Abstract
Hair cells are sensory receptors for the auditory and vestibular system in vertebrates. The transcription factor Atoh1 is both necessary and sufficient for the differentiation of hair cells, and is strongly upregulated during hair-cell regeneration in nonmammalian vertebrates. To identify genes involved in hair cell development and function, we performed RNA-seq profiling of purified Atoh1-expressing hair cells from the neonatal mouse cochlea. We identified >600 enriched transcripts in cochlear hair cells, of which 90% have not been previously shown to be expressed in hair cells. We identified 233 of these hair cell genes as candidates to be directly regulated by Atoh1 based on the presence of Atoh1 binding sites in their regulatory regions and by analyzing Atoh1 ChIP-seq datasets from the cerebellum and small intestine. We confirmed 10 of these genes as being direct Atoh1 targets in the cochlea by ChIP-PCR. The identification of candidate Atoh1 target genes is a first step in identifying gene regulatory networks for hair-cell development and may inform future studies on the potential role of Atoh1 in mammalian hair cell regeneration.
Keywords: Atoh1, cochlea, hair cells, inner ear
Introduction
Atoh1 is the first transcription factor to be expressed in hair cells, and is essential for hair-cell development. Previous studies of Atoh1 mutant mice have revealed multiple functions of Atoh1 in the genesis, survival, maturation, and function of hair cells (Bermingham et al., 1999; Chen et al., 2002; Woods et al., 2004; Pan et al., 2012; Yang et al., 2012b; Cai et al., 2013). Overexpression of Atoh1 in immature rodent inner ears can induce ectopic hair cells in both sensory and nonsensory regions of the cochlea (Zheng and Gao, 2000; Woods et al., 2004), suggesting the sufficiency of Atoh1 for hair-cell formation in parts of the inner ear. However, the ability of Atoh1 to induce new hair cells in the cochlea declines precipitously with age (Liu et al., 2012; Yang et al., 2012a), although the reasons for this decline are currently not known.
Although Atoh1 is both necessary and sufficient for hair-cell development, the precise molecular mechanism by which Atoh1 mediates hair-cell genesis is unknown. A very small number of Atoh1 targets have been identified by expression profiling of tissues or cell lines (Krizhanovsky et al., 2006; Scheffer et al., 2007a,b). Genome-wide studies have also identified Atoh1 targets in the nervous system and intestine (Klisch et al., 2011; Kim et al., 2014). A previous study combined Atoh1 ChIP-seq (to identify Atoh1 binding sites) together with histone-seq (to identify global H3K4 methylation status), and RNA-seq (to compare expression profiles of wild-type and Atoh1-null cerebella; Klisch et al., 2011). The resultant Atoh1 “targetome” suggests that Atoh1 regulates the expression of genes responsible for diverse biological processes, including cell proliferation, differentiation, migration, and metabolism. This study also pinpointed an extended E-box-containing sequence termed AtEAM as a consensus binding site for Atoh1 (Klisch et al., 2011). A second strategy combining the cerebellar Atoh1 targetome with microarray data from the dorsal spinal cord identified several additional Atoh1 targets specific for dorsal spinal cord interneurons (Lai et al., 2011).
The small number of hair cells in the cochlea has militated against identification of Atoh1 target genes in hair cells by ChIP-seq. However, the success of Atoh1 target identification in the dorsal spinal cord suggests a strategy of hair cell RNA-seq combined with ChIP-seq data from other tissues may allow the identification of some Atoh1 targets in hair cells. We used RNA-sequencing to identify transcripts in Atoh1-expressing cells from the neonatal mouse cochlea and found 614 genes enriched over tenfold in Atoh1-expressing cells. We performed an in situ hybridization screen to validate the expression of 60 of these enriched genes, of which 34 showed specific hair cell expression. We searched for the Atoh1-binding sites in 10 of the validated genes and verified direct Atoh1 binding in these gene loci by ChIP-PCR. These Atoh1 targets may be useful tools in the assembly of a hair cell gene regulatory network and may allow us to understand why the ability of Atoh1 to induce hair-cell transdifferentiation declines with age.
Materials and Methods
Experimental animals.
Atoh1 −/− (MGI: Atoh1tm1Hzo), Atoh1A1GFP/A1GFP (MGI: Atoh1tm4.1Hzo), and Atoh1flox/flox (MGI: Atoh1tm3Hzo) mice were generated as previously described (Ben-Arie et al., 1997; Shroyer et al., 2007; Rose et al., 2009). Atoh1-CreERT2 (MGI: Tg(Atoh1-cre/Esr1*)14Fsh; (Machold and Fishell, 2005) and R26R-YFP (MGI: Gt(ROSA)26Sortm1(EYFP)Cos; (Srinivas et al., 2001) transgenic lines were obtained from Jackson Laboratories. Genotyping was performed by PCR using the following primers: for different Atoh1 alleles, Atoh1-forward (ACG CAC TTC ATC ACT GGC), Atoh1-reverse (GGC ACT GGC TTC TCT TGG), and Neo-forward (GCA TCG CCT TCT ATC GCC) yield a 600 bp wild-type allele band and a 400 bp null allele band. HA-forward (GCG ATG ATG GCA CAG AAG G) and HA-reverse (GAA GGG CAT TTG GTT GTC TCA G) yield a 1 kb Atoh1 EGFP-tagged allele band and a 350 bp floxed allele band. For Atoh1-CreERT2, Cre1F (GCC TGC ATT ACC GGT CGA TGC AAC GA),and Cre1R (GTG GCA GAT GGC GCG GCA ACA CCA TT) yield a 700 bp band. For R26R-YFP, oIMR0316 (GGA GCG GGA GAA ATG GAT ATG), oIMR0883 (AAA GTC GCT CTG AGT TGT TAT), and oIMR4982 (AAG ACC GCG AAG AGT TTG TC) yield a 320 bp YFP+ band. To generate the inducible Atoh1 conditional knock-out (CKO) mice, Atoh1-CreERT2; Atoh1± males were crossed with Atoh1flox/flox; R26R-YFP homozygous females. One dose of 2 mg tamoxifen and 2 mg progesterone was administered to pregnant females at embryonic day (E)17.5 by oral gavage. Progesterone was coadministered to prevent late fetal abortions (Nakamura et al., 2006). Tamoxifen and progesterone were dissolved together in peanut oil, both at a concentration of 20 mg/ml. The genotypes of embryos or newborn pups from these crosses were determined as above. The Baylor College of Medicine Institutional Animal Care and Use committee approved all animal experiments.
Hair-cell purification.
Whole inner ears were dissected from homozygous P0 Atoh1A1GFP/A1GFP mice and incubated in Ca2+, Mg2+-free (CMF) PBS. The cochleae were isolated and the spiral ganglia and Reissner's membrane removed to expose the organ of Corti. Isolated cochleae were washed with CMF-PBS and then incubated in 0.1% Trypsin-EDTA (Sigma-Aldrich) diluted in CMF-PBS for 10 min at 37°C. The trypsin solution was removed and the tissue rinsed in DMEM with 5% fetal calf serum (FCS). The tissue was then gently triturated with a 1000 μl pipette tip in CMF-PBS containing 5% FCS for ∼100 times to generate a single-cell suspension. Hair cells were purified on a BD FACSAria cell-sorting flow cytometer using a 100 μm nozzle and 488 nm excitation. Gates were set each time using small number of cells from the same sample to identify the viable cells and GFP fluorescence. GFP+ and GFP− cells were individually collected in CMF-PBS with 5% FCS. Sorted cells were spun down at 1000 × g for 10 min at 4°C. The supernatant was carefully removed and pellets were suspended in cell lysis buffer from an RNeasy Plus Micro kit (Qiagen) and stored at −80°C for future RNA extraction.
qPCR.
Total RNA was extracted from fluorescence-activated cell sorting (FACS)-purified cells using an RNeasy Plus Micro kit (Qiagen). cDNA was generated using Superscript III Reverse Transcriptase (Invitrogen). Quantitative PCR (qPCR) was performed with Master SYBR Green Kit (Applied Biosystems) on a Step One Plus real-time PCR system (Applied Biosystems). Each reaction was performed in triplicate. Relative quantification of gene expression was analyzed by the ΔΔCT method (Livak and Schmittgen, 2001) using the ribosomal gene L19 as an internal control. Gene-specific primer sets used for qPCR were as follows: Atoh1-F (ATG CAC GGG CTG AAC CA) and Atoh1-R (TCG TTG TTG AAG GAC GGG ATA); L19-F (GGT CTG GTT GGA TCC CAA TG), and L19-R (CCC GGG AAT GGA CAG TCA).
RNA-sequencing.
RNA-seq libraries of FACS purified cells were generated as previously described (Lott et al., 2011). Total RNA was extracted from FACS-purified cells (∼100,000 cells for each library) using an RNeasy Plus Micro kit (Qiagen). mRNA purification, RNA fragmentation, first strand and second strand cDNA synthesis, adaptor ligation, and PCR amplification were performed using the Illumina mRNA-Seq Sample Prep Kit. SPRI beads (Ampure XP, Beckman) were used in each purification step after RNA fragmentation for size selection. Duplicate libraries were made for GFP+ cells and GFP− cells. All libraries were analyzed for quality and concentration using an Agilent Bioanalyzer. A sample from each library was also cloned into the TOPO-Blunt vector. Ten clones were randomly picked and sequenced to verify that ribosomal RNA was depleted from the libraries. Sequencing was performed at the Genomic and RNA Profiling Core in Baylor College of Medicine using Illumina HiSeq2000 100bp Paired-End Platform. Fastq files of paired-end reads and BigWig files have been deposited in the NCBI GEO database, Accession No. GSE65633.
RNA-seq data analysis.
RNA-seq reads were mapped to the mouse reference genome (mm9) using TopHat (Trapnell et al., 2009). Read counts per gene were obtained by counting the number of reads, which overlap the exons of genes defined by RefGene (NCBI). Read counts were normalized to a library size and differentially expressed genes were identified by performing a negative binomial test using the DESeq package (Anders and Huber, 2010). P values were adjusted using the Benjamini and Hochberg (1995) multiple testing procedure. Genes with adjusted p values <1 × 10−10 were marked as significant.
Prediction of Atoh1 binding sites.
To predict Atoh1 binding sites, the MACS package (Zhang et al., 2008) was used to select the top 5000 Atoh1 binding sites from ChIP-Seq data obtained from the neonatal cerebellum (Klisch et al., 2011). MEME-ChIP (Machanick and Bailey, 2011) was used to analyze the flanking sequences of the top 5000 sites, from which 87 motifs were selected that were present >10 times in these 5000 sites. Conserved promoter or enhancer regions within 5 kb of the transcriptional start site of each gene were identified from the UCSC browser conservation track and these regions were searched for the 87 Atoh1 binding motifs.
RNA probe synthesis.
Primer sets for each candidate gene were selected to target a 500–700 bp DNA fragment in a single exon of each gene for screen. A T7 RNA polymerase sequence (5′-GGATCCTAATACGACTCACTATAGGGAG-3′) was added to the 5′ end of each reverse primer. Mouse genomic DNA was used as the template for PCR. The PCR product of the correct size was purified with a PCR Purification Kit (Qiagen). Purified DNA was used as the template for RNA probe synthesis with T7 polymerase (Promega) using standard protocols (Stern, 1998).
In situ hybridization.
