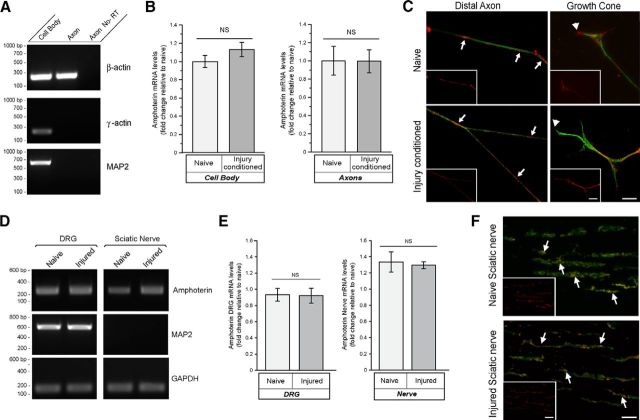
Because the levels of amphoterin protein increased in the axons with injury conditioning, we investigated whether intraaxonal amphoterin mRNA levels or aggregation might change with axotomy. We used axonal isolates as in Figure 1 that were validated for purity based on absence γ-actin and MAP2 mRNAs, but showed clear amplification of β-actin mRNA (Fig. 3A). We were not able to detect any significant differences in amphoterin mRNA levels comparing the axonal compartment of naive versus injury-conditioned neuron cultures by RT-qPCR (Fig. 3B). Similarly, the RT-qPCR showed no significant differences comparing the cell body compartment mRNA levels of the naive versus injury-conditioned neuron cultures (Fig. 3B). Axonal amphoterin mRNA FISH signal intensity also appeared similar between the injury-conditioned and naive DRG cultures (Fig. 3C). Amphoterin mRNA FISH signals showed granular profiles, as seen with other axonally transported mRNAs, and there was no apparent difference in these profiles with injury conditioning. RT-qPCR analyses of DRG and sciatic nerve samples showed that the amphoterin mRNA levels were comparable to the GAPDH control bands in naive and 7 d post injury in vivo (Fig. 3D). RT-qPCR confirmed no significant changes in amphoterin mRNA in the sciatic nerve or L4–L5 DRGs after crush injury (Fig. 3E). Consistent with the axonal nature of the neuronal processes traversing the sciatic nerve (Zheng et al., 2001), no MAP2 mRNA was detected in the sciatic nerve samples, but there was a clear signal in the DRGs where the neuronal cell bodies reside (Fig. 3D). Given that the axonal levels of amphoterin mRNA do not appear to change in vitro or in vivo after axotomy, an intraaxonal increase in translation of amphoterin mRNA after injury could account for the increase in axonal amphoterin protein demonstrated in Figure 1.
Figure 3.
Axonal levels of amphoterin mRNA do not change with injury. A, B, Neuronal cultures prepared from 7 d injury-conditioned versus naive DRGs were used for fractionation of cell body versus axonal RNA. RT-qPCR shows β-actin in both cell body and axonal preparations, but γ-actin and MAP2 RT-qPCR products were only detected in the cell body preparations (A). By RT-qPCR, amphoterin mRNA levels show no significant differences comparing cell body preparations or axonal preparations from the naive versus injury-conditioned neurons (B). C, Representative exposure-matched images of FISH/IF for amphoterin mRNA (red) and NF protein (green). Large panels show merged images and insets show only the RNA signal. There are overall comparable signals for amphoterin mRNA in axon shaft and growth cones of the naive versus injury-conditioned DRG neurons. Axonal amphoterin mRNA appears granular in the axon shaft (arrows) and growth cone (arrowheads) Scale bars, 10 μm. D, E, Representative RT-qPCR for naive versus 7 d postsciatic nerve crush injury for L4–L5 DRGs and sciatic nerve proximal to crush site. GAPDH was used as a loading control and the absence of MAP2 mRNA amplification from nerve samples confirms axonal nature of these nerve RNA preparations (D). RT-qPCR showed no significant alterations in amphoterin mRNA in the L4–L5 DRGs or sciatic nerve in vivo comparing the naive versus 7 d axotomized samples (E). F, Representative confocal XYZ images of FISH/IF signals for naive and 7 d post-crush-injury sciatic nerve are shown (matched for laser power, PMT gain/offset). Amphoterin mRNA is displayed in red and NF protein is shown in green, with merged mRNA and NF in main images and mRNA only for insets. Intraaxonal amphoterin mRNA (arrows) is seen in both the naive and injured nerve, with no apparent differences in abundance between the two. Scale bars, 10 μm.
A 60 nt 3′-UTR element confers axonal localization for amphoterin mRNA
We showed previously that the 3′-UTR of amphoterin mRNA is sufficient for localization of heterologous mRNAs into sensory axons, both in culture and in vivo (Donnelly et al., 2013). To determine whether a subregion of this 3′-UTR might be responsible for axonal localization, we transfected DRG neurons with GFPMYR plus varying lengths of the amphoterin UTR and used FISH to test for axonal localizing capability. DRGs expressing GFPMYR plus full 3′-UTR of amphoterin (nt 797–1238; GFPMYR3′-amph797- 1238) showed robust intraaxonal GFP mRNA FISH signal (Fig. 4Aa). With GFPMYR constructs plus 3′-UTR consisting of amphoterin nt 993–1238 or 797–993 (GFPMYR3′-amph993–1238 and GFPMYR3′-amph797–993, respectively), axonal signals for the antisense GFP mRNA probe were no different from the sense GFP mRNA probe in this exposure-matched image series (Fig. 4Ab,Ac vs Ae). However, intraaxonal GFP signals were clearly seen when amphoterin nt 738–797 was included in the reporter construct 3′-UTR (GFPMYR3′-amph738–797; Fig. 4Ad). Quantification of the axonal GFP RNA signals over multiple transfection experiments showed significantly greater pixel intensity in axons for GFPMYR3′-amph738–797 and GFPMYR3′-amph738–1238 compared with GFPMYR3′-amph797–993 and GFPMYR3′-amph993–1238 (Fig. 4B). When quantified across multiple culture preparations, these normalized axon FISH signal intensities show some background fluorescence for the GFPMYR3′-amph797–993 and GFPMYR3′-amph993–1238-transfected neurons, as we have reported previously for GFP reporter RNAs without any localizing elements (Ben-Yaakov et al., 2012; Merianda et al., 2013b). Together, these data indicate that the most proximal 60 nt of amphoterin's 3′-UTR contains a motif that is both necessary and sufficient for axonal localization in DRG neurons.
Figure 4.
