Abstract
Deadwood is an important biodiversity hotspot in forest ecosystems. While saproxylic insects and wood-inhabiting fungi have been studied extensively, little is known about deadwood-inhabiting bacteria. The study we present is among the first to compare bacterial diversity and community structure of deadwood under field conditions. We therefore compared deadwood logs of two temperate forest tree species Fagus sylvatica and Picea abies using 16S rDNA pyrosequencing to identify changes in bacterial diversity and community structure at different stages of decay in forest plots under different management regimes. Alphaproteobacteria, Acidobacteria and Actinobacteria were the dominant taxonomic groups in both tree species. There were no differences in bacterial OTU richness between deadwood of Fagus sylvatica and Picea abies. Bacteria from the order Rhizobiales became more abundant during the intermediate and advanced stages of decay, accounting for up to 25% of the entire bacterial community in such logs. The most dominant OTU was taxonomically assigned to the genus Methylovirgula, which was recently described in a woodblock experiment of Fagus sylvatica. Besides tree species we were able to demonstrate that deadwood physico-chemical properties, in particular remaining mass, relative wood moisture, pH, and C/N ratio serve as drivers of community composition of deadwood-inhabiting bacteria.
Deadwood is an important structural component in forest ecosystems. It provides shelter and nutrition to various organisms, primarily fungi and saproxylic insects1,2,3. It also partakes in numerous ecosystem functions4, including carbon sequestration and nutrient cycling5,6,7,8. Many investigations have focused on the diversity and composition of fungal communities, their roles in wood decomposition9,10, and their interactions with different forest management regimes11,12. Conversely, the role of prokaryotes in deadwood and related ecosystem processes has only been examined in a few case studies such as the investigation into bacterial communities in deadwood of an East Asian pine species13 and the presence of coexisting bacteria with Hypholoma fasciculare sampled in seven tree stumps by Valášková et al.14. Substrate properties such as nutrient and water content have been shown to strongly influence wood colonization by microbes10,15. Greaves16 first developed a concept concerning a functional classification of wood-inhabiting bacteria: with (1) bacteria that affect permeability but do not cause losses in material strength, (2) bacteria that attack wood structures, (3) bacteria that act as integral synergistically members of the total microflora and (4) the “passive” bacteria, which may act as antagonists to other bacteria. Waterlogged and oxygen-depleted wood is mainly degraded by bacteria17 through a process that may be so slow that trees remain intact long enough to think of salvaging submerged logs18. However, they may be inefficient as wood decayers on their own, though bacteria, especially members of the Actinobacteria are expected to be among the early initial colonizers of deadwood and to permeate or even degrade its lignified cell walls via secretion of cellulases19,20,21. The composition of the primary wood-inhabiting bacterial communities may also be a consequence of the composition of the surrounding soil bacterial communities22. Associations among saprotrophic or wood-decaying Basidiomycota and bacteria in deadwood have been examined in several studies and reviews published over the last three decades21,23,24,25. Both antagonistic25,26 and mutualistic interactions27,28 have been observed during wood decomposition processes. A recent network analysis revealed non-random co-occurrence patterns of bacterial nitrogenase-encoding nifH genes with fungal sporocarps on deadwood logs of Fagus sylvatica and Picea abies29. This finding provided further evidence of potential mutualistic interactions between fungi and methylotrophic N-fixing bacteria that consume methanol, which is a by-product of enzymatic lignin degradation27,30,31. Hervé et al.32 investigated changes in bacterial community structure over time in deadwood inoculated with the lignin-degrading fungus Phanerochaete chrysosporium. They discovered variations in bacterial community composition across time to be incidental, but also identified members of the Burkholderiaceae to be always present in the mycosphere.
Apart from these studies, little is known about the community dynamics of wood-inhabiting bacteria during wood decay in temperate ecosystems. The study we here present is among the first to investigate bacterial diversity and community structure in deadwood under field conditions and applying deep 16S rDNA metabarcoding. Specifically, it compares the bacteria in deadwood logs of two common temperate timber tree species grown in geographic proximity, the deciduous Fagus sylvatica and the conifer Picea abies, at different stages of decay and under different forest management. Deciduous and coniferous woods have very different physico-chemical properties33,34. We anticipated that the structures of the bacterial communities would depend strongly on the properties of the deadwood and the identity of the tree species from which it derived. The primary objective of this study was therefore to determine which of these wood properties correspond or determine the composition and diversity of deadwood-inhabiting bacterial communities, and to identify the key players in the bacterial communities in the two deadwood types on at least the family level. In relation to a previously conducted study on the distribution of nifH genes in deadwood29, we additionally assumed that N-fixing bacteria from the order Rhizobiales were more abundant during the intermediate stages of wood decay, when fungal sporocarp richness is known to be highest and the provision of nitrogen is crucial29.
Results
Wood properties
In addition to assigning each log to a decay class, we determined their C and N contents, the concentrations of both elements per unit wood density (g/cm3), and also their relative wood moisture contents and pH values (Tables S2a and S2b). The C/N ratio in Picea logs ranged from 629.7 ± 48.4 to 422.9 ± 54.4 and was thus substantially greater and more variable than that in Fagus logs (376.9 ± 11.5 to 193.7 ± 15.6). In logs of both tree species, the C/N ratio decreased slightly with increasing decay class. This decrease was largely due to an increase in the logs' N concentration as they decayed (from 0.13 to 0.25% on F. sylvatica and 0.08 to 0.14% on P. abies, Table S2a). Due to the significant decrease of wood density (from 0.5 to 0.2g/cm3 on F. sylvatica and 0.35 to 0.15g/cm3 on P. abies), we also observed a decrease in total N and C content per density unit (Table S2a). There was no significant difference in wood moisture between the two tree species, but the moisture contents of both species' logs increased significantly with the extent of decay, from 52.2% ± 5.7 to 155.2% ± 9.1 in F. sylvatica logs and 48.7% ± 11.6 to 163.1% ± 24.6 in P. abies. Finally, the pH of F. sylvatica logs was significantly higher (P < 0.001) than that of P. abies logs; this trend was independent of the logs' state of decay.
Sequence data analysis
In total, 125,183 reads were obtained from the 47 amplicon libraries by 454 pyrosequencing of the deadwood samples. Sequences were initially quality checked, trimmed (115,750 sequences), normalized per sample (1,837 reads per sample) and screened for potential chimeras. CD-HIT clustering of the remaining 73,099 sequences (discarding potential chimeras) yielded 7,388 OTUs at a 97% cutoff, 5,016 of which were singletons, 807 doubletons, and 368 tripletons. A total of 85 OTUs (among them 56 singletons) stemming from Cyanobacteria related to chloroplasts were also removed from the dataset.