Heads of neonatal mouse pups were fixed in 4% paraformaldehyde in PBS overnight at 4°C, cryoprotected in 30% sucrose in PBS at 4°C, embedded in OCT compound (Sakura Finetek), and cryosectioned at 14 μm. The in situ hybridization procedure was modified from previous protocols (Harland, 1991; Birren et al., 1993; Groves et al., 1995). Sections were fixed in 4% paraformaldehyde in PBS, pH 7.2, for 10 min at room temperature, followed by three 5 min washes in DEPC-treated PBS. The sections were treated with 1 μg/ml proteinase K in DEPC-PBS for 5 min at room temperature, followed by three 5 min washes in DEPC-PBS and refixation in 4% paraformaldehyde in PBS, pH 7.2, for 10 min at room temperature. Sections were acetylated in 0.25% acetic anhydride in 0.1 m triethanolamine, pH 8.0, for 10 min at room temperature, followed by three 5 min washes in DEPC-PBS. Slides were incubated in hybridization buffer (50% Formamide, 5× SSC, 50 μg/ml Yeast tRNA, 100 μg/ml heparin, 1× Denhardt's Solution, 0.1% Tween 20, 0.1% CHAPS, 5 mm EDTA) for 1–2 h at 65°C. One-hundred microliters of digoxygenin-labeled probe (1 mg/ml) was added to each slide and the slides covered with glass coverslips. The slides were incubated in a chamber humidified with 5× SSC, 50% formamide at 65°C overnight. Coverslips were removed by rinsing in 0.2× SSC and the slides washed in 0.2× SSC at 65°C for 1 h. The slides were then washed in 0.2× SSC for 5 min at room temperature, followed by another 5 min wash in 0.1% Tween 20 in PBS (PTw). The slides were blocked in 10% lamb serum in PTw at room temperature for 1 h and then stained with anti-digoxygenin-alkaline phosphatase antibody (1:2000) for 1–3 h at room temperature in a humidified chamber. The slides were then washed three times for 5 min each in PTw and equilibrated with freshly made alkaline phosphatase buffer (100 mm Tris, pH 9.5, 50 mm MgCl2, 100 mm NaCl, 0.1% Tween 20) for 10 min. The slides were developed in alkaline phosphatase buffer containing 0.33 mg/ml NBT and 0.18 mg/ml BCIP in the dark at room temperature until the purple reaction product had developed to a satisfactory degree. The reaction was stopped by washing the slides in PBS three times for 15 min each, followed by fixation in 4% paraformaldehyde in PBS, pH 7.2, for 30 min. The slides were then rinsed and mounted in 80% glycerol in PBS.
Chromatin immunoprecipitation.
Cochleae were dissected from P0 Atoh1A1GFP/GFP pups and stored in DMEM with 5% FBS. Cochleae were incubated with 500 μm thermolysin in DMEM for 30–45 min at 37°C and washed in DMEM with 5% FBS. The sensory epithelium from each cochlea was dissected out and pooled together in PBS. Eight sensory epithelia were collected for each sample and centrifuged at 470 × g for 10 min at 4°C. The pooled sensory epithelia were cross-linked with 500 μl PBS containing 13.5 μl of 36.5% formaldehyde solution for 20 min; the sample was vortexed every 5 min during the incubation. The fixation was then quenched with 57 μl of 1.25 m glycine for 5 min. The cross-linked tissue was centrifuged at 470 × g for 10 min at 4°C with soft deceleration. The supernatant was then removed, and the pellet was washed with 500 μl of ice-cold PBS three times. The PBS was removed without disturbing the pellet and the sample snap frozen in liquid nitrogen, and stored at −80°C.
Chromatin immunoprecipitation was performed using the “micro-ChIP” protocol of Dahl and Collas (2009) with some modification. Cross-linked sensory epithelia in the tube were lysed in 120 μl lysis buffer (50 mm Tris-HCl, pH 8.0, 10 mm EDTA, 1% SDS with fresh 1% protease inhibitor and 1 mm PMSF), and incubate for 5 min on ice. The sample was sonicated using a Bioruptor (Diagenode) programmed for 30 s on, 30 s off, for 15 cycles, with vortexing after every five cycles. Four-hundred microliters of RIPA ChIP buffer (10 mm Tris-HCl, pH 7.5, 140 mm NaCl, 1 mm EDTA, 0.5 mm EGTA, 1% Triton X-100, 0.1% SDS, 0.1% Na-deoxycholate, 1% protease inhibitor and 1 mm PMSF) was added to the tube. The sample was then mixed and centrifuged at 12,000 × g for 10 min at 4°C. Supernatant (450 μl) was collected into a new tube and the pellet re-extracted with another 400 μl of RIPA ChIP buffer. To precipitate Atoh1GFP-bound DNA, 10 μl of blocking magnetic beads and 10 μl of anti-GFP magnetic beads (Chromotek, GFP-Trap-M) were washed twice with dilution buffer (1.0 m Tris, pH 7.5, 150 mm NaCl, 1 mm EDTA). The ChIP sample solution (400 μl) was added to 400 μl dilution buffer containing the blocking magnetic beads. The sample mixture was then incubated for 30 min at 4°C. After incubation, the tube was placed on a magnetic rack, the beads were captured, and the solution was extracted and mixed with anti-GFP magnetic beads. This mixture was vortexed and incubated for 2 h at 4°C with moderate shaking. The bead-containing solution was placed on the ice-cold magnetic rack to capture the beads and the beads then washed three times with 1 ml ice-cold RIPA buffer at 4°C with moderate shaking for 10 min each. After the final wash, 1 ml TE buffer was added to the beads and incubated for 10 min at 4°C with moderate shaking. After removal of TE, 100 μl of a 10% solution of Chelex-100 beads was added and the sample vortexed for 10 s and boiled for 10 min. One microliter of 25 mg/ml proteinase K was added to the sample, vortexed, and incubated for 40 min at 56°C, with vortexing every 10 min followed by boiling again for 10 min. The sample was cooled to room temperature, centrifuged for 10 s and the supernatant transferred to a chilled clean tube. The sample was then stored at −20°C for up to 1 week or used directly for ChIP-PCR with appropriate primers. As a negative control, GFP ChIP was performed as above from a wild-type mouse.
Results
RNA-seq analysis identifies transcripts enriched in Atoh1-expressing hair cells
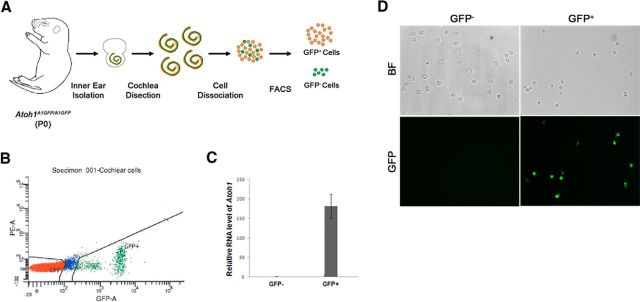
We used FACS on cells isolated from Atoh1A1GFP/A1GFP knock-in mice (Rose et al., 2009) to purify differentiating hair cells that express Atoh1 (Fig. 1A). Cochleae were dissected from newborn (P0) Atoh1A1GFP/A1GFP mice, dissociated into single cells, and GFP+ and GFP− cells were isolated by FACS sorting based on the intensity of GFP fluorescence (Fig. 1B). At P0 we saw no observable apical-basal differences in GFP intensity in the intact cochleae of Atoh1A1GFP/A1GFP mice (Cai et al., 2013). To confirm Atoh1-GFP expression, both sorted populations were fixed and GFP fluorescence was examined under a microscope. In the GFP+ population, ∼ 90% expressed levels of GFP that were detectable by eye and by costaining with antibodies to Myosin6. No GFP+ cells were detected in the GFP− population (Fig. 1C). To further examine Atoh1 expression level in the sorted cell populations, we extracted total RNA from both GFP+ and GFP− populations and performed quantitative reverse-transcriptase PCR to measure the relative mRNA level of Atoh1. Atoh1 transcripts were highly enriched in the GFP+ population by almost 180-fold (Fig. 1D).
Figure 1.
A, Schematic diagram of the purification of GFP cells from the neonatal Atoh1A1GFP/A1GFP mouse cochlea. B, Sample FACS profile showing the distribution of GFP-expressing cells (green). C, Q-PCR analysis of GFP-positive and -negative cells sorted from the Atoh1A1GFP/A1GFP cochlea shows an almost 180-fold enrichment of Atoh1 mRNA in the GFP+ population. D, Fluorescence and bright field images of the GFP-positive and -negative cell populations.
To identify genes that are enriched in differentiating hair cells, we compared the transcripts in GFP+ and GFP− cells from P0 Atoh1A1GFP/A1GFP mice using RNA-sequencing. We obtained between 98,000,000 and 189,000,000 reads for each sample, with 83–85% of the paired reads mapping correctly to the reference genome. Two biological replicates of each population showed high reproducibility (r = 0.98 for the GFP+ populations and 0.96 for the GFP− populations). By comparing the transcripts between GFP+ and GFP− populations, we identified 614 transcripts that have over tenfold enrichment in the GFP+ cell population (p < 1.0E-30). We also identified 329 transcripts that were downregulated over tenfold in the GFP+ population (data not shown). Of the 614 upregulated genes, 57 have been previously identified as hair cell-specific genes (Table 1), 82 have been identified in adult inner or outer hair cells (Liu et al., 2014), and 74 have also been identified as candidate hair cell genes based on the behavior of chick homologues of these genes during utricle regeneration (Ku et al., 2014). Finally, we also identified 22 enriched transcripts in the GFP+ population that are known deafness genes (Cabp2, Cib2, Cldn14, Dfnb59, Gipc3, Gxcr1, Gxcr2, Ildr1, Lhfpl5, Msrb3, Myo3a, Myo6, Myo7a, Pdzd7, Pou4f3, Ptprq, Smpx, Tmc1, Tmie, Ush1 g, Ush2a).
Table 1.