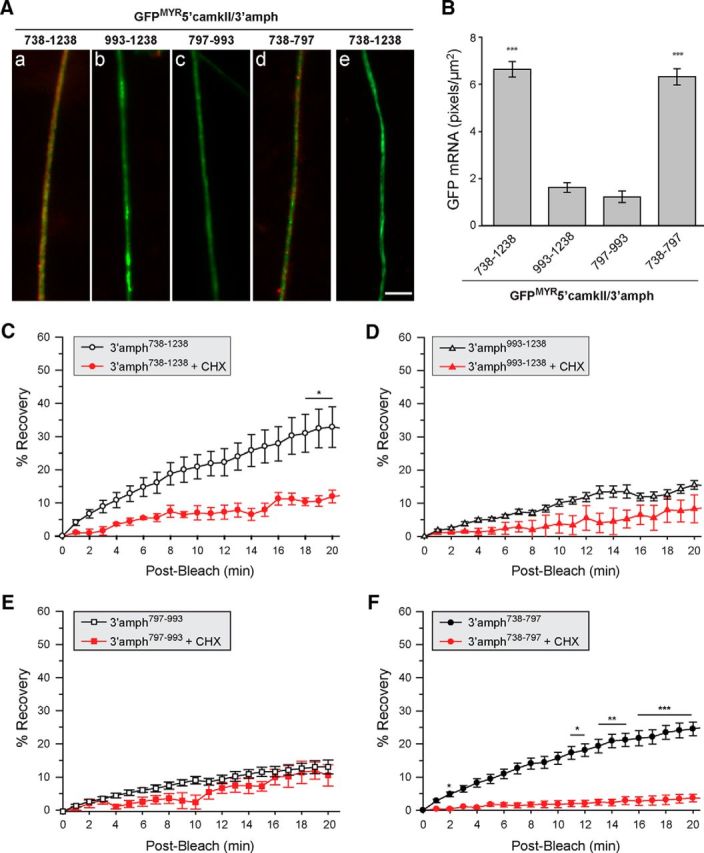
A 60 nt segment of amphoterin 3′-UTR is sufficient for axonal localization in DRG neurons. A, Representative exposure-matched FISH/IF images for axons of neurons transfected with indicated GFPMYR-5′camkII/3′-amph constructs. GFP mRNA is shown in red and NF protein in green (antisense probes are shown in Aa–Ad and sense cRNA probe is shown in Ae). Quantifications of GFP RNA FISH signals in DRG axons across multiple experiments are shown in B as average ± SEM (n ≥ 30 over three independent experiments; ***p ≤ 0.001 by Student's t test). Scale bar, 10 μm. C–F, Postbleach signals from FRAP studies for distal axons of DRG neurons transfected with the same GFPMYR constructs as in A are shown. In each case, the ROI was at least 400 μm from the cell body. Average signal intensity normalized to prebleach and postbleach signal for each experiment is shown; error bars indicate the SEM of normalized data (n ≥ 7 over at least 3 independent experiments). CHX, Cultures pretreated with 150 μg/ml cycloheximide for 20 min before photobleaching (n ≥ 4 over at least 3 independent experiments; *p ≤ 0.05, **p ≤ 0.01, and ***p ≤ 0.001 for indicated time points compared with t = 0 min postbleach by repeated-measures ANOVA with Bonferroni post hoc comparisons).
To determine whether this axonally localizing segment of amphoterin's 3′-UTR could also support intraaxonal translation, we used FRAP of transfected DRG neurons. The myristoylation motif (MYR) in the GFP was shown to markedly decrease diffusion of newly synthesized protein in neuronal processes (Aakalu et al., 2001); translation dependence for fluorescent recovery of such diffusion-limited GFP in axons has been illustrated by application of protein synthesis inhibitors before FRAP imaging (Yudin et al., 2008). As described previously (Donnelly et al., 2013), GFPMYR3′-amph738–1238 showed robust recovery after photobleaching comparing time lapse sequence with postbleach (t = 0) signal intensity, but no significant recovery was seen when protein synthesis was inhibited using cycloheximide (Fig. 4C). Consistent with the data shown in Figure 4, A and B, DRGs transfected with GFPMYR3′-amph797–993 or GFPMYR3′-amph993–1238 showed no significant increase in axonal GFP signals over the 20 min postbleach interval (Fig. 4D,E). There were also no significant differences between the control and cycloheximide-treated cultures at each time point for GFPMYR3′-amph797–993 or GFPMYR3′-amph993–1238 (Fig. 4D,E), indicating that the modestly increasing axonal GFP signals were the result of GFP mRNA translation. Conversely, axonal signals for neurons expressing GFPMYR with amphoterin nt 738–797 (GFPMYR3′-amph738–797) showed recovery after photobleaching comparable to the neurons expressing GFPMYR3′-amph738–1238 (Fig. 4A,D). Similar to the GFPMYR3′-amph738–1238-expressing neurons, the fluorescence recovery for GFP signals in GFPMYR3′-amph738–797 was blocked by the translation inhibitor cycloheximide (Fig. 4F). Therefore, the axonally localized GFPMYR3′-amph738–797 can be translated comparable to reporter carrying the full length of the amphoterin 3′-UTR.
Amphoterin UTR sequences are sufficient for axonal localization and translational regulation in vivo
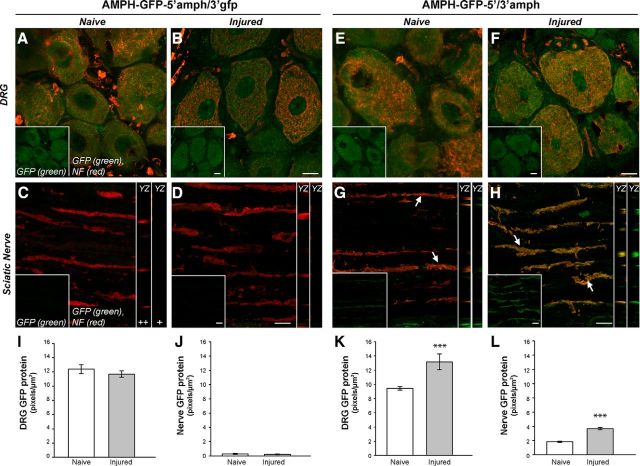
Because studies in Figure 3 showed amphoterin mRNA localization in sciatic nerve axons, we investigated whether the UTR sequences of amphoterin might be sufficient for axonal localization in vivo. For this, we generated LV to express an amphoterin-AcGFP fusion protein (AMPH-GFP) with amphoterin 5′-UTR and amphoterin or GFP 3′-UTRs (LV-AMPH-GFP-5′/3′-amph and LV-AMPH-GFP-5′amph/3′gfp, respectively). We have shown previously that the 5′ and 3′-UTRs of GFP have no axonally localizing activity and reporter mRNAs carrying these are restricted to the cell body (Merianda et al., 2013a and 2013b). Seven days after injection of LV into L4–L5 nerve roots bilaterally, animals were subjected to unilateral sciatic nerve crush. The AMPH-GFP-5′/3′-amph- and AMPH-GFP-5′amph/3′gfp-expressing animals showed robust GFP fluorescence in the neuronal cell bodies of the L4–L5 DRGs (Fig. 5A,B, E,F). GFP signals seen at the injection site were also comparable between the AMPH-GFP-5′/3′-amph and AMPH-GFP-5′amph/3′gfp expressing animals (data not shown). For the distal nerve, only the AMPH-GFP-5′/3′-amph-expressing animals, and not the AMPH-GFP-5′amph/3′gfp-expressing animals, showed GFP signals (Fig. 5C,D, G,H). Orthogonal sections showed that GFP signal in the AMPH-GFP-5′/3′-amph-expressing animals was clearly intraaxonal (Fig. 5G,H, right). Intra-axonal GFP signals were clearly present in the uninjured nerves of the AMPH-GFP-5′/3′-amph-expressing animals, but at much lower levels than the 7 d nerve crush samples (Fig. 5G vs H). Analyses of multiple samples for colocalization of NF and GFP signals showed a Pearson's coefficient of 0.63 ± 0.18 for AMPH-GFP-5′/3′-amph compared with 0.043 ± 0.27 for AMPH-GFP-5′amph/3′gfp expressing animals (n = 16 each).
