Abstract
A sheet of choroid plexus epithelial cells extends into each cerebral ventricle and secretes signaling factors into the CSF. To evaluate whether differences in the CSF proteome across ventricles arise, in part, from regional differences in choroid plexus gene expression, we defined the transcriptome of lateral ventricle (telencephalic) versus fourth ventricle (hindbrain) choroid plexus. We find that positional identities of mouse, macaque, and human choroid plexi derive from gene expression domains that parallel their axial tissues of origin. We then show that molecular heterogeneity between telencephalic and hindbrain choroid plexi contributes to region-specific, age-dependent protein secretion in vitro. Transcriptome analysis of FACS-purified choroid plexus epithelial cells also predicts their cell-type-specific secretome. Spatial domains with distinct protein expression profiles were observed within each choroid plexus. We propose that regional differences between choroid plexi contribute to dynamic signaling gradients across the mammalian cerebroventricular system.
Keywords: cerebrospinal fluid, choroid plexus, next-generation sequencing
Introduction
The choroid plexus-CSF (ChP-CSF) system is essential for the development and maintenance of the vertebrate brain (Johanson et al., 2008; Lowery and Sive, 2009; Lehtinen and Walsh, 2011; Lehtinen et al., 2013). ChP-distributed factors actively regulate proliferation and survival of neural stem cells (X. Huang et al., 2009; Lehtinen et al., 2011). However, little is known about the regulation of the CSF proteome and whether regionalization across choroid plexi contributes to differential protein secretion.
The choroid plexi, located in each brain ventricle, consist of epithelial cells enveloping a stromal, fenestrated capillary network (Sturrock, 1979; Wilting and Christ, 1989). ChP epithelial cells secrete CSF, and tight junctions between cells form the blood-CSF barrier, preventing diffusion of systemic signals into CSF (Damkier et al., 2013). The choroid plexi differentiate from the roof plate (Currle et al, 2005; Emerich et al., 2005) and can be identified by early papillarity by mouse embryonic day (E)11–E12 (Sturrock, 1979). The hindbrain (myelencephalic) ChP (hChP) of the fourth ventricle forms first, followed by the bilateral development of the telencephalic ChP (tChP) of the lateral ventricles, and finally, the diencephalic ChP of the third ventricle (Kappers, 1955; Dziegielewska et al., 2001; Currle et al., 2005). Cell lineages originating from the hindbrain roof plate contribute to the hChP (Awatramani et al., 2003; Currle et al., 2005; Hunter and Dymecki, 2007). However, it is not known whether derivation of tChP versus hChP from separate lineages defines their positional identity, leading to ChP tissues that are functionally distinct in their protein secretion or receptivity. The possibility that each ChP produces a regionalized CSF would enable a powerful means to deliver region-specific signals to distinct populations of neural stem cells.
Although CSF flow directed from the lateral toward the fourth ventricle allows mixing of CSF, previous studies have suggested that CSF composition is regionalized between ventricles (Cavanagh et al., 1983; Zappaterra et al., 2007). Recent gene expression studies provide insight into the general ChP secretome, but these studies have relied on intact, in toto tChP and have not investigated tissue regionalization (Marques et al., 2011; Liddelow et al., 2012, 2013). Although epithelial cells represent the predominant cell type in the ChP (Keep and Jones, 1990), the ChP is vascularized, and vascular cells represent important sources of secreted signals in the nervous system (Palmer et al., 2000; Tavazoie et al., 2008; Kokovay et al., 2010; Delgado et al., 2014). Parsing the various ChP cell types, the regionalized secretomes of tChP versus hChP, and ultimately their contribution to the CSF proteome, would impart greater insight to the availability of extrinsic signals during brain development.
We demonstrate that the positional identities of the telencephalic and hindbrain choroid plexi derive from anterior–posterior gene expression domains that parallel their axial tissues of origin in the rodent and primate brain. We define the embryonic ChP secretome by assaying FACS-purified ChP epithelial cells alongside in toto ChP, and uncover molecular heterogeneity between choroid plexi that contributes to regional ChP-protein secretion in an age-dependent manner, coupling the choroid plexi to the production of signal gradients across the ventricular system.
Materials and Methods
Animals.
Timed pregnant CD-1 dams were obtained from Charles River Laboratories. mT/mG/Foxj1Cre+/− mice were obtained through heterozygous breedings (Muzumdar et al., 2007; Zhang et al., 2007). Ttr::RFP mice were obtained through heterozygous breeding of C57BL/6J × CBA/J mice outbred onto CD-1 mice (Kwon and Hadjantonakis, 2009). All animal experimentation was performed under protocols approved by the IACUCs of Boston Children's Hospital (BCH) and UC Irvine. Tissues were collected from both male and female embryos.
Fluorescence-activated cell sorting.
Ttr::RFP embryos were checked for genotype under a fluorescence microscope. Ttr::RFP-negative littermates were collected and processed in parallel to set the signal threshold for the RFP gate. tChP and hChP tissues were dissected and collected separately, and digested in type II collagenase (Life Technologies) in HBSS for 15–20 min with occasional tapping followed by TrypLE Express (Life Technologies) for 15–20 min. Dissociated cells were filtered through a 40 μm mesh strainer. Fluorescence-activated cell sorting (FACS) was performed with dead-cell exclusion by the green LIVE/DEAD Fixable Stain Kit (Life Technologies). We excluded debris and dead cells by side-scatter area and forward-scatter area, and single-cell selection by forward-scatter height and forward-scatter area before analyzing for RFP expression. RFP-positive cells comprised ∼30–50% and ∼15–20% of all single cells in the tChP and hChP, respectively; post-sort purity was >92%.
RNA sequencing and analysis.
