Abstract
Introduction
Flow cytometry has been around for over 40 years, but only recently has the opportunity arisen to move into the high-throughput domain. The technology is now available and is highly competitive with imaging tools under the right conditions. Flow cytometry has, however, been a technology that has focused on its unique ability to study single cells and appropriate analytical tools are readily available to handle this traditional role of the technology.
Areas covered
Expansion of flow cytometry to a high-throughput (HT) and high-content technology requires both advances in hardware and analytical tools. The historical perspective of flow cytometry operation as well as how the field has changed and what the key changes have been discussed. The authors provide a background and compelling arguments for moving toward HT flow, where there are many innovative opportunities. With alternative approaches now available for flow cytometry, there will be a considerable number of new applications. These opportunities show strong capability for drug screening and functional studies with cells in suspension.
Expert opinion
There is no doubt that HT flow is a rich technology awaiting acceptance by the pharmaceutical community. It can provide a powerful phenotypic analytical toolset that has the capacity to change many current approaches to HT screening. The previous restrictions on the technology, based on its reduced capacity for sample throughput, are no longer a major issue. Overcoming this barrier has transformed a mature technology into one that can focus on systems biology questions not previously considered possible.
Keywords: automated classification, drug screening, flow cytometry, high throughput, pathway analysis, phenotypic classification
1. Introduction
The power of flow cytometry lies in its unique ability to acquire measurements on every single cell without interference from sample background or other cells, and evaluate each one for many different functional parameters, all in microseconds. The fastest cell sorters can process around 100,000 cells/second, and the fastest analyzers about 70,000 cells/second. Therefore, these instruments offer far greater cell-analysis rates than any imaging system available today, including all the high-throughput HCS instruments so readily used for screening. The major limitations of flow cytometry are the need to keep cells in suspension and the very limited access to spatial information. This is an obvious disadvantage if the model cells of interest are those attached to culture dishes.
Flow cytometry (FC) does not have a long tradition of being employed as a high-throughput cell-analysis technology, where the term “high-throughput” implies thousands to tens of thousands of samples per day. In fact, most common applications of FC analysis process only 50 to 150 samples on any given day. Thus for researchers familiar only with traditional applications of flow cytometry and conventional hardware, this technology would not be associated with a high sample-analysis rate.
However, this notion is not correct. The modern FC instruments are suited to high-throughput studies, and the availability of high-throughput cytometry (HT) may be one of the most exciting technological opportunities for the fields of study employing a systems biology approach. With laboratory automation and robotics, it is now possible to process significantly larger numbers of samples per day on a single instrument. Subsequent analyses of multiple populations of cells to quantify various phenotypes can be accomplished in almost real time. However, there are significant differences in how these studies are performed using HT devices, as opposed to traditional flow cytometry. Additionally, it has to be recognized, issues are that HT flow cytometry is still an emerging technology; therefore the practical issues facing the users are very significant.
Flow cytometry is a tool that allows analysis of single cells producing data that are inherently quantitative in nature. The original attempts to study single cells, in the 1930s, involved simple tools focused on counting individual blood cells [1]. Subsequently, the flow cytometry pioneers introduced [2,3] sensitive photodetectors to separate cells and particles and attempted to evaluate cellular content by measuring cellular absorbance [4] or by other electronic means [5]. In the early 1950s, Wallace Coulter proposed a highly accurate cell counter based on measurement of impedance that changed the world of cellular analysis [6,7]. This technology was the basis for Mack Fulwyler’s invention of the cell sorter, the first true flow cytometer in today’s sense of the word. When Fulwyler designed his cell sorter in 1965, little did he know that the fundamental technology of his instrument, which was able to measure just a couple of samples a day [8], in less than 50 years would be the basis for automated systems with the ability to process a thousand samples an hour [9]. As flow cytometry significantly increased its capabilities, so too were the applications expanded, from cellular impedance measurement to one-color fluorescence[10], all the way to studies employing 17 or more fluorescently-labeled biomarkers [11]. Thus it can be argued that modern flow cytometry can acquire by far the highest content among single-cell analysis tools. More recent developments in flow cytometry hardware allowed the amount of functional data per cell to increase well beyond any assay available for imaging modalities; this will be discussed below [12].
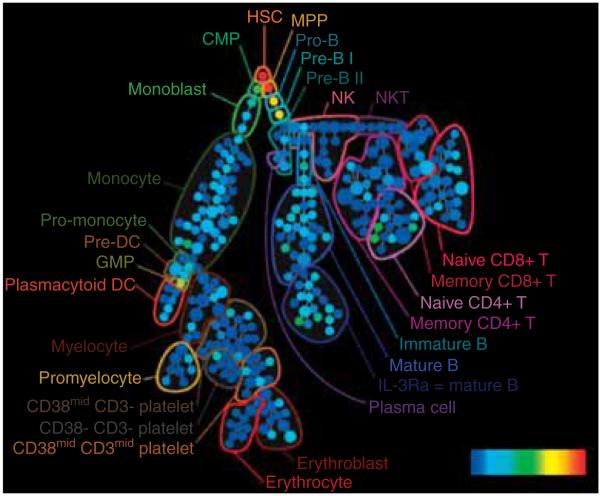
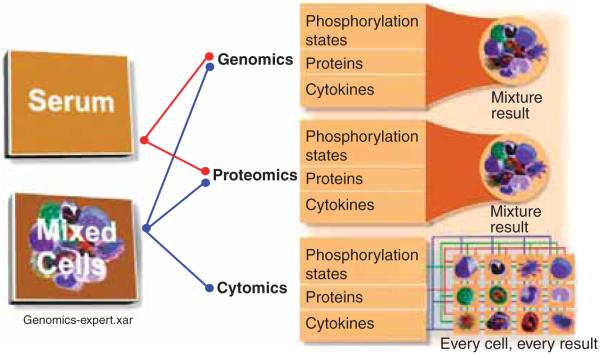
What sets flow cytometry apart from other technologies is the ability to study, measure, and analyze heterogeneous populations of cells one cell at a time. While genomic and proteomic approaches are powerful systems biology tools, they also have their disadvantages owing to the fact that samples are processed and analyzed in bulk. However, if mixed cells (serum, tissue, etc.) are the starting materials, the specific source of detected biological response is unclear. Proteins found in mixed-cell samples could be derived from any, some, or all of the present cell populations. On the other hand, FC is able to differentiate various cell types [13-22]. This ability to classify individual cells into populations defined on a basis of phenotypic differences is a strong suit of cytometry and enables this approach to study complex mixtures of cells without loss of information. Figure 1 illustrates this issue. In most cases, the separation and classification of cells is achieved using unique surface markers signifying well-understood functional properties of cells.
Figure 1.
The major advantage of the cytomics (i.e., single-cell analysis) approach is the ability to interrogate single cells and to separate functionally and phenotypically distinctive populations of cells during the process of statistical analysis of the entire biological system. Every defined population can be studied separately or in the context of functional relations to other populations.
Reproduced from a poster presented at the NIH Common Fund Single Cell Analysis Workshop, April 17 – 18, 2012 [115] by the University Cytometry Laboratories with their permission.
However, flow cytometry is not restricted to cellular surface-antigen expression. Functional characteristics are also well within the capacity of the technology and facilitate the differentiation of functional subsets of cellular populations [23-44].
2. The traditional data-analysis techniques of flow cytometry
The traditional flow cytometry analysis pipeline involves fluorescence-based detection of descriptors of cell function or molecular content and an interactive, operator-guided data-analysis component that uses 1- or 2-D visualization formats that allow “gating” (selection of cellular populations of interest) and computation of summary statistics. The data processing can be performed by an FC operator using various dedicated cytometry-analysis packages. One of the most important aspects of this technology is a well-established standardization of data structure, described by the Flow Cytometry Standard (FCS), that was established 3 decades ago by the International Society for Advancement of Cytometry (ISAC), and has been recognized by all FC hardware manufacturers [45]. This standard was further enhanced to FCS 2.0 in 1990 [46], and again in 1997 to FCS 3.0 [47], with a minor revision to FCS 3.1 in 2010 [48]. The next-generation version of the standard is currently in development, and will cover not only the data representation but also many important aspects of processing an analysis, such as gating, mathematical transformations, etc. [49]. The existence of the standard leads to a thriving market of third-party data analysis packages. Commercial programs such as FCS Express™ (De Novo Software, Los Angeles, CA), FlowJo™ (Tree Star, Ashland, OR), WinList (Verity Software House, Topsham, Maine), Kaluza™ (Beckman-Coulter, Brea, CA), and others can be used with data obtained from various FC instruments. Additionally, a number of free tools written for users of R language for scientific computing are available within Bioconductor library Table 1 (see above).