Previously characterized hair cell genes identified in RNA-seq of P1 Atoh1-GFP+ cells
| Gene | Expression in GFP+ cells (RPKM) | Expression in GFP− cells (RPKM) | Fold-change (GFP+ vs GFP−) | p | References |
|---|---|---|---|---|---|
| Atoh1 | 19042.084 | 40.151 | 474.258 | 6.23E-130 | Bermingham et al., 1999; Chen et al., 2002; Cai et al., 2013 |
| Atp7b | 1270.502 | 64.270 | 19.768 | 5.07E-43 | Ding et al., 2011 |
| Barhl1 | 561.825 | 1.982 | 12.810 | 2.54E-73 | Li et al., 2002 |
| Bdnf | 156.249 | 0.500 | 283.453 | 6.73E-44 | Ylikoski et al., 1993; Wheeler et al., 1994 |
| Cacnb2 | 3669.813 | 288.711 | 312.498 | 5.67E-37 | Neef et al., 2009 |
| Calb2 | 9535.022 | 754.472 | 12.711 | 1.41E-35 | Dechesne et al., 1991, 1993 |
| Calm1 | 200561.183 | 12606.473 | 12.638 | 1.84E-42 | Walker et al., 1993 |
| Cdh23 | 45229.825 | 347.607 | 15.909 | 8.06E-27 | Müller, 2008 |
| Celsr3 | 3879.836 | 63.198 | 61.391 | 6.77E-74 | Shima et al., 2002 |
| Chrm4 | 594.397 | 33.833 | 17.568 | 1.21E-34 | Maison et al., 2010 |
| Chrna10 | 25097.049 | 49.883 | 503.115 | 6.37E-127 | Elgoyhen et al., 2001 |
| Chrna9 | 12030.115 | 17.392 | 691.694 | 2.97E-86 | Elgoyhen et al., 1994 |
| Cldn14 | 446.603 | 5.849 | 76.361 | 1.95E-54 | Ben-Yosef et al., 2003 |
| Cldn9 | 23743.783 | 1413.451 | 16.798 | 4.54E-44 | Nunes et al., 2006 |
| Ctbp2 | 2032.242 | 8.167 | 70.452 | 1.72E-31 | Schmitz et al., 2000 |
| Dll1 | 4063.849 | 57.682 | 239.864 | 8.02E-78 | Morrison et al., 1999 |
| Dll3 | 14095.426 | 58.764 | 119.103 | 4.46E-111 | Hartman et al., 2007 |
| Foxj1 | 10620.589 | 89.172 | 133.781 | 3.26E-93 | Yu et al., 2011 |
| Fscn2 | 8200.816 | 61.300 | 205.116 | 3.45E-45 | Shin et al., 2010 |
| Gfi1 | 10651.143 | 51.927 | 52.077 | 5.78E-105 | Wallis et al., 2003 |
| Gpr98 | 17877.609 | 343.293 | 505.351 | 1.37E-37 | McGee et al., 2006 |
| Grxcr1 | 10736.947 | 21.247 | 584.267 | 1.63E-126 | Odeh et al., 2010 |
| Grxcr2 | 8326.395 | 14.251 | 19.827 | 2.45E-127 | Imtiaz et al., 2014 |
| Hes6 | 27504.001 | 1387.183 | 75.974 | 6.40E-47 | Qian et al., 2006 |
| Jag2 | 41818.936 | 550.437 | 573.769 | 2.72E-58 | Lanford et al., 1999 |
| Lhfpl5 | 32615.662 | 56.845 | 452.087 | 5.35E-135 | Longo-Guess et al., 2005 |
| Lhx3 | 14148.409 | 31.296 | 21.718 | 5.58E-128 | Hertzano et al., 2007 |
| Lmo1 | 12269.408 | 564.951 | 277.373 | 2.62E-49 | Deng et al., 2006 |
| Loxhd1 | 6198.799 | 22.348 | 22.931 | 4.21E-81 | Grillet et al., 2009 |
| Mcoln3 | 4954.974 | 216.084 | 618.731 | 1.38E-51 | Di Palma et al., 2002 |
| Myo3a | 7421.246 | 11.994 | 374.468 | 1.51E-128 | Walsh et al., 2002 |
| Myo3b | 10718.111 | 28.622 | 15.678 | 2.62E-123 | Merritt et al., 2012 |
| Myo6 | 176638.988 | 11266.614 | 49.712 | 1.76E-43 | Avraham et al., 1997 |
| Myo7a | 74294.144 | 1494.476 | 380.593 | 1.83E-37 | Hasson et al., 1995 |
| Nhlh1 | 9522.399 | 25.020 | 259.873 | 4.27E-121 | Krüger et al., 2006 |
| Pcp4 | 26486.479 | 101.921 | 602.423 | 1.04E-115 | Thomas et al., 2003 |
| Pou4f3 | 25774.460 | 42.785 | 331.022 | 2.01E-76 | Erkman et al., 1996; Xiang et al., 1998 |
| Ptprq | 32675.674 | 98.711 | 98.435 | 2.41E-71 | Goodyear et al., 2003 |
| Pvalb | 23332.031 | 237.031 | 13.086 | 6.02E-88 | Pack and Slepecky, 1995 |
| Pvrl1 | 21596.648 | 1650.421 | 17.679 | 8.31E-38 | Togashi et al., 2011 |
| Rab15 | 32493.128 | 1837.926 | 18.510 | 2.82E-46 | Lai et al., 2011 |
| Rassf4 | 14720.011 | 795.253 | 10.701 | 5.97E-48 | Lai et al., 2011 |
| Selm | 26344.158 | 2461.804 | 275.918 | 4.66E-32 | Lai et al., 2011 |
| Smpx | 11162.484 | 40.456 | 195.793 | 2.51E-113 | Yoon et al., 2011 |
| Sstr2 | 480.310 | 2.453 | 161.149 | 5.35E-66 | Bodmer et al., 2012 |
| Strc | 11791.437 | 14.699 | 802.190 | 3.93E-55 | Verpy et al., 2001 |
| Tmc1 | 1173.027 | 98.556 | 11.902 | 8.21E-33 | Kawashima et al., 2011 |
| Tmc2 | 1621.727 | 3.260 | 497.489 | 5.12E-103 | Kawashima et al., 2011 |
| Tmie | 4513.184 | 369.072 | 12.228 | 1.03E-35 | Mitchem et al., 2002 |
| Tomt | 9470.009 | 143.013 | 66.218 | 2.92E-78 | Ahmed et al., 2008 |
| Ush1g | 815.769 | 5.931 | 137.532 | 3.25E-60 | Kikkawa et al., 2003 |
| Ush2a | 32496.762 | 48.641 | 668.099 | 2.46E-40 | Pearsall et al., 2002 |
List of known hair cell genes whose transcripts are enriched in neonatal Atoh1-GFP+ cells. The gene name is indicated, together with the expression and fold-change in expression level (RPKM) compared with GFP− cells and the calculated p value for difference between the GFP+ and GFP− populations.
Genes enriched in Atoh1-expressing hair cells are involved in biological processes associated with sensory organ and neuronal development
To analyze the function of the genes enriched in Atoh1-expressing cells, we made use of the gene ontology (GO) database DAVID (Huang et al., 2009a,b). We focused on 313 genes that were enriched by at least tenfold and showed transcript levels at >3000 RPKM. GO analysis suggests several of these genes are involved in biological process associated with sensory organ development (Table 2; 24 genes, p = 2.11E-10), as well as molecular functions that are known to be critical for hair cell development, such as cytoskeletal protein binding (20 genes, p = 2.68E-04), channel activity (17 genes, p = 0.001439), and motor activity (11 genes, p = 0.001123). It is known that sensory hair cells share some properties with neurons, such as synapse formation, synaptic vesicle release, and the regulation of membrane potential. Accordingly, we identified a number of genes associated with neuron differentiation in Atoh1-expressing cells (30 genes, p = 4.92E-11) and genes known to be synapse components (23 genes, p = 2.05E-06).
Table 2.
Gene ontology analysis of genes identified in RNA-seq of P1 Atoh1-GFP+ cells
| GO category | GO term | No. of genes | Genes |
|---|---|---|---|
| Biological process | Neuron differentiation | 30 | Myo7a, Uchl1, Jag2, Atoh1, Mcoln3, Lhx3, Pou4f3, Rtn4rl2, Gfi1, Nefl, Kndc1, Ush2a, Cdh23, Tomt, Myo6, Fscn2, Kif5c, Celsr3, Dll1, Gpr98, Sall3, Bbs1, Lhfpl5, 6530402F18Rik, 2610109H07Rik, Chrnb2, Ngfr, Slitrk6, Wnt7a, Grk1 |
| Sensory organ development | 24 | Tomt, Myo6, Fscn2, Cryab, Myo7a, Tmie, Jag2, Dll1, Pax2, Prph2, Prox1, Gpr98, Ptprq, Lhfpl5, Atoh1, Pvrl1, Chrna9, Mcoln3, Pou4f3, Gfi1, Chrna10, Ush2a, Cdh23, Grk1 | |
| Molecular function | Calcium ion binding | 32 | Spock2, Clstn3, Jag2, Cacnb2, Dlk2, Cdh1, Calb2, Necab2, Mmp24, Syp, Pvalb, Pcp4, Pls1, Cib2, S100a1, Cdh23, Dtna, Tesc, Hpcal1, Celsr3, Dll1, Actn3, Anxa4, Gas6, S100a13, Gpr98, Pkd2l1, Chga, Umodl1, Cadps2, Dll4, Calm1 |
| Cytoskeletal protein binding | 20 | Obscn, Myo6, Baiap2l2, Fscn2, Myo3a, Myo7a, Myo3b, Lmo7, Actn3, Rph3a, Gpr98, Pacsin1, Fhod3, Myo16, Pls1, AI428936, Lmod1, Eps8l2, Ush2a, Tmod1 | |
| Channel activity | 17 | Scn1b, Cacnb2, Pkd2l1, Kcnmb2, Kcna10, Accn4, Chrna9, Mcoln3, P2rx3, Kcnh6, Kcnf1, Scn11a, Chrnb2, Kcnh2, Chrna1, Chrna10, Chrng | |
| Motor activity | 11 | Myo6, Kif27, Myo3a, Myo7a, Myo3b, Kif5c, Myo16, Kif19a, Dynlrb2, Kif21b, Dnaic2 | |
| Cell component | Cell junction | 24 | Gpr156, Ica1, Cldn9, Dlgap3, Rimbp2, Ctnnd2, Lmo7, Cdh1, Rims2, Rph3a, Calb2, Syp, Cbln1, Pvrl1, Cadps2, Pcp4, Snph, Chrnb2, Ssx2ip, Tjp3, Chrna1, Chrna10, Chrng, Dtna |
| Synapse | 23 | Gpr156, Rab3b, Ica1, Cplx1, Myo6, Dlgap3, Clstn3, Myo7a, Rimbp2, Rims2, Rph3a, Syp, Cbln1, Chrna9, Cadps2, Snph, Chrnb2, Chrna1, Chrna10, Nefm, Chrng, Cdh23, Dtna |
Gene ontology analysis of the 313 genes whose transcripts showed high ranks in both expression and fold-change in our RNA-seq result (expression level >3000 RPKM, fold-change >10). A representative number of GO categories and terms are shown with >10 genes per category.
In situ validation of genes enriched in Atoh1-expressing cochlear cells
We performed an in situ hybridization screen in the P0 mouse cochlea to validate the expression of the genes enriched in Atoh1-expressing cochlear cells. We focused on the 313 genes that showed high ranks in both expression and fold-change in our RNA-seq result (expression level >3000 RPKM, fold-change >10). We chose an expression of RPKM>3000 as an arbitrary filter based on our empirical correlation of RPKM values with a positive signal by in situ hybridization. We designed RNA probes for 60 genes from this list, and performed in situ hybridization on sections of neonatal mouse cochlea to validate their expression. Surprisingly, only 34 of 60 genes showed specific expression in cochlear hair cells (Table 3; Fig. 2A,B). Although the remaining 26 genes were expressed in hair cells, we observed additional sites of expression such as supporting cells, Kölliker's organ, or the outer sulcus. Some of the validated hair-cell-specific genes were previously shown to be expressed in the inner ear hair cells or associated with hair-cell development and hearing-defect phenotypes, such as Lhx3 (Hertzano et al., 2007; Rajab et al., 2008), Lhfpl5 (Kalay et al., 2006), and Srrm4 (Nakano et al., 2012). However, many of the genes in our screen have never been studied in the inner ear before (Table 3; Fig. 2B). These include the phospholipid binding protein Annexin A4, the transcription factor and tumor suppressor Castor (Casz1; Charpentier et al., 2013), RBM24, an RNA-binding protein implicated in Notch-Delta signaling (Maragh et al., 2014), the Notch receptor modifying enzyme Mfng, and the lysosome regulatory gene melanoregulin (Mreg). Some genes associated with cytoskeletal binding and motor activity are specifically expressed in cochlear hair cells, such as Eps8l2, Kif21b, and Pacsin1 (Table 3; Fig. 2A,B). Several ion channels also show a hair-cell-restricted expression pattern, such as Chrna9, Chrna10, Kcnh6, and Scn11a (Table 3; Fig. 2A,B). Because Atoh1 is expressed in all differentiating hair cells in both the auditory and vestibular end organs, we also checked the expression of our validated genes in the vestibular organs. All the genes we found to be specifically expressed in cochlear hair cells also show expression in vestibular hair cells at neonatal stages (Table 3; Fig. 2C).
Table 3.