Each sequenced sample comprised tissue or cells isolated from the tChP or hChP of E18.5 mouse embryos and pooled across a single litter. Total RNA was isolated from whole ChP or FACS-purified ChP epithelial cells by using the RNeasy Micro Kit (Qiagen) and converted to cDNA and preamplified using the Ovation RNA-seq System V2 (NuGEN) following the manufacturer's protocol. cDNA was converted to Illumina paired-end sequencing libraries following the standard protocol (TruSeq v2) and sequenced on a Illumina HiSeq 2000 instrument to a depth of ∼20–60 million pass-filter reads per library, after standard quality control filters. RNA-seq data have been deposited in the NCBI Gene Expression Omnibus database (GEO; www.ncbi.nlm.nih.gov/geo) with the GEO series accession number GSE66312. The 50 base paired-end reads were mapped to the UCSC mm9 mouse reference genome and “known genes” transcriptome using TopHat v2, and fragments per kilobase per million reads (FPKM) values were estimated using cufflinks v2 (Trapnell et al., 2012). We then used the HTSeq and DESeq software packages to perform library normalization, principle component analysis (PCA), and differential expression testing (Anders and Huber, 2010; Anders et al., 2013). Because the tChP and hChP samples came from the same biological replicates, differential expression was modeled as either a paired-sample single-factor design or a multifactor design contrasting both region (tChP vs hChP) and cell type (whole ChP vs purified epithelial cells). Hierarchical clustering and heatmap generation were performed in R on the library-normalized, variance-stabilized read counts from DESeq, using Pearson correlation and complete linkage. For heatmap display purposes, genes were scaled across conditions to have a mean of zero and 1 standard deviation. Genes selected for hierarchical clustering had a false discovery rate (FDR; Benjamini-Hochberg adjusted p value) <0.05. Gene Ontology term enrichment analysis was done with DAVID v6.7 (http://david.abcc.ncifcrf.gov/; D. W. Huang et al., 2009a,b), comparing tChP- and hChP-enriched genes with a minimum twofold expression difference (FDR < 0.05) to the full mouse transcriptome. The statistic reported for the annotation clusters, the enrichment score, is the geometric mean of the enrichment p values for the annotation terms in each cluster, in −log10 scale.
Macaque and human brain samples.
Tissue samples for molecular analyses were available through the following sources: (1) Rhesus macaque samples were collected from fresh brain specimens or brain slabs preserved in RNAlater from the Sestan Laboratory at Yale University, (2) human samples were obtained from frozen brain specimens from the Sestan Laboratory at Yale University, and (3) human samples were obtained under an IRB approved protocol at BCH. Samples were processed as follows: fresh macaque brains were chilled on ice for 15–30 min before sectioning. Brains were placed ventral side up onto a chilled aluminum plate (1 cm thick) on ice. ChP samples were collected from both lateral and fourth ventricles. Samples were snap frozen in isopentane (JT Baker)/dry ice at −30 to −40°C and stored at −80°C. Fresh frozen human samples (from Yale University) were microscopically inspected, and ChP samples were dissected from lateral and fourth ventricles using a dental drill (AnyXing, 300D) and a Lindemann Bone Cutter H162A.11.016 or diamond disk saw (Dental Burs; r = 11 mm) on a 1-cm-thick aluminum plate over dry ice. Dissected tissue samples were stored at −80°C before further processing. Human tissue specimens from BCH were reviewed by a neuropathologist (H.G.W.L.), and nonidentifying information was recorded for each specimen. Tissue sections were stained with hematoxylin and eosin (H&E) according to standard methods. RNA was extracted using miRNeasy Micro kit (Qiagen), reverse transcribed (Promega), and subjected to qRT-PCR using Taqman probes purchased from Life Technologies (see Table 2). All tissues were handled in accordance with ethical guidelines and regulations for the research use of human brain tissue set forth by the NIH (http://bioethics.od.nih.gov/humantissue.html). All fresh nonhuman primate samples were collected in accordance with a protocol approved by Yale University's Committee on Animal Research and NIH guidelines.
Table 2.
qRT-PCR measurement of genes detected by RNA-seq to be differentially expressed between tChP and hChP
| Gene | Taqman probe | tChP fold exp. | hChP fold exp. | p |
|---|---|---|---|---|
| Upregulated in tChP | ||||
| Bmp2 | Mm01340178_m1 | 1.000 | 0.647 ± 0.054 | 0.056 |
| Bmp15 | Mm00437797_m1 | 1.000 | 0.383 ± 0.100 | 0.015 |
| Cdh26 | Mm01212671_m1 | 1.000 | 0.401 ± 0.088 | 0.012 |
| Cntn4 | Mm00476065_m1 | 1.000 | 0.084 ± 0.011 | 0.037 |
| Emx2a | Mm00550241_m1 | 1.000 | 0.014 ± 0.0084 | <0.001 |
| Fbln5 | Mm00488601_m1 | 1.000 | 0.719 ± 0.222 | 0.306 |
| Fezf1 | Mm01164970_g1 | 1.000 | 0.043 ± 0.010 | 0.058 |
| Fezf2 | Mm01320618_g1 | 1.000 | 0.024 ± 0.013 | 0.046 |
| Lepr | Mm00440181_m1 | 1.000 | 0.473 ± 0.088 | 0.012 |
| Mc3r | Mm00434876_s1 | 1.000 | 0.237 ± 0.051 | 0.021 |
| Otx1a | Mm01616047_m1 | 1.000 | 0.075 ± 0.016 | 0.003 |
| Rdh10 | Mm01227200_m1 | 1.000 | 0.543 ± 0.035 | <0.001 |
| Six3a | Mm01237639_m1 | 1.000 | 0.534 ± 0.117 | 0.043 |
| Slit3 | Mm01326974_m1 | 1.000 | 0.345 ± 0.118 | 0.032 |
| Trf | Mm00446715_m1 | 1.000 | 0.879 ± 0.023 | 0.025 |
| Wnt8b | Mm00442108_g1 | 1.000 | 0.005 ± 0.001 | 0.003 |
| Upregulated in hChP | ||||
| Abca4 | Mm00492035_m1 | 1.000 | 3.052 ± 0.759 | 0.044 |
| En2a | Mm00438710_m1 | 1.000 | 167.44 ± 19.86 | 0.001 |
| Gata6 | Mm00802636_m1 | 1.000 | 1.862 ± 0.249 | 0.017 |
| Gdf7 | Mm00807130_m1 | 1.000 | 306.962 ± 12.906 | <0.001 |
| HoxA2a | Mm00439361_m1 | 1.000 | 1058.461 ± 179.043 | 0.004 |
| Igf2 | Mm00439565_g1 | 1.000 | 1.318 ± 0.057 | 0.004 |
| Igsf5 | Mm00511211_m1 | 1.000 | 3.242 ± 0.859 | 0.027 |
| Meis1a | Mm00487664_m1 | 1.000 | 30.208 ± 6.161 | <0.001 |
| Mstn | Mm01254559_m1 | 1.000 | 7.481 ± 2.005 | 0.018 |
| Osmr | Mm01307326_m1 | 1.000 | 2.882 ± 0.712 | 0.046 |
| Penkb | Mm01212875_m1 | 1.000 | 78.753 ± 9.654 | <0.001 |
| Shhb | Mm00436528_m1 | 1.000 | 198.548 ± 50.537 | 0.008 |
| Slc4a10 | Mm00473806_m1 | 1.000 | 3.001 ± 0.637 | 0.022 |
| Sod3b | Mm00448831_s1 | 1.000 | 5.203 ± 1.459 | 0.035 |
| Wisp1 | Mm01200484_m1 | 1.000 | 4.200 ± 0.695 | 0.015 |
| Wnt5a | Mm00437347_m1 | 1.000 | 5.558 ± 0.187 | <0.001 |
| Similarly expressed in telencephalic and hindbrain ChP | ||||
| Cst3 | Mm00438347_m1 | 1.000 | 1.021 ± 0.041 | 0.792 |
| Igfbp2 | Mm00492632_m1 | 1.000 | 1.014 ± 0.246 | 0.866 |
| Ttrb | Mm00443267_m1 | 1.000 | 0.902 ± 0.042 | 0.074 |
DAVID was used to functionally annotate differentially expressed genes, and candidates from three categories (transcription factors, secreted factors, and cell-surface receptors) were chosen for validation. Fold expression (Fold exp.) values are represented as mean ± SEM with tChP = 1.000. Statistical analysis was performed using two-tailed t test.
atChP/hChP fold-change graphed in Figure 3.
bqRT-PCR values graphed in Figure 6.