Table 1.
Flow cytometry data analysis libraries for R/Bioconductor environment. While most of these do require a moderately high level of proficiency in R language, they provide functionality not available in off-the-shelf flow cytometry software packages.
| Bioconductor package | Functionality | Authors | Ref. |
|---|---|---|---|
| Basic computational infrastructure | |||
| flowCore | S4 data structures and basic functions to read/write and process flow cytometry data files |
Byron Ellis, Perry Haaland, Florian Hahne, Nolwenn Le Meur, Nishant Gopalakrishnan |
[97,98] |
| plateCore | Basic S4 data structures and routines for analyzing flow cytometry samples collected from multiwall plates |
Errol Strain, Florian Hahne, and Perry Haaland | [99] |
| flowStats | Methods and functions to analyze flow cytometry data beyond the basic infrastructure provided by the flowCore package |
Florian Hahne, Nishant Gopalakrishnan, Alireza Hadj Khodabakhshi, Chao-Jen Wong, Kyongryun Lee |
[100] |
| flowUtils | Functions to import gates, transformations, and compensations defined in compliance with Gating-ML standard |
Nishant Gopalakrishnan, Florian Hahne, Byron Ellis, Robert Gentleman, Mark Dalphin, Nolwenn Le Meur, Barclay Purcell |
[101] |
| flowFlowJo | Importation of basic flowJo workspaces into BioConductor environment | John J. Gosink | [102] |
| ncdfFlow | netCDF storage-based methods and functions for manipulation of flow cytometry data |
Mike Jiang, Greg Finak, Nishant Gopalakrishnan | [103] |
| Processing, clustering, gating, automated analysis, and quality control | |||
| Logicle | Generalized biexponential transform for cytometry data | Wayne Moore, David Parks | [80] |
| flowMeans | Non-parametric clustering for flow cytometry | Nima Aghaeepour | [84] |
| flowClust | Robust model-based clustering using a t-mixture model with Box–Cox transformation |
Raphael Gottardo, Kenneth Lo | [85] |
| samSPECTRAL | Data reduction and spectral clustering for analysis of high-throughput flow cytometry data |
Habil Zare and Parisa Shooshtari | [104] |
| flowMerge | Merging of mixture components for model-based automated gating | Greg Finak, Raphael Gottardo | [105] |
| flowType | Automated phenotyping | Nima Aghaeepour | [106] |
| flowTrans | Maximum likelihood estimation of parameters for flow cytometry data transformation |
Greg Finak, Juan Manuel-Perez, Raphael Gottardo | [87] |
| flowFP | Fingerprint generation for multidimensional flow cytometry data | Herb Holyst, Wade Rogers | [88] |
| flowPhyto | Automated computational methods for analysis of marine biology data obtained from high-throughput systems such as SeaFlow |
David M. Schruth and Francois Ribalet | [107] |
| flowQ | Quality control tools for flow cytometry data | Robert Gentleman, Florian Hahne, J. Kettman, Nolwenn Le Meur, Nishant Gopalakrishnan |
[108] |
| QUALIFIER | Quality control tools for gated flow cytometry data | Mike Jiang, Greg Finak, Raphael Gottardo | [109] |
| RchyOptimyx | Automated determination of the minimal sets of markers Able to identify a target population to a desired level of purity |
Adrin Jalali, Nima Aghaeepour | [110] |
| FlowAND | FlowAnd is a flow cytometric tool for systematic and efficient analysis of large multidimensional datasets |
Anna-Maria Lahesmaa-Korpinen, Ping Chen, Erkka Valo, Javier Nunez Fontarnau, Sampsa Hautaniemi |
[111] |
| Visualization and interactive analysis | |||
| flowViz | Visualization tools for flow cytometry data (histograms, dot plots, density plots, etc.) |
Byron Ellis, Robert Gentleman, Florian Hahne, Nolwenn Le Meur, Deepayan Sarkar |
[112] |
| iFlow | Visualization and exploratory analysis of flow cytometry data | Kyongryun Lee, Florian Hahne, Deepayan Sarkar | [113] |
| flowPlots | Graphical displays with embedded statistical tests for flow cytometry | Natalie Hawkins, Steve Self | [114] |
| SPADE | Processing and visualization of multidimensional FC data using spanning-tree progression of density-normalized events (SPADE) |
Michael Linderman, Erin Simonds, Zach Bjornson, Peng Qiu |
[72] |
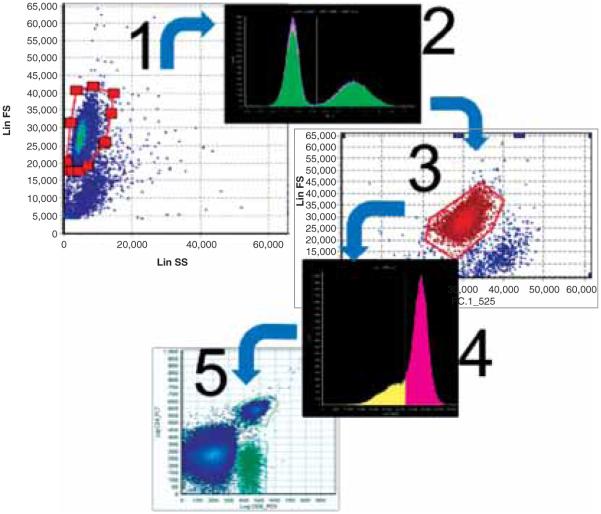
All these tools use a conceptually similar process, which starts with so-called compensation (linear unmixing of raw fluorescence readouts using information available from single-stained controls), followed by visualization of the compensated values in the form of 2-D dot plots or density plots that enable creation of gates (manually defined regions of interest) in a cascading manner (see Figure 2).
Figure 2.
Traditional flow cytometry analyzes one sample at a time. Due to the interactivity required for visualization and gating, this approach is relatively slow and cumbersome. In the provided example four gating steps were defined. The first and the third gates are polygonal and are defined in two-dimensional spaces formed by FC-measured variables. The steps numbered 2 and 4 use only one variable and simple thresholding to separate cells demonstrating high intensity of fluorescenceinspecificbands.
Some specific applications of flow cytometry require more specialized approaches. For example, Gemstone™ (Verity Software House, Topsham, ME) uses Markov-chain modeling to define the origin and final differentiation state of a person’s white blood cells. This technique enables a physician to determine the differentiation state of cell types that might be predictive of particular clinical conditions, or a clinical scientist to better understand the outcome of patients for whom flow cytometry is being used to monitor changes in cellular phenotypes [50,51].
3. High-throughput flow cytometry
3.1 The emergence of the technology
While there is no authoritative threshold of throughput for a high-throughput screening (HTS) technology, some fundamental premises are generally accepted. First, assays are carried out in 96-, 384-, or 1536-well plates that are usually set up and manipulated within the context of an automation system. Second, screening practitioners usually define throughput in terms of the “number of compounds” that can be screened per day. If single wells are the basis for reporting the results of screening, most HTS operations would expect at least 10,000 to 50,000 collectable events daily; some technologies claim far higher efficiency, such as up to 108 enzyme-based reactions per day [52].
The technology of high-throughput flow cytometry combines the very high cell-analysis rate – a capability of any FC instrumentation – with speedy sample processing and access to common HTS sample-handling formats such as multi-well plates.
It is not uncommon for a technological concept to be envisioned many years prior to its emergence as a practical capability. In the case of HTFC, the key ideas of barcoding samples [53], using complex file structure for storing data representations of multiple samples [54], and the use of time-of-measurement as a parameter for separating multiple continuously measured samples [55] were all proposed over 20 years ago, and all are now fundamental components of HTFC domain.
Although the carousels for traditional analysis tubes have been available for many FC instrument providing a possibility of automated analysis, the key peripheral for making HTFC a reality came through the integration of robotic sampling into the FC process by Sklar’s group [56-58]. This was transformational for the field – the Sklar’s technology allowed analysis of hundreds of samples arranged in standard multi-well plates in a high-speed, orderly fashion. The advantage of Sklar’s device is that it allows sampling of small volumes (~ 1 µL). In contrast, most current sampling systems available for commercial flow cytometers sample 25 – 250 µL – volumes far greater than normally available for the majority of HT assays. Integration with robust robotic technology effects sampling for 96-, 384- or 1536-well plates.