Summary of hair cell genes validated by in situ hybridization
| Expression | Fold-change | P0 vestibule | Gradient (high to low) | Published HC gene | ChIP-seq | Atoh1 direct targets | Expression in Atoh1-CKO | |
|---|---|---|---|---|---|---|---|---|
| Anxa4 | 58650.61 | 74.378 | HC | Base to apex | No | Intestine | Yes | Decreased |
| Atoh1 | 19042.08 | 474.258 | HC | Apex to base | Yes | Cerebellum, intestine | Yes | Decreased |
| Calb2 | 9535.022 | 12.638 | HC | Not observed | Yes | Cerebellum | ND | ND |
| Casz1 | 9969.698 | 85.483 | HC | Not observed | No | Cerebellum, intestine | ND | ND |
| Chrna10 | 25097.05 | 503.115 | HC | Base to apex | Yes | Intestine | Yes | ND |
| Chrna9 | 12030.11 | 691.694 | HC | Not observed | Yes | None | ND | ND |
| Eps8l2 | 15530.19 | 64.052 | HC | Base to apex | Yes | Intestine | ND | ND |
| Grxcr2 | 8326.395 | 584.267 | HC | Not observed | Yes | None | ND | No change |
| Kcnh6 | 12791.54 | 154.773 | HC | Not observed | No | Intestine | ND | ND |
| Kif21b | 8369.936 | 11.176 | HC | Base to apex | No | Cerebellum, intestine | ND | ND |
| Lhfpl5 | 32615.66 | 573.769 | HC | Not observed | Yes | Intestine | ND | ND |
| Lhx3 | 14148.41 | 452.087 | HC | Not observed | Yes | Intestine | ND | Decreased |
| Mfng | 10493.52 | 76.080 | HC | Not observed | No | Cerebellum, intestine | ND | Decreased |
| Mgat5b | 22037.06 | 214.706 | HC | Not observed | No | Cerebellum, intestine | Yes | No change |
| Mreg | 16063.03 | 139.686 | HC | Base to apex | No | Cerebellum, intestine | Yes | No change |
| Mycl1 | 26009.33 | 41.131 | HC | Not observed | Yes | Cerebellum, intestine | ND | ND |
| Myo3b | 10718.11 | 374.468 | HC | Base to apex | Yes | Cerebellum, intestine | ND | ND |
| Naca | 10408.7 | 24.968 | ND | Base to apex | No | Cerebellum, intestine | ND | ND |
| Pacsin1 | 4302.44 | 40.762 | HC | Base to apex | No | Cerebellum, intestine | ND | ND |
| Pcp4 | 26486.48 | 259.873 | HC | Base to apex | Yes | Cerebellum | Yes | No change |
| Ptgir | 4722.92 | 133.553 | No | Base to apex | No | None | ND | ND |
| Ptprq | 32675.67 | 331.022 | HC | Base to apex | Yes | None | ND | No change |
| Rab11fip1 | 3025.383 | 89.836 | HC | Not observed | No | Intestine | ND | ND |
| Rasd2 | 127825.8 | 507.234 | HC | Not observed | No | Cerebellum, intestine | Yes | Decreased |
| Rbm24 | 16001.61 | 229.243 | HC | Not observed | No | Intestine | Yes | Decreased |
| Scn11a | 11604.04 | 797.871 | HC | Apex to base | Yes | Cerebellum | Yes | Decreased |
| Sema5b | 32913.11 | 29.366 | ND | Base to apex | No | Cerebellum, intestine | ND | ND |
| Slc6a11 | 13126.97 | 309.753 | HC | Not observed | No | Cerebellum, intestine | ND | No change |
| Smpx | 11162.48 | 275.918 | HC | Not observed | Yes | Cerebellum | ND | ND |
| Srrm4 | 8899.872 | 164.984 | HC | Apex to base | Yes | Cerebellum, intestine | Yes | Decreased |
| Stard10 | 30766.48 | 25.747 | HC | Not observed | No | Intestine | ND | ND |
| Thsd7b | 28906.74 | 298.100 | HC | Not observed | No | Intestine | ND | No change |
| Ttc21a | 16745.86 | 329.525 | HC | Not observed | No | Cerebellum, intestine | ND | ND |
| Umodl1 | 10437.93 | 331.222 | HC | Not observed | No | Cerebellum, intestine | ND | ND |
Summary of genes whose specific expression in hair cells was validated by in situ hybridization. Gene name, expression level (RPKM), and fold-change over GFP− cells are indicated, together with expression pattern in the cochlea or vestibular system. If a transcript showed a gradient of expression in the cochlea, this is indicated. The table also lists whether each gene was identified as an Atoh1 target by ChIP-seq in either the cerebellum (Klisch et al., 2011) or small intestine (Kim et al., 2014), as well as whether the gene was shown to be a direct target of Atoh1 in the cochlea by ChIP-PCR (Fig. 5). Finally, the expression of direct Atoh1 target genes was measured in vivo 24 h after deletion of Atoh1 (Fig. 5).
HC, Hair cell; IHC, inner hair cell; ND, not determined.
Figure 2.
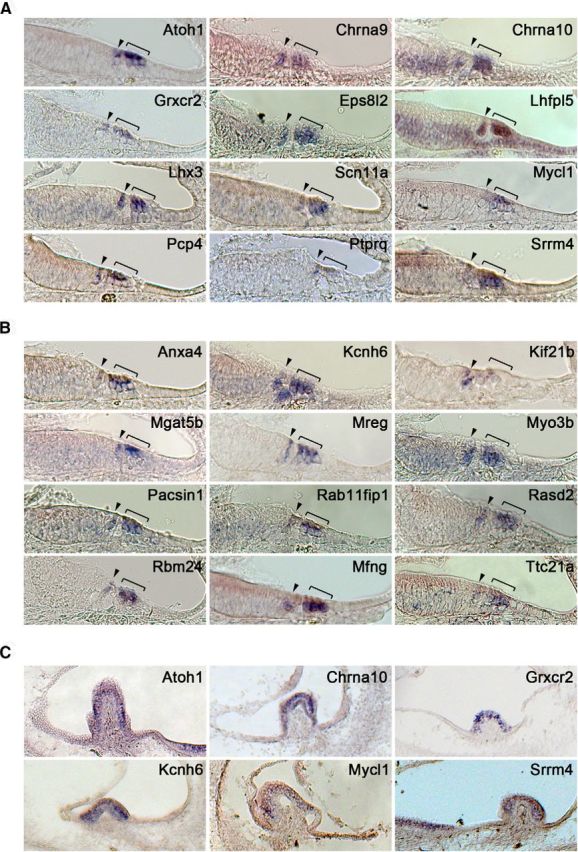
In situ hybridization validation of transcripts enriched in Atoh1-GFP+ cells. A, Representative sample of 12 genes whose expression has previously been reported in hair cells. Images are taken from the basal turn of a neonatal mouse cochlea. B, Representative sample of 12 genes whose expression has not previously been reported in hair cells. Images are taken from the basal turn of a neonatal mouse cochlea. Brackets mark the outer hair cell region; arrowhead marks the inner hair cell region. C, Expression of sample of enriched transcripts in the cristae of the neonatal mouse vestibular system.
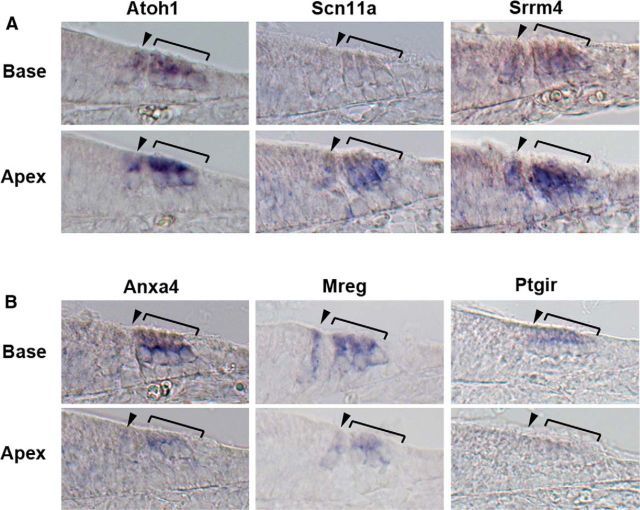
Atoh1 expression initiates in the basal region of the cochlea at E13.5 and spreads along the cochlear duct to the apex (Chen et al., 2002; Cai et al., 2013). However, by the day of birth, Atoh1 expression is beginning to be downregulated in the hair cells in the basal turn of the cochlea, and this downregulation continues for several days along the length of the cochlea to the apex (Groves et al., 2013). Many of the genes in our screen also show a gradient of expression in hair cells along the apical-basal axis of the cochlear duct (Table 3; Fig. 3). Some genes are expressed more strongly in apical hair cells than in the basal ones at P0 in a similar gradient to Atoh1, such as Srrm4 and Scn11a (Fig. 3). These genes are likely to be regulated by Atoh1 (although they may not necessarily be direct targets) and like Atoh1 are downregulated as hair cells mature. In contrast, some genes, such as Chrna10 and Mreg, showed an opposite gradient, with higher expression in basal hair cells compared with apical hair cells (Fig. 3). It is likely that these genes reflect the ongoing basal-apical wave of differentiation of hair cells and represent markers of maturing hair cells. Alternatively, it is possible that some of the genes showing differential apical-basal expression maintain this differential expression as the cochlea matures and might, for example, be involved in regulating tonotopic differences in hair cells along the cochlear duct.
Figure 3.
Examples of hair cell transcripts expressed in gradients along the neonatal cochlea. A, Atoh1, Scn11a, and Srrm4 are expressed more strongly in the apex than the base of the cochlea, consistent with their being downregulated as hair cells mature. B, Anxa4, Mreg, and Ptgir are expressed more strongly in the base than the apex, consistent with their upregulation and maintenance as hair cells mature. Brackets mark the outer hair cell region; arrowhead marks the inner hair cell region.
Identification of Atoh1 direct target genes in neonatal cochlear hair cells
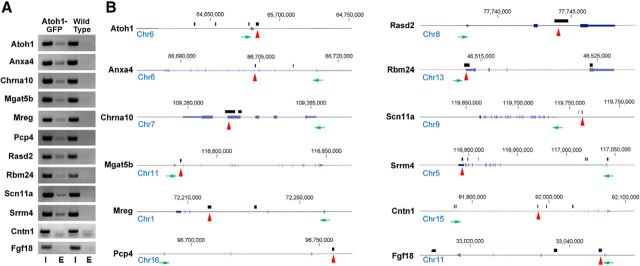
Although Atoh1 is both necessary and sufficient for hair cell development, little is known about the molecular function of Atoh1 in hair cells and very few genes have been identified as direct targets of Atoh1 in hair cells. To select potential Atoh1 targets, we cross-referenced our validated hair-cell-specific genes from the P0 cochlea with Atoh1 ChIP-seq datasets from mouse cerebellum and intestine (Klisch et al., 2011; Kim et al., 2014). We found that genes for 233 of 313 hair-cell-enriched transcripts contain Atoh1 binding regions within 10 kb of the transcriptional start site in either cerebellar or intestinal ChIP-seq experiments, or have AtEAM binding sites within 5 kb of their transcriptional start site (Klisch et al., 2011). We designed specific primers for the binding locus of nine candidate genes (Anxa4, Chrna10, Mgat5b, Mreg, Pcp4, Rasd2, Rbm24, Scn11a, and Srrm4), as well as Atoh1, as it is known to regulate its own expression by binding to an autoregulatory enhancer (Helms et al., 2000; Groves et al., 2013; Jarman and Groves, 2013). We isolated sensory epithelia of P0 Atoh1A1GFP/A1GFP cochleae and performed chromatin immunoprecipitation with camelid nanobodies against the Atoh1-GFP fusion protein. By PCR amplification of the immunoprecipitated DNA, we verified all 10 genes to contain upstream or downstream elements that are directly bound by Atoh1 in the neonatal cochlear sensory epithelium. We also analyzed two genes, Fgf18 and Cntn1 that have been shown to be direct targets of Atoh1 in the cerebellum (Klisch et al., 2011), but which are not expressed at significant levels in cochlear hair cells. Neither gene region gave a positive result in our ChIP-PCR experiments (Fig. 4). The nine newly identified Atoh1 direct targets include genes that have been previously reported to be essential for cochlear development, such as Chrna10 and Srrm4 (Simmons and Morley, 2011; Nakano et al., 2012), and genes that have not previously been described in ear development, such as Anxa4, Mgat5b, Mreg, Pcp4, Rasd2, Rbm24, and Scn11a (Fig. 4).
Figure 4.
A, Verification of direct targets of Atoh1 by ChIP-PCR. Ten genes that were identified as candidate Atoh1 targets (including Atoh1 itself as a positive control) were analyzed by chromatin immunoprecipitation from sensory epithelium dissected from neonatal Atoh1A1GFP/A1GFP mice. Wild-type mice were used as a negative control. In addition, ChIP-PCR for two genes (Fgf18 and Cntn1) that are directly regulated by Atoh1 in the cerebellum but are not expressed in hair cells were also used as a negative control. PCR amplification of candidate Atoh1 binding regions was performed as described in Materials and Methods. Input DNA and experimental lanes are indicated by I and E, respectively. B, Schematic diagram showing the chromosomal location and structure of each gene, the direction of transcription (green arrows), and the location of Atoh1 binding sites (black bars) identified by ChIP-Seq from cerebellum (Atoh1, Mgat5b, Mreg, Pcp4, Rasd2, Scn11a, Srrm4, Cntn1, Fgf18; Klisch et al., 2011) or intestine (Anxa4, Chrna10, Rbm24; Kim et al., 2014). Regions used for ChIP-PCR in A are shown with red arrows.