Quantitative PCR.
Expression of genes in mouse, macaque, and human tissues (except HOXA2) were measured by Taqman qPCR. Taqman gene expression probes were purchased from Life Technologies, using 18S as an internal control, and run on a StepOne Plus instrument (Applied Biosystems). Expression of HOXA2 in macaque and human tissues were measured by SYBR-green qPCR using the following forward (F) and reverse (R) primer probes: HOXA2 (F): GAAGAAGGCGGCCAAGAAAA; HOXA2 (R): GCTGTGTGTTGGTGTAAGCA; GAPDH (F): ACAACTTTGGTATCGTGGAAGG; GAPDH (R): GCCATCACGCCACAGTTTC. qPCR studies were performed using at least three biological replicates, and statistical analyses were performed using two-tailed t tests. Results are reported as mean ± SEM.
Microarrays.
Whole tChP and hChP were isolated from three biological replicates (pooled litters) and total RNA was isolated using the TRIzol reagent (Life Technologies), followed by DNA cleanup using the Turbo DNA-free Kit (Life Technologies). Polyadenylated mRNA was purified and cDNA was generated and in vitro transcribed according to Illumina's protocols. The amplified, biotin-labeled cRNA was hybridized to the Illumina MouseWG-6 v2.0 Expression BeadChip array. Microarray data were analyzed using the Partek Genomics Suite software and cross-referenced to the RNA-seq data in Excel.
Mass spectrometry.
Protein (30 μg) from either native CSF, collected as described previously (Zappaterra et al., 2013), or ChP-conditioned medium, was reduced in 0.1 m DTT and filtered twice with a 30 kDa spin filter (Millipore). Samples were alkylated, washed, and digested overnight with 2 μg trypsin (Promega) at 37°C. Digested proteins were eluted, acidified, desalted, and dried. Each sample was reconstituted and analyzed by a nanoLC system (Eksigent) equipped with LC-chip system (cHiPLCnanoflex, Eksigent) coupled online to a Q-Exactive mass spectrometer (Thermo Scientific). Peptides were separated by a linear gradient with a gradient length of 180 min. Before submitting for database search, the Thermo “raw” files were converted into the Mascot generic format (MGF-files) using the ProteoWizard software tool. All tandem mass spectrometry (MS/MS) data were analyzed using Mascot (v2.3.01, Matrix Science). The Mascot searches were performed using a concatenated mouse protein sequence database (UNIPROT.MOUSE, downloaded June 2014). Protein sequence database was searched with tryptic cleavage specificity, a fragment ion mass tolerance of 20 mμ and a parent ion tolerance of 10 ppm. Scaffold (version Scaffold_4.2.1, Proteome Software) was used to validate MS/MS-based peptide and protein identifications, resulting in an overall peptide and protein FDR of 1%. The subsequent output gave protein IDs with spectral counts corresponding to unique peptides identified for the protein per sample. Proteins were accepted as present if identified in any of the biological replicates.
Choroid plexus-conditioned medium.
tChP and hChP were dissected, cultured in neurobasal medium (Life Technologies) supplemented with penicillin-streptomycin and glutamine, 10 μl medium per ventricle of ChP for 24 h, centrifuged at 10,000 × g at 4°C for 10 min, and stored at −80°C.
Antibodies.
The following antibodies were purchased: Aquaporin-1 (Millipore and Santa Cruz Biotechnology), Cystatin C (Millipore), EC-SOD (Millipore), EMX2 (Santa Cruz Biotechnology), GDF7 (Millipore), GFP (Abcam), HOXA2 (Sigma-Aldrich), MEIS1 (Abcam), OTX1 (Abcam), PECAM1 (BD PharMingen), Proenkephalin (Thermo Scientific), Transthyretin (Dako), HRP-conjugated Transferrin (Immunology Consultants Laboratory).
Immunostaining.
Paraffin sections (5 μm) of brains were dehydrated, and antigen retrieval was performed using Antigen Unmasking Solution (Vector Laboratories). Sections were blocked and permeabilized (0.04% Tween20 in PBS, 5% serum), followed by antibody incubation. Cryosections (14 μm) of brains were blocked, permeabilized, and incubated with antibodies. All tissues were counterstained with Hoechst 33258.
Western blot quantification.
Blots were exposed on Bio-Rad chemidoc, and images captured were converted to 8 bit files. Intensities were quantified using ImageJ, and relative intensities were calculated based on the loading control from the same blot. Statistical analysis was performed on the relative intensities using two-tailed t test. Results are reported as mean ± SEM.
ELISA.
Shh concentrations in ChP-conditioned medium were measured using the Mouse Shh N-Terminus Quantikine ELISA kit (R&D Systems) per the manufacturer's instructions. tChP- and hChP-conditioned media were measured by BCA protein assay kit (Pierce), and samples with equal total protein were assayed by ELISA.
Biochemical assay.
Relative SOD activity was measured in diluted rat CSF samples (1:10) using the Superoxide Dismutase Assay Kit (Cayman Chemical) according to the manufacturer's instructions.