3.2 Multifactorial high-throughput FC
The concept of HT cellular screening implies that a great number of samples will be processed, but does not indicate whether the measurements should represent responses of cells exposed to thousands of compounds, or to just a few compounds characterized by very large number of variables describing cellular responses in multiple relevant conditions. Multifactorial flow cytometry, which focuses on the latter model, may potentially have a greater impact on systems biology than the traditionally understood throughput-focused screening. It is important to underline the difference between traditional multiparametric flow cytometry and multifactorial flow cytometry. The increase in number of simultaneously measured biomarkers conveying functional parameters of cells underscores multiparamertic cytometry. However, it is the availability of automation and high throughput that allowed for rapid collection of data from biological samples exposed to various environments or perturbants (drugs or growth factors). Therefore, multifactorial data describe a complex response pattern that a heterogeneous population may demonstrate. An example of such a multifactorial flow cytometry study performed by Nolan’s group is summarized in Figure 3 (figure is based on personal communication). In this example, more than 2000 dose–response curves characterizing 14 populations exposed to a set of activating molecules and various drugs were generated from cells arranged in a single 96-well plate.
Figure 3.
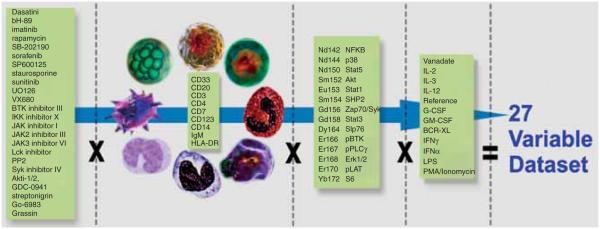
The multifactorial experimental design demonstrated in this figure, based on a study from Bodenmiller (personal communication), allows simultaneous study of responses demonstrated by nine populations of cells to a number of drugs in the presence of 14 activators. Over 30 functional parameters based on molecular markers of phosphorylation were measured for every one of the 14 cell types in the system.
It is also crucial to note that multifactorial flow cytometry differs in some important aspects from other multifactorial analysis techniques, such as image-based HCS. The techniques utilizing flow employ labels (usually fluorescence-based) which directly indicate the presence of certain molecular markers or physiological features. Imaging systems can simultaneously collect only a very limited number of spectrally distinctive fluorescence intensities. A typical image-based study employs three fluorescence markers, whereas studies utilizing a flow cytometry system often make use of 10 markers and more. On the other hand, a huge number of secondary parameters can be derived from just a few image-based features [59] For example, using just one fluorescent label, an automated microscope system can derive various descriptors of location, texture, or shape [60]. These concepts are employed in techniques such as location proteomics [61]. Often the image-based descriptors can be used to characterize very complex biological responses that are not necessarily easy to summarize by any single molecular biomarker.
The employed descriptors in these advanced imaging approaches, such as image texture in the nuclear region or other exotic feature, may not be directly linked to any well-defined molecular biomarker but this not mean that they are less powerful. For example, in the study by Neumann et al. the researchers were able to pinpoint involvement of over 500 genes in the cell-division process, using millions of images characterized by hundreds of various image features [62]. If one attempted to repeat such a study using molecular markers for every one of the studied genes, one would need 500 well-defined molecular targets for labeling.
Consequently, image-based single-cell analysis excels if cell function or observed perturbation is described in a fuzzy way (for instance, by change in shape during the differentiation). It is not an accident that one of the most common applications in HCS is measurement of protein translocation, where temporal–spatial relationships are key components of these functional probes. However, performing image-based measurements requires computationally complex algorithms; the image-based features used to characterize cells are often not orthogonal and the data have to be further reduced. The biological complexity is addressed by combining many image features using statistical machine-learning techniques [63].
In contrast to imaging methods, multifactorial flow cytometry benefits directly from access to the very high dimensionality of biological information. Flow cytometry relies on labeled molecular biomarkers and is capable of quantifying many of them simultaneously. Therefore, highly complex biological systems comprising large number of cellular populations with many different functions can be studied in FC by exposing the sample to various external factors, such as drugs in many concentrations or activators, or measurements at multiple time points. For every combination of factors, an independent high-content FC experiment may be performed. The addition of HT ability of modern FC means that a complete combinatorial set may be measured and quantified within minutes. This is a blend of speed and content unavailable to any other single-cell analysis technique.
Although the number of factors is limited only by the experimental design and the plate format used, the number of simultaneously measured molecular features depends on the detection technology employed. Owing to the spectral overlap between fluorescence labels and to the practical problems of staining biochemistry, fluorescence-based detection in flow has not exceeded 17 bands, with the exception of approaches such as multispectral cytometry of Raman-based detection that may be able to expand this number further [64-67]. Recently, in a radical departure from optical detection, Tanner et al. proposed utilization of mass spectroscopy-based measurements in flow cytometry [68,69], in which isotopes of heavy metals are the conjugated molecules linked to specific antibodies. This approach pushes the number of available functional descriptors much higher.
4. Flow cytometry data processing
4.1 The computational dilemma
The combination of multifactorial design of flow experiments and throughput produces data sets that cannot be interpreted using traditional flow cytometry software. High throughput is driven by quick processing of many samples, either by sequential analysis using robotic samplers, or by barcoding and multiplexing, increases the number of simultaneously available functional parameters acquired for every measured cell and produces data sets of a high order. The HTFC data sets are too large and complex for methods which typically produce simple spreadsheet-like outputs, and are practically inaccessible for rapid interpretation by flow cytometer operators and researchers.
As mentioned before, flow cytometry data processing and statistical analysis traditionally assumed that only a single file (representing a single biological sample) is analyzed at a given time and that the phenotypes of interest are defined by flow cytometer operators who create cascading set of gates. Therefore, visualization and analysis of data representing multiple samples requires a repetitive process. Consequently, information extracted from one sample could not be easily correlated with measurements performed on other samples. It is also clear that within the described operator-driven format, the task of statistical analysis of thousands of samples could take several hours or even days. This is why HT flow cytometry requires a change in approach.
Some flow cytometry analysis software vendors responded to the challenge by providing batch-analysis utility. However, the batch-processing function delivers a very limited solution given the biological differences between samples and the fact that traditional flow cytometry data analysis requires interactivity for defining populations and subpopulations of interest. The problem with traditional analytical tools is the lack of capacity to capture the entire assay or present the results in a fast, simple way, and researchers should not have an expectation that they must wait days for results. The real purpose of performing HT assays is to create large-scale comparisons of samples, drugs, activators, or cell types. The processes must be viewed as a system approach, not as a sample approach. Addressing the data complexity of multifactorial flow cytometry data required the concerted use of bioinformatics and machine-learning tools. Some of the proposed solutions are summarized below.
4.2 Storage, analysis, and mining of high-content FC data
Data for dozens of parameters collected on millions of cells introduce many challenges at the stages of processing and visualization. Storage, annotation, and exchange of highly structured multifactorial data sets require specialty custom-tailored informatics solutions designed with FC in mind. One such approach is the Cytobank system developed by a team at Stanford University. Cytobank captures all the important metadata describing multifactorial high-content FC data and combines the information about experimental design with sophisticated data-presentation techniques and statistical analysis of FC measurements. The key to the successful use of tools like Cytobank is careful documentation of the experimental design to provide accurate details regarding antibodies, conjugates, reagents, optical filters employed, etc. Cytobank also includes some sophisticated data-processing capabilities, such as automated extraction of response curves and interpretation of cellular barcodes [70]. Further knowledge deduction for the creation of networks based on Bayesian learning from HT flow cytometry has been shown to be possible [71]. Cytobank can also use advanced visualization tools such as the SPADE algorithm [72], which assists in displaying and identifying large numbers of simultaneously studied cell phenotypes (see Figure 4). The Cytobank system was chosen to power the FlowRepository, an ISAC-backed project to build a public repository of cytometry data to promote reproducibility, exchangeability, and peer review of FC experiments. An example of an advanced analysis enabled by this system is the construction of “Markov neighborhoods” for each variable based on a variety of dependencies and which performs structured learning using a constrained search [73].
Figure 4.
This figure demonstrates the SPADE algorithm as applied to hematopoietic cells. This technology packages data into regions of similarity so that it is possible to see the strength and range of interactions within a complex system. SPADE uses a color- and shape-coded process to allow the investigator to get an overview of a very complex system.
Reproduced from [95] with permission of the American Association for the Advancement of Science.