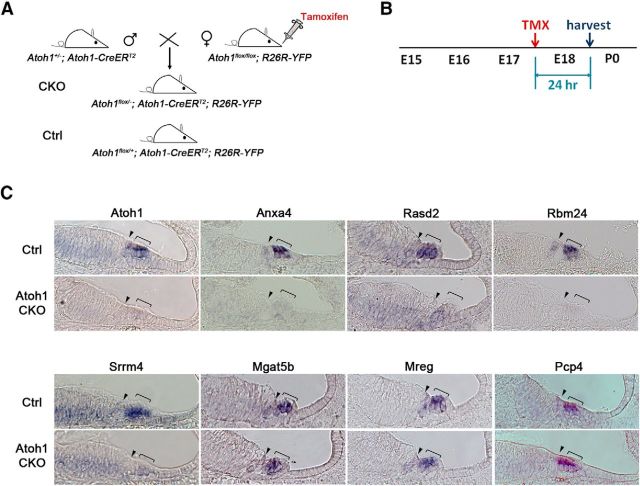
We have previously shown that Atoh1 protein is quite unstable in cochlear hair cells, as conditional deletion of Atoh1 using the Cre-Lox system leads to hair cell death 12 h after the onset of Cre-mediated recombination (Cai et al., 2013). We tested to what extent our verified Atoh1 target genes were sensitive to the loss of Atoh1 by conditionally deleting Atoh1 from hair cells at E17.5 as previously described (Cai et al., 2013). We crossed Atoh1±; Atoh1-CreERT2 males with Atoh1flox/flox; R26R-YFP female mice and administered tamoxifen and progesterone to female mice on the 17th day of pregnancy by oral gavage to generate Atoh1 CKO (Atoh1flox/−; Atoh1-CreERT2; R26R-YFP) and control (Atoh1flox/+; Atoh1-CreERT2; R26R-YFP) animals. We collected embryos 24 h later, and examined the expression of seven verified Atoh1 target genes, as well as Atoh1 itself as a positive control. Four genes (Anxa4, Rasd2, Rbm24, and Srrm4) were rapidly and completely downregulated in hair cells 24 h after deletion of Atoh1 (Fig. 5). However, three other genes (Mgat5b, Mreg, and Pcp4) continued to be expressed in Atoh1 CKO hair cells after 24 h at levels that were indistinguishable from their control counterparts. This persistence may simply reflect the stability of mRNA transcripts for these genes, or may indicate that these genes require Atoh1 for their initiation but not maintenance, due to the action of other transcription factors that can maintain transcription even after Atoh1 is downregulated.
Figure 5.
Some, but not all Atoh1 target genes are rapidly downregulated after deletion of Atoh1. A, Breeding scheme to generate Atoh1-CKO mice. Female Atoh1flox/flox; R26R-YFP mice were mated with Atoh1±; Atoh1-CreERT2 males to generate mice that carried the CreERT2 allele, one copy of the R26R-YFP Cre reporter and either an Atoh1 flox/+ (50%) or Atoh1 flox/− (50%) allele. Use of the R26R-YFP Cre reporter provides a readout of the leakiness, speed, and efficiency of the system. B, Pregnant dams received one pulse of tamoxifen/progesterone (TMX) at E17.5, followed by kill 24 h later. C, Four of seven genes tested (Anxa4, Rasd2, Rbm24, and Srrm4) showed rapid downregulation of transcripts 24 h after tamoxifen administration. However, three other genes (Mgat5b, Mreg, and Pcp4) were still expressed 24 h after tamoxifen administration. Brackets mark the outer hair cell region; arrowhead marks the inner hair cell region.
Although Atoh1 is necessary and to some degree sufficient for hair cell development in the inner ear, its function is transient as it is downregulated shortly after birth (Groves et al., 2013). However, some of the genes we have verified to be direct targets of Atoh1, such as Chrna10, continue to be expressed in mature outer hair cells after Atoh1 ceases to be expressed (Elgoyhen et al., 2001), and were expressed strongly in basal regions of the cochlea even as Atoh1 itself was being downregulated (Fig. 3). It is likely that Atoh1 participates in the assembly of transcriptional complexes at these loci, which remain active after Atoh1 expression ceases. For example, the Pou4f3 gene, which is expressed in hair cells throughout life (Erkman et al., 1996; Xiang et al., 1998), has 5′ regulatory regions that are directly bound and regulated by Atoh1 (Masuda et al., 2011) but these regions also contain sites for other transcription factors such as Etv4, N-Myc, and Ets2 (Ikeda et al., 2015). It is possible that at least three of our verified Atoh1 targets, Mgat5b, Mreg, and Pcp4 may be regulated in this way, as they continued to be expressed in the hair cells of Atoh1 CKO mice (Fig. 3). Alternatively, it is also possible that the transcripts for these genes are quite stable and remain detectable in hair cells at least 24 h after loss of Atoh1 protein.
Discussion
We present one of the first RNA-seq studies to characterize purified cell populations from the mammalian cochlea. We were able to obtain high quality RNA-seq libraries from 100,000 purified Atoh1-GFP+ hair cells that gave excellent paired-end mapping of sequenced reads. A number of previous studies have produced valuable gene expression information from microarray profiling of different populations from the inner ear (Sato et al., 2009; Hertzano et al., 2011; Sinkkonen et al., 2011; Son et al., 2012). Our study, together with other studies such as the recently described SHIELD database (shield.hms.harvard.edu) suggest that RNA-seq analysis of purified cell populations can be successfully accomplished with small numbers of inner ear hair cells; for example, we have recently been able to obtain RNA-seq data of similar quality from as few as 10,000 cells purified from the cochlea using standard RNA-seq library preparation reagents (M. L. Basch and A. K. Groves, unpublished observations).
We found 614 transcripts to be significantly enriched in hair cells, and of these at least 10% have been reported previously to be specifically expressed in hair cells. From our initial list of 614 genes, we selected 313 genes that were enriched >10-fold in Atoh1+ cells, and which gave expression values of >3000 RPKM. Strikingly, when we analyzed 60 genes from this list by in situ hybridization, only ∼50% (34 genes) showed exclusive and specific expression in hair cells. We note that one of our criteria for selecting genes for validation was the fold-change between Atoh1-GFP+ hair cells and all other cell types in the ear. As such, it follows that a high degree of transcript enrichment in hair cells does not preclude that the transcript in question is present in other cell types in the ear; simply, that it is expressed at lower levels. This emphasizes that validation of RNA-seq data by analysis of mRNA in situ expression is a crucial step in the identification and confirmation of truly cell-type specific transcripts, even in cell populations enriched to >90% purity.
Our transcriptomic analysis was performed on hair cells purified from the newborn mouse cochlea. At this age, almost all hair cells have differentiated in the organ of Corti, although they continue to mature over the next 14 d until the onset of hearing at ∼2 weeks of age. This maturation is reflected in many ways, for example, the elaboration of the stereociliary hair bundle (Frolenkov et al., 2004), the acquisition of functional mechanotransduction apparatus (Lelli et al., 2009), mature ion currents, and the divergence in morphology and function of inner and outer hair cells (Belyantseva et al., 2000). Some genes are specifically expressed in either inner or outer hair cells from an early age, for example, Fgf8 is restricted to inner hair cells shortly after the onset of Atoh1 expression (Jacques et al., 2007), and we observed Kif21b expression exclusively in inner hair cells at P1 (Fig. 2B). However, other genes, such as Chrna9, begin to be expressed in all cochlear hair cells before becoming restricted to outer hair cells (Elgoyhen et al., 1994; Liu et al., 2014; Fig. 2A), which may reflect radial (neural–abneural) gradients of differentiation signals. It will be of great interest to determine the transcriptional regulators that establish hair-cell-specific patterns of gene expression, either in development or during hair cell maturation.
At present, very little is known about changes in gene expression that accompany and regulate the maturation of hair cells. Our data suggest that at least some components of mature hair cells are already expressed in neonatal hair cells. First, gene ontology analysis of significantly enriched hair cell genes in our study identified cohorts of genes involved in motor activity, channel activity and calcium binding, synapse formation and function, in addition to genes associated with neural or sensory organ development (Table 2). Second, we were able to detect transcripts for some previously characterized components of the hair bundle (Shin et al., 2013), such as Apba1, Calb2, Eps8l2, Fscn2, Hspa4l, Myo3b, and Stard10 in addition to the unconventional myosins Myo6 and Myo7a that are expressed early in differentiating hair cells. Third, we compared our data to a recent study that used Affymetrix microarrays to identify the transcriptomes of 2000 individually isolated inner and outer hair cells from 25- to 30-d-old mice (Liu et al., 2014). Comparison of these transcriptomes to our neonatal hair cell data suggest that >80 genes expressed in neonatal hair cells continue to be expressed in mature inner or outer hair cells. Finally, we were also able to identify transcripts for 22 genes identified in syndromic or nonsyndromic forms of deafness.
The challenge and utility of identifying Atoh1 targets in hair cells
Atoh1 plays a central role in the differentiation and survival of hair cells (Jarman and Groves, 2013), and has been proposed as a candidate for gene therapy to restore hair cells lost through injury or aging (Baker et al., 2009; Husseman and Raphael, 2009). Discovering the direct transcriptional targets of Atoh1 is key to understanding its function and possible reasons for its reduced efficacy in promoting hair-cell formation in the aging ear (Kelly et al., 2012; Liu et al., 2012). However, genome-wide analysis of Atoh1 binding sites in hair cells by techniques, such as ChIP-seq, is confounded by the relatively small numbers of hair cells per cochlea and the fact that hair cells represent a small fraction of the total cell types in the cochlear duct. In contrast, it has been possible to obtain ChIP-seq data from Atoh1-expressing cerebellar granule cells (Klisch et al., 2011) and intestinal secretory cells (Kim et al., 2014). In an attempt to identify new Atoh1 targets in hair cells, we interrogated the transcriptome of cochlear hair cells obtained by RNA-sequencing to identify loci that are strongly and specifically expressed in Atoh1+ hair cells and which have enhancers containing Atoh1 consensus binding sites (AtEAMs; Klisch et al., 2011), or which have been shown to be occupied by Atoh1 in the cerebellum or gut by ChIP-seq. This approach allowed us to identify 233 candidate Atoh1 target genes, of which 68 were common to our hair cell transcriptome and to ChIP-seq analyses from the cerebellum and gut. We verified 10 genes, including Atoh1 itself, as strong candidates to be direct targets of Atoh1. Previous studies have also identified several other direct targets of Atoh1 in hair cells, such as Pou4f3 (Masuda et al., 2011; Ikeda et al., 2015), Selm, Rassf4, and Rab15 (Lai et al., 2011).
Atoh1 can regulate a large and diverse array of genes in different cell types with functions including transcriptional control, cell-cycle regulation, cell migration, metabolic control, and even housekeeping functions (Klisch et al., 2011; Lai et al., 2011). These processes are not common to all Atoh1-expressing cells, for example, cerebellar granule neuron precursors are both migratory and proliferative, whereas hair cells are not. Clearly then, the cellular context in which Atoh1 functions will determine its selection of targets. Although Atoh1 is expressed in all inner ear hair cells, it is not known whether Atoh1 has distinct targets or overlapping targets in different types of hair cell, for example, auditory versus vestibular hair cells, inner hair cells versus outer hair cells, or type I versus type II vestibular hair cells, or instead simply regulates common, generic aspects of hair-cell development. All 10 of our validated Atoh1 targets showed comparable expression in inner and outer hair cells, and were also expressed in vestibular hair cells (Table 3); although the expression of many more validated Atoh1 targets will need to be tested to determine whether this trend is maintained. It is also possible that Atoh1 establishes a generic pattern of hair-cell gene expression, with later transcription factors modulating these targets in a hair-cell-specific manner after Atoh1 expression is downregulated.
Progress in identifying direct targets of Atoh1 may shed light on the function of this gene in regeneration as well as normal hair-cell differentiation. Ectopic expression of Atoh1 in the inner ear, either in vivo or in vitro, can lead to the formation of ectopic hair cells (Zheng and Gao, 2000; Kawamoto et al., 2003; Izumikawa et al., 2005; Gubbels et al., 2008; Kelly et al., 2012; Liu et al., 2012). Moreover, Atoh1 is rapidly upregulated in supporting cells of nonmammalian vertebrates as a consequence of hair cell loss (Cafaro et al., 2007; Ma et al., 2008; Ma and Raible, 2009; Lewis et al., 2012). However, recent studies in mice suggest that the ability of inner ear cells to transdifferentiate into hair cells in the presence of ectopic Atoh1 declines with age (Kelly et al., 2012; Liu et al., 2012). There are a number of possible explanations for this decline, for example, the presence or absence of post-translational modifications of Atoh1 itself (Mulvaney and Dabdoub, 2012), an age-dependent upregulation of inhibitors of Atoh1, such as Id HLH factors, or an age-dependent decline in transcription factors or cofactors that cooperate with Atoh1 to activate hair cell genes (Ikeda et al., 2015). Finally, it is possible that direct epigenetic modification of Atoh1 target genes renders them unavailable for transcription, even in the presence of exogenously expressed Atoh1 (Groves et al., 2013). Our identification of direct Atoh1 targets in hair cells in the present study may allow the testing of these hypotheses.