Results
Transcriptome analysis of whole-choroid plexi and purified choroid plexus epithelial cells
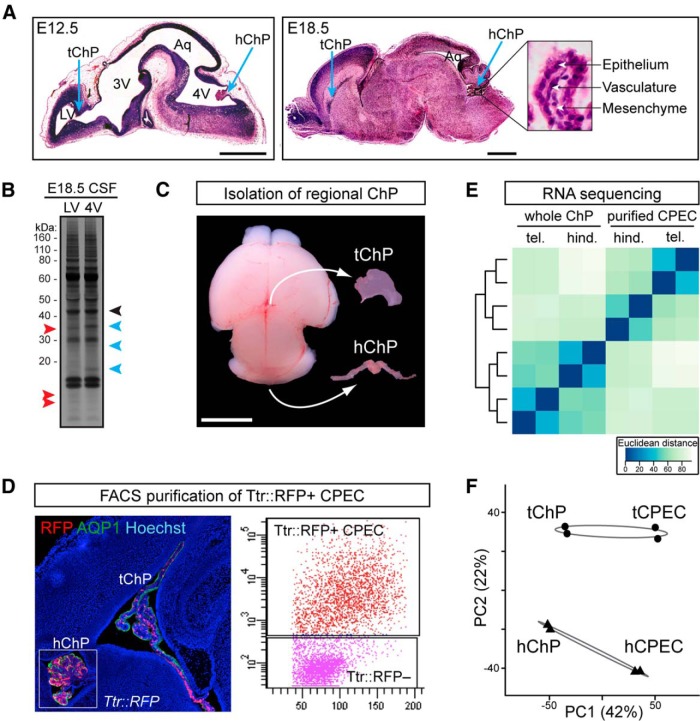
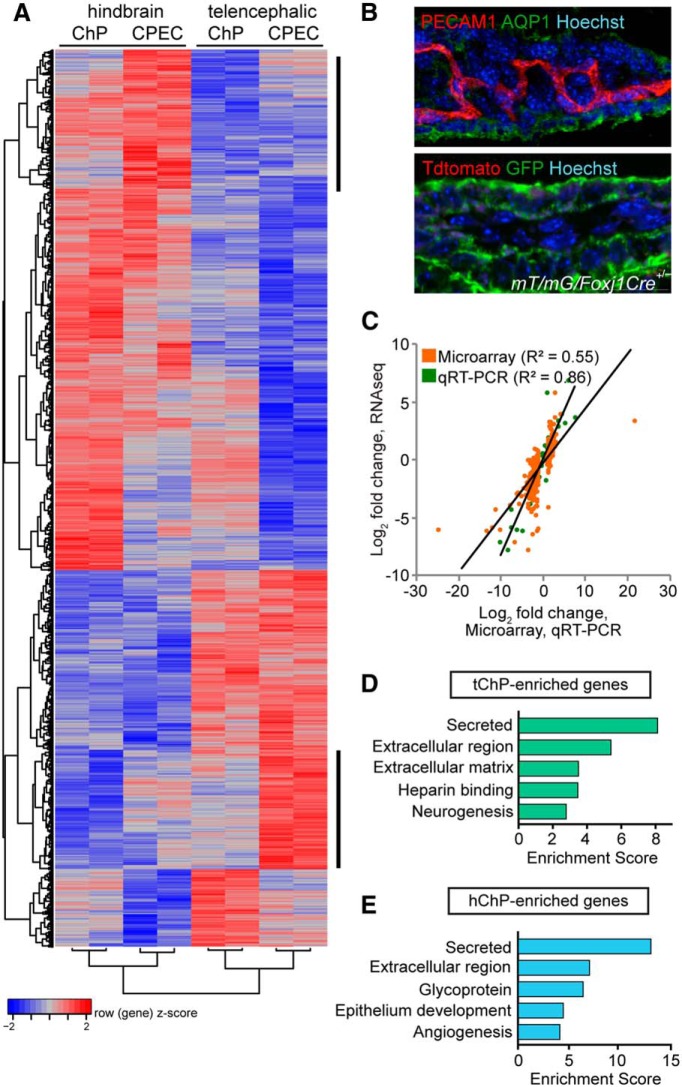
Because ChP-secreted signals may be a source of regionalized CSF in the lateral and fourth ventricles (Fig. 1A,B), we performed next-generation RNA sequencing (RNA-seq) analyses of telencephalic and hindbrain choroid plexi to identify transcriptional signatures that define positional heterogeneity between these tissues. First, RNA-seq of whole tChP and hChP was performed at E18.5 in mice (Fig. 1C), when proliferation of the choroid plexi nears completion (Hunter and Dymecki, 2007), peak CSF protein concentration is observed (see Fig. 6G; Lehtinen et al., 2011), and the ventricular system nears maturation, with distinct ventricular compartments and flow (Johanson et al., 2008). Second, because whole ChP consists of epithelial, vascular, and mesenchymal cells (Fig. 1A), transcriptome profiling of the bulk ChP is likely to miss smaller but significant cell-type-specific expression characteristics. Therefore, we performed parallel RNA-seq of purified ChP epithelial cells (CPEC), the cells responsible for CSF secretion, isolated from Transthyretin::RFP (Ttr::RFP) transgenic mice (Kwon and Hadjantonakis, 2009) by FACS (Fig. 1D). Of note, Ttr::RFP expression is mosaic across ChP epithelial cells (Fig. 1D). However, this mosaic expression, which becomes increasingly sparse with age, appears spatially random within the ChP epithelium and does not correlate with actual Transthyretin (TTR) expression as tested by immunohistochemistry or in situ hybridization (data not shown). Combining these datasets, we filtered and aligned 50 bp paired-end reads to the reference mouse genome and transcriptome (UCSC mm9), estimated gene expression levels, and performed sample-to-sample correlation analysis (Fig. 1E), as well as PCA (Fig. 1F) on the normalized read counts of ∼20,000 detected genes. PCA confirmed that the majority of variance between samples corresponded to the two variables of interest, cell type and ChP regional identity (Fig. 1F), suggesting that significant transcriptome-wide expression patterns distinguish both the constituent cell types of the whole ChP, as well as the positional identity of tChP versus hChP. In addition, both PCA and sample-to-sample correlation analysis (Fig. 1E,F) showed that these effects were greater than the individual variability across two biological replicates. We therefore proceeded to perform differential expression analysis using a paired sample, two-factor model contrasting both region (tChP vs hChP) and tissue type (whole ChP vs purified epithelial cells), identifying a total of 1699 differentially expressed genes at a threshold of twofold expression difference and a 5% FDR, including 684 genes displaying regional heterogeneity between tChP and hChP (Fig. 2A; Table 1) and 526 genes enriched in purified epithelial cells that we hypothesized encode a significant fraction of the ChP secretome. Furthermore, 256 gene expression differences between tChP and hChP were only detected in the purified epithelial cells, demonstrating the importance of cell-type-specific transcriptional profiling (Fig. 2B). Altogether, these data strongly support our hypothesis that regional gene expression differences in the ChP contribute to differential protein secretion into the CSF.
Figure 1.