4.3 Current approaches for advanced analysis of flow cytometry data
The sine qua non for automated analysis of high-throughput flow cytometry data is the availability and practical implementation of modern pattern-recognition, machine-learning, and statistical procedures to interpret, process, and analyze flow cytometry data sets. Without access to a statistical analysis toolkit, flow cytometry practitioners are limited to manual data analysis, which is not scalable and cannot be extended to HT experiments and resultant data models. Owing to biological and instrumental variability, a simple batch-processing approach that re-applies an operator’s manual analysis steps to a large number of flow cytometry files does not work well in practice and cannot be recommended. Therefore, the success of automation in FC data processing is intimately connected to progress in the development of easy-to-use scientific computational programs for general statistical analysis.
The early work on application of machine learning-inspired methods in FC dates back to the early 2000s [74-76]. State-of-the-art classification techniques such as neural networks and support vector machines were demonstrated to perform well when applied to flow cytometry data sets. Similar approaches utilizing advanced pattern-recognition tools were subsequently applied successfully by other researchers [77,78]. Tools such as Logicle Transformation [79] were embedded within the field and have now been widely applied [80]. However, the initial lack of a unified development environment for FC data analysis severely hindered progress and limited the impact of modern pattern-recognition methods on automation of FC data processing. Only recently has modern statistical methodology finally become available to all cytometry practitioners willing to invest a reasonable amount of time in learning the basics of statistics and programming.
This methodology became possible due to the efforts of the bioinformatics community, which faced a similar set of problems dealing with large data sets and the demand for immense data collection and analysis throughput. The push toward standardization, data exchangeability, and ability to peer-review the procedure, techniques, and algorithms used resulted in the adoption of a popular open-source statistical programming language know as R for development and deployment of bioinformatics processing tools. R was an ideal choice – it is a dialect of the powerful S language created by John Chambers at Bell Labs, which has been used by statisticians since the 1980s. However, in contrast to S, R is an open-source project initiated by Ross Ihaka and Robert Gentleman at the University of Auckland, New Zealand. The openness of R and the involvement of one of its creators in the field of bioinformatics were important factors leading to the Bioconductor initiative.
The Bioconductor project, which aimed at coordinating R-based bioinformatics-related data-analysis packages for biologists, was initiated in 2001 at the Dana Farber Cancer Institute [81]. The 1.0 release of Bioconductor contained only 15 packages, none of which related to cytometry. By 2011 the number of bioinformatics and computational biology packages grew to 467 and included a number of cytometry-related toolkits that are now widely utilized to build custom HTFC data-processing pipelines. Among these packages, several directly address the problems of subjectivity, lack of reproducibility, and time consumption inherent in manual FC analysis, and provide modern statistical methods for observer-independent analysis and classification. The aptly named flowCore package provides a basis for most of the other cytometry-related Bioconductor tools [82]. It supplies standardized ways of reading and saving structured FC data and provides a model for the representation of FC data as R objects. flowCore merges the functionality of the older R-packages prada and rflowcyt. The package uses the flowFrame class to represent single FC data sets functionally equivalent to a standard FCS file (FCS version 2.0, 3.0, and LMD-style files are supported), and the flowSet class to represent a set of logically connected FC experiments (for instance, arranged in a multi-well plate, or as a set of test tubes). All the traditionally employed data transformations (such as log, biexponential, arcsinh, etc.) are provided, as well as specialized tools to apply gates of various types and shape (polygon, ellipsoid, rectangle, etc.). The package works in conjunction with flowViz, which offers traditional modes of FC data visualization such as histograms, density plots, and dot plots [83]. The package flowMeans provides an extension to the popular k-means algorithm specifically tuned for dealing with non-spherical cell populations [84]. flowClust, another important R/Bioconductor toolkit, implements advanced model-based clustering that uses multivariate t-mixture models and the Box–Cox transformation [85,86].
flowMerge further extends the flowClust methodology, while flowType makes use of intensity thresholding, k-means, flow-Means, and flowClust to partition every fluorescence marker into “positive” and “negative” populations in order to produce automated phenotype tabulation. With a large number of available data transformations, it is imperative to select the transformation in accordance with best practices and to maintain well-defined transformation protocols. The R/Bioconductor package flowTrans offers help and assistance in the search for optimal transformations for subsequent data processing and visualization [87]. flowFP, developed by Holyst and Rogers [88,89], allows the user to generate a fingerprint-like description of the multivariate probability distribution functions representing FC data. Another important tool for high-throughput cytometry, flowQ, was developed to automate the process of quality control, which is crucial for HT experiments involving measurements of hundreds or even thousands of samples. The flowQ Bioconductor package provides various QC functions ranging from a simple check on number of cells measured in every well of the analyzed plates to determination of boundary effects, discovery of abnormal flow rates, or sudden jumps in measurement intensities. flowQ also provides automated reporting capability.
Since high-throughput cytometry experiments often require simultaneous rather than sequential processing of large numbers of files, the traditional data representation used by the cytometry community, and also provided by flowCore, may not be adequate, since it keeps the entire flowFrame object in computer memory for data manipulation and processing. A solution to this problem provided by the package ncdfFlowSet utilizes netCDF (network common data form) tools. Owing to the availability of compression and chunking features, netCDF is an ideal format for storage of large HT cytometry experiments. The specialized package plateCore specifically addresses the issue of handling flow cytometry data organized in 96- or 384-well plate formats and allows seamless integration of the previously mentioned tools with plate-based HT cytometry [90].
SPADE (already mentioned in the context of Cytobank) is another important algorithm available in the Bioconductor repository. The SPADE algorithm, which was developed by Peng Qiu in Sylvia Plevritis’ lab in collaboration with Garry P. Nolan’s laboratory, utilizes a spanning-tree representation of density-normalized cellular populations to visualize phenotypes of cells in complex systems (Figure 4). The resultant trees are much easier to interpret than a cascade of 2-D projections, especially for multifactorial high-dimensionality data sets such as those obtained using the CyTOF system [12,72].
Although the bulk of the described efforts in modern HTFC analysis are related to development of basic building blocks for automated data-processing pipelines, the important issue of interfacing the automated processing algorithms is also being addressed by the FC community [91,92]. iFlow is a GUI linking R/Bioconductor packages and allows even inexperienced users to explore the available functions and tools provided by Bioconductor [93]. Many of the Bioconductor packages discussed above are listed in Table 1.
The users who feel intimidated by the requirement to program in R/Bioconductor environment in order to build processing pipelines for multifactorial HT cytometry assays and who would like to use a ready-made off-the-shelf solution regrettably do not have a wide choice. Currently there are no commercially available FC processing packages which out-of-box fully embrace multifactorial formats and allow quick processing of large number of samples, keeping the metadata and relationships between samples intact. However, R-code can be used within general-use biodata-automated processing platform such as Pipeline Pilot (Accelrys, Inc., San Diego, CA) or freely available GenePattern from Broad Institute [94].
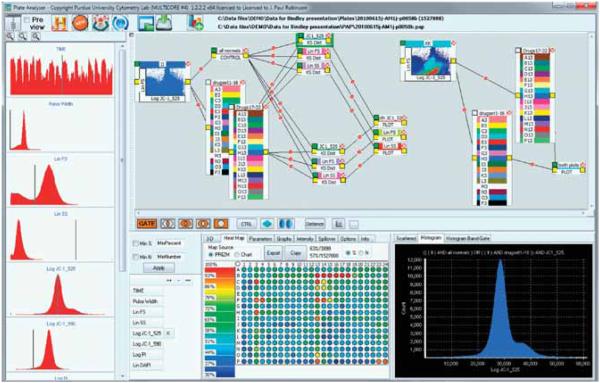
The only fully integrated multifactorial flow cytometry processing system known to us is the PlateAnalyzer platform developed in Robinson’s lab [95,96]. The package was designed employing the concept of visual programming, which can be summarized as the process of programming by manipulating graphical objects instead of writing textual code. The idea dates back to the early 1970s and David Canfield Smith’s Pygmalion programming environment, which used an icon-based programming paradigm in which the user created, modified, and linked together small pictorial objects, with defined operators and computational actions. The concept of a graphical design canvas allowing the use of independent pictorial “modules” and connecting “pipes” fits very well with the openness and complexity of multifactorial HT cytometry assay development. The scientific and engineering community is already using visual programming in tools such as LabVIEW, visualization package AVS, or machine-learning environment Orange.
PlateAnalyzer is de facto a visual programming toolkit allowing an FC analyst to combine various processing steps, control the input for the algorithms, and apply various processing operators graphically, interacting with the design canvas, rather than encoding the required processing steps in a language such as R (see Figure 5). In the PlateAnalyzer system, small icons or boxes represent programmatic entities (fragments of functional code), and lines (or pipes) connecting these objects allow for the flow of information and indicate relationships between operators.