Notes
Supplemental material for this article is available at https://www.bcm.edu/research/labs/andrew-groves/publications. Seven data tables and a legend document describe qPCR and PCR primers, a list of all 614 hair cell-enriched genes, 233 candidate Atoh1 target genes, and comparisons of our hair cell-enriched genes with known deafness genes, genes identified in adult hair cells and genes identified in chick utricle hair cells, and expression levels of 19218 transcripts in Atoh1-GFP+ hair cells at P1. This material has not been peer reviewed.
Footnotes
This project was supported in part by The Genomic and RNA Profiling Core and the RNA In Situ Core at Baylor College of Medicine with the expert assistance of Drs Lisa D. White and Cecilia Ljunberg, funding from NIH Grant P30HD024064 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development, The Cytometry and Cell Sorting Core at Baylor College of Medicine with the expert assistance of Joel M. Sederstrom, funding from NIH Grants P30 AI036211, P30 CA125123, and S10 RR024574, NIH Grants DC006185 and DC011657 to A.K.G., Department of Defense Grant DOD W81XWH-11-2-004 to A.K.G., and H.Y.Z. is an investigator of the Howard Hughes Medical Institute. We thank Hongyuan Zhang, Alyssa Crowder, and Huiling Li for excellent technical assistance and members of the Groves laboratory for their advice and comments; David He for advice and sharing data with us; and Ying-Wooi Wan for help in handling sequencing data.
The authors declare no competing financial interests.
References
- Ahmed ZM, Masmoudi S, Kalay E, Belyantseva IA, Mosrati MA, Collin RW, Riazuddin S, Hmani-Aifa M, Venselaar H, Kawar MN, Tlili A, van der Zwaag B, Khan SY, Ayadi L, Riazuddin SA, Morell RJ, Griffith AJ, Charfedine I, Caylan R, Oostrik J, et al. Mutations of LRTOMT, a fusion gene with alternative reading frames, cause nonsyndromic deafness in humans. Nat Genet. 2008;40:1335–1340. doi: 10.1038/ng.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avraham KB, Hasson T, Sobe T, Balsara B, Testa JR, Skvorak AB, Morton CC, Copeland NG, Jenkins NA. Characterization of unconventional MYO6, the human homologue of the gene responsible for deafness in Snell's waltzer mice. Hum Mol Genet. 1997;6:1225–1231. doi: 10.1093/hmg/6.8.1225. [DOI] [PubMed] [Google Scholar]
- Baker K, Brough DE, Staecker H. Repair of the vestibular system via adenovector delivery of Atoh1: a potential treatment for balance disorders. Adv Otorhinolaryngol. 2009;66:52–63. doi: 10.1159/000218207. [DOI] [PubMed] [Google Scholar]
- Belyantseva IA, Adler HJ, Curi R, Frolenkov GI, Kachar B. Expression and localization of prestin and the sugar transporter GLUT-5 during development of electromotility in cochlear outer hair cells. J Neurosci. 2000;20:RC116. doi: 10.1523/JNEUROSCI.20-24-j0002.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Arie N, Bellen HJ, Armstrong DL, McCall AE, Gordadze PR, Guo Q, Matzuk MM, Zoghbi HY. Math1 is essential for genesis of cerebellar granule neurons. Nature. 1997;390:169–172. doi: 10.1038/36579. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- Ben-Yosef T, Belyantseva IA, Saunders TL, Hughes ED, Kawamoto K, Van Itallie CM, Beyer LA, Halsey K, Gardner DJ, Wilcox ER, Rasmussen J, Anderson JM, Dolan DF, Forge A, Raphael Y, Camper SA, Friedman TB. Claudin 14 knockout mice, a model for autosomal recessive deafness DFNB29, are deaf due to cochlear hair cell degeneration. Hum Mol Genet. 2003;12:2049–2061. doi: 10.1093/hmg/ddg210. [DOI] [PubMed] [Google Scholar]
- Bermingham NA, Hassan BA, Price SD, Vollrath MA, Ben-Arie N, Eatock RA, Bellen HJ, Lysakowski A, Zoghbi HY. Math1: an essential gene for the generation of inner ear hair cells. Science. 1999;284:1837–1841. doi: 10.1126/science.284.5421.1837. [DOI] [PubMed] [Google Scholar]
- Birren SJ, Lo L, Anderson DJ. Sympathetic neuroblasts undergo a developmental switch in trophic dependence. Development. 1993;119:597–610. doi: 10.1242/dev.119.3.597. [DOI] [PubMed] [Google Scholar]
- Bodmer D, Brand Y, Radojevic V. Somatostatin receptor types 1 and 2 in the developing mammalian cochlea. Dev Neurosci. 2012;34:342–353. doi: 10.1159/000341291. [DOI] [PubMed] [Google Scholar]
- Cafaro J, Lee GS, Stone JS. Atoh1 expression defines activated progenitors and differentiating hair cells during avian hair cell regeneration. Dev Dyn. 2007;236:156–170. doi: 10.1002/dvdy.21023. [DOI] [PubMed] [Google Scholar]
- Cai T, Seymour ML, Zhang H, Pereira FA, Groves AK. Conditional deletion of Atoh1 reveals distinct critical periods for survival and function of hair cells in the organ of Corti. J Neurosci. 2013;33:10110–10122. doi: 10.1523/JNEUROSCI.5606-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier MS, Taylor JM, Conlon FL. The CASZ1/Egfl7 transcriptional pathway is required for RhoA expression in vascular endothelial cells. Small GTPases. 2013;4:231–235. doi: 10.4161/sgtp.26849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Johnson JE, Zoghbi HY, Segil N. The role of Math1 in inner ear development: uncoupling the establishment of the sensory primordium from hair cell fate determination. Development. 2002;129:2495–2505. doi: 10.1242/dev.129.10.2495. [DOI] [PubMed] [Google Scholar]
- Dahl JA, Collas P. MicroChIP: chromatin immunoprecipitation for small cell numbers. Methods Mol Biol. 2009;567:59–74. doi: 10.1007/978-1-60327-414-2_4. [DOI] [PubMed] [Google Scholar]
- Dechesne CJ, Winsky L, Moniot B, Raymond J. Localization of calretinin mRNA in rat and guinea pig inner ear by in situ hybridization using radioactive and non-radioactive probes. Hear Res. 1993;69:91–97. doi: 10.1016/0378-5955(93)90096-J. [DOI] [PubMed] [Google Scholar]
- Dechesne CJ, Winsky L, Kim HN, Goping G, Vu TD, Wenthold RJ, Jacobowitz DM. Identification and ultrastructural localization of a calretinin-like calcium-binding protein (protein 10) in the guinea pig and rat inner ear. Brain Res. 1991;560:139–148. doi: 10.1016/0006-8993(91)91224-O. [DOI] [PubMed] [Google Scholar]
- Deng M, Pan L, Xie X, Gan L. Differential expression of LIM domain-only (LMO) genes in the developing mouse inner ear. Gene Expr Patterns. 2006;6:857–863. doi: 10.1016/j.modgep.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Di Palma F, Belyantseva IA, Kim HJ, Vogt TF, Kachar B, Noben-Trauth K. Mutations in Mcoln3 associated with deafness and pigmentation defects in varitint-waddler (Va) mice. Proc Natl Acad Sci U S A. 2002;99:14994–14999. doi: 10.1073/pnas.222425399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding D, He J, Allman BL, Yu D, Jiang H, Seigel GM, Salvi RJ. Cisplatin ototoxicity in rat cochlear organotypic cultures. Hear Res. 2011;282:196–203. doi: 10.1016/j.heares.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgoyhen AB, Johnson DS, Boulter J, Vetter DE, Heinemann S. Alpha 9: an acetylcholine receptor with novel pharmacological properties expressed in rat cochlear hair cells. Cell. 1994;79:705–715. doi: 10.1016/0092-8674(94)90555-X. [DOI] [PubMed] [Google Scholar]
- Elgoyhen AB, Vetter DE, Katz E, Rothlin CV, Heinemann SF, Boulter J. alpha10: a determinant of nicotinic cholinergic receptor function in mammalian vestibular and cochlear mechanosensory hair cells. Proc Natl Acad Sci U S A. 2001;98:3501–3506. doi: 10.1073/pnas.051622798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkman L, McEvilly RJ, Luo L, Ryan AK, Hooshmand F, O'Connell SM, Keithley EM, Rapaport DH, Ryan AF, Rosenfeld MG. Role of transcription factors Brn-3.1 and Brn-3.2 in auditory and visual system development. Nature. 1996;381:603–606. doi: 10.1038/381603a0. [DOI] [PubMed] [Google Scholar]
- Frolenkov GI, Belyantseva IA, Friedman TB, Griffith AJ. Genetic insights into the morphogenesis of inner ear hair cells. Nat Rev Genet. 2004;5:489–498. doi: 10.1038/nrg1377. [DOI] [PubMed] [Google Scholar]
- Goodyear RJ, Legan PK, Wright MB, Marcotti W, Oganesian A, Coats SA, Booth CJ, Kros CJ, Seifert RA, Bowen-Pope DF, Richardson GP. A receptor-like inositol lipid phosphatase is required for the maturation of developing cochlear hair bundles. J Neurosci. 2003;23:9208–9219. doi: 10.1523/JNEUROSCI.23-27-09208.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillet N, Schwander M, Hildebrand MS, Sczaniecka A, Kolatkar A, Velasco J, Webster JA, Kahrizi K, Najmabadi H, Kimberling WJ, Stephan D, Bahlo M, Wiltshire T, Tarantino LM, Kuhn P, Smith RJ, Müller U. Mutations in LOXHD1, an evolutionarily conserved stereociliary protein, disrupt hair cell function in mice and cause progressive hearing loss in humans. Am J Hum Genet. 2009;85:328–337. doi: 10.1016/j.ajhg.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves AK, George KM, Tissier-Seta JP, Engel JD, Brunet JF, Anderson DJ. Differential regulation of transcription factor gene expression and phenotypic markers in developing sympathetic neurons. Development. 1995;121:887–901. doi: 10.1242/dev.121.3.887. [DOI] [PubMed] [Google Scholar]
- Groves AK, Zhang KD, Fekete DM. The genetics of hair cell development and regeneration. Annu Rev Neurosci. 2013;36:361–381. doi: 10.1146/annurev-neuro-062012-170309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubbels SP, Woessner DW, Mitchell JC, Ricci AJ, Brigande JV. Functional auditory hair cells produced in the mammalian cochlea by in utero gene transfer. Nature. 2008;455:537–541. doi: 10.1038/nature07265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harland RM. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol. 1991;36:685–695. doi: 10.1016/S0091-679X(08)60307-6. [DOI] [PubMed] [Google Scholar]
- Hartman BH, Hayashi T, Nelson BR, Bermingham-McDonogh O, Reh TA. Dll3 is expressed in developing hair cells in the mammalian cochlea. Dev Dyn. 2007;236:2875–2883. doi: 10.1002/dvdy.21307. [DOI] [PubMed] [Google Scholar]
- Hasson T, Heintzelman MB, Santos-Sacchi J, Corey DP, Mooseker MS. Expression in cochlea and retina of myosin VIIa, the gene product defective in Usher syndrome type 1B. Proc Natl Acad Sci U S A. 1995;92:9815–9819. doi: 10.1073/pnas.92.21.9815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms AW, Abney AL, Ben-Arie N, Zoghbi HY, Johnson JE. Autoregulation and multiple enhancers control Math1 expression in the developing nervous system. Development. 2000;127:1185–1196. doi: 10.1242/dev.127.6.1185. [DOI] [PubMed] [Google Scholar]
- Hertzano R, Dror AA, Montcouquiol M, Ahmed ZM, Ellsworth B, Camper S, Friedman TB, Kelley MW, Avraham KB. Lhx3, a LIM domain transcription factor, is regulated by Pou4f3 in the auditory but not in the vestibular system. Eur J Neurosci. 2007;25:999–1005. doi: 10.1111/j.1460-9568.2007.05332.x. [DOI] [PubMed] [Google Scholar]