Next-generation mRNA sequencing reveals regional heterogeneity between telencephalic and hindbrain choroid plexi. A, Left, H&E staining of sagittal E12.5 mouse brain. Right, H&E staining of sagittal E18.5 mouse brain and hChP (inset). Patency of lateral ventricle (LV), third ventricle (3V), and fourth ventricle (4V) changes dramatically with development, and the Aqueduct of Sylvius (Aq) constricts prior to birth. Blue arrows indicate tChP and hChP. Scale bar, 2 mm. High-magnification of hChP in E18.5 (inset) shows ChP structure with epithelial cells enveloping a core of vascular and mesenchymal cells. B, Silver stain of LV and 4V CSF, E18.5 mouse. Although some proteins are present at similar levels in the CSF of both ventricles (black arrowhead), others are more enriched in the CSF of the LV (red arrowhead) or of the 4V (blue arrowhead), suggesting regional differences in CSF protein composition between ventricles despite potential mixing between compartments due to CSF flow. C, Whole tChP and hChP were dissected from wild-type and Ttr::RFP transgenic mice at E18.5. Scale bar, 2 mm. D, ChP from Ttr::RFP mice were dissociated and FACS-sorted to purify the CPECs. Left, Sagittal view of tChP and hChP (inset) from E18.5 Ttr::RFP transgenic mouse. Sections were immunostained with anti-AQP1 (green) and Hoechst (blue), and imaged along with native RFP (red). Right, Representative FACS plot showing the purification of RFP+ CPEC. E, F, RNA was isolated from either whole ChP or purified CPEC from two biological replicates of each (embryonic litters) and processed for RNA sequencing. E, Euclidean distance matrix and clustering dendrogram indicating the correlation across all eight samples. F, PCA of gene expression estimates, with circles representing tChP and triangles representing hChP, and ellipses indicating the 99% confidence interval for the SE of the centroid for each region. Both PCA and clustering indicate that a large proportion of variance (42%) is attributed to tissue type (whole ChP vs purified CPEC), whereas a smaller but highly significant proportion of variance (22%) is due to regional gene expression differences between tChP and hChP, with both effects strongly outweighing the variability between biological replicates.
Figure 6.
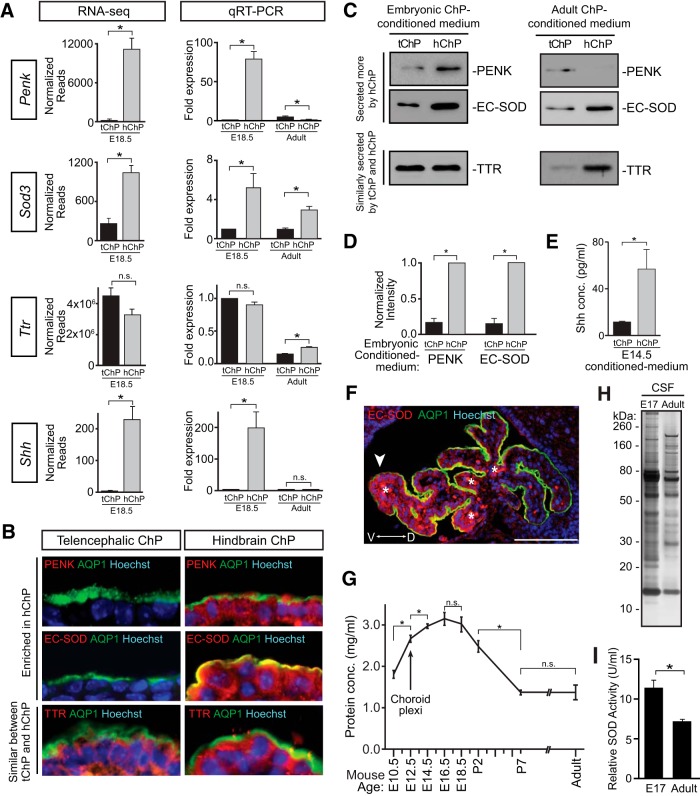
Differential protein expression and secretion between telencephalic and hindbrain choroid plexi establish anterior–posterior gradients in an age-dependent manner. A, Left, RNA-seq normalized read counts from DESeq analysis (Penk: tChP = 170.21 ± 296.22, hChP = 11132.09 ± 1815.991, FDR = 3.26 × 10−19; Sod3: tChP = 260.48 ± 85.500, hChP = 1039.99 ± 96.738, FDR = 4.55 × 10−12; Ttr: tChP = 4455,127.72 ± 514,497.268, hChP = 3239,035.52 ± 322,273.113, FDR = 3.42 × 10−01; Shh: tChP = 3.93 ± 2.097, hChP = 229.19 ± 40.774, FDR = 1.55 × 10−13). Right, qRT-PCR validation of differentially expressed genes Penk, Sod3, Shh, and similarly expressed gene Ttr at E18.5 represented as mean ± SEM, with tChP expression = 1.00. (Penk tChP, 1.00, Penk hChP, 78.75 ± 9.65, n = 4, t test, p = 0.0002; Sod3 tChP, 1.00, Sod3 hChP, 5.20 ± 1.46, n = 4, t test, p = 0.035; Ttr tChP, 1.00, Ttr hChP, 0.90 ± 0.04, n = 4, t test, p = 0.074; Shh tChP, 1.00, Shh hChP, 198.55 ± 50.53, n = 4, t test, p = 0.008; Table 2). qRT-PCR analyses in adult ChP demonstrates overall downregulation of Penk, Sod3, Ttr, and Shh gene expression. Additional comparisons between tChP and hChP in adults reveals that Penk is more abundantly expressed in adult tChP than hChP, the pattern of differential Sod3 expression between adult tChP and hChP is maintained, and Ttr is more abundantly expressed by adult hChP than tChP. Adult fold expression is represented as mean ± SEM, with E18.5 tChP expression = 1.00. (Adult Penk tChP, 4.824 ± 1.194, Adult Penk hChP, 0.818 ± 0.312, n = 3, t test, p = 0.031; Adult Sod3 tChP, 0.996 ± 0.134, Adult Sod3 hChP, 2.940 ± 0.362, n = 3, t test, p = 0.007; Adult Ttr tChP, 0.147 ± 0.012, Adult Ttr hChP, 0.249 ± 0.005, n = 3, t test, p = 0.001; Adult Shh tChP, 0.010 ± 0.004, Shh hChP, 0.018 ± 0.002, n = 5, t test, p = 0.130). B, Immunostaining of E16.5 ChP with anti-PENK, anti-EC-SOD, anti-TTR, anti-AQP1, and Hoechst. Top, Expression of PENK (red) was enriched in hChP epithelium. Middle, Expression of EC-SOD (red) was enriched in hChP epithelium. Bottom, Expression of TTR (red) was similar in tChP and hChP epithelia. C, Left, E16.5 mouse tChP and hChP were cultured for 24 h in equal volumes of base medium per ChP. Equal volumes of ChP-conditioned medium were immunoblotted with antibodies for PENK, EC-SOD, and TTR. Secretion of PENK and EC-SOD was greater by hChP than tChP. Secretion of TTR was similar between tChP and hChP. Right, Adult mouse tChP and hChP were cultured and analyzed as described above. Secretion of PENK was greater by tChP than hChP, secretion of EC-SOD was greater by adult hChP than tChP, and secretion of TTR was greater by adult hChP than tChP. D, Quantification of embryonic PENK and EC-SOD immunoblots normalized to TTR, shown in C. Data are represented as mean ± SEM (PENK tChP = 0.167 ± 0.056; hChP = 1.0, n = 4, t test, p < 0.0001; EC-SOD tChP = 0.146 ± 0.073; hChP = 1.0, n = 3, t test, p < 0.001). E, E14.5 mouse tChP and hChP were cultured for 24 h in equal volumes of base medium per ChP. Equal volumes of ChP-conditioned medium were measured for Shh concentration by ELISA, represented as mean concentration (pg/ml) ± SEM (tChP = 11.58 ± 0.65, hChP = 56.83 ± 16.79; n = 3, t test, p = 0.027). F, Immunostaining of hChP with anti-EC-SOD reveals cellular gradient of enriched expression. Section taken at level of gestational day 18 sagittal plate 10 from Prenatal