Figure 5.
Screenshot of PlateAnalyzer, an interactive analysis system for multifactorial FC data. The GUI uses a pipeline metaphor to enable even inexperienced users access to sophisticated embedded visual-programming tools.
Reproduced from [12] with permission of International Drug Discovery and Russell Publishing.
In the case of PlateAnalyzer, the user composes a data cytometry data processing pipeline (termed a logic map), by creating connections between boxes indicating various inputs, outputs, and operators. These trees represent a series of processes of data reduction and manipulation steps that can be applied to the data set of interest. The innovative aspect of PlateAnalyzer processing is that not only does it allow for visual pipeline building, but also all the designed pipelines can be utilized in a sequential or parallel fashion. In other words, a typical HT assay or screen that leads to collection of hundreds of data sets is processed virtually in real time, since the data sets flow through the processing pipeline in parallel. This is possible because data reduction and processing in multifactorial flow cytomery is a so-called embarrassingly parallel problem, i.e., a computational task in which there exists no dependency between parallel subtasks (that is, processing of subsamples exposed to a given combination of factors). This property also means that PlateAnalyzer is easily portable to a grid-computing format, which can address the issue of processing very large screens or data mining complex databases of cytometry assays.
5. Future applications/new opportunities
HT flow cytometry is a transformational technology that opens up new opportunities for systems biology, especially in a multifactorial version. In the last couple of years, the number of samples that can be processed in modern FC devices has grown from just a few per hour to thousands. At the same time, the complexity of the data has also increased owing to the increasing number of simultaneously measured markers as well as to the use of multifactorial experiment-design formats. As outlined here, an increasing number of tools are being actively developed to process, manipulate, visualize, store, and mine these data sets. With the availability of cellular barcoding technology and the introduction of new high-content measurement techniques such as mass spectrometry-based FC, the well-established field of cytometry is likely to expand its role in systems biology. This will enable large-scale studies of signaling networks and regulatory pathways, discovery of new molecular targets, and quantification of cellular responses to various activators of function, drugs, and stress [73].
6. Expert opinion
There is little doubt that the paradigm is changing in the world of flow cytometry. As practiced for almost five decades the technology has been useful, is successful, and makes a significant contribution to current needs. However, this traditional technology fails to serve the needs of a systems approach to large-scale biology. This approach demands the development of new assay designs, new reagents, some new detection opportunities, and most definitely new analytical approaches. A few possibilities have shown significant promise in moving toward these goals. Some are already available but in limited use. The complications in moving flow cytometry into the very-high-content and high-throughput domain are significant. HT flow cannot be performed without automated preparatory tools, which are commonly used in the image-screening world, but not as yet in the flow world. This situation has to change. Without these automated processes, it is not possible to establish large-scale experiments with acceptable levels of quality control. The same robotic instrumentation currently used in screening labs works for HT flow cytometry equally well and should be adopted.
Detection instrumentation is changing as well. For several years, a few groups have had the capacity to collect 15 or more fluorescent probes simultaneously using the most advanced instruments available. In addition, hyperspectral tools are also advancing and have the possibility to expand the capacity of current instruments. The one major expansion in the field has been the CyTOF-based analysis using isotopes of heavy metals instead of fluorescent probes. Without the need to deal with fluorescence compensation and with the capability to operate in a virtual digital mode, this technology is extremely powerful, yet immature.
As these new technologies mature, the opportunities for unique application will increase. For example, the CyTOF has a clear-cut advantage for the analysis of multiple populations like bone marrow or blood, where the relationships between cell types may be complex. For less complicated applications such as drug screening, HT flow cytometry is an amazingly successful tool that is sure to see growth as the advantages become more obvious. Flow cytometry has been a technology neatly packaged for decades. The covers are coming off the package as this powerful tool creates new opportunities and enters the world of systems biology. If we constantly think of flow cytometry as a tool with current limitations and only capable of extracting a few populations of cells and identify an extracellular antigen or provide the status of the cell cycle of a population, we will not see the new opportunities. The next generation of flow cytometry capabilities has stretched the limits of complexity in defining cellular relationships as well as evaluating complicated drug–cell interactions.
Article highlights.
Flow cytometry can be performed in high throughput as well as a very high content mode.
Data analysis for such data sets, while complex, can be performed in a rapid and robust way.
Advanced classification solutions are plentiful and can be modified to be applied to very large flow cytometry data sets in a semi-automated fashion.
Automated preparation of assays is a key aspect of high-throughput flow cytometry and is required for the throughput and quality control.
HT flow now has the capacity to be an effective systems biology tool as very large assay systems can be developed and run in a very short time window.
Very complex analysis is possible with HT flow particularly when you integrate the entire assay and use new tools that can be applied in parallel.
Acknowledgements
The authors thank G Lawler for proofreading the manuscript.
Declaration of Interest
The authors declare support in the form of NIH grants 1R56AI089511 and 1R33CA140084 and funding from Beckman Coulter Corp.
Bibliography
Papers of special note have been highlighted as either of interest (•) or of considerable interest (••) to readers.
- 1.Moldavan A. Photo-electric technique for the counting of microscopical cells. Science. 1934;80:188–9. doi: 10.1126/science.80.2069.188. [DOI] [PubMed] [Google Scholar]
- 2.Gucker FT, Jr, O’Konski CT. Electronic methods of counting aerosol particles. Chem Rev. 1949;44:373–3. doi: 10.1021/cr60138a009. [DOI] [PubMed] [Google Scholar]
- 3.Gucker FT, Jr, O’Konski CT, Pickard HB, Pitts JN., Jr A photoelectronic counter for colloidal particles. J Am Chem Soc. 1947;69:2422–2. doi: 10.1021/ja01202a053. [DOI] [PubMed] [Google Scholar]
- 4.Caspersson TO. Uber den chemischen Aufbau der Strukturen des Zellkernes. Skand Arch Physiol. 1936;73 [Google Scholar]
- 5.Crosland-Taylor PJ. A device for counting small particles suspended in fluid through a tube. Nature. 1953;171:37–8. doi: 10.1038/171037b0. [DOI] [PubMed] [Google Scholar]
- 6.Coulter WH. Means for counting particles suspended in a fluid. US2656508(1953)
- 7.Coulter WH. High speed automatic blood cell counter and cell size analyzer. Proc Natl Electron Conf. 1956;12:1034–42. [Google Scholar]
- 8.Fulwyler MJ. Electronic separation of biological cells by volume. Science. 1965;150:910–11. doi: 10.1126/science.150.3698.910. [DOI] [PubMed] [Google Scholar]
- •. The invention of the cell sorter by Fulwyler was the definitive transformation for the creation of a new approach to single cell analysis.
- 9.Edwards BS, Kuckuck F, Sklar LA. Plug flow cytometry: an automated coupling device for rapid sequential flow cytometric sample analysis. Cytometry. 1999;37:156–9. [PubMed] [Google Scholar]
- 10.Dittrich W, Gohde W. Impulsfluorometrie bei Einzelzellen in Suspensionen. Zeitschrift fur Naturforschung. 1969;24b:360–1. [PubMed] [Google Scholar]
- 11.Perfetto SP, Chattopadhyay PK, Roederer M. Seventeen-colour flow cytometry: unravelling the immune system. Nat Rev Immunol. 2004;4:648–55. doi: 10.1038/nri1416. [DOI] [PubMed] [Google Scholar]
- •. Moving the barriers constantly by increasing the number of simultaneous fluorochromes analyzed. This proved that very high content flow was possible.
- 12.Bendall SC, Simonds EF, Qiu P, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–96. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••. The first paper demonstrating very high content flow data using mass cytometry – a technology sure to be transformative in many areas of single cell analysis. This technology increases the number of measurable parameters well beyond 40 – 50 defined markers.