In total, 61,831 sequence reads distributed to 1,180 OTUs were retained for analysis. Taxonomic assignments were achieved for 99.75% of these filtered OTUs (61,776 sequences (99.9%)) at the phylum level (including proteobacterial classes). 1,056 (58,498 sequences (94.6%)), 901 (53,732 sequences (86.9%)) and 633 (39,645 (64.1%)) OTUs were classified at the level of order, family, and genus level, respectively.
Bacterial 16S rDNA diversity, richness and relationship with wood parameters
Species richness across all samples ranged from 105 to 378 OTUs with an average of 258 ± 56. We did not observe any significant differences in mean OTU richness between the two tree species (P = 0.52) (Fig. S2). One-way analyses of the variance in mean bacterial species richness for different decay classes revealed an increase from decay class 1 to 3 in Fagus logs (Fig. S2): the species richness in decay class 1 logs (191.6 ± 21.1) was significantly lower than in decay class 3 (291.9 ± 19.4). We did not detect any significant variation in OTU richness in Picea logs, although the species richness in the two later stages of decay (271 ± 17) was slightly higher than that in stages 1 and 2 (254 ± 14, Fig. S2). This increase is reflected in the observed correlations between OTU richness and the remaining mass of the logs (which provides a measure of their extent of decay). Bacterial OTU richness correlated significantly and positively with the extent of wood decay (P < 0.0005, R2 = 0.45) in Fagus logs but not in Picea logs (P = 0.25, R2 = 0.06) (Fig. S3).
The mean relative abundance of dominant bacterial phyla (including proteobacterial classes) did not differ greatly between the two tree species (Kronafiles SK1, SK2, SK3 in supplementary information). Alphaproteobacteria were dominant on both Fagus (38.9 ± 3.1%) and Picea logs (41.6 ± 2.1%); other abundant phyla included Acidobacteria (14.9 ± 2% and 20.9 ± 0.7%) and Actinobacteria (10.5 ± 0.7% and 11.4 ± 0.1%). However, there were significant differences in the relative abundances of these phyla between decay classes in logs of the same tree species (Fig. 1, Table S3). The relative contribution of Alphaproteobacteria increased from decay class 1 to 4 in both Fagus and Picea logs (from 28.1 ± 3% to 50.7 ± 1.9% and 30.4 ± 3.7% to 44.1 ± 1.5%, respectively) (Tab. S3). The relative abundances of Acidobacteria were consistent across decay classes but the contribution of Firmicutes decreased significantly from decay class 1 to 4 in both Fagus (9.6 ± 3.2% to 0.6 ± 0.2%) and Picea (5.2 ± 0.8% to 0.4 ± 0.2%) logs.
Figure 1. Relative abundances of phylogenetic groups (bacterial phyla including proteobacterial classes) in deadwood from two species (Fagus sylvatica = FASY, Picea abies = PIAB) in different stages of decay (decay classes 1–4).
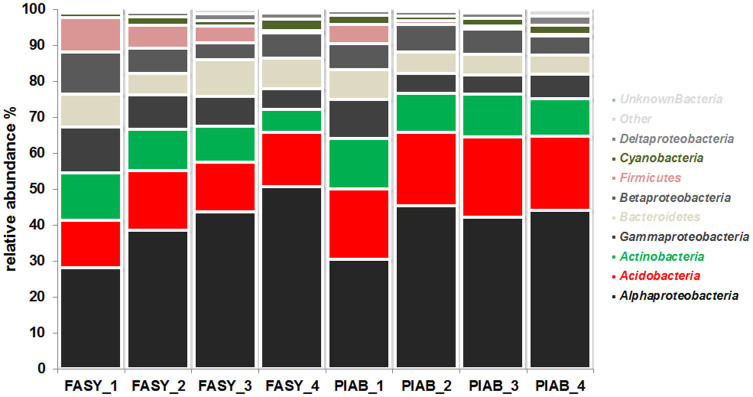
OTUs that could not be taxonomically assigned at the phylum/subphylum level are reported as “Others” and comprise ≤ 0.006% of all sequences. The category “other” also includes all OTUs with <1.5% relative sequence abundance.
These shifts of relative abundances of the dominant phyla have further been observed performing rank abundance correlations (Table S4). This analysis revealed that the remaining mass per log had a significantly negative impact on the abundances of Alphaproteobacteria and Deltaproteobacteria (on F. sylvatica and P. abies), and Cyanobacteria (on F. sylvatica). In turn, Actinobacteria, Firmicutes (both on F. sylvatica and P. abies), Gammaproteobacteria (on logs of and F. sylvatica) and Bacteroidetes (on logs of P. abies) significantly decreased during wood decay. Analogous results were observed for the impact of relative wood moisture on the abundances of the respective phyla, since this parameter significantly increases with mass loss (see Table S2b). The abundances of Alphaproteobacteria (on Fagus sylvatica) and Deltaproteobacteria (on both tree species) correlated negatively and significantly to C/N in contrast to Firmicutes. Members of this phylum were also found to be positively and significantly impacted by higher pH on logs of Picea abies, whereas Acidobacteria correlated negatively and significantly to pH on logs of Fagus sylvatica (Table S4).
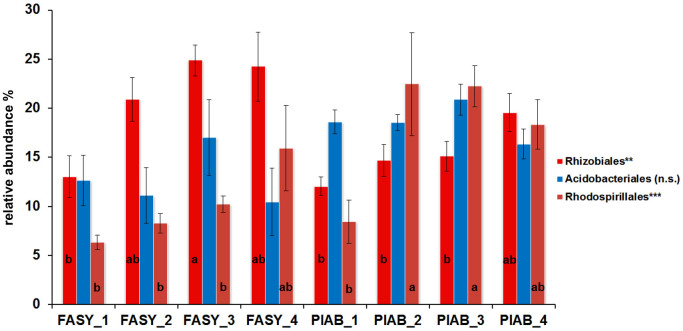
At the order level, members of the Rhizobiales were dominant in Fagus logs (22%), followed by Acidobacteriales (13%) and Rhodospirillales (11%) (Kronafiles SK1 and SK3 in supplementary information, Fig. 2). In Picea logs, Rhodospirillales (20%) were dominant, followed by Acidobacteriales (18%) and Rhizobiales (17%). As shown in Fig. 2, we also observed a significant increase of Rhizobiales from decay class one to three in Fagus logs (15.4 ± 5.8% to 26.9 ± 10.1%). Similarly, for Picea deadwood, the contribution of Rhizobiales went from 14.4 ± 6.4% in decay class 1 to 19.9 ± 6.6% in decay class 4. The most abundant OTU in our dataset appeared to be affiliated with the methanotrophic Methylovirgula genus from the family Beijerinckiaceae. In addition, there were another 5 highly abundant OTUs that were assigned to methanotrophic bacteria of the genera Methyloferula, Methylocella and Methylocystis. Pearson rank correlations revealed that Methylovirgula abundance had a significant negative correlation with the C/N ratio in F. sylvatica logs (R2 = 0.38, P = 0.04) (Table S5). We also found that the abundance of Methylovirgula and Methyloferula correlated significantly and negatively with the remaining log mass but positively with the logs' moisture content (Table S5). No such correlations were observed for Picea logs. Fig. S4 further clearly illustrates the increase in the relative abundance of this dominant OTU as decay proceeds.