Mouse Brain Atlas (Schambra, 2008). Arrowhead indicates region of enhanced EC-SOD expression along the ventral region of the hChP. Asterisks mark ChP vasculature. Double-headed arrow orients along dorsal (D)–ventral (V) axis. Scale bar, 100 μm. G, Total CSF protein concentration over the course of mouse embryonic and postnatal development represented as mean ± SEM (E10.5, 1.8 ± 0.09, n = 6; E12.5, 2.67 ± 0.08, n = 7; E14.5, 2.97 ± 0.06, n = 4; E16.5, 3.15 ± 0.16, n = 6; E18.5/P0, 3.01 ± 0.18, n = 7; P2, 2.48 ± 0.14, n = 3; P7, 1.37 ± 0.06, n = 6; adult, 1.37 ± 0.12, n = 3). CSF protein concentration increases following development of choroid plexi at E11–E12 (indicated by arrow; E10.5 vs E12.5, t test, p = 0.00003; E12.5 vs E14.5, t test, p = 0.031), declines postnatally (P2 vs P7, t test, p = 0.00004; and remains stable between P7 and adulthood, t test, p = 0.984). H, Silver stain of 1 μl of E17 and adult rat CSF. Total CSF protein complexity decreases in adulthood. I, Relative SOD activity decreases in rat CSF with age (E17 CSF, 11.3 ± 0.7; Adult CSF, 7.1 ± 0.1; n = 3, t test, p < 0.05).
Figure 2.
Transcriptional heterogeneity reveals regional choroid plexus identity. A, Hierarchical clustering of all ∼2500 genes showing significant regional differential expression between hChP (1426 genes) and tChP (1102 genes) at FDR < 0.05. Among these, 684 genes showed at least a two-fold regional enrichment. Red and blue indicate relatively higher and lower expression, with genes independently scaled to a mean of zero and 1 SD for display purposes. Vertical bars at the right of the heatmap highlight clusters of epithelial cell-specific or -enhanced regional expression differences. B, Top, Section from E18.5 mouse immunostained with anti-PECAM1 (red), anti-AQP1 (green), and Hoechst (blue). PECAM1 expression was enriched in the vascular component of the ChP. Bottom, Section from E16.5 mT/mG/FoxJ1Cre+/− reporter mice, which show recombination (GFP-positive staining cells) in nearly all ChP epithelial cells, were immunostained with anti-GFP (green) and Hoechst (blue). FOXJ1 expression was enriched in purified ChP epithelial cells. C, Linear regression of log2 fold-change in expression values for tChP- and hChP-enriched genes by RNA-seq, compared with gene expression microarray (orange) or qRT-PCR (green) reveals positive correlations in fold-change values between the three technologies, providing high confidence in our RNA-seq sample size, data quality, and analysis. D, E, Gene ontology analysis of tChP- and hChP-enriched genes compared with the reference mouse transcriptome reveals significant overrepresentation of genes encoding putative secreted factors. The top five functional clusters for each ChP are plotted (FDR < 0.05).
Table 1.
Differentially expressed genes characterize the positional identities of tChP and hChP
| Gene | Category | Fold-change | FDR |
|---|---|---|---|
| 25 most enriched genes in tChP | |||
| Dmrt1 | Transcription factor | Infiniteb | 0.03102 |
| Dmrt3 | Transcription factor | 209.31 | 0.00000 |
| Emx2 | Transcription factor | 119.65 | 0.00000 |
| Dmrta2 | Transcription factor | 101.97 | 0.00000 |
| Alx3 | Transcription factor | 71.29 | 0.00021 |
| Six3a | Transcription factor | 56.86 | 0.00000 |
| Six3os1 | Noncoding RNA | 24.20 | 0.00000 |
| Rspo2 | Secreted | 21.88 | 0.00000 |
| Vit | Extracellular matrix | 21.43 | 0.00000 |
| Chodl | Membrane component | 20.52 | 0.00000 |
| Emx2os | Noncoding RNA | 16.99 | 0.00000 |
| Nrn1 | Cell surface? | 16.30 | 0.00000 |
| Fezf1a | Transcription factor | 15.58 | 0.00170 |
| Gm19277 | Predicted gene | 13.75 | 0.00020 |
| Wnt8ba | Secreted | 12.98 | 0.00040 |
| Kcnip1 | Ion transport | 12.19 | 0.00000 |
| Gm11413 | Predicted gene | 11.82 | 0.00074 |
| Vstm2l | Secreted | 11.23 | 0.00105 |
| 4930412O13Rik | Predicted gene | 10.48 | 0.00032 |
| Otx1a | Transcription factor | 10.44 | 0.00000 |
| Nmur2 | Receptor | 9.84 | 0.00000 |
| Fgf17 | Secreted | 9.72 | 0.03804 |
| Fezf2a | Transcription factor | 9.12 | 0.01878 |
| Mc3ra | Receptor | 8.26 | 0.00000 |
| Klhl14 | Membrane component | 8.11 | 0.00006 |
| 25 most enriched genes in hChP | |||
| Hoxa3 | Transcription factor | Infiniteb | 0.00000 |
| Hoxb2 | Transcription factor | Infiniteb | 0.00001 |
| Hoxb3 | Transcription factor | Infiniteb | 0.00000 |
| Gdf7a | Secreted | 213.59 | 0.00000 |
| Irx2 | Transcription factor | 213.21 | 0.00000 |
| Lmx1b | Transcription factor | 173.13 | 0.00000 |
| Hoxa2a | Transcription factor | 136.73 | 0.00000 |
| Msx3 | Transcription factor | 91.55 | 0.01816 |
| Gm20554 | Predicted gene | 80.09 | 0.00634 |
| Tacr3 | Receptor | 68.80 | 0.00000 |
| Meis1a | Transcription factor | 67.70 | 0.00000 |
| Penka | Secreted | 65.40 | 0.00000 |
| C130021I20Rik | Non-coding RNA | 65.26 | 0.00000 |
| Hoxa1 | Transcription factor | 60.79 | 0.00071 |
| Shha | Secreted | 58.32 | 0.00000 |
| Lrrtm4 | Plasma membrane | 46.18 | 0.02512 |
| Gbp11 | Extracellular vesicular exosome | 43.50 | 0.00002 |
| Cnpy1 | Regulator of FGF signaling | 36.45 | 0.00028 |
| Wnt6 | Secreted | 27.53 | 0.00173 |
| Gfra1 | Receptor | 26.30 | 0.00000 |
| Thsd4 | Extracellular matrix | 22.39 | 0.00022 |
| Fibin | Secreted | 21.87 | 0.01517 |
| En2a | Transcription factor | 19.43 | 0.00000 |
| Tmem90b | Plasma membrane | 19.04 | 0.02612 |
| Adamts16 | Extracellular matrix | 17.53 | 0.00000 |
The top 50 genes with the greatest expression differences between tChP and hChP, defined by fold-change of expression, are listed along with their functional category. Fold-change was calculated from DESeq normalized read counts. In total, there are 149 genes upregulated in tChP and 322 genes upregulated in hChP (fold-change ≥2, FDR < 0.05).