- 13.Loken MR, Leibson PJ, Schreiber H. Heritable and non-heritable variations in the phenotype of myeloma cells as detected by a fluorescence-activated cell sorter. J Histochem Cytochem. 1979;27:1647–8. doi: 10.1177/27.12.521620. [DOI] [PubMed] [Google Scholar]
- 14.Boss MA, Delia D, Robinson JB, Greaves MF. Differentiation-linked expression of cell surface markers on HL60 leukemic cells. Blood. 1980;56:910–16. [PubMed] [Google Scholar]
- 15.Barlogie B, Maddox AM, Johnston DA, et al. Quantitative cytology in leukemia research. Blood Cells. 1983;9:35–55. [PubMed] [Google Scholar]
- 16.Lanier LL, Le AM, Phillips JH, et al. Subpopulations of human natural killer cells defined by expression of the Leu-7 (HNK-1) and Leu-11 (NK-15) antigens. J Immunol. 1983;131:1789–1796. [PubMed] [Google Scholar]
- 17.Scollay R, Shortman K. Thymocyte subpopulations: an experimental review, including flow cytometric cross-correlations between the major murine thymocyte markers. Thymus. 1983;5:245–95. [PubMed] [Google Scholar]
- 18.Drexler HG, Gignac SM, Minowada J. Routine immunophenotyping of acute leukaemias. Blut. 1988;57:327–39. doi: 10.1007/BF00320752. [DOI] [PubMed] [Google Scholar]
- 19.Patel SS, Duby AD, Thiele DL, Lipsky PE. Phenotypic and functional characterizations of human T cell clones. J Immunol. 1988;141:3726–36. [PubMed] [Google Scholar]
- 20.Wasserman K, Subklewe M, Pothoff G, et al. Expression of surface markers on alveolar macrophages from symptomatic patients with HIV infection as detected by flow cytometry. Chest. 1994;105:1324–34. doi: 10.1378/chest.105.5.1324. [DOI] [PubMed] [Google Scholar]
- 21.Spiekermann K, Emmendoerffer A, Elsner J, et al. Altered surface marker expression and function of G-CSF-induced neutrophils from test subjects and patients under chemotherapy. Br J Haematol. 1994;87:31–8. doi: 10.1111/j.1365-2141.1994.tb04866.x. [DOI] [PubMed] [Google Scholar]
- 22.Kearsey JA, Stadnyk AW. Isolation and characterization of highly purified rat intestinal intraepithelial lymphocytes. J Immunol Methods. 1996;194:35–48. doi: 10.1016/0022-1759(96)00052-x. [DOI] [PubMed] [Google Scholar]
- 23.Hulett HR, Bonner WA, Barrett J, et al. Automated separation of mammalian cells as a function of intracellular fluorescence. Science. 1969;166:747–7. [PubMed] [Google Scholar]
- 24.Parry MF, Root RK, Metcalf JA, et al. Myeloperoxidase deficiency: prevalence and clinical significance. Ann Intern Med. 1981;95:293–301. doi: 10.7326/0003-4819-95-3-293. [DOI] [PubMed] [Google Scholar]
- 25.Anderson DC, Schmalstieg FC, Arnaout MA, et al. Abnormalities of polymorphonuclear leukocyte function associated with a heritable deficiency of high molecular weight surface glycoproteins (GP138): common relationship to diminished cell adherence. J Clin Invest. 1984;74:536–51. doi: 10.1172/JCI111451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bassoe C-F, Bjerknes R. The effect of serum opsonins on the phagocytosis of Staphylococcus aureus and zymosan particles, measured by flow cytometry. Acta Pathol Microbiol Immunol Scand [C] 1984;92:51–8. doi: 10.1111/j.1699-0463.1984.tb00051.x. [DOI] [PubMed] [Google Scholar]
- 27.Wolber RA, Duque RE, Robinson JP, Oberman HA. Oxidative product formation in irradiated neutrophils. A flow cytometric analysis. Transfusion. 1987;27:167–70. doi: 10.1046/j.1537-2995.1987.27287150192.x. [DOI] [PubMed] [Google Scholar]
- 28.Kay NE, Perri RT. Natural killer function in B-chronic lymphocytic leukemia. Nouvelle Revue Francaise d’Hematologie. 1988;30:343–5. [PubMed] [Google Scholar]
- 29.Falk RJ, Terrell RS, Charles LA, Jennette JC. Anti-neutrophil cytoplasmic autoantibodies induce neutrophils to degranulate and produce oxygen radicals in vitro. Proc Natl Acad Sci USA. 1990;87:4115–19. doi: 10.1073/pnas.87.11.4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuen AR, Sikic BI. Multidrug resistance in lymphomas. J Clin Oncol. 1994;12:2453–9. doi: 10.1200/JCO.1994.12.11.2453. [DOI] [PubMed] [Google Scholar]
- 31.Keh D, Gerlach M, Kurer I, et al. The effects of nitric oxide (NO) on platelet membrane receptor expression during activation with human alpha-thrombin. Blood Coagul Fibrinolysis. 1996;7:615–24. doi: 10.1097/00001721-199609000-00007. [DOI] [PubMed] [Google Scholar]
- 32.Kotowicz K, Callard RE, Friedrich K, et al. Biological activity of IL-4 and IL-13 on human endothelial cells: functional evidence that both cytokines act through the same receptor. Int Immunol. 1996;8:1915–1925. doi: 10.1093/intimm/8.12.1915. [DOI] [PubMed] [Google Scholar]
- 33.Broxterman HJ, Schuurhuis GJ, Lankelma J, et al. Highly sensitive and specific detection of P-glycoprotein function for haematological and solid tumour cells using a novel nucleic acid stain. Br J Cancer. 1997;76:1029–34. doi: 10.1038/bjc.1997.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katoh N, Hirano S, Kishimoto S, Yasuno H. Calcium channel blockers suppress the contact hypersensitivity reaction (CHR) by inhibiting antigen transport and presentation by epidermal Langerhans cells in mice. Clin Exp Immunol. 1997;108:302–8. doi: 10.1046/j.1365-2249.1997.d01-1024.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knight CJ, Panesar M, Wright C, et al. Altered platelet function detected by flow cytometry. Effects of coronary artery disease and age. Arteriosclerosis Thromb Vasc Biol. 1997;17:2044–53. doi: 10.1161/01.atv.17.10.2044. [DOI] [PubMed] [Google Scholar]
- 36.Amann S, Reinke C, Valet G, et al. Flow-cytometric investigation of cellular metabolism during oxidative stress and the effect of tocopherol. Int J Vitam Nutr Res. 1999;69:356–61. doi: 10.1024/0300-9831.69.5.356. [DOI] [PubMed] [Google Scholar]
- 37.Fulcher D, Wong S. Carboxyfluorescein succinimidyl ester-based proliferative assays for assessment of T cell function in the diagnostic laboratory. Immunol Cell Biol. 1999;77:559–64. doi: 10.1046/j.1440-1711.1999.00870.x. [DOI] [PubMed] [Google Scholar]
- 38.Puskas F, Gergely P, Jr, Banki K, Perl A. Stimulation of the pentose phosphate pathway and glutathione levels by dehydroascorbate, the oxidized form of vitamin C. Federation Am Soc Exp Biol J. 2000;14:1352–61. doi: 10.1096/fj.14.10.1352. [DOI] [PubMed] [Google Scholar]
- 39.Han Y, Bu LM, Ji X, et al. Modulation of multidrug resistance by andrographolid in a HCT-8/5-FU multidrug-resistant colorectal cancer cell line. Chin J Dig Dis. 2005;6:82–6. doi: 10.1111/j.1443-9573.2005.00197.x. [DOI] [PubMed] [Google Scholar]
- 40.Lopez JP, Wang-Rodriguez J, Chang C, et al. Gefitinib inhibition of drug resistance to doxorubicin by inactivating ABCG2 in thyroid cancer cell lines. Arch Otolaryngol Head Neck Surg. 2007;133:1022–7. doi: 10.1001/archotol.133.10.1022. [DOI] [PubMed] [Google Scholar]
- 41.Lu J, Wang XY, Tang WH. Glutamine attenuates nitric oxide synthase expression and mitochondria membrane potential decrease in interleukin-1beta-activated rat hepatocytes. Eur J Nutr. 2009;48:333–9. doi: 10.1007/s00394-009-0018-x. [DOI] [PubMed] [Google Scholar]
- 42.Liu JT, Guo X, Ma WJ, et al. Mitochondrial function is altered in articular chondrocytes of an endemic osteoarthritis, Kashin-Beck disease. Osteoarthritis Cartilage. 2010;18:1218–26. doi: 10.1016/j.joca.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 43.McElnea EM, Quill B, Docherty NG, et al. Oxidative stress, mitochondrial dysfunction and calcium overload in human lamina cribrosa cells from glaucoma donors. Mol Vis. 2011;17:1182–91. [PMC free article] [PubMed] [Google Scholar]
- 44.Paino IM, Miranda JC, Marzocchi-Machado CM, et al. Phagocytosis and nitric oxide levels in rheumatic inflammatory states in elderly women. J Clin Lab Anal. 2011;25:47–51. doi: 10.1002/jcla.20429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murphy RF, Chused TM. A proposal for a flow cytometric data file standard. Cytometry. 1984;5:553–5. doi: 10.1002/cyto.990050521. [DOI] [PubMed] [Google Scholar]
- •. The fundamental paper established base standards, for flow cytometry. This key innovation transformed the field by establishing file standards something the HT image screening world has failed to do.