Figure 5.
Amphoterin 3′-UTR is needed for axonal localization in vivo. A–H, Representative, exposure-matched confocal images of L4–L5 DRGs and distal sciatic nerve for animals transduced with LV encoding amphoterin-GFP fusion protein plus 5′-UTR of amphoterin and 3′-UTRs of GFP or amphoterin are shown [AMPH-GFP-5′amph/3′gfp (A–D) and AMPH-GFP-5′/3′-amph (E–H), respectively), with NF protein in red and GFP in green. All samples are 14 d post-LV transduction of L4–L5 nerve roots; naive (A, C, E, G) and 7 d after sciatic nerve crush (B, D, F, H) samples are shown. The top shows confocal XYZ images of L4–L5 DRGs, with merged channels in main panel and GFP signal only in the insets; bottom shows XYZ projections of sciatic nerve as merge of GFP and NF signals in the main panels and GFP only in the inset (lower left of each panel). YZ orthogonal projections of GFP + NF (++) and GFP only (+) are shown as strip images adjacent to each sciatic nerve image in C. GFP signals are seen in the DRGs cell bodies expressing either AMPH-GFP-5′amph/3′gfp (A, B) or AMPH-GFP-5′/3′-amph (C, D). However, the sciatic nerve only shows GFP signals in AMPH-GFP-5′/3′-amph-expressing animals (arrows), with a clear increase after nerve crush injury (G, H) Scale bars: DRG panels, 25 μm; sciatic nerve panels, 10 μm; orthogonal projections are at same scale as main panels. I–L, Quantification of GFP signal intensity is shown for DRG (I, K) and sciatic nerves (J, L) for AMPH-GFP-5′amph/3′gfp-expressing animals (I, J) and AMPH-GFP-5′/3′-amph-expressing animals (K, L). Values represent mean intensity ± SEM from images matched for exposure, gain, offset, and after processing (n ≥ 30 from at least three separate experiments; **p ≤ 0.01; ***p ≤ 0.001 by Student's t test).
Quantitation across multiple animals showed a significant increase in GFP signals in the DRG and distal nerves of the AMPH-GFP-5′/3′-amph-expressing animals after nerve crush compared with the uninjured animals (Fig. 5K,L). There was no change in GFP signals in the DRGS of the AMPH-GFP-5′amph/3′gfp-expressing animals with nerve crush injury (Fig. 5I). These data indicate that the nontranslated regions of amphoterin mRNA provide for both axonal mRNA targeting and translational regulation after nerve injury.
Axonally generated amphoterin protein increases axonal growth in DRG neurons
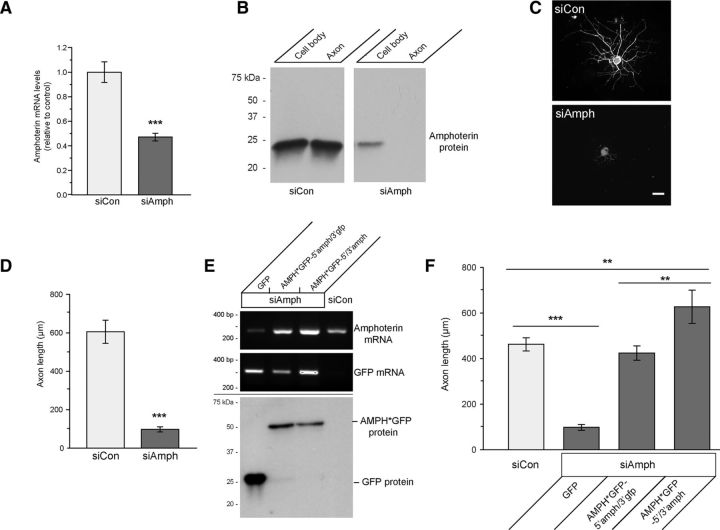
Exogenous amphoterin protein has been shown to increase neurite outgrowth (Huttunen and Rauvala, 2004; Hock et al., 2007 and references within). To determine whether amphoterin expression might contribute to axonal growth in the adult DRG neurons used here, we used siRNA to deplete amphoterin in cultured DRG neurons. DRGs transfected with siRNA targeting amphoterin's coding sequence (siAmph) showed a significant depletion of amphoterin mRNA and protein (Fig. 6A,B). siAmph-transfected cultures also showed markedly reduced axon length compared with those transfected with nontargeting control siRNA (Fig. 6C,D).
Figure 6.
Amphoterin depletion decreases axonal outgrowth. A, B, siAmph mRNA-transfected DRG neuron cultures show significantly decreased amphoterin mRNA by RT-qPCR compared with nontargeting control (siCon) (A); there is also a clear decrease in amphoterin protein with siAmph compared with siCon in cell body and axonal preparations by immunoblotting (B). C, D, Representative images for NF immunofluorescence for DRG neurons transfected with siCon and siAmph (C); quantitation of the longest axon per neuron over multiple transfection experiments is also shown (D) for siAmph versus siCon; there is a marked reduction in axon length with depletion of amphoterin (***p ≤ 0.001 by Student's t test for n ≥ 30 over 3 independent experiments). Scale bar, 100 μm. E, F, siRNA-resistant Amphoterin-GFP constructs (AMPH*GFP) was used to test for potential off target effects of the siAmph. Cell-body-restricted (AMPH*GFP-5′amph/3′gfp) or axonally targeted (AMPH*GFP-5′/3′-amph) constructs were used to distinguish rescue effects of axonally synthesized AMPH*GFP. RT-qPCR shows rescue of amphoterin mRNA levels upon expression of AMPH*GFP-5′amph/3′gfp and AMPH*GFP-5′/3′-amph (E, top and middle, respectively). AMPH*GFP protein expression is comparable between the siAmph + AMPH*GFP-5′amph/3′gfp and siAmph + AMPH*GFP-5′/3′-amph transfected cultures (E, bottom). Length of longest axon in siAmph + GFP, AMPH*GFP-5′amph/3gfp, and AMPH*GFP-5′/3′-amph cotransfected cultures is shown in F. Both AMPH*GFP-5′amph/3′gfp and the AMPH*GFP-5′/3′-amph transfection rescue the axon growth deficit of siAmph transfection; however, axon lengths in the cultures expressing the axonally targeted AMPH*GFP-5′/3′-amph exceed those in control cultures (n ≥ 30 over 3 independent experiments; **p ≤ 0.01 and ***p ≤ 0.001 by Student's t test).