aDifferential expression of genes validated by qRT-PCR (Table 2).
bTranscript was undetected in one region.
Transcriptional heterogeneity between choroid plexi
To validate differentially expressed genes, we first compared the results to an independent expression microarray experiment performed on three additional biological replicates of whole tChP versus hChP (Fig. 2C). Due to the greater sensitivity and dynamic range of RNA-seq, fewer genes were detected and identified as differentially expressed by microarray. However, among 477 candidates detected by both approaches, the direction of change was concordant, with a high correlation of fold-change values across platforms (R2 = 0.55). Differential gene expression was further verified by targeted qRT-PCR of 35 candidate genes in functional categories of interest, including transcription factors, cell surface receptors, and secreted factors (Figs. 2C, 3A, 6A; Table 2), with all genes displaying expression trends consistent with RNA-seq results (R2 = 0.86). Finally, we also verified candidates by inspection of in situ hybridization data from publically available resources, including GenePaint and Allen Brain Atlas to visually confirm gene expression in ChP epithelium (data not shown).
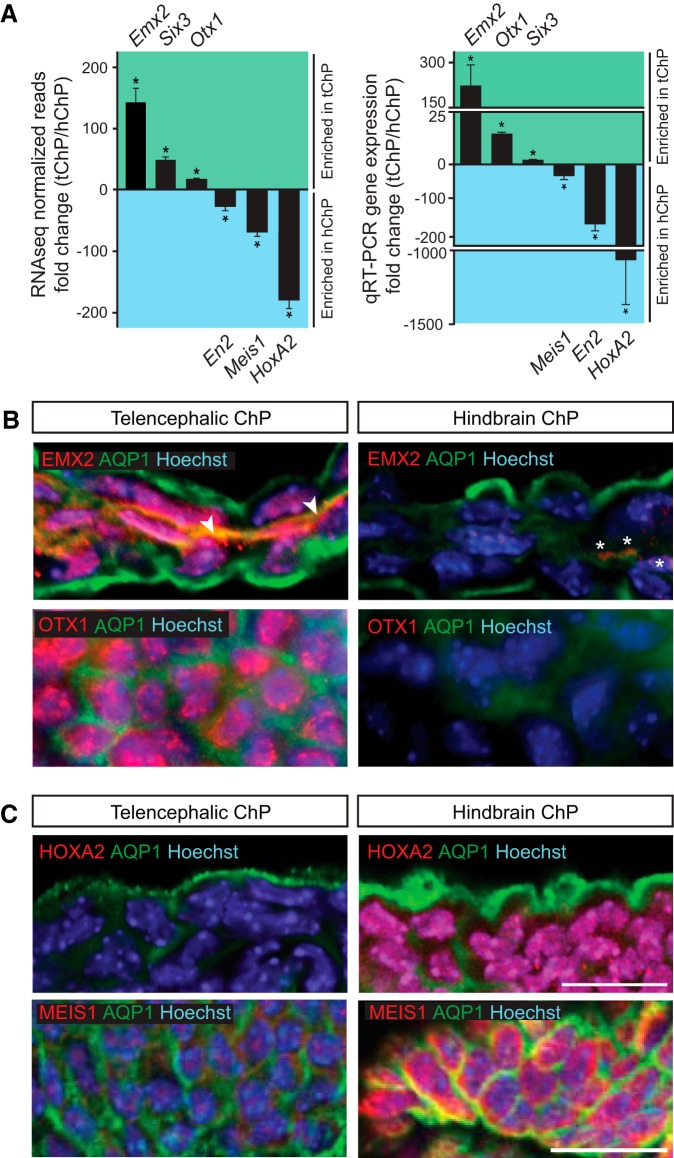
Figure 3.
Transcriptional heterogeneity elucidates distinct positional identities for choroid plexi. A, Left, Cufflinks-estimated RNA-seq FPKM counts from ChP (n = 4) for differentially expressed transcription factors represented as mean fold-change (tChP/hChP) ± SEM (Emx2, 142.67 ± 23.23, t test, p = 0.0009; Six3, 48.61 ± 4.75, t test, p < 0.0001; Otx1, 17.067 ± 2.10, t test, p = 0.0003; En2, −29.69 ± 3.56, t test, p < 0.0001; Meis1, −67.99 ± 7.18, t test, p = 0.0002; HoxA2, −177.50 ± 13.47, t test, p < 0.0001). Right, qRT-PCR validation of differentially expressed transcription factors in whole dissected tChP and hChP from E18.5 mouse represented as mean fold-change (tChP/hChP) ± SEM (Emx2, 205.40 ± 85.98, t test, p < 0.0001; Otx1, 14.36 ± 0.621, t test, p = 0.0061; Six3, 1.993 ± 0.113, t test, p = 0.0085; Meis1, −33.85 ± 9.57, t test, p = 0.0016; En2, −167.34 ± 19.42, t test, p = 0.0011; HoxA2, −1065.464 ± 302.98, t test, p = 0.0308). B, Immunostaining of E16.5 ChP with anti-EMX2 (red, top) and anti-OTX1 (red, bottom) antibodies reveals higher EMX2 and OTX1 expression in tChP than hChP. Also shown: anti-AQP1 (green) staining for apical surface of choroid plexus epithelium and Hoechst (blue). EMX2 staining of the vasculature marked by an arrowhead, weak EMX2 signal detected in hChP, indicated with asterisks. C, Immunostaining of E16.5 ChP with anti-HOXA2 (red, top) and anti-MEIS1 (red, bottom; whole mount ChP) antibodies reveals higher HOXA2 and MEIS1 expression in hChP than tChP. Also shown: anti-AQP1 (green) and Hoechst (blue). Scale bar, 20 μm.