- 46.Dean PN, Bagwell CB, Lindmo T, et al. Data file standard for flow cytometry. Data File Standards Committee of the Society for Analytical Cytology. Cytometry. 1990;11:323–32. doi: 10.1002/cyto.990110303. [DOI] [PubMed] [Google Scholar]
- 47.Seamer LC, Bagwell CB, Barden L, et al. Proposed new data file standard for flow cytometry, version FCS 3.0. Cytometry. 1997;28:118–22. doi: 10.1002/(sici)1097-0320(19970601)28:2<118::aid-cyto3>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 48.Spidlen J, Moore W, Parks D, et al. Data File Standard for Flow Cytometry, version FCS 3.1. Cytometry A. 2010;77:97–100. doi: 10.1002/cyto.a.20825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spidlen J, Shooshtari P, Kollmann TR, Brinkman RR. Flow cytometry data standards. BMC Res Notes. 2011;4:50–2011. doi: 10.1186/1756-0500-4-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Telford WG, Komoriya A, Packard BZ, Bagwell CB. Multiparametric analysis of apoptosis by flow cytometry. Methods Mol Biol (Clifton, N.J.) 2011;699:203–27. doi: 10.1007/978-1-61737-950-5_10. [DOI] [PubMed] [Google Scholar]
- 51.Bagwell CB. Breaking the dimensionality barrier. Methods Mol Biol (Clifton, N.J.) 2011;699:31–51. doi: 10.1007/978-1-61737-950-5_2. [DOI] [PubMed] [Google Scholar]
- •. The first directly applicable toolset for transforming flow cytometry data into phenotypically useful patterns for clinical interpretation.
- 52.Agresti JJ, Antipov E, Abate AR, et al. Ultrahigh-throughput screening in drop-based microfluidics for directed evolution. Proc Natl Acad Sci USA. 2010;107:4004–9. doi: 10.1073/pnas.0910781107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robinson JP, Maguire DJ, King G, et al. Integration of a barcode reader with a commercial flow cytometer. Cytometry. 1992;13:193–7. doi: 10.1002/cyto.990130214. [DOI] [PubMed] [Google Scholar]
- 54.Robinson JP, Durack G, Kelley S. An innovation in flow cytometry data collection & analysis producing a correlated multiple sample analysis in a single file. Cytometry. 1991;12:82–90. doi: 10.1002/cyto.990120112. [DOI] [PubMed] [Google Scholar]
- ••. The first demonstration of large-scale file management in flow cytometry by the creation of combined files representing entire assays. This concept is basic to all high-throughput flow analysis.
- 55.Durack G, Lawler G, Kelley S, et al. Time interval gating for analysis of cell function using flow cytometry. Cytometry. 1991;12:701–6. doi: 10.1002/cyto.990120803. [DOI] [PubMed] [Google Scholar]
- 56.Edwards BS, Kuckuck FW, Prossnitz ER, et al. HTPS flow cytometry: a novel platform for automated high throughput drug discovery and characterization. J Biomol Screen. 2001;6:83–90. doi: 10.1177/108705710100600204. [DOI] [PubMed] [Google Scholar]
- 57.Kuckuck FW, Edwards BS, Sklar LA. High throughput flow cytometry. Cytometry. 2001;44:83–90. [PubMed] [Google Scholar]
- 58.Young SM, Bologa C, Prossnitz ER, et al. High-throughput screening with HyperCyt flow cytometry to detect small molecule formylpeptide receptor ligands. J Biomol Screen. 2005;10:374–82. doi: 10.1177/1087057105274532. [DOI] [PubMed] [Google Scholar]
- 59.Held M, Schmitz MHA, Fischer B, et al. CellCognition: time-resolved phenotype annotation in high-throughput live cell imaging. Nat Methods. 2010;7:747–54. doi: 10.1038/nmeth.1486. [DOI] [PubMed] [Google Scholar]
- 60.Peng T, Bonamy GMC, Glory-Afshar E, et al. Determining the distribution of probes between different subcellular locations through automated unmixing of subcellular patterns. Proc Natl Acad Sci USA. 2010;107:2944–9. doi: 10.1073/pnas.0912090107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen X, Velliste M, Murphy RF. Automated interpretation of subcellular patterns in fluorescence microscope images for location proteomics. Cytometry A. 2006;69:631–40. doi: 10.1002/cyto.a.20280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Neumann B, Walter T, Jean-Karim H. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464:721–7. doi: 10.1038/nature08869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Loo LH, Wu LF, Altschuler SJ. Image-based multivariate profiling of drug responses from single cells. Nat Methods. 2007;4:445–53. doi: 10.1038/nmeth1032. [DOI] [PubMed] [Google Scholar]
- 64.Robinson JP. Multispectral cytometry: the next generation. Biophotonics Int. 2004;2004:36–40. [Google Scholar]
- •. Increasing the dimensionality of flow cytometry using a 32 channel PMT array allowed the application of spectral deconvolution processes to very high content data.
- 65.Gregori G, Patsekin V, Rajwa B, et al. Hyperspectral cytometry at the single-cell level using a 32-channel photodetector. Cytometry A. 2012;81:35–44. doi: 10.1002/cyto.a.21120. [DOI] [PubMed] [Google Scholar]
- 66.Watson DA, Gaskill DF, Brown LO, et al. Spectral measurements of large particles by flow cytometry. Cytometry A. 2009;75:460–4. doi: 10.1002/cyto.a.20706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goddard G, Martin JC, Naivar M, et al. Single particle high resolution spectral analysis flow cytometry. Cytometry A. 2006;69:842–51. doi: 10.1002/cyto.a.20320. [DOI] [PubMed] [Google Scholar]
- 68.Ornatsky O, Bandura D, Baranov V, et al. Highly multiparametric analysis by mass cytometry. J Immunol Methods. 2010;361:1–20. doi: 10.1016/j.jim.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 69.Ornatsky O, Baranov VI, Bandura DR, et al. Multiple cellular antigen detection by ICP-MS. J Immunol Methods. 2006;308:68–76. doi: 10.1016/j.jim.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 70.Krutzik PO, Clutter MR, Trejo A, Nolan GP. Fluorescent cell barcoding for multiplex flow cytometry. Curr Protoc Cytom Chapter 6:Unit-2011. doi: 10.1002/0471142956.cy0631s55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••. A key paper that demonstrated that one could increase the dimensionality of data in flow without a vast number of new fluorescent probes.
- 71.Linderman MD, Bruggner R, Athalye V, et al. High-throughput Bayesian network learning using heterogeneous multicore computers; Proceedings of the 24th ACM International Conference on Supercomputing; 2010. pp. 95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Qiu P, Simonds EF, Bendall SC, et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol. 2011;29(10):886–91. doi: 10.1038/nbt.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••. A very advanced approach to visualizing huge datasets of flow data resultant from mass cytometry driven systems. The SPADE approach allows the user to identify subsets of cells that share particular relationships in a graphical format.