Figure 2. Relative abundances of the three dominant phylogenetic groups (bacterial orders) in deadwood of the two studied tree species (Fagus sylvatica = FASY, Picea abies = PIAB) in different stages of decay (decay classes 1–4).
Differences between decay classes and tree species were analyzed by employing one-way analysis of variance and Fisher's Least Significant Difference (LSD) post hoc test (ns = not significant, * P < 0.05, ** P < 0.01, *** P < 0.001).
Effect of forest management on bacterial richness
Our results also revealed a significant impact of forest management regimes on bacterial OTU richness. The analysis of forest management regimes' influence was based on separated data for Fagus and Picea logs in the respective forest plots. When considering Fagus logs in spruce plots, and vice versa (Fig. S5), we observed that the highest OTU richness in Fagus logs was in the unmanaged plots (P < 0.001). While the Picea logs with the greatest OTU richness were those in managed spruce forests (269.2 ± 14.5), their richness was not significantly different to that for spruce deadwood in managed beech forests (248.2 ± 17.1) or unmanaged beech forests (266.8 ± 12.1). We also found that the richness of bacterial OTUs in Fagus logs was negatively associated with land use intensity (Fig. S6) by calculating its correlations with the intensity indices SMI (R2 = 0.31, P = 0.02) and ForMI (R2 = 0.21, P = 0.07).
Bacterial community structure and variation with progressing wood decay
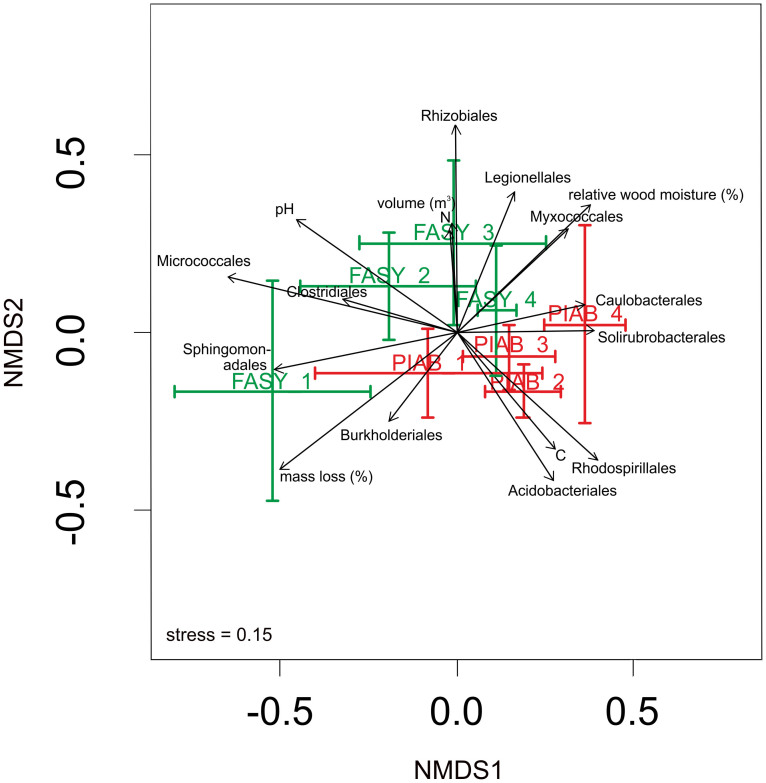
As revealed by perMANOVA and NMDS tree species and decay classes significantly explained the observed variation in bacterial community structure (P = 0.0001), while forest management type did not (Table 1, Fig. 3). The importance of tree species was further confirmed with one-way ANOSIM using either relative abundance (Bray-Curtis or Euclidean) or presence/absence (Jaccard or Sörensen) data (Table S6; always P = 0.001). Furthermore, physico-chemical properties of the different types of deadwood were found to correlate significantly with bacterial community structure (as presented by dominant bacterial order; Fig. 3). Specifically, the decay class, remaining mass, volume, density, relative wood moisture, pH, C/N ratio and the concentrations and contents of C and N in the logs contributed significantly (P = 0.043-0.0001) to the observed variation in bacterial community structure in both tree species (Table 2). At the tree species level, bacterial community structure correlated significantly with decay class, remaining mass, wood density, C/N ratio and C content. Wood volume, C concentrations, N and C content and pH were important in Fagus deadwood, while the C/N ratio contributed significantly to explaining the variation in bacterial community structure in Picea deadwood (Table 2).
Table 1. Results of perMANOVA analysis of the Bray-Curtis dissimilarities for bacterial OTU community structure in relation to tree species, decay class (assigned based on the remaining mass of the log in question), management regime, and their interaction, Df = degrees of freedom; SS = sum of squares; MS = mean sum of squares; Pseudo-F = F value by permutation. Bold face indicates statistical significance (P < 0.05); P-values are based on 9999 permutations (i.e. the lowest possible P-value is 0.0001).
| Df | SS | F | R2 | P | |
|---|---|---|---|---|---|
| Tree species | 1 | 0.8052 | 5.6484 | 0.09982 | 0.0001 |
| Decay class | 1 | 0.6076 | 4.2622 | 0.07532 | 0.0001 |
| Management type | 2 | 0.3967 | 1.3914 | 0.04918 | 0.0527 |
| Tree species x Decay class | 1 | 0.2377 | 1.6674 | 0.02947 | 0.0315 |
| Tree species x Management type | 2 | 0.4011 | 1.4068 | 0.04972 | 0.0466 |
| Decay class x Management type | 2 | 0.3474 | 1.2186 | 0.04307 | 0.1492 |
| Tree species x Decay class x Management type | 2 | 0.2815 | 0.9872 | 0.03489 | 0.4543 |
| Residuals | 35 | 4.9895 | 0.61853 | ||
| Total | 46 | 8.0667 | 1 |
Figure 3. Two-dimensional non-metric multidimensional scaling (NMDS) ordination plots of bacterial community structure across the different tree species at each stage of decay (FASY1-4, PIAB1–4).