- Hertzano R, Elkon R, Kurima K, Morrisson A, Chan SL, Sallin M, Biedlingmaier A, Darling DS, Griffith AJ, Eisenman DJ, Strome SE. Cell type-specific transcriptome analysis reveals a major role for Zeb1 and miR-200b in mouse inner ear morphogenesis. PLoS Genet. 2011;7:e1002309. doi: 10.1371/journal.pgen.1002309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Husseman J, Raphael Y. Gene therapy in the inner ear using adenovirus vectors. Adv Otorhinolaryngol. 2009;66:37–51. doi: 10.1159/000218206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda R, Pak K, Chavez E, Ryan AF. Transcription factors with conserved binding sites near ATOH1 on the POU4F3 gene enhance the induction of cochlear hair cells. Mol Neurobiol. 2015;51:672–684. doi: 10.1007/s12035-014-8801-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imtiaz A, Kohrman DC, Naz S. A frameshift mutation in GRXCR2 causes recessively inherited hearing loss. Hum Mutat. 2014;35:618–624. doi: 10.1002/humu.22545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumikawa M, Minoda R, Kawamoto K, Abrashkin KA, Swiderski DL, Dolan DF, Brough DE, Raphael Y. Auditory hair cell replacement and hearing improvement by Atoh1 gene therapy in deaf mammals. Nat Med. 2005;11:271–276. doi: 10.1038/nm1193. [DOI] [PubMed] [Google Scholar]
- Jacques BE, Montcouquiol ME, Layman EM, Lewandoski M, Kelley MW. Fgf8 induces pillar cell fate and regulates cellular patterning in the mammalian cochlea. Development. 2007;134:3021–3029. doi: 10.1242/dev.02874. [DOI] [PubMed] [Google Scholar]
- Jarman AP, Groves AK. The role of atonal transcription factors in the development of mechanosensitive cells. Semin Cell Dev Biol. 2013;24:438–447. doi: 10.1016/j.semcdb.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalay E, Li Y, Uzumcu A, Uyguner O, Collin RW, Caylan R, Ulubil-Emiroglu M, Kersten FF, Hafiz G, van Wijk E, Kayserili H, Rohmann E, Wagenstaller J, Hoefsloot LH, Strom TM, Nürnberg G, Baserer N, den Hollander AI, Cremers FP, Cremers CW, et al. Mutations in the lipoma HMGIC fusion partner-like 5 (LHFPL5) gene cause autosomal recessive nonsyndromic hearing loss. Hum Mutat. 2006;27:633–639. doi: 10.1002/humu.20368. [DOI] [PubMed] [Google Scholar]
- Kawamoto K, Ishimoto S, Minoda R, Brough DE, Raphael Y. Math1 gene transfer generates new cochlear hair cells in mature guinea pigs in vivo. J Neurosci. 2003;23:4395–4400. doi: 10.1523/JNEUROSCI.23-11-04395.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima Y, Géléoc GS, Kurima K, Labay V, Lelli A, Asai Y, Makishima T, Wu DK, Della Santina CC, Holt JR, Griffith AJ. Mechanotransduction in mouse inner ear hair cells requires transmembrane channel-like genes. J Clin Invest. 2011;121:4796–4809. doi: 10.1172/JCI60405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly MC, Chang Q, Pan A, Lin X, Chen P. Atoh1 directs the formation of sensory mosaics and induces cell proliferation in the postnatal mammalian cochlea in vivo. J Neurosci. 2012;32:6699–6710. doi: 10.1523/JNEUROSCI.5420-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikkawa Y, Shitara H, Wakana S, Kohara Y, Takada T, Okamoto M, Taya C, Kamiya K, Yoshikawa Y, Tokano H, Kitamura K, Shimizu K, Wakabayashi Y, Shiroishi T, Kominami R, Yonekawa H. Mutations in a new scaffold protein Sans cause deafness in Jackson shaker mice. Hum Mol Genet. 2003;12:453–461. doi: 10.1093/hmg/ddg042. [DOI] [PubMed] [Google Scholar]
- Kim TH, Li F, Ferreiro-Neira I, Ho LL, Luyten A, Nalapareddy K, Long H, Verzi M, Shivdasani RA. Broadly permissive intestinal chromatin underlies lateral inhibition and cell plasticity. Nature. 2014;506:511–515. doi: 10.1038/nature12903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klisch TJ, Xi Y, Flora A, Wang L, Li W, Zoghbi HY. In vivo Atoh1 targetome reveals how a proneural transcription factor regulates cerebellar development. Proc Natl Acad Sci U S A. 2011;108:3288–3293. doi: 10.1073/pnas.1100230108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizhanovsky V, Soreq L, Kliminski V, Ben-Arie N. Math1 target genes are enriched with evolutionarily conserved clustered E-box binding sites. J Mol Neurosci. 2006;28:211–229. doi: 10.1385/JMN:28:2:211. [DOI] [PubMed] [Google Scholar]
- Krüger M, Schmid T, Krüger S, Bober E, Braun T. Functional redundancy of NSCL-1 and NeuroD during development of the petrosal and vestibulocochlear ganglia. Eur J Neurosci. 2006;24:1581–1590. doi: 10.1111/j.1460-9568.2006.05051.x. [DOI] [PubMed] [Google Scholar]
- Ku YC, Renaud NA, Veile RA, Helms C, Voelker CC, Warchol ME, Lovett M. The transcriptome of utricle hair cell regeneration in the avian inner ear. J Neurosci. 2014;34:3523–3535. doi: 10.1523/JNEUROSCI.2606-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai HC, Klisch TJ, Roberts R, Zoghbi HY, Johnson JE. In vivo neuronal subtype-specific targets of Atoh1 (Math1) in dorsal spinal cord. J Neurosci. 2011;31:10859–10871. doi: 10.1523/JNEUROSCI.0445-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanford PJ, Lan Y, Jiang R, Lindsell C, Weinmaster G, Gridley T, Kelley MW. Notch signalling pathway mediates hair cell development in mammalian cochlea. Nat Genet. 1999;21:289–292. doi: 10.1038/6804. [DOI] [PubMed] [Google Scholar]
- Lelli A, Asai Y, Forge A, Holt JR, Géléoc GS. Tonotopic gradient in the developmental acquisition of sensory transduction in outer hair cells of the mouse cochlea. J Neurophysiol. 2009;101:2961–2973. doi: 10.1152/jn.00136.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis RM, Hume CR, Stone JS. Atoh1 expression and function during auditory hair cell regeneration in post-hatch chickens. Hear Res. 2012;289:74–85. doi: 10.1016/j.heares.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Price SM, Cahill H, Ryugo DK, Shen MM, Xiang M. Hearing loss caused by progressive degeneration of cochlear hair cells in mice deficient for the Barhl1 homeobox gene. Development. 2002;129:3523–3532. doi: 10.1242/dev.129.14.3523. [DOI] [PubMed] [Google Scholar]
- Liu H, Pecka JL, Zhang Q, Soukup GA, Beisel KW, He DZ. Characterization of transcriptomes of cochlear inner and outer hair cells. J Neurosci. 2014;34:11085–11095. doi: 10.1523/JNEUROSCI.1690-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Dearman JA, Cox BC, Walters BJ, Zhang L, Ayrault O, Zindy F, Gan L, Roussel MF, Zuo J. Age-dependent in vivo conversion of mouse cochlear pillar and deiters' cells to immature hair cells by atoh1 ectopic expression. J Neurosci. 2012;32:6600–6610. doi: 10.1523/JNEUROSCI.0818-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−δδC(T) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Longo-Guess CM, Gagnon LH, Cook SA, Wu J, Zheng QY, Johnson KR. A missense mutation in the previously undescribed gene Tmhs underlies deafness in hurry-scurry (hscy) mice. Proc Natl Acad Sci U S A. 2005;102:7894–7899. doi: 10.1073/pnas.0500760102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lott SE, Villalta JE, Schroth GP, Luo S, Tonkin LA, Eisen MB. Noncanonical compensation of zygotic X transcription in early Drosophila melanogaster development revealed through single-embryo RNA-seq. PLoS Biol. 2011;9:e1000590. doi: 10.1371/journal.pbio.1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma EY, Raible DW. Signaling pathways regulating zebrafish lateral line development. Curr Biol. 2009;19:R381–R386. doi: 10.1016/j.cub.2009.03.057. [DOI] [PubMed] [Google Scholar]
- Ma EY, Rubel EW, Raible DW. Notch signaling regulates the extent of hair cell regeneration in the zebrafish lateral line. J Neurosci. 2008;28:2261–2273. doi: 10.1523/JNEUROSCI.4372-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machanick P, Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011;27:1696–1697. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machold R, Fishell G. Math1 is expressed in temporally discrete pools of cerebellar rhombic-lip neural progenitors. Neuron. 2005;48:17–24. doi: 10.1016/j.neuron.2005.08.028. [DOI] [PubMed] [Google Scholar]
- Maison SF, Liu XP, Vetter DE, Eatock RA, Nathanson NM, Wess J, Liberman MC. Muscarinic signaling in the cochlea: presynaptic and postsynaptic effects on efferent feedback and afferent excitability. J Neurosci. 2010;30:6751–6762. doi: 10.1523/JNEUROSCI.5080-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maragh S, Miller RA, Bessling SL, Wang G, Hook PW, McCallion AS. Rbm24a and rbm24b are required for normal somitogenesis. PLoS One. 2014;9:e105460. doi: 10.1371/journal.pone.0105460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda M, Dulon D, Pak K, Mullen LM, Li Y, Erkman L, Ryan AF. Regulation of POU4F3 gene expression in hair cells by 5′ DNA in mice. Neuroscience. 2011;197:48–64. doi: 10.1016/j.neuroscience.2011.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee J, Goodyear RJ, McMillan DR, Stauffer EA, Holt JR, Locke KG, Birch DG, Legan PK, White PC, Walsh EJ, Richardson GP. The very large G-protein-coupled receptor VLGR1: a component of the ankle link complex required for the normal development of auditory hair bundles. J Neurosci. 2006;26:6543–6553. doi: 10.1523/JNEUROSCI.0693-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt RC, Manor U, Salles FT, Grati M, Dose AC, Unrath WC, Quintero OA, Yengo CM, Kachar B. Myosin IIIB uses an actin-binding motif in its espin-1 cargo to reach the tips of actin protrusions. Curr Biol. 2012;22:320–325. doi: 10.1016/j.cub.2011.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchem KL, Hibbard E, Beyer LA, Bosom K, Dootz GA, Dolan DF, Johnson KR, Raphael Y, Kohrman DC. Mutation of the novel gene Tmie results in sensory cell defects in the inner ear of spinner, a mouse model of human hearing loss DFNB6. Hum Mol Genet. 2002;11:1887–1898. doi: 10.1093/hmg/11.16.1887. [DOI] [PubMed] [Google Scholar]
- Morrison A, Hodgetts C, Gossler A, Hrabé de Angelis M, Lewis J. Expression of Delta1 and Serrate1 (Jagged1) in the mouse inner ear. Mech Dev. 1999;84:169–172. doi: 10.1016/S0925-4773(99)00066-0. [DOI] [PubMed] [Google Scholar]
- Müller U. Cadherins and mechanotransduction by hair cells. Curr Opin Cell Biol. 2008;20:557–566. doi: 10.1016/j.ceb.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvaney J, Dabdoub A. Atoh1, an essential transcription factor in neurogenesis and intestinal and inner ear development: function, regulation, and context dependency. J Assoc Res Otolaryngol. 2012;13:281–293. doi: 10.1007/s10162-012-0317-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura E, Nguyen MT, Mackem S. Kinetics of tamoxifen-regulated Cre activity in mice using a cartilage-specific CreERT to assay temporal activity windows along the proximodistal limb skeleton. Dev Dyn. 2006;235:2603–2612. doi: 10.1002/dvdy.20892. [DOI] [PubMed] [Google Scholar]