- 73.Sachs K, Itani S, Carlisle J, et al. Learning signaling network structures with sparsely distributed data. J Comput Biol. 2009;16:201–12. doi: 10.1089/cmb.2008.07TT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Davey HM, Jones A, Shaw AD, Kell DB. Variable selection and multivariate methods for the identification of microorganisms by flow cytometry. Cytometry. 1999;35:162–8. doi: 10.1002/(sici)1097-0320(19990201)35:2<162::aid-cyto8>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 75.Boddy L, Wilkins MF, Morris CW. Pattern recognition in flow cytometry. Cytometry. 2001;44:195–209. doi: 10.1002/1097-0320(20010701)44:3<195::aid-cyto1112>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 76.Wilkins MF, Hardy SA, Boddy L, Morris CW. Comparison of five clustering algorithms to classify phytoplankton from flow cytometry data. Cytometry. 2001;44:210–17. doi: 10.1002/1097-0320(20010701)44:3<210::aid-cyto1113>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 77.Rajwa B, Venkatapathi M, Ragheb K, et al. Automated classification of bacterial particles in flow by multiangle scatter measurement and support vector machine classifier. Cytometry A. 2008;73:369–79. doi: 10.1002/cyto.a.20515. [DOI] [PubMed] [Google Scholar]
- 78.Quinn J, Fisher PW, Capocasale RJ, et al. A statistical pattern recognition approach for determining cellular viability and lineage phenotype in cultured cells and murine bone marrow. Cytometry A. 2007;71:612–24. doi: 10.1002/cyto.a.20416. [DOI] [PubMed] [Google Scholar]
- 79.Parks DR, Roederer M, Moore WA. A new "Logicle" display method avoids deceptive effects of logarithmic scaling for low signals and compensated data. Cytometry A. 2006;69:541–51. doi: 10.1002/cyto.a.20258. [DOI] [PubMed] [Google Scholar]
- 80.Moore WA, Parks DR. Update for the logicle data scale including operational code implementations. Cytometry A. 2012;81A:273–7. doi: 10.1002/cyto.a.22030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hahne F, LeMeur N, Brinkman RR, et al. flowCore: a Bioconductor package for high throughput flow cytometry. BMC Bioinform. 2009;10:106. doi: 10.1186/1471-2105-10-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sarkar D, Le MN, Gentleman R. Using flowViz to visualize flow cytometry data. Bioinformatics. 2008;24:878–9. doi: 10.1093/bioinformatics/btn021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Aghaeepour N, Nikolic R, Hoos HH, Brinkman RR. Rapid cell population identification in flow cytometry data. Cytometry A. 2011;79:6–13. doi: 10.1002/cyto.a.21007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lo K, Hahne F, Brinkman RR, Gottardo R. flowClust: a Bioconductor package for automated gating of flow cytometry data. BMC Bioinform. 2009;10:145. doi: 10.1186/1471-2105-10-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lo K, Brinkman RR, Gottardo R. Automated gating of flow cytometry data via robust model-based clustering. Cytometry A. 2008;73:321–32. doi: 10.1002/cyto.a.20531. [DOI] [PubMed] [Google Scholar]
- 87.Finak G, Perez JM, Weng A, Gottardo R. Optimizing transformations for automated, high throughput analysis of flow cytometry data. BMC Bioinform. 11:546–2010. doi: 10.1186/1471-2105-11-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rogers WT, Holyst HA. FlowFP: A Bioconductor Package for Fingerprinting Flow Cytometric Data. Adv Bioinform. 2009;2009:11. doi: 10.1155/2009/193947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rogers WT, Moser AR, Holyst HA, et al. Cytometric fingerprinting: quantitative characterization of multivariate distributions. Cytometry A. 2008;73:430–41. doi: 10.1002/cyto.a.20545. [DOI] [PubMed] [Google Scholar]
- 90.Bruckner S, Wang L, Yuan R, et al. Flow-based combinatorial antibody profiling: an integrated approach to cell characterization. Methods Mol Biol (Clifton, N.J.) 2011;699:97–118. doi: 10.1007/978-1-61737-950-5_6. [DOI] [PubMed] [Google Scholar]
- 91.Hammer MM, Kotecha N, Irish JM, et al. WebFlow: a software package for high-throughput analysis of flow cytometry data. Assay Drug Dev Technol. 2009;7:44–55. doi: 10.1089/adt.2008.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shulman N, Bellew M, Snelling G, et al. Development of an automated analysis system for data from flow cytometric intracellular cytokine staining assays from clinical vaccine trials. Cytometry A. 2008;73:847–56. doi: 10.1002/cyto.a.20600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lee K, Hahne F, Sarkar D, Gentleman R. iFlow: A Graphical User Interface for Flow Cytometry Tools in Bioconductor. Adv Bioinform. 2009;2009:3. doi: 10.1155/2009/103839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Reich M, Liefeld T, Gould J, et al. GenePattern 2.0. Nat Genet. 2006;38:500–1. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 95.Robinson JP, Davisson VJ, Narayanan PK. Hight Content-High Throughput drug discovery: Flow cytometry rises to meet the challenge. Int Drug Discov. 2011:10–14. [Google Scholar]
- ••. Demonstration of the application of both HT sample collection capacity integrated with a unique robust and rapid multifactorial analytical toolset.
- 96.Robinson JP, Patsekin V, Rajwa B, et al. High throughput flow cytometry: speeding up the process has increased the technology’s utility in systems biology. Genet Eng Biotechnol News. 2011;15:20–2. [Google Scholar]
- 97.Hahne F, LeMeur N, Brinkman RR, et al. Flowcore: a Bioconductor package for high throughput flow cytometry. BMC Bioinform. 2009;10:106–6. doi: 10.1186/1471-2105-10-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hahne F, Arlt D, Sauermann M, et al. Statistical methods and software for the analysis of highthroughput reverse genetic assays using flow cytometry readouts. Genome Biol. 2006;7:R77. doi: 10.1186/gb-2006-7-8-r77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Strain E, Hahne F, Brinkman RR, Haaland P. Analysis of high-throughput flow cytometry data using plateCore. Adv Bioinformatics. 2009;2009:10. doi: 10.1155/2009/356141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hahne F, Gopalakrishnan N, Khodabakhshi AH, et al. flowStats: statistical methods for the analysis of flow cytometry data. 2012 Available from: http://www.bioconductor.org/packages/2.9/bioc/html/flowStats.html.
- 101.Gopalakrishnan N, Hahne F, Ellis B, et al. Flowutils: utilities for flow cytometry. 2012 Available from: http://www.bioconductor.org/packages/2.9/bioc/html/flowUtils.html.
- 102.Gosink JJ, Means GD, Rees WA, et al. Bridging the divide between manual gating and bioinformatics with the bioconductor package flowFlowJo. Adv Bioinformatics. 2009;2009:8. doi: 10.1155/2009/809469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jiang M, Finak G, Gopalakrishnan N. ncdfFlow: a package that provides Ncdf based storage for flow cytometry data. 2012 Available from: http://www.bioconductor.org/packages/devel/bioc/html/ncdfFlow.html.
- 104.Zare H, Shooshtari P, Gupta A, Brinkman RR. Data reduction for spectral clustering to analyze high throughput flow cytometry data. BMC Bioinform. 2010;11:403–3. doi: 10.1186/1471-2105-11-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Finak G, Bashashati A, Brinkman R, Gottardo R. Merging mixture components for cell population identification in flow cytometry. Adv Bioinform. 2009;2009:12. doi: 10.1155/2009/247646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Aghaeepour N, Nikolic R, Hoos HH, Brinkman RR. Rapid cell population identification in flow cytometry data. Cytometry A. 2011;79:6–13. doi: 10.1002/cyto.a.21007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ribalet F, Schruth DM, Armbrust EV. flowPhyto: enabling automated analysis of microscopic algae from continuous flow cytometric data. Bioinformatics (Oxford, England) 2011;27:732–3. doi: 10.1093/bioinformatics/btr003. [DOI] [PubMed] [Google Scholar]
- 108.Le Meur N, Rossini A, Gasparetto M, et al. Data quality assessment of ungated flow cytometry data in high throughput experiments. Cytometry A. 2007;71:393–403. doi: 10.1002/cyto.a.20396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jiang M, Finak G. QUALIFIER: quality assessment of gated flow cytometry. doi: 10.1186/1471-2105-13-252. Available from: http://www.bioconductor.org/ packages/release/bioc/vignettes/QUALIFIER/inst/doc/QUALIFIER.pdf. [DOI] [PMC free article] [PubMed]
- 110.Aghaeepour N, Jalali A. RchyOptimyx:gating hierarchy optimization for flow cytometry. doi: 10.1002/cyto.a.22209. Available from: http://www.bioconductor.org/packages/release/bioc/vignettes/RchyOptimyx/inst/doc/RchyOptimyx.pdf. [DOI] [PMC free article] [PubMed]
- 111.Lahesmaa-Korpinen A-M, Chen P, Valo V, et al. FlowAND. 2012 Available from: http://csbi.ltdk.helsinki.fi/flowand/index.html.
- 112.Sarkar D, Le Meur N, Gentleman R. Using flowViz to visualize flow cytometry data. Bioinformatics (Oxford, England) 2008;24:878–9. doi: 10.1093/bioinformatics/btn021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lee K, Hahne F, Sarkar D, Gentleman R. iFlow: a graphical user interface for flow cytometry tools in bioconductor. Adv Bioinform. 2009;2009:3. doi: 10.1155/2009/103839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hawkins N, Self S. Flowplots: flowPlots: analysis plots and data class for gated flow cytometry data. 2012 Available from: http://www.bioconductor.org/packages/devel/bioc/html/flowPlots.html.
- 115.NIH Common Fund Single Cell Analysis Workshop. 2012 Apr 17-18; Available from: https://www.blsmeetings.net/singlecell2012/agenda.cfm.