Plots show centroids within a single decay stage, bars represent one SD along both NMDS axes. Statistical significances (R2 and P-values) are based on Goodness-of-fit statistics for environmental variables and bacterial order abundances per sample.
Table 2. Goodness-of-fit statistics (R2) for parameters fitted to the non-metric multidimensional scaling (NMDS) ordination of bacterial community structure. The significance estimates were based on 999 permutations. Significant factors (Bonferroni corrected P < 0.05) are indicated in bold. Marginally significant variables (Bonferroni corrected P < 0.10) are indicated in italics. Data on remaining mass%, density, corrected N, and C content are auto-correlated (compare Table S2).
| Fagus and Picea | Fagus | Picea | ||||
|---|---|---|---|---|---|---|
| R2 | P | R2 | P | R2 | P | |
| decay class | 0.5257 | 0.001 | 0.6087 | 0.001 | 0.5205 | 0.001 |
| remaining mass% | 0.6449 | 0.001 | 0.7669 | 0.001 | 0.5999 | 0.001 |
| volume (m3) | 0.1519 | 0.025 | 0.3149 | 0.027 | 0.0806 | 0.308 |
| wood density (g/cm3) | 0.7144 | 0.001 | 0.7536 | 0.001 | 0.6133 | 0.001 |
| rel. wood moisture% | 0.2959 | 0.001 | 0.4438 | 0.004 | 0.3654 | 0.013 |
| C% | 0.3011 | 0.001 | 0.2357 | 0.073 | 0.1944 | 0.126 |
| N% | 0.1352 | 0.043 | 0.1989 | 0.103 | 0.204 | 0.112 |
| C/N | 0.1932 | 0.008 | 0.1917 | 0.109 | 0.2071 | 0.09 |
| C (g/cm3) | 0.7175 | 0.001 | 0.7569 | 0.001 | 0.6174 | 0.001 |
| N (g/cm3) | 0.4622 | 0.001 | 0.3728 | 0.011 | 0.1601 | 0.163 |
| pH | 0.493 | 0.001 | 0.3439 | 0.015 | 0.1438 | 0.197 |
Similarity percentage analysis (SIMPER) for bacterial families revealed contrasting patterns for the two tree species/decay classes. In Fagus logs, members of Acidobacteriaceae (Acidobacteria) explained roughly 17% of the community variation among the 4 decay classes (Fig. S7). Burkholderiaceae (Betaproteobacteria), which accounted for 5% of the total bacteria on Fagus logs, explained roughly 9% of the total variation across different decay classes. This family appeared to be dominant in decay class 1 logs. The opposite pattern was observed for the family Beijerinckiaceae, which explained 8.5% of the community variation and which was almost absent in the earliest stage of decay but became much more abundant as decay progressed (Fig. S7). This family is represented by the most abundant OTU, which was assigned to Methylovirgula. In Picea logs, members of the Acetobacteraceae (Alphaproteobacteria) explained 20% of the total community variation across all decay classes. They were most dominant in decay class 2 logs. Acidobacteriaceae were consistently present at all decay stages and explained 10% of the community variation. They were the dominant bacterial family at all decay stages. Burkholderiaceae and Beijerinckiaceae explained 7 and 6.6% of the community variation, respectively, but there were no clear trends in their abundance comparable to those observed for Fagus logs.
Discussion
We here present one of the first field studies on 16S rDNA bacterial community structure in deadwood of two Central European tree species using 454 pyrosequencing that provides an extensive insight into the diversity and composition of deadwood dwelling microorganisms on a rather underexplored substrate. Using a very comprehensive dataset of physico-chemical wood properties, we were able to identify key wood properties that correlate with bacterial community structure and abundances of dominant bacterial phyla (including proteobacterial classes). In line with the results of our recent study on the distribution of nifH genes29, we found that members of the order Rhizobiales became significantly more abundant during the intermediate to advanced stages of decay, indicating that they may play an important ecological role and contribute significantly to N-cycling.
We did not observe any difference in bacterial richness between the two tree species but did find that richness potentially increased as decay progressed (on logs of Fagus sylvatica). It is difficult to put these findings into context due to the absence of comparable data sets. However, the steady increase in bacterial richness as decay proceeds is consistent with results on fungal species richness obtained using the molecular techniques employed in this work9,35.
Our study also revealed a significant impact of forest management on bacterial OTU richness, which was significantly higher in Fagus logs in the unmanaged beech forest plots than in Fagus and Picea deadwood in the respective managed plots. This suggests that the bacterial communities in unmanaged forests are more species-rich, which may be due to a higher level of substrate continuity arising from the absence of wood extraction12. When comparing Fagus logs in managed beech stands to their counterparts in managed spruce stands (and vice versa for Picea logs), we also discovered a significant impact of the surrounding stand structures on the bacterial richness. Nacke et al.36, who studied soil 16S rDNA diversity in grassland and forest plots using the same experimental platform also detected a significantly higher Shannon diversity index in unmanaged beech plots than managed beech and managed spruce plots. In contrast to Purahong et al.12 who observed a higher fungal diversity in Fagus logs in managed beech stands than in Picea logs in managed spruce stands, we did not find such a clear pattern for bacterial OTU richness.
Alphaproteobacteria, followed by Acidobacteria and Actinobacteria were the dominant phyla in this study. The relative abundances of these phyla are similar to those observed by Nacke et al.36 in forest soils and in a separate study on decayed wood samples14. The relative abundance of Acidobacteria in Picea logs (20.9%) was higher than that in Fagus (14.9%). A previous study using a clone library sequencing approach13 similarly revealed members of the Proteobacteria to be dominant on Keteleeria evelyniana deadwood, followed by Actinobacteria and Acidobacteria. In contrast, Bacteroidetes was the second most abundant phylum (rather than Acidobacteria or Actinobacteria) across a range of samples studied in a different set of wood colonization experiments22. The most abundant OTU in this study was assigned to Mucilaginibacter (Bacteroidetes), a genus that has been shown to contain degraders of pectin and xylan37. This genus was also represented by the 13th most abundant OTU in our dataset. Its abundance in Fagus logs increased continuously from 14.6% to 26.7% from decay classes 1 to 4 but in Picea logs it decreased from 25.3% to 15.2% with increasing decay stage. In contrast to the findings of Hervé et al.32, the genus Dyella of the Xanthomonadaceae only contributed marginally (0.02%) to the total sequence dataset.
Although Actinobacteria were expected to be among the dominant important early colonizers of deadwood21, they contributed only 10.5% and 11.4% of the total bacterial richness in logs of Fagus and Picea, respectively. However, we found that their relative abundance decreased significantly with progressing wood decay confirming their potential role in the early colonization and decomposition of dead wood logs. We assume that their dominant detection in culture-based studies may arise because of culturing conditions favoring them and because of fast germination of the dormant spores under conducive culturing conditions.