Although amphoterin protein was reduced in the axonal compartment of the DRGs transfected with siAmph (Fig. 6B), the siRNAs obviously do not selectively target the axonal cohort of amphoterin mRNA. To address the potential role(s) for the axonally synthesized amphoterin protein in axonal growth, we used transfection with “siRNA-resistant” amphoterin-GFP fusion protein constructs (AMPH*GFP) in an attempt to rescue any growth deficits from amphoterin depletion. For this, we restricted the AMPH*GFP-5′amph mRNA to the cell body using the 3′-UTR of GFP mRNA (AMPH*GFP-5′amph/3′gfp) or targeted the mRNA into axons using the 3′-UTR of amphoterin mRNA (AMPH*GFP-5′/3′-amph). In siAmph-transfected cultures, cotransfection with AMPH*GFP-5′amph/3′gfp and AMPH*-5′/3′-amph generated approximately equivalent levels of mRNA and protein expressed from these constructs in DRG neurons (Fig. 6E). The decrease in axonal growth with the siAmph transfection was rescued by both the cell-body-restricted AMPH*GFP-5′amph/3′gfp and axonally targeted AMPH*GFP-5′/3′-amph. However, axons in amphoterin-depleted neurons cotransfected with axonally targeted AMPH*GFP-5′/3′-amph were consistently much longer than those in neurons cotransfected with the cell-body-restricted AMPH*GFP-5′amph/3′gfp (Fig. 6F). We next compared cell-body-restricted AMPH*GFP-5′amph/3′gfp to axonally targeted AMPH*GFP-5′amph/3′-amph738–797, where we could distinguish the endogenous amphoterin from AMPH*GFP. There were no differences in expression of the endogenous amphoterin, indicating that neither the cell-body-restricted or axonally targeted AMPH*GFP resulted in an compensatory alterations in expression of endogenous amphoterin in these DRG cultures (data not shown). We cannot exclude the possibility that trafficking of some AMPH*GFP from the cell body in the siAmph-transfected cultures may account for the rescue seen with the restricted AMPH*GFP-5′amph/3′gfp mRNA expression. In culture preparations, axonal signals for AMPH-GFP were seen in replated neurons overexpressing the cell-body-restricted AMPH-GFP-5′amph/3′gfp constructs (data not shown). This suggests that some cell-body-synthesized amphoterin protein can traffic into axons, at least over the few-hundred-micron distances that axons extend in vitro.
Because both the axonally targeted and cell-body-restricted AMPH*GFP constructs appeared to rescue the axon growth deficit seen with depletion of endogenous amphoterin, we investigated whether DRG axonal growth might be altered with simple overexpression of cell-body-restricted versus axonally targeted AMPH-GFP. Transfection with AMPH-GFP carrying the amphoterin 5′-UTR and nontargeting GFP 3′-UTR (AMPH-GFP-5′amph/3′gfp) did not alter growth above control GFP transfection. Conversely, expression of AMPH-GFP-5′/3′-amph738–1238 nearly doubled the axon length (Fig. 7A,B). In situ hybridization showed approximately equivalent AMPH-GFP mRNA levels in cell bodies of AMPH-GFP-5′/3′-amph738–1238 and AMPH-GFP-5′amph/3′gfp-transfected cultures; however, AMPH-GFP mRNA was only seen in the axons of the AMPH-GFP-5′/3′-amph738–1238-expressing DRGs, not the AMPH-GFP-5′amph/3′gfp-expressing DRGs (data not shown). On analysis of additional AMPH-GFP constructs with varying 5′ and 3′-UTRs, AMPH-GFP mRNA only localized into axons when nt 738–797 from amphoterin mRNA was included in the 3′-UTR (Fig. 7C–E), emphasizing the functional role of this 60 nt segment in targeting amphoterin mRNA into axons. It should be noted that the 3′Amph738–797 containing mRNAs would use the downstream SV40 poly-adenylation site in the pcDNA3.1 used for cloning here, so these mRNAs would have a 3′-UTR of at least 188 nt.
Figure 7.
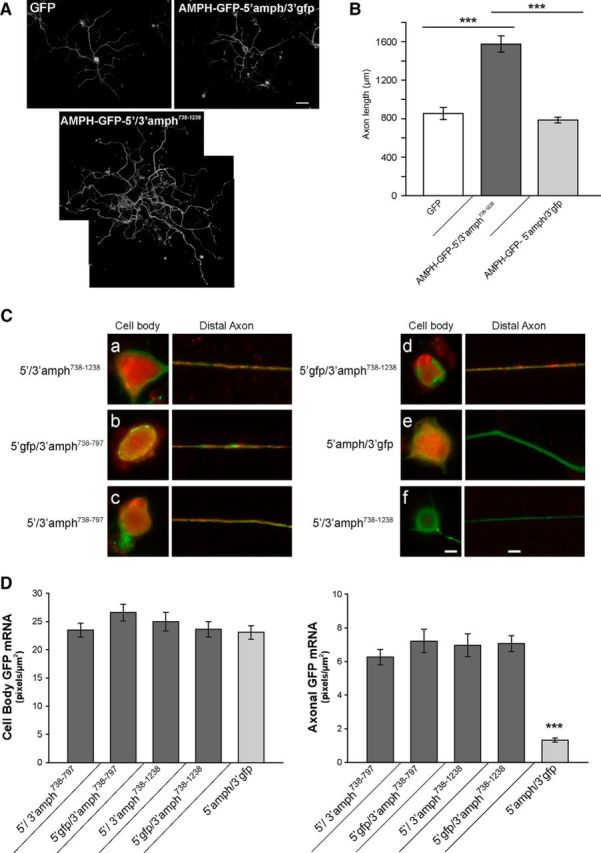
Overexpression of axonally targeted amphoterin mRNA increases axon growth. A, B, Representative montage images of NF-H-immunostained DRG neurons are shown in A for transfections with GFP, AMPH-GFP-5′/3′-amph738–1238, AMPH-GFP-5′amph/3′gfp. Average length of the longest axon ± SEM for the neurons expressing GFP, AMPH-GFP-5′/3′-amph738–1238, AMPH-GFP-5′amph/3′gfp is shown in B (n ≥ 30 neurons in 3 separate experiments; ***p ≤ 0.001 by Student's t test). Scale bar, 100 μm. C, D, Exposure-matched FISH/IF images for cell body (left) and distal axon (right) of DRG neurons transfected with AMPH-GFP plus the indicated 5′ and 3′-UTRs are shown in C. GFP mRNA is shown red and NF protein is shown in green (a–e show antisense GFP probes and f shows sense GFP probe). Axonal GFP mRNA is only seen for AMPH-GFP that includes nt 738–797 from amphoterin mRNA 3′-UTR. Quantitation of GFP mRNA intensity in DRG axons and cell body is shown as average ± SEM for cell bodies and axons as indicated (D). There are no significant differences in cell body GFP RNA levels between the different UTR-containing constructs. All constructs containing amphoterin nt 738–797 and 738–1238 show significantly higher GFP mRNA in axons than the AMPH-GFP-5′amph/3′gfp. The axonal levels for AMPH-GFP-5′amph/3′gfp are comparable to those for GFPMYR3′-amph797–993 and GFPMYR3′-amph993–1238 shown in Figure 3F (n ≥ 30 processes in three experiments; ***p ≤ 0.001 by Student's t test). Scale bars: cell body, 20 μm; distal axon, 10 μm.