- Nakano Y, Jahan I, Bonde G, Sun X, Hildebrand MS, Engelhardt JF, Smith RJ, Cornell RA, Fritzsch B, Bánfi B. A mutation in the Srrm4 gene causes alternative splicing defects and deafness in the Bronx Waltzer mouse. PLoS Genet. 2012;8:e1002966. doi: 10.1371/journal.pgen.1002966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef J, Gehrt A, Bulankina AV, Meyer AC, Riedel D, Gregg RG, Strenzke N, Moser T. The Ca2+ channel subunit β2 regulates Ca2+ channel abundance and function in inner hair cells and is required for hearing. J Neurosci. 2009;29:10730–10740. doi: 10.1523/JNEUROSCI.1577-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes FD, Lopez LN, Lin HW, Davies C, Azevedo RB, Gow A, Kachar B. Distinct subdomain organization and molecular composition of a tight junction with adherens junction features. J Cell Sci. 2006;119:4819–4827. doi: 10.1242/jcs.03233. [DOI] [PubMed] [Google Scholar]
- Odeh H, Hunker KL, Belyantseva IA, Azaiez H, Avenarius MR, Zheng L, Peters LM, Gagnon LH, Hagiwara N, Skynner MJ, Brilliant MH, Allen ND, Riazuddin S, Johnson KR, Raphael Y, Najmabadi H, Friedman TB, Bartles JR, Smith RJ, Kohrman DC. Mutations in Grxcr1 are the basis for inner ear dysfunction in the pirouette mouse. Am J Hum Genet. 2010;86:148–160. doi: 10.1016/j.ajhg.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pack AK, Slepecky NB. Cytoskeletal and calcium-binding proteins in the mammalian organ of Corti: cell type-specific proteins displaying longitudinal and radial gradients. Hear Res. 1995;91:119–135. doi: 10.1016/0378-5955(95)00173-5. [DOI] [PubMed] [Google Scholar]
- Pan N, Jahan I, Kersigo J, Duncan JS, Kopecky B, Fritzsch B. A novel Atoh1 “self-terminating” mouse model reveals the necessity of proper Atoh1 level and duration for hair cell differentiation and viability. PLoS One. 2012;7:e30358. doi: 10.1371/journal.pone.0030358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearsall N, Bhattacharya G, Wisecarver J, Adams J, Cosgrove D, Kimberling W. Usherin expression is highly conserved in mouse and human tissues. Hear Res. 2002;174:55–63. doi: 10.1016/S0378-5955(02)00635-4. [DOI] [PubMed] [Google Scholar]
- Qian D, Radde-Gallwitz K, Kelly M, Tyrberg B, Kim J, Gao WQ, Chen P. Basic helix-loop-helix gene Hes6 delineates the sensory hair cell lineage in the inner ear. Dev Dyn. 2006;235:1689–1700. doi: 10.1002/dvdy.20736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajab A, Kelberman D, de Castro SC, Biebermann H, Shaikh H, Pearce K, Hall CM, Shaikh G, Gerrelli D, Grueters A, Krude H, Dattani MT. Novel mutations in LHX3 are associated with hypopituitarism and sensorineural hearing loss. Hum Mol Genet. 2008;17:2150–2159. doi: 10.1093/hmg/ddn114. [DOI] [PubMed] [Google Scholar]
- Rose MF, Ren J, Ahmad KA, Chao HT, Klisch TJ, Flora A, Greer JJ, Zoghbi HY. Math1 is essential for the development of hindbrain neurons critical for perinatal breathing. Neuron. 2009;64:341–354. doi: 10.1016/j.neuron.2009.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Doi K, Hibino H, Kubo T. Analysis of gene expression profiles along the tonotopic map of mouse cochlea by cDNA microarrays. Acta Otolaryngol Suppl. 2009;562:12–17. doi: 10.1080/00016480902926464. [DOI] [PubMed] [Google Scholar]
- Scheffer D, Sage C, Corey DP, Pingault V. Gene expression profiling identifies Hes6 as a transcriptional target of ATOH1 in cochlear hair cells. FEBS Lett. 2007a;581:4651–4656. doi: 10.1016/j.febslet.2007.08.059. [DOI] [PubMed] [Google Scholar]
- Scheffer D, Sage C, Plazas PV, Huang M, Wedemeyer C, Zhang DS, Chen ZY, Elgoyhen AB, Corey DP, Pingault V. The α1 subunit of nicotinic acetylcholine receptors in the inner ear: transcriptional regulation by ATOH1 and co-expression with the gamma subunit in hair cells. J Neurochem. 2007b;103:2651–2664. doi: 10.1111/j.1471-4159.2007.04980.x. [DOI] [PubMed] [Google Scholar]
- Schmitz F, Königstorfer A, Südhof TC. RIBEYE, a component of synaptic ribbons: a protein's journey through evolution provides insight into synaptic ribbon function. Neuron. 2000;28:857–872. doi: 10.1016/S0896-6273(00)00159-8. [DOI] [PubMed] [Google Scholar]
- Shima Y, Copeland NG, Gilbert DJ, Jenkins NA, Chisaka O, Takeichi M, Uemura T. Differential expression of the seven-pass transmembrane cadherin genes Celsr1–3 and distribution of the Celsr2 protein during mouse development. Dev Dyn. 2002;223:321–332. doi: 10.1002/dvdy.10054. [DOI] [PubMed] [Google Scholar]
- Shin JB, Longo-Guess CM, Gagnon LH, Saylor KW, Dumont RA, Spinelli KJ, Pagana JM, Wilmarth PA, David LL, Gillespie PG, Johnson KR. The R109H variant of fascin-2, a developmentally regulated actin crosslinker in hair-cell stereocilia, underlies early-onset hearing loss of DBA/2J mice. J Neurosci. 2010;30:9683–9694. doi: 10.1523/JNEUROSCI.1541-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JB, Krey JF, Hassan A, Metlagel Z, Tauscher AN, Pagana JM, Sherman NE, Jeffery ED, Spinelli KJ, Zhao H, Wilmarth PA, Choi D, David LL, Auer M, Barr-Gillespie PG. Molecular architecture of the chick vestibular hair bundle. Nat Neurosci. 2013;16:365–374. doi: 10.1038/nn.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroyer NF, Helmrath MA, Wang VY, Antalffy B, Henning SJ, Zoghbi HY. Intestine-specific ablation of mouse atonal homolog 1 (Math1) reveals a role in cellular homeostasis. Gastroenterology. 2007;132:2478–2488. doi: 10.1053/j.gastro.2007.03.047. [DOI] [PubMed] [Google Scholar]
- Simmons DD, Morley BJ. Spatial and temporal expression patterns of nicotinic acetylcholine alpha9 and alpha10 subunits in the embryonic and early postnatal inner ear. Neuroscience. 2011;194:326–336. doi: 10.1016/j.neuroscience.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkkonen ST, Chai R, Jan TA, Hartman BH, Laske RD, Gahlen F, Sinkkonen W, Cheng AG, Oshima K, Heller S. Intrinsic regenerative potential of murine cochlear supporting cells. Sci Rep. 2011;1:26. doi: 10.1038/srep00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son EJ, Wu L, Yoon H, Kim S, Choi JY, Bok J. Developmental gene expression profiling along the tonotopic axis of the mouse cochlea. PLoS One. 2012;7:e40735. doi: 10.1371/journal.pone.0040735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern CD. Detection of multiple gene products simultaneously by in situ hybridization and immunohistochemistry in whole mounts of avian embryos. Curr Top Dev Biol. 1998;36:223–243. doi: 10.1016/s0070-2153(08)60505-0. [DOI] [PubMed] [Google Scholar]
- Thomas S, Thiery E, Aflalo R, Vayssettes C, Verney C, Berthuy I, Créau N. PCP4 is highly expressed in ectoderm and particularly in neuroectoderm derivatives during mouse embryogenesis. Gene Expr Patterns. 2003;3:93–97. doi: 10.1016/S1567-133X(02)00100-X. [DOI] [PubMed] [Google Scholar]
- Togashi H, Kominami K, Waseda M, Komura H, Miyoshi J, Takeichi M, Takai Y. Nectins establish a checkerboard-like cellular pattern in the auditory epithelium. Science. 2011;333:1144–1147. doi: 10.1126/science.1208467. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verpy E, Masmoudi S, Zwaenepoel I, Leibovici M, Hutchin TP, Del Castillo I, Nouaille S, Blanchard S, Lainé S, Popot JL, Moreno F, Mueller RF, Petit C. Mutations in a new gene encoding a protein of the hair bundle cause non-syndromic deafness at the DFNB16 locus. Nat Genet. 2001;29:345–349. doi: 10.1038/ng726. [DOI] [PubMed] [Google Scholar]
- Walker RG, Hudspeth AJ, Gillespie PG. Calmodulin and calmodulin-binding proteins in hair bundles. Proc Natl Acad Sci U S A. 1993;90:2807–2811. doi: 10.1073/pnas.90.7.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis D, Hamblen M, Zhou Y, Venken KJ, Schumacher A, Grimes HL, Zoghbi HY, Orkin SH, Bellen HJ. The zinc finger transcription factor Gfi1, implicated in lymphomagenesis, is required for inner ear hair cell differentiation and survival. Development. 2003;130:221–232. doi: 10.1242/dev.00190. [DOI] [PubMed] [Google Scholar]
- Walsh T, Walsh V, Vreugde S, Hertzano R, Shahin H, Haika S, Lee MK, Kanaan M, King MC, Avraham KB. From flies' eyes to our ears: mutations in a human class III myosin cause progressive nonsyndromic hearing loss DFNB30. Proc Natl Acad Sci U S A. 2002;99:7518–7523. doi: 10.1073/pnas.102091699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler EF, Bothwell M, Schecterson LC, von Bartheld CS. Expression of BDNF and NT-3 mRNA in hair cells of the organ of Corti: quantitative analysis in developing rats. Hear Res. 1994;73:46–56. doi: 10.1016/0378-5955(94)90281-X. [DOI] [PubMed] [Google Scholar]
- Woods C, Montcouquiol M, Kelley MW. Math1 regulates development of the sensory epithelium in the mammalian cochlea. Nat Neurosci. 2004;7:1310–1318. doi: 10.1038/nn1349. [DOI] [PubMed] [Google Scholar]
- Xiang M, Gao WQ, Hasson T, Shin JJ. Requirement for Brn-3c in maturation and survival, but not in fate determination of inner ear hair cells. Development. 1998;125:3935–3946. doi: 10.1242/dev.125.20.3935. [DOI] [PubMed] [Google Scholar]
- Yang J, Bouvron S, Lv P, Chi F, Yamoah EN. Functional features of trans-differentiated hair cells mediated by Atoh1 reveals a primordial mechanism. J Neurosci. 2012a;32:3712–3725. doi: 10.1523/JNEUROSCI.6093-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SM, Chen W, Guo WW, Jia S, Sun JH, Liu HZ, Young WY, He DZ. Regeneration of stereocilia of hair cells by forced Atoh1 expression in the adult mammalian cochlea. PLoS One. 2012b;7:e46355. doi: 10.1371/journal.pone.0046355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylikoski J, Pirvola U, Moshnyakov M, Palgi J, Arumäe U, Saarma M. Expression patterns of neurotrophin and their receptor mRNAs in the rat inner ear. Hear Res. 1993;65:69–78. doi: 10.1016/0378-5955(93)90202-C. [DOI] [PubMed] [Google Scholar]
- Yoon H, Lee DJ, Kim MH, Bok J. Identification of genes concordantly expressed with Atoh1 during inner ear development. Anat Cell Biol. 2011;44:69–78. doi: 10.5115/acb.2011.44.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Lau D, Ng CP, Roy S. Cilia-driven fluid flow as an epigenetic cue for otolith biomineralization on sensory hair cells of the inner ear. Development. 2011;138:487–494. doi: 10.1242/dev.057752. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng JL, Gao WQ. Overexpression of Math1 induces robust production of extra hair cells in postnatal rat inner ears. Nat Neurosci. 2000;3:580–586. doi: 10.1038/75753. [DOI] [PubMed] [Google Scholar]