In contrast to soils, where pH might serve as predictor for bacterial community structure and as a determinant for relative abundances of dominant bacterial phyla38,39 we did not observe such an important impact of pH in deadwood. In fact, we found negative correlations for Acidobacteria (on P. abies) and Alphaproteobacteria (on F. sylvatica), which have also been reported for soils36,39, but the rather small variance of pH between different wood species and its independency from decay classes does not substantially explain the shifts of the relative abundances of the dominant phyla. Our results rather reveal that the remaining mass or the respective decay class could be used as a potential predictor for the shifts in bacterial abundances in deadwood. However, since they are determined by significant variations of the wood physico-chemical properties (e.g. C and N concentration, relative wood moisture, density; compare Tables S2a and S2b), we assume that variations in the abundances of phyla are rather determined by a combination of wood properties than by single parameters, such as pH, alone.
In line with our assumptions, Rhizobiales was the dominant order in the intermediate and advanced decay classes. In fact, they accounted for almost 25% of the total sequence abundance in F. sylvatica logs of decay class 3. The relative contribution of Rhizobiales in the wood block experiments of Folman et al.25 and de Boer and van der Wal24 was also around 25% even though the total number of wood-inhabiting bacteria in that study was reduced by the bactericidal effects of the white-rot fungus Hypholoma fasciculare. The high abundance of Rhizobiales in that case was attributed to a mutualistic/predatory interaction since they were not detected on wood blocks without the fungus. The hypothesis that wood-degrading fungi meet their N requirements by associating with N-fixing bacteria was first raised by Cowling and Merrill40. This hypothesis has evolved over time, and microbial N-fixation in deadwood has since been demonstrated in several studies conducted over the last few decades41,42,43,44. This hypothesis was further corroborated by the detection of methanotrophic bacteria in deadwood that utilize methane as their sole carbon source45 and possess the nifH gene encoding the enzyme dinitrogenase reductase, which is important for dinitrogen fixation in some taxa31,46. Lenhart et al.47 identified eight saprotrophic fungi that produce significant amounts of methane under oxic conditions in the absence of methanogenic archaea, further supporting the possibility of mutualistic interactions between fungi and methanotrophic N-fixers.
Interestingly, the most abundant OTU in our dataset was taxonomically affiliated to the genus Methylovirgula, whose type strains Vorobev et al.31 isolated from the beech woodblocks studied by Folman et al.25. These methylotrophic bacteria are specialized to utilize methanol as their sole carbon source. Partial sequences of the nifH gene from these strains provided evidence of their N-fixing potential. We found that the abundance of Methylovirgula in Fagus logs correlated significantly and negatively with the C/N ratio (Table S5). Together with the preferential occurrence of this genus during the advanced stages of decay (Fig. S4), this finding supports the hypothesis that wood-decaying fungi interact mutualistically with certain bacterial taxa while suppressing or disfavoring others21,24. This is further supported by the results of Brunner and Kimmins42, who studied deadwood from two coniferous tree species in various stages of decay and found that nitrogenase activity was highest in decay classes 3 and 4. Their data indicated that the total rate of N fixation in such logs was up to 2.1kg N*ha−1*a−1.
Tree species was found as the main factor shaping the bacterial community structure in deadwood. This finding is comparable with the results of previous studies on bacterial community structure in forest soils beneath different tree species36,48. We also found that decay class significantly contributed to the shift in the bacterial community structure unlike the forest management regime. This is entirely consistent with our previous observations on the distribution of nifH genes within the bacterial communities in the same logs29. The observed effect of the tree species mainly attributed to the wood physico-chemical properties (Table 2 and the corresponding attributes in Table S2a). Among these parameters we were able to identify pH, C and N availability, and wood moisture as the main determinants corresponding to drivers of the bacterial community structure in deadwood. These findings can also be compared to the results of recent studies that verified the impact of soil chemical properties (especially pH) on the corresponding bacterial36,39,48 and fungal49 communities.
This paper describes the bacterial diversity and community structure in deadwood of two tree species. The results presented in this study elucidate the functional traits of specific bacterial taxa involved in wood degradation. We have demonstrated that the deadwood bacterial community structures are influenced mainly by the trees species and specifically by wood's physico-chemical properties, where the most important drivers were remaining mass, density, pH, water content and C/N ratio. We also found that members of the order Rhizobiales were dominant in the studied deadwood logs, accounting for up to 25% of all bacteria present during the intermediate and late stages of decay. The most abundant OTU in our data set was assigned to the genus Methylovirgula, which has been shown to contain the nifH gene. This further supports the hypothesis that microbial N-fixation plays an important role in wood decomposition.
Methods
Experimental design, deadwood selection and physico-chemical properties
The study was conducted on plots of the German Biodiversity Exploratories50 located in the “Schwäbische Alb” UNESCO Biosphere Reserve in southwestern Germany according to the sampling scheme displayed in Hoppe et al.29. We surveyed deadwood logs in nine very intensively investigated 1ha plots (VIPs), with three plots representing three different forest types and management regimes, respectively: (i) unmanaged beech forests, where timber harvesting stopped several decades (20–70 years) ago, (ii) managed beech forests dominated by Fagus sylvatica and (iii) managed spruce forests dominated by Picea abies, which in both cases are characterised by uniform tree species composition, forest structure and site conditions. In April 2009, a set of 48 logs, equally representing the two tree species (24 logs per tree species (P. abies and F. sylvatica) located on the forest floor were randomly selected and their properties (length, diameter, tree species, e.g.) were characterized (see Table S1). The logs were selected to ensure that some of the Fagus logs were located in Picea-dominated plots and vice versa. In June 2009, 3–7 wood samples were collected from each log (according to log length; compare Purahong et al.12 and supplementary information) using a cordless Makita BDF451 drill (Makita, Anja, Japan) equipped with a 2 × 42cm wood auger as described by Hoppe et al.29 (further details provided in supplementary information; Fig S1). The upper surface layer and bark of the deadwood was removed to avoid external bacterial contamination. Prior to analysis, the wood samples were weighed, dried at 60°C to constant mass, and reweighed. The concentrations of C and N in wood samples were determined by total combustion using a Truspec elemental analyzer (Leco, St. Joseph, MI, USA). The samples' densities and relative wood moisture contents were calculated based on their dry masses. Sample pH values were determined by shaking 1g of dried wood in 10mL of distilled water for 120 minutes and measuring the pH of the resulting aqueous extract. Each deadwood log was assigned to one of 4 decay classes based on its remaining mass (%) by k-means cluster analysis as described by Kahl et al.5 and Hoppe et al.29. Higher decay classes corresponded to more extensive decay (Tables S2a and S7).