Discussion
The results of an increasing number of studies point to the role of localized protein synthesis for development, maintenance, and regeneration of mammalian axons (Jung et al., 2012; Perry and Fainzilber, 2014). The work presented here indicates that translation of axonal amphoterin mRNA is enhanced after injury conditioning. A 60 nt element in amphoterin's 3′-UTR is responsible for its localization in cultured sensory neurons and in vivo. Localization of neuronal mRNAs has most often been ascribed to structures in their UTRs, with the 3′-UTR more frequently containing localizing activities than the 5′-UTR (Gomes et al., 2014). Similar to our findings with amphoterin mRNA, localized translational regulation after injury has been documented for a few axonal mRNAs, including Importin β1, RanBP1, and Stat3α (Hanz et al., 2003; Yudin et al., 2008; Ben-Yaakov et al., 2012). However, translational activation of these mRNAs is linked to the initial injury response with increased axoplasmic Ca2+. Moreover, their protein products are linked to retrograde cell body responses rather than affecting localized growth of the axons directly, as we see with amphoterin. Therefore, different trans-acting factors are likely responsible for the translational control of amphoterin mRNA in the DRG axons. With the constitutive transport of amphoterin mRNA into the PNS axons studied here, identifying the nature of those regulatory factors and mechanisms for their translation-promoting activities may provide future tools to manipulate axon growth. miRNAs have been implicated in posttranscriptional regulation of some axonal mRNAs, but we were not able to detect any miRNA recognition sequences in nt 738–797 of amphoterin mRNA, where we have demonstrated that a functional motif(s) resides.
Several studies have linked amphoterin to neural growth, showing that exogenous amphoterin protein increases neurite outgrowth from neuron-like cell lines, embryonic stem cells, and primary neurons (Merenmies et al., 1991; Hori et al., 1995; Huttunen et al., 1999; Huttunen et al., 2002; Saleh et al., 2013). Regulation of cell migration by amphoterin has also been implicated in tumor invasiveness (Taguchi et al., 2000; Riuzzi et al., 2006). Amphoterin mRNA was shown to localize to the cell periphery of C6 glioma cells, but the functional significance of this localization was not tested (Punnonen et al., 1999; Fages et al., 2000). Because the axonally targeted AMPH-GFP-5′/3′-amph mRNA increases axon growth well above that seen with a cell-body-restricted AMPH-GFP-5′amph/3′gfp, our data indicate that locally synthesized amphoterin protein functions as a growth-associated protein.
Amphoterin was initially identified as a 30 kDa heparin-binding protein from brain that supports neurite outgrowth. Sequencing of the amphoterin cDNA predicted a dipolar protein with N-terminal basic and C-terminal acidic domains typical of HMG proteins (Merenmies et al., 1991). As an HMG protein, amphoterin localizes to the nucleus and mice lacking this HMG protein show early postnatal lethality due to hypoglycemia (Calogero et al., 1999). Amphoterin levels are highest in embryonic cells, but expression is maintained into adulthood (Rauvala and Pihlaskari, 1987) and it appears ubiquitously expressed across different cell types (Hock et al., 2007). In the nucleus, HMG proteins bind to the minor groove of DNA, where they were thought to assist with organization of chromatin and nucleosomes (Wolffe, 1999). However, amphoterin/HMGB1 knock-out mice show altered transcriptional responses to glucocorticoids, indicating that the nuclear amphoterin protein contributes to more specific transcriptional regulation than would be predicted from a general chromatin-binding protein (Calogero et al., 1999). Interestingly, cytoplasmic amphoterin protein can be secreted through nonclassical pathways from living cells (Gardella et al., 2002) or released from dying cells (Scaffidi et al., 2002; Bell et al., 2006). This extracellular amphoterin protein has an entirely different function than its interactions with nucleic acids.
Hyperacetylation of amphoterin in monocytes and macrophages is associated with its translocation from the nucleus (Bonaldi et al., 2003). In macrophages, LPS- and interferon-induced activation of Jak and Stat1 is needed for amphoterin's hyperacetylation (Lu et al., 2014). Both secreted amphoterin protein and that released from dying cells have been implicated as proinflammatory factors (Lotze and Tracey, 2005). This could also be true of the neuronal amphoterin protein because the nuclear amphoterin protein rapidly redistributes to the neuronal cytoplasm during ischemia (Faraco et al., 2007). Injection of exogenous amphoterin into brain triggers the expression of inflammatory cytokines and increased sensitivity to ischemia (Faraco et al., 2007) and rats treated with glycyrrhizin, an inhibitor of amphoterin, are partially protected from brain injury (Gong et al., 2014). Here, the amphoterin protein shows both cytoplasmic and nuclear localization in neuronal cell body, but the majority of the cell body protein becomes cytoplasmic in injury-conditioned neurons (Fig. 3A). Failure of the cell-body-restricted AMPH-GFP-5′amph/3′gfp to show increased AMPH-GFP protein expression in the DRG after axotomy (Fig. 5) suggests that the shift from nuclear to cytoplasmic amphoterin predominance in the injury-conditioned neuronal perikaryon is not due to translational regulation of amphoterin mRNA in the cell body as we see in axons. This could point to two distinct cohorts of amphoterin protein that the DRG neuron can draw from, with the distinction being driven by the axonal localization and translational control of amphoterin mRNA versus protein trafficking. It is intriguing to speculate whether localizing amphoterin mRNA represents a general cellular mechanism to overcome the nuclear targeting of amphoterin protein. Nonetheless, we cannot rule out the possibility that some of the intraxonal amphoterin protein derives from cell body protein transport/trafficking based on our studies. Future work will be needed in vivo, where the transport rates across the distances separating the distal injured axon and cell body can help to distinguish localized synthesis and protein trafficking.
Along the cell surface, amphoterin elicits intracellular actions through binding to Toll-Like Receptors 2 and 4 (TLR2 and TLR4, respectively) and/or receptor for Advanced Glycation End-products (RAGE) (Lotze and Tracey, 2005). Amphoterin was shown to be upregulated in the injured rodent spinal cord (Kawabata et al., 2010). This increase in amphoterin levels precedes the induction of proinflammatory cytokines in the cord (Chen et al., 2011). Several lines of evidence support that amphoterin's proinflammatory actions are generated by TLR whereas amphoterin's growth-promoting activities are generated by RAGE (Lotze and Tracey, 2005). However, RAGE, TLR2, and TLR4 also show increased levels in the injured spinal cord (Kawabata et al., 2010; Chen et al., 2011), so it is not clear whether these proinflammatory effects of amphoterin after CNS injury are mediated by TLRs or RAGE. In reptiles and fish, in contrast to mammals, the injured spinal cord can spontaneously regenerate and this may provide some insight. Geckos were shown to express two HMGB1 isoforms, gHMGB1a (paralog of rat amphoterin/HMGB1) and gHMGB1b (Dong et al., 2013). Similar to mammals, gHMGB1b is upregulated after spinal cord injury. gHMGB1a and gHMGB1b bind to RAGE, but not TLR4 or TLR2 (Dong et al., 2013), which suggests that RAGE drives the effects of amphoterin in the spinal cord injury for geckos. HMGB1 is similarly upregulated in the injured zebrafish spinal cord and depletion of the HMGB1 decreases axon regeneration (Fang et al., 2014). The axonally generated rat amphoterin protein enhances axon growth from PNS neurons. Therefore, axonal amphoterin behaves similarly to HMGB1 in the injured spinal cord of the regeneration-competent gecko and zebrafish.