DNA isolation, PCR and pyrosequencing
Total community DNA was isolated from 1g of each previously homogenized wood sample using a modified CTAB-protocol51 as described by Hoppe et al.29. All DNA extracts from the wood subsamples of the 48 logs were pooled into a composite extract prior to PCR amplification. In total, 47 16S rDNA gene amplicon libraries were obtained while no amplified product was found from one sample of Fagus sylvatica. For the amplicon library production we used fusion primers designed with pyrosequencing primer B, a barcode and the primer 341f52 as a forward primer and pyrosequencing primer A and the 907r53 to amplify the V3-V5 region of the eubacterial 16S rDNA gene. The primers were barcoded with a set of 10nt MID-barcodes provided by Roche (Roche Applied Science, Mannheim, Germany). For each composite DNA extract the amplicon libraries was amplified separately by PCR in triplicate 50µl reaction mixtures containing 25 µl 2x GoTaq Green Mastermix (Promega, Madison, WI, USA), 25 µM of each primer and approximately 20 ng template DNA. PCR was performed with an initial denaturation period of 1 min at 98°C followed by 30 cycles of 95°C for 45 s, 57°C for 45 s, and 72°C for 1 min 30s, then a final elongation step at 72°C for 10min. After checking the quality of the PCR products by separation on a 1.5% agarose gel, the replicates were pooled and purified by gel extraction using the QIAquick Gel Extraction Kit (QIAGEN, Hilden, Germany). The purified DNA was quantified using a fluorescence spectrophotometer (Cary Eclipse, Agilent Technologies, Waldbronn, Germany). An equimolar mixture of each library was subjected to unidirectional pyrosequencing from the 907r ends of the amplicons, using a 454 Titanium amplicon sequencing kit and a Genome Sequencer FLX 454 System (454 Life Sciences/Roche Applied Biosystems, Mannheim, Germany) at the department of Soil Ecology, UFZ.
Bioinformatic Analysis
We performed multiple levels of sequence quality filtering. The 454 bacterial 16S sequences were extracted based on 100% barcode similarity. Sequences were trimmed from barcodes and sized to a minimum length of 350nt to cover the V4-V5 region of the 16S rRNA gene using mothur54. Sequence reads with an average quality score of <20, bases and homo-polymers of >8 bases were removed. Unique good quality sequences from the dataset were filtered and checked for chimeras using the uchime algorithm, as implemented in mothur. To avoid sampling size effects, the number of reads per sample was normalized to 1,837 for each data set by randomly subsampling to the lowest number of reads among samples.
The revised complete sequence dataset was then clustered and assigned to OTUs using CD-HIT-EST of CD-HIT version -4.5.455 at a 97% threshold of pairwise sequence similarity. We used GAST (global alignment for sequence taxonomy)56 to taxonomically assign the OTUs against the arb-silva database 11457 as downloaded in August 2013.
Statistical analysis
We performed a procrustes analysis58 using the protest function in vegan59 to test the impact of excluding the rare taxa (singletons, doubletons, tripletons). Procrustes analysis indicated that removing of rare taxa had no effect on the results obtained (procrustes correlation R = 0.94, P = 0.001), so singletons, doubletons, and tripletons were discarded prior to further analysis. OTUs stemming from Cyanobacteria related to chloroplasts were also discarded (85 OTUs incl. 56 singletons).
The effects of tree species, decay class and forest management type on the 16S OTU community structure in the sampled logs were analyzed by perMANOVA. ANOVA was used to assess the influence of tree species and decay class on bacterial OTU richness and the two richness estimators. We performed one-way analysis of variance (ANOVA) to identify significant (P < 0.05) differences between mean OTU richness in association with the respective tree species. ANOVA was coupled with Shapiro-Wilk's W test for normality and Levene's test for equality of group variances. In addition, we assessed the impact of forest management intensity on bacterial diversity using two land use intensity indices, SMI (Silvicultural Management Intensity indicator)60 and ForMi (Forest Management Intensity index)61, that were developed in parallel within the Biodiversity Exploratories research community. Pearson Rank correlations between the relative abundances of dominant bacterial phyla (including proteobacterial subphyla) and selected wood properties were performed in PAST63. One-way analysis of similarities (ANOSIM) calculations based on four commonly used distance measures in conjunction with data on OTU abundance and presence/absence was performed in PAST63 to test for significant differences in bacterial community structures and compositions among different tree species and decay classes, respectively. Assessments of statistical significance were based on 999 permutations and P values were Bonferroni-corrected. NMDS was conducted in R to investigate the influence of the following wood physico-chemical parameters on bacterial community structure: decay classes, the concentrations of the macronutrients C and N, relative wood moisture, wood density, remaining mass, and pH. Goodness-of-fit statistics (R2) for environmental variables fitted to the NMDS ordinations of the bacterial communities were calculated using the envfit function of the “vegan” package, with P values being based on 999 permutations60. Similarity Percentage analysis (SIMPER) was performed in PAST62 to identify the important bacterial families responsible for the observed clustering of samples.
Author Contributions
B.H., D.K., T.K., J.B. and F.B. conceived and designed the study; B.H., T.A. and T.K. performed the field and laboratory work; B.H. and T.W. analyzed the data; B.H. and T.W. wrote the paper. All authors reviewed and edited the manuscript and also provided input for the discussion.
Additional information
Data accessibility: The raw sequence data are available from the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/Traces/study/) under experiment SRX589509. Corresponding MIDs and metadata are provided in Table S7.
Supplementary Material
Supplementary Information
Acknowledgments
Our work was funded in part by contributing projects to the DFG Priority Program 1374 on “Infrastructure-Biodiversity-Exploratories” (KR 3587/1-1, KR 3587/3-2, BA 2821/9-2, BU 941/17-1, HO 1961/5-1, HO 1961/5-2). We thank the managers of the three Exploratories, Swen Renner, Sonja Gockel, Kerstin Wiesner and Martin Gorke for their work in maintaining the plot and project infrastructure; Simone Pfeiffer and Christiane Fischer giving support through the central office, Michael Owonibi for managing the central data base, and Markus Fischer, Eduard Linsenmair, Dominik Hessenmöller, Jens Nieschulze, Daniel Prati, Ingo Schöning, Ernst-Detlef Schulze, Wolfgang W. Weisser and the late Elisabeth Kalko for their role in setting up the Biodiversity Exploratories project. Field work permits were issued by the responsible state environmental offices of Baden-Württemberg (according to § 72 BbgNatSchG). The funders had no role in the study design, data collection and analysis, decision to publish, or the preparation of the manuscript. Julia Moll and Renate Rudloff are thanked for helpful comments or laboratory support. Sigrid Härtling, and Beatrix Schnabel performed the 454 sequencing at the Department of Soil Ecology. Christina Lachmann helped with bioinformatics.
References
- Harmon M. E. et al. Ecology of coarse woody debris in temperate ecosystems. Adv. Ecol. Res. 15, 133–302 (1986). [Google Scholar]
- Lassauce A., Paillet Y., Jactel H. & Bouget C. Deadwood as a surrogate for forest biodiversity: Meta-analysis of correlations between deadwood volume and species richness of saproxylic organisms. Ecol. Indic. 11, 1027–1039 (2011). [Google Scholar]
- Stokland J. N., Siitonen J. & Jonsson B. G. Biodiversity in Dead Wood. [1–509] (Cambridge University Press, Cambridge, 2012). [Google Scholar]
- Cornwell W. K. et al. Plant traits and wood fates across the globe: rotted, burned, or consumed? Glob. Change Biol. 15, 2431–2449 (2009). [Google Scholar]
- Kahl T., Mund M., Bauhus J. & Schulze E. D. Dissolved organic carbon from European beech logs: Patterns of input to and retention by surface soil. Ecoscience 19, 1–10 (2012). [Google Scholar]
- Litton C. M., Raich J. W. & Ryan M. G. Carbon allocation in forest ecosystems. Glob. Change Biol. 13, 2089–2109 (2007). [Google Scholar]
- Herrmann S. & Bauhus J. Effects of moisture, temperature and decomposition stage on respirational carbon loss from coarse woody debris (CWD) of important European tree species. Scand. J. For. Res. 28, 346–357 (2012). [Google Scholar]
- Chambers J. Q., Higuchi N., Schimel J. P., Ferreira L. V. & Melack J. M. Decomposition and carbon cycling of dead trees in tropical forests of the central Amazon. Oecologia 122, 380–388 (2000). [DOI] [PubMed] [Google Scholar]
- Kubartova A., Ottosson E., Dahlberg A. & Stenlid J. Patterns of fungal communities among and within decaying logs, revealed by 454 sequencing. Mol. Ecol. 21, 4514–4532 (2012). [DOI] [PubMed] [Google Scholar]
- Rajala T., Peltoniemi M., Pennanen T. & Makipaa R. Fungal community dynamics in relation to substrate quality of decaying Norway spruce (Picea abies [L.] Karst.) logs in boreal forests. FEMS Microbiol. Ecol. 81, 494–505 (2012). [DOI] [PubMed] [Google Scholar]
- Müller J., Engel H. & Blaschke M. Assemblages of wood-inhabiting fungi related to silvicultural management intensity in beech forests in southern Germany. Eur. J. For. Res. 126, 513–527 (2007). [Google Scholar]
- Purahong W. et al. Changes within a single land-use category alter microbial diversity and community structure: Molecular evidence from wood-inhabiting fungi in forest ecosystems. J. Environ. Manage. 139, 109–119 (2014). [DOI] [PubMed] [Google Scholar]
- Zhang H. B., Yang M. X. & Tu R. Unexpectedly high bacterial diversity in decaying wood of a conifer as revealed by a molecular method. Int. Biodeterior. Biodegradation 62, 471–474 (2008). [Google Scholar]
- Valaskova V., de Boer W., Klein Gunnewiek P. J. A., Pospisek M. & Baldrian P. Phylogenetic composition and properties of bacteria coexisting with the fungus Hypholoma fasciculare in decaying wood. ISME J. 3, 1218–1221 (2009). [DOI] [PubMed] [Google Scholar]
- Volkenant K. Totholz als Lebensraum von Mycozönosen im fortschreitenden Zersetzungsprozess - Eine Chronosequenzstudie an Fagus sylvatica-Totholz im Nationalpark Kellerwald-Edersee. Dissertation thesis, Universität Kassel, Berichte des Forschungszentrums Waldökosysteme. (2007).
- Greaves H. The bacterial factor in wood decay. Wood Sci. Technol. 5, 6–16 (1971). [Google Scholar]
- Kirker G. T., Prewitt M. L., Diehl W. J. & Diehl S. V. Community analysis of preservative-treated southern pine (Pinus spp.) using terminal restriction fragment length polymorphism (T-RFLP) analysis. Part 2: Bacteria field study. Holzforschung 66, 529–535 (2012). [Google Scholar]
- Tenenbaum D. J. Underwater logging: Submarine rediscovers lost wood. Environm. Health. Persp. 112, 892–895 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel G. & Nilsson T. [Developments in the study of soft rot and bacterial decay.]. Forest Products biotechnology. [Bruce A., , Palfreyman J. W., eds. (eds)] [37–62] (Taylor & Francis: London 1998). [Google Scholar]
- Lynd L. R., Weimer P. J., van Zyl W. H. & Pretorius I. S. Microbial cellulose utilization: Fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 66, 506–577 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen C. A. Bacterial associations with decaying wood: A review. Int. Biodeterior. Biodegradation 37, 101–107 (1996). [Google Scholar]
- Sun H., Terhonen E., Kasanen R. & Asiegbu F. O. Diversity and community structure of primary wood-inhabiting bacteria in boreal forest. Geomicrobiol. J. 31, 315–324 (2013). [Google Scholar]
- Blanchette R. A. & Shaw C. G. Associations among bacteria, yeasts, and basidiomycetes during wood decay. Phytopathology 68, 631–637(1978). [Google Scholar]
- de Boer W., van der Wal A. [Interactions between saprotrophic basidiomycetes and bacteria.]. Ecology of saprotrophic basidiomycetes. [Boddy L., , Franklin J. C., , van West P., eds. (eds.)] [143–153] (Mycological Society Symposia Series, vol. 28, Academic Press, London, 2008). [Google Scholar]
- Folman L. B., Klein Gunnewiek P. J. A., Boddy L. & de Boer W. Impact of white-rot fungi on numbers and community composition of bacteria colonizing beech wood from forest soil. FEMS Microbiol. Ecol. 63, 181–191 (2008). [DOI] [PubMed] [Google Scholar]
- de Boer W. et al. Mechanism of antibacterial activity of the white-rot fungus Hypholoma fasciculare colonizing wood. Can. J. Microbiol. 56, 380–388 (2010). [DOI] [PubMed] [Google Scholar]
- Merrill W. & Cowling E. B. Role of nitrogen in wood deterioration - amount and distribution of nitrogen in fungi. Phytopathology 56, 1083–1090 (1966). [Google Scholar]
- Lindahl B. D. & Finlay R. D. Activities of chitinolytic enzymes during primary and secondary colonization of wood by basidiomycetous fungi. New Phytol. 169, 389–397 (2006). [DOI] [PubMed] [Google Scholar]
- Hoppe B. et al. Network analysis reveals ecological links between N-fixing bacteria and wood-decaying fungi. PLoS ONE e88141. 10.1371/journal.pone.0088141 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedysh S., Horz H.-P., Dunfield P. & Liesack W. A novel pmoA lineage represented by the acidophilic methanotrophic bacterium Methylocapsa acidophila B2. Arch. Microbiol. 177, 117–121 (2001). [DOI] [PubMed] [Google Scholar]