RAGE signaling has been implicated in neurite growth and regeneration. Transgenic mice expressing dominant-negative RAGE in neurons show significantly decreased myelinated fiber density and nerve conduction velocities at 21 d after sciatic nerve crush compared with their wild-type littermates (Rong et al., 2004b). The administration of soluble RAGE or antibodies to amphoterin similarly reduces sensory and motor function recovery after nerve crush injury (Rong et al., 2004a). Exogenous amphoterin also augments neurotrophic factor driven axon growth from adult DRG neurons and this effect requires expression of RAGE (Saleh et al., 2013). RAGE has been shown to localize to distal neurites and its downstream signaling activates pathways associated with neurite growth (Huttunen et al., 2002; Rong et al., 2004a; Kim et al., 2012; Saleh et al., 2013). Therefore, RAGE is well positioned to exert the growth-promoting effects of the locally synthesized amphoterin that we have studied here. Future studies will be needed to determine whether axonally generated amphoterin activates RAGE in an autocrine or paracrine fashion.
In summary, our studies point to a growth-promoting effect of the intraaxonally synthesized amphoterin protein. The redistribution of cell body amphoterin from nucleus to cytoplasm after axotomy versus translational upregulation to generate amphoterin protein in distal axons suggests that the neuron can draw from two different sources for this protein. This could be quite consistent with the multifunctional nature of amphoterin, providing both proinflammatory and growth-promoting activities by controlling when and where amphoterin is presented. Amphoterin has been suggested as causative for hypersensitivity after spinal nerve ligation (Shibasaki et al., 2010) and neuronal death after CNS injury (Faraco et al., 2007; Kawabata et al., 2010; Chen et al., 2011; Gong et al., 2014). Distinct spatial and temporal regulation of the different amphoterin sources in the neuron may contribute to these activities that seem contradictory to the proregenerative activities that we and others have seen. Conversely, Niemi et al. (2013) recently implicated macrophage infiltration of the DRGs in the increased axon regeneration seen after the same PNS injury conditioning that we have used here. Release of amphoterin from DRG neurons by relocalization from the nucleus to cytoplasm could participate in recruitment of macrophages. Therefore, both cohorts of neuronal amphoterin protein could effectively contribute to the increased PNS regeneration in injury conditioning. Future work will be needed to dissect the functions of these different subcellular sources of the neuronal amphoterin protein.
Footnotes
The initial experiments in this work were funded by the Paralyzed Veterans of America Spinal Cord Research Foundation (Grant 2520 to T.T.M.). Subsequent work was funded by the National Institutes of Health (Grant R01-NS041596 to J.L.T. and Grants P20-GM103446 and R21-NS085691 to S.Y.), the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (J.L.T. and A.B.), The Academy of Finland (H.R.), and the Sigrid Jusélius Foundation (H.R.). P.B.V. is supported by the National Institutes of Health (Grant R25-GM066526). J.L.T. is supported by the South Carolina SmartState Endowed Chairs Program. The eGFPMYR5′camkII plasmid was graciously provided by Erin Schuman.
The authors declare no competing financial interests.
References
- Aakalu G, Smith WB, Nguyen N, Jiang C, Schuman EM. Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron. 2001;30:489–502. doi: 10.1016/S0896-6273(01)00295-1. [DOI] [PubMed] [Google Scholar]
- Andreassi C, Zimmermann C, Mitter R, Fusco S, De Vita S, Saiardi A, Riccio A. An NGF-responsive element targets myo-inositol monophosphatase-1 mRNA to sympathetic neuron axons. Nat Neurosci. 2010;13:291–301. doi: 10.1038/nn.2486. [DOI] [PubMed] [Google Scholar]
- Bell CW, Jiang W, Reich CF, 3rd, Pisetsky DS. The extracellular release of HMGB1 during apoptotic cell death. Am J Physiol Cell Physiol. 2006;291:C1318–C1325. doi: 10.1152/ajpcell.00616.2005. [DOI] [PubMed] [Google Scholar]
- Ben-Yaakov K, Dagan SY, Segal-Ruder Y, Shalem O, Vuppalanchi D, Willis DE, Yudin D, Rishal I, Rother F, Bader M, Blesch A, Pilpel Y, Twiss JL, Fainzilber M. Axonal transcription factors signal retrogradely in lesioned peripheral nerve. EMBO J. 2012;31:1350–1363. doi: 10.1038/emboj.2011.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blesch A. Lentiviral and MLV based retroviral vectors for ex vivo and in vivo gene transfer. Methods. 2004;33:164–172. doi: 10.1016/j.ymeth.2003.11.005. [DOI] [PubMed] [Google Scholar]
- Bonaldi T, Talamo F, Scaffidi P, Ferrera D, Porto A, Bachi A, Rubartelli A, Agresti A, Bianchi ME. Monocytic cells hyperacetylate chromatin protein HMGB1 to redirect it towards secretion. EMBO J. 2003;22:5551–5560. doi: 10.1093/emboj/cdg516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calogero S, Grassi F, Aguzzi A, Voigtländer T, Ferrier P, Ferrari S, Bianchi ME. The lack of chromosomal protein Hmg1 does not disrupt cell growth but causes lethal hypoglycaemia in newborn mice. Nat Genet. 1999;22:276–280. doi: 10.1038/10338. [DOI] [PubMed] [Google Scholar]
- Chen KB, Uchida K, Nakajima H, Yayama T, Hirai T, Rodriguez Guerrero A, Kobayashi S, Ma WY, Liu SY, Zhu P, Baba H. High-mobility group box-1 and its receptors contribute to proinflammatory response in the acute phase of spinal cord injury in rats. Spine. 2011;36:2122–2129. doi: 10.1097/BRS.0b013e318203941c. [DOI] [PubMed] [Google Scholar]
- Dong Y, Gu Y, Huan Y, Wang Y, Liu Y, Liu M, Ding F, Gu X, Wang Y. HMGB1 protein does not mediate the inflammatory response in spontaneous spinal cord regeneration: a hint for CNS regeneration. J Biol Chem. 2013;288:18204–18218. doi: 10.1074/jbc.M113.463810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly CJ, Willis DE, Xu M, Tep C, Jiang C, Yoo S, Schanen NC, Kirn-Safran CB, van Minnen J, English A, Yoon SO, Bassell GJ, Twiss JL. Limited availability of ZBP1 restricts axonal mRNA localization and nerve regeneration capacity. EMBO J. 2011;30:4665–4677. doi: 10.1038/emboj.2011.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly CJ, Park M, Spillane M, Yoo S, Pacheco A, Gomes C, Vuppalanchi D, McDonald M, Kim HH, Kim HK, Merianda TT, Gallo G, Twiss JL. Axonally synthesized beta-actin and GAP-43 proteins support distinct modes of axonal growth. J Neurosci. 2013;33:3311–3322. doi: 10.1523/JNEUROSCI.1722-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fages C, Nolo R, Huttunen HJ, Eskelinen E, Rauvala H. Regulation of cell migration by amphoterin. J Cell Sci. 2000;113:611–620. doi: 10.1242/jcs.113.4.611. [DOI] [PubMed] [Google Scholar]
- Fang P, Pan HC, Lin SL, Zhang WQ, Rauvala H, Schachner M, Shen YQ. HMGB1 contributes to regeneration after spinal cord injury in adult zebrafish. Mol Neurobiol. 2014;49:472–483. doi: 10.1007/s12035-013-8533-4. [DOI] [PubMed] [Google Scholar]
- Faraco G, Fossati S, Bianchi ME, Patrone M, Pedrazzi M, Sparatore B, Moroni F, Chiarugi A. High mobility group box 1 protein is released by neural cells upon different stresses and worsens ischemic neurodegeneration in vitro and in vivo. J Neurochem. 2007;103:590–603. doi: 10.1111/j.1471-4159.2007.04788.x. [DOI] [PubMed] [Google Scholar]
- Gardella S, Andrei C, Ferrera D, Lotti LV, Torrisi MR, Bianchi ME, Rubartelli A. The nuclear protein HMGB1 is secreted by monocytes via a non-classical, vesicle-mediated secretory pathway. EMBO Rep. 2002;3:995–1001. doi: 10.1093/embo-reports/kvf198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes C, Merianda TT, Lee SJ, Yoo S, Twiss JL. Molecular determinants of the axonal mRNA transcriptome. Dev Neurobiol. 2014;74:218–232. doi: 10.1002/dneu.22123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong G, Xiang L, Yuan L, Hu L, Wu W, Cai L, Yin L, Dong H. Protective effect of glycyrrhizin, a direct HMGB1 inhibitor, on focal cerebral ischemia/reperfusion-induced inflammation, oxidative stress, and apoptosis in rats. PLoS One. 2014;9:e89450. doi: 10.1371/journal.pone.0089450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanz S, Perlson E, Willis D, Zheng JQ, Massarwa R, Huerta JJ, Koltzenburg M, Kohler M, van-Minnen J, Twiss JL, Fainzilber M. Axoplasmic importins enable retrograde injury signaling in lesioned nerve. Neuron. 2003;40:1095–1104. doi: 10.1016/S0896-6273(03)00770-0. [DOI] [PubMed] [Google Scholar]
- Hock R, Furusawa T, Ueda T, Bustin M. HMG chromosomal proteins in development and disease. Trends Cell Biol. 2007;17:72–79. doi: 10.1016/j.tcb.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori O, Brett J, Slattery T, Cao R, Zhang J, Chen JX, Nagashima M, Lundh ER, Vijay S, Nitecki D. The receptor for advanced glycation end products (RAGE) is a cellular binding site for amphoterin: mediation of neurite outgrowth and co-expression of rage and amphoterin in the developing nervous system. J Biol Chem. 1995;270:25752–25761. doi: 10.1074/jbc.270.43.25752. [DOI] [PubMed] [Google Scholar]
- Huttunen HJ, Rauvala H. Amphoterin as an extracellular regulator of cell motility: from discovery to disease. J Intern Med. 2004;255:351–366. doi: 10.1111/j.1365-2796.2003.01301.x. [DOI] [PubMed] [Google Scholar]
- Huttunen HJ, Fages C, Rauvala H. Receptor for advanced glycation end products (RAGE)-mediated neurite outgrowth and activation of NF-kappaB require the cytoplasmic domain of the receptor but different downstream signaling pathways. J Biol Chem. 1999;274:19919–19924. doi: 10.1074/jbc.274.28.19919. [DOI] [PubMed] [Google Scholar]
- Huttunen HJ, Kuja-Panula J, Rauvala H. Receptor for advanced glycation end products (RAGE) signaling induces CREB-dependent chromogranin expression during neuronal differentiation. J Biol Chem. 2002;277:38635–38646. doi: 10.1074/jbc.M202515200. [DOI] [PubMed] [Google Scholar]
- Jung H, Yoon BC, Holt CE. Axonal mRNA localization and local protein synthesis in nervous system assembly, maintenance and repair. Nat Rev Neurosci. 2012;13:308–324. doi: 10.1038/nrn3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H, Gkogkas CG, Sonenberg N, Holt CE. Remote control of gene function by local translation. Cell. 2014;157:26–40. doi: 10.1016/j.cell.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabata H, Setoguchi T, Yone K, Souda M, Yoshida H, Kawahara K, Maruyama I, Komiya S. High mobility group box 1 is upregulated after spinal cord injury and is associated with neuronal cell apoptosis. Spine. 2010;35:1109–1115. doi: 10.1097/BRS.0b013e3181bd14b6. [DOI] [PubMed] [Google Scholar]
- Kim J, Wan CK, J O'Carroll S, Shaikh SB, Nicholson LF. The role of receptor for advanced glycation end products (RAGE) in neuronal differentiation. J Neurosci Res. 2012;90:1136–1147. doi: 10.1002/jnr.23014. [DOI] [PubMed] [Google Scholar]
- Liu-Yesucevitz L, Bassell GJ, Gitler AD, Hart AC, Klann E, Richter JD, Warren ST, Wolozin B. Local RNA translation at the synapse and in disease. J Neurosci. 2011;31:16086–16093. doi: 10.1523/JNEUROSCI.4105-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotze MT, Tracey KJ. High-mobility group box 1 protein (HMGB1): nuclear weapon in the immune arsenal. Nat Rev Immunol. 2005;5:331–342. doi: 10.1038/nri1594. [DOI] [PubMed] [Google Scholar]
- Lu B, Antoine DJ, Kwan K, Lundbäck P, Wähämaa H, Schierbeck H, Robinson M, Van Zoelen MA, Yang H, Li J, Erlandsson-Harris H, Chavan SS, Wang H, Andersson U, Tracey KJ. JAK/STAT1 signaling promotes HMGB1 hyperacetylation and nuclear translocation. Proc Natl Acad Sci U S A. 2014;111:3068–3073. doi: 10.1073/pnas.1316925111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merenmies J, Pihlaskari R, Laitinen J, Wartiovaara J, Rauvala H. 30-kDa heparin-binding protein of brain (amphoterin) involved in neurite outgrowth: amino acid sequence and localization in the filopodia of the advancing plasma membrane. J Biol Chem. 1991;266:16722–16729. [PubMed] [Google Scholar]
- Merianda TT, Vuppalanchi D, Yoo S, Blesch A, Twiss JL. Axonal transport of neural membrane protein 35 mRNA increases axon growth. J Cell Sci. 2013a;126:90–102. doi: 10.1242/jcs.107268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merianda TT, Gomes C, Yoo S, Vuppalanchi D, Twiss JL. Axonal localization of neuritin/CPG15 mRNA in neuronal populations through distinct 5′ and 3′-UTR elements. J Neurosci. 2013b;33:13735–13742. doi: 10.1523/JNEUROSCI.0962-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minis A, Dahary D, Manor O, Leshkowitz D, Pilpel Y, Yaron A. Subcellular transcriptomics-dissection of the mRNA composition in the axonal compartment of sensory neurons. Dev Neurobiol. 2014;74:365–381. doi: 10.1002/dneu.22140. [DOI] [PubMed] [Google Scholar]