- Vorobev A. V. et al. Methylovirgula ligni gen. nov., sp nov., an obligately acidophilic, facultatively methylotrophic bacterium with a highly divergent mxaF gene. Int. J. Syst. Evol. Microbiol. 59, 2538–2545 (2009). [DOI] [PubMed] [Google Scholar]
- Hervé V., Le Roux X., Uroz S., Gelhaye E. & Frey-Klett P. Diversity and structure of bacterial communities associated with Phanerochaete chrysosporium during wood decay. Environm. Microbiol. 16, 2238–2252 (2014). [DOI] [PubMed] [Google Scholar]
- Carlquist S. Comparative wood anatomy: Systematic, ecological, and evolutionary aspects of dicotyledon wood. [1–448] (Springer, Berlin-Heidelberg, 2001) [Google Scholar]
- Fengel D. & Wegener G. Wood: chemistry, ultrastructure, reactions [1–613] (Walter de Gruyter, New York, 1983). [Google Scholar]
- Ovaskainen O. et al. Combining high-throughput sequencing with fruit body surveys reveals contrasting life-history strategies in fungi. ISME J. 7, 1696–1709 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacke H. et al. Pyrosequencing-based assessment of bacterial community structure along different management types in German forest and grassland soils. PLoS ONE e17000. 10.1371/journal.pone.0017000 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankratov T. A., Tindall B. J., Liesack W. & Dedysh S. N. Mucilaginibacter paludis gen. nov., sp. nov. and Mucilaginibacter gracilis sp. nov., pectin-, xylan- and laminarin-degrading members of the family Sphingobacteriaceae from acidic Sphagnum peat bog. Int. J. Syst. Evol. Microbiol. 57, 2349–2354 (2007). [DOI] [PubMed] [Google Scholar]
- Baker K. L. et al. Environmental and spatial characterisation of bacterial community composition in soil to inform sampling strategies. Soil Biol. Biochem. 41, 2292–2298 (2009). [Google Scholar]
- Lauber C. L., Hamady M., Knight R. & Fierer N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environm. Microbiol. 75, 5111–5120 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowling E. B. & Merrill W. Nitrogen in wood and its role in wood deterioration. Can. J. Bot. 44, 1539–1554 (1966). [Google Scholar]
- Aho P. E., Seidler R. J., Evans H. J. & Raju P. N. Distribution, enumeration, and identification of Nitrogen-fixing bacteria associated with decay in living white fir trees. Phytopathology 64, 1413–1420 (1974). [Google Scholar]
- Brunner A. & Kimmins J. P. Nitrogen fixation in coarse woody debris of Thuja plicata and Tsuga heterophylla forests on northern Vancouver Island. Can. J. For. Res. 33, 1670–1682 (2003). [Google Scholar]
- Seidler R. J., Aho P. E., Evans H. J. & Raju P. N. Nitrogen fixation by bacterial isolates from decay in living white fir trees [Abies concolor (Gord and Glend) Lindl]. J. Gen. Microbiol. 73, 413–416 (1972). [Google Scholar]
- Spano S. D., Jurgensen M. F., Larsen M. J. & Harvey A. E. Nitrogen-fixing bacteria in Douglas fir residue decayed by Fomitopsis pinicola. Plant Soil 68, 117–123 (1982). [Google Scholar]
- Hanson R. S. & Hanson T. E. Methanotrophic bacteria. Microbiol. Rev. 60, 439–471 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobev A. V. et al. Methyloferula stellata gen. nov., sp. nov., an acidophilic, obligately methanotrophic bacterium that possesses only a soluble methane monooxygenase. Int. J. Syst. Evol. Microbiol. 61, 2456–2463 (2011). [DOI] [PubMed] [Google Scholar]
- Lenhart K. et al. Evidence for methane production by saprotrophic fungi. Nat. Commun. 10.1038/ncomms2049 (2012). [DOI] [PubMed] [Google Scholar]
- Hackl E., Zechmeister-Boltenstern S., Bodrossy L. & Sessitsch A. Comparison of diversities and compositions of bacterial populations inhabiting natural forest soils. Appl. Environm. Microbiol. 70, 5057–5065 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wubet T. et al. Differences in soil fungal communities between European beech (Fagus sylvatica L.) dominated forests are related to soil and understory vegetation. PLoS ONE e47500. 10.1371/journal.pone.0047500 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M. et al. Implementing large-scale and long-term functional biodiversity research: The Biodiversity Exploratories. Basic Appl. Ecol. 11, 473–485 (2010). [Google Scholar]
- Doyle J. J. & Doyle J. L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15 (1987). [Google Scholar]
- Muyzer G., Dewaal E. C. & Uitterlinden A. G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environm. Microbiol. 59, 695–700 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D. J. [16S/23S rRNA sequencing]. Nucleic acid techniques in bacterial systematics. [Stackebrandt E., , Goodfellow M., eds. (eds)] [125–175] (Wiley: New York, 1991). [Google Scholar]
- Schloss P. D. et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environm. Microbiol. 75, 7537–7541 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. & Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 (2006). [DOI] [PubMed] [Google Scholar]
- Huse S. M. et al. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. Plos Genetics e1000255. 10.1371/journal.pgen.1000255 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, 590–596 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peres-Neto P. R. & Jackson D. A. How well do multivariate data sets match? The advantages of a Procrustean superimposition approach over the Mantel test. Oecologia 129, 169–178 (2001). [DOI] [PubMed] [Google Scholar]
- Oksanen J. Multivariate analysis of ecological communities in R: vegan tutorial. (2013). at <http://cc.oulu.fi/~jarioksa/opetus/metodi/vegantutor.pdf> (accessed: August 20th 2014).
- Schall P. & Ammer C. How to quantify forest management intensity in Central European forests. Eur. J.For. Res. 132, 379–396 (2013). [Google Scholar]
- Kahl T. & Bauhus J. An index of forest management intensity based on assessment of harvested tree volume, tree species composition and dead wood origin. Nat. Conserv. 7, 15–27 (2014). [Google Scholar]
- Hammer Ø., Harper D. A. T. & Ryan P. D. PAST: Paleontological statistics software package for education and data analysis. Palaeontologia Electronica 4, 1–9 (2001). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information