- Niemi JP, DeFrancesco-Lisowitz A, Roldán-Hernández L, Lindborg JA, Mandell D, Zigmond RE. A critical role for macrophages near axotomized neuronal cell bodies in stimulating nerve regeneration. J Neurosci. 2013;33:16236–16248. doi: 10.1523/JNEUROSCI.3319-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry RB, Fainzilber M. Local translation in neuronal processes–in vivo tests of a “heretical hypothesis.”. Dev Neurobiol. 2014;74:210–217. doi: 10.1002/dneu.22115. [DOI] [PubMed] [Google Scholar]
- Punnonen EL, Fages C, Wartiovaara J, Rauvala H. Ultrastructural localization of beta-actin and amphoterin mRNA in cultured cells: application of tyramide signal amplification and comparison of detection methods. J Histochem Cytochem. 1999;47:99–112. doi: 10.1177/002215549904700111. [DOI] [PubMed] [Google Scholar]
- Rauvala H, Pihlaskari R. Isolation and some characteristics of an adhesive factor of brain that enhances neurite outgrowth in central neurons. J Biol Chem. 1987;262:16625–16635. [PubMed] [Google Scholar]
- Riuzzi F, Sorci G, Donato R. The amphoterin (HMGB1)/receptor for advanced glycation end products (RAGE) pair modulates myoblast proliferation, apoptosis, adhesiveness, migration, and invasiveness: functional inactivation of RAGE in L6 myoblasts results in tumor formation in vivo. J Biol Chem. 2006;281:8242–8253. doi: 10.1074/jbc.M509436200. [DOI] [PubMed] [Google Scholar]
- Rong LL, Trojaborg W, Qu W, Kostov K, Yan SD, Gooch C, Szabolcs M, Hays AP, Schmidt AM. Antagonism of RAGE suppresses peripheral nerve regeneration. FASEB J. 2004a;18:1812–1817. doi: 10.1096/fj.04-1899com. [DOI] [PubMed] [Google Scholar]
- Rong LL, Yan SF, Wendt T, Hans D, Pachydaki S, Bucciarelli LG, Adebayo A, Qu W, Lu Y, Kostov K, Lalla E, Yan SD, Gooch C, Szabolcs M, Trojaborg W, Hays AP, Schmidt AM. RAGE modulates peripheral nerve regeneration via recruitment of both inflammatory and axonal outgrowth pathways. FASEB J. 2004b;18:1818–1825. doi: 10.1096/fj.04-1900com. [DOI] [PubMed] [Google Scholar]
- Rouhiainen A, Kuja-Panula J, Wilkman E, Pakkanen J, Stenfors J, Tuominen RK, Lepäntalo M, Carpén O, Parkkinen J, Rauvala H. Regulation of monocyte migration by amphoterin (HMGB1) Blood. 2004;104:1174–1182. doi: 10.1182/blood-2003-10-3536. [DOI] [PubMed] [Google Scholar]
- Saleh A, Smith DR, Tessler L, Mateo AR, Martens C, Schartner E, Van der Ploeg R, Toth C, Zochodne DW, Fernyhough P. Receptor for advanced glycation end-products (RAGE) activates divergent signaling pathways to augment neurite outgrowth of adult sensory neurons. Exp Neurol. 2013;249:149–159. doi: 10.1016/j.expneurol.2013.08.018. [DOI] [PubMed] [Google Scholar]
- Scaffidi P, Misteli T, Bianchi ME. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature. 2002;418:191–195. doi: 10.1038/nature00858. [DOI] [PubMed] [Google Scholar]
- Shibasaki M, Sasaki M, Miura M, Mizukoshi K, Ueno H, Hashimoto S, Tanaka Y, Amaya F. Induction of high mobility group box-1 in dorsal root ganglion contributes to pain hypersensitivity after peripheral nerve injury. Pain. 2010;149:514–521. doi: 10.1016/j.pain.2010.03.023. [DOI] [PubMed] [Google Scholar]
- Smith DS, Skene JH. A transcription-dependent switch controls competence of adult neurons for distinct modes of axon growth. J Neurosci. 1997;17:646–658. doi: 10.1523/JNEUROSCI.17-02-00646.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi A, Blood DC, del Toro G, Canet A, Lee DC, Qu W, Tanji N, Lu Y, Lalla E, Fu C, Hofmann MA, Kislinger T, Ingram M, Lu A, Tanaka H, Hori O, Ogawa S, Stern DM, Schmidt AM. Blockade of RAGE-amphoterin signalling suppresses tumour growth and metastases. Nature. 2000;405:354–360. doi: 10.1038/35012626. [DOI] [PubMed] [Google Scholar]
- Taylor AM, Berchtold NC, Perreau VM, Tu CH, Li Jeon N, Cotman CW. Axonal mRNA in uninjured and regenerating cortical mammalian axons. J Neurosci. 2009;29:4697–4707. doi: 10.1523/JNEUROSCI.6130-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twiss JL, Smith DS, Chang B, Shooter EM. Translational control of ribosomal protein L4 is required for rapid neurite extension. Neurobiol Dis. 2000;7:416–428. doi: 10.1006/nbdi.2000.0293. [DOI] [PubMed] [Google Scholar]
- Vuppalanchi D, Coleman J, Yoo S, Merianda TT, Yadhati AG, Hossain J, Blesch A, Willis DE, Twiss JL. Conserved 3′-untranslated region sequences direct subcellular localization of chaperone protein mRNAs in neurons. J Biol Chem. 2010;285:18025–18038. doi: 10.1074/jbc.M109.061333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis DE, van Niekerk EA, Sasaki Y, Mesngon M, Merianda TT, Williams GG, Kendall M, Smith DS, Bassell GJ, Twiss JL. Extracellular stimuli specifically regulate localized levels of individual neuronal mRNAs. J Cell Biol. 2007;178:965–980. doi: 10.1083/jcb.200703209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis D, Li KW, Zheng JQ, Chang JH, Smit AB, Smit A, Kelly T, Merianda TT, Sylvester J, van Minnen J, Twiss JL. Differential transport and local translation of cytoskeletal, injury-response, and neurodegeneration protein mRNAs in axons. J Neurosci. 2005;25:778–791. doi: 10.1523/JNEUROSCI.4235-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe AP. Architectural regulations and Hmg1. Nat Genet. 1999;22:215–217. doi: 10.1038/10267. [DOI] [PubMed] [Google Scholar]
- Yoo S, Kim HH, Kim P, Donnelly CJ, Kalinski AL, Vuppalanchi D, Park M, Lee SJ, Merianda TT, Perrone-Bizzozero NI, Twiss JL. A HuD-ZBP1 ribonucleoprotein complex localizes GAP-43 mRNA into axons through its 3′ untranslated region AU-rich regulatory element. J Neurochem. 2013;126:792–804. doi: 10.1111/jnc.12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudin D, Hanz S, Yoo S, Iavnilovitch E, Willis D, Gradus T, Vuppalanchi D, Segal-Ruder Y, Ben-Yaakov K, Hieda M, Yoneda Y, Twiss JL, Fainzilber M. Localized regulation of axonal RanGTPase controls retrograde injury signaling in peripheral nerve. Neuron. 2008;59:241–252. doi: 10.1016/j.neuron.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng JQ, Kelly TK, Chang B, Ryazantsev S, Rajasekaran AK, Martin KC, Twiss JL. A functional role for intraaxonal protein synthesis during axonal regeneration from adult sensory neurons. J Neurosci. 2001;21:9291–9303. doi: 10.1523/JNEUROSCI.21-23-09291.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]