We next performed gene ontology annotation clustering of the most regionally differentially expressed genes to determine the functions of the most highly enriched subset of genes in either the tChP or hChP (Fig. 2D,E). Secreted factors were the top enriched annotations for both tChP- and hChP-enriched genes (enrichment scores: tChP, 8.12; hChP, 13.31), supporting our hypothesis that regional heterogeneity of gene expression within the ChP may significantly contribute to differential protein secretion into the CSF. Although there are fewer enriched genes in tChP compared with hChP (149 genes vs 322 genes), a greater proportion of the tChP-enriched genes fall in the secreted functional category (30 genes of 149 vs 57 genes of 322).
Conserved positional identities between telencephalic and hindbrain choroid plexi
Our differential gene expression findings reveal that tChP and hChP are transcriptionally heterogeneous tissues. We further hypothesized that these differences, at least in part, arise from the choroid plexi retaining memories of their developmental origins from distinct positions along the rostral-caudal axis of the CNS. Consistent with this hypothesis, we found that the tChP is enriched for markers of the posterior half of the telencephalon [e.g., prosomeres (p)3/4] including Emx2, Otx1, and Six3 (Puelles and Rubenstein, 1993), which we verified by qRT-PCR (Fig. 3A; Table 2). We further confirmed the regionalized protein expression pattern of EMX2 and OTX1 by immunostaining (Fig. 3B). EMX2 was found in subsets of tChP epithelial cells, consistent with the previous observation that Emx2 mutant mice lack the tChP (Mallamaci et al., 2000). OTX1 was observed more uniformly in nearly all tChP epithelial cells examined (Fig. 3B).
The vertebrate Hox genes play a critical role in patterning the primary and secondary axes of the developing embryo (Philippidou and Dasen, 2013) and the hChP exhibits a Hox gene cluster signature including En2, HoxA, and HoxB genes (Table 1). We confirmed the differential expression of these genes, as well as Meis1, by qRT-PCR (Fig. 3A; Table 2). By immunostaining, HOXA2 and MEIS1 were found to be more highly expressed in hChP than tChP (Fig. 3C). Both transcription factors localized to subsets of hChP epithelial cells. Together, these data reflect that at the transcriptome level, distinct lineages of cells originate from specific rhombomeres and arrive at the hChP, as previously suggested by genetic lineage tracing studies (Awatramani et al., 2003; Hunter and Dymecki, 2007).
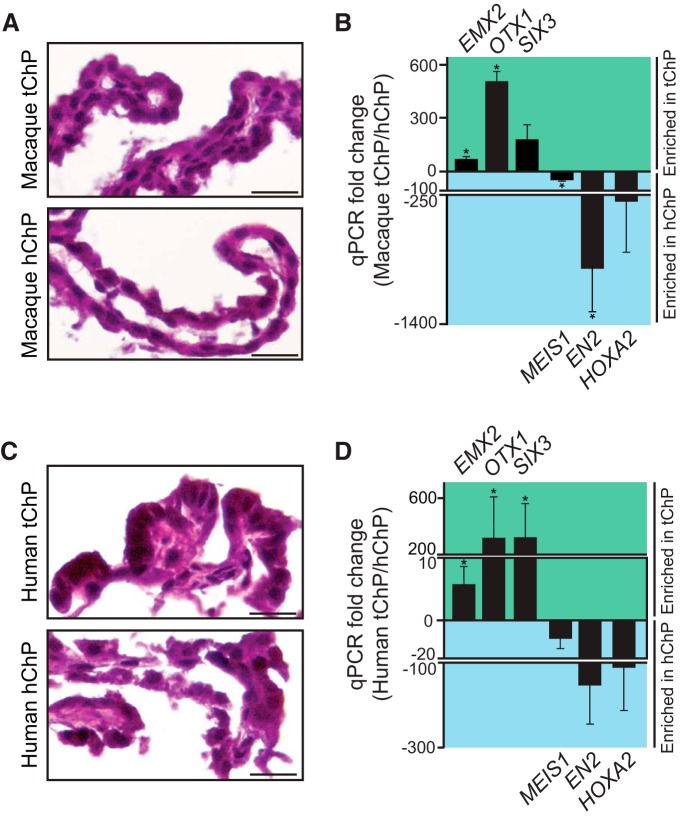
We next tested whether the regional identity of tChP and hChP we observed in mouse is conserved in macaque and human brain. Normal macaque choroid plexi from animals spanning E60 to 13 years old (Fig. 4A) were analyzed for markers of ChP positional identity by qRT-PCR (Fig. 4B). EMX2, OTX1, and SIX3 expression were enriched in tChP compared with hChP [mean fold-change (tChP/hChP) ± SEM; EMX2, 69.310 ± 15.467, p = 0.019; OTX1, 509.212 ± 54.897, p = 0.0004; SIX3, 179.401 ± 83.648 (not detected in 1 of 3 hChPs tested), p = 0.287], whereas MEIS1, EN2, and HOXA2 expression were enriched in hChP compared with tChP [mean fold-change (tChP/hChP) ± SEM; MEIS1, −64.673 ± 9.168, p < 0.0001; EN2, −666.360 ± 586.365, p < 0.0001; HOXA2, −269.846 ± 269.145, p = 0.171; Fig. 4B]. Similarly, paired samples of normal human choroid plexi from five individuals 11 d, 5, 13, 36, and 44 years of age (Fig. 4C), were analyzed by qRT-PCR for expression of positional identity genes (Fig. 4D). Conserved among primate ChP, expression of EMX2, OTX1, and SIX3 were enriched in the human tChP [mean fold-change (tChP/hChP) ± SEM; EMX2, 5.540 ± 2.953, p = 0.0007; OTX1, 308.947 ± 294.251, p < 0.0001; SIX3, 312.103 ± 242.809, p < 0.0001; Fig. 4D]; whereas expression of MEIS1, EN2, and HOXA2 were enriched in the human hChP [mean fold-change (tChP/hChP) ± SEM; MEIS1, −9.070 ± 5.420, p = 0.174; EN2, −150.898 ± 95.988, p = 0.157; HOXA2, −113.994 ± 100.118, p = 0.292; Fig. 4D]. All macaque and human tissue samples were confirmed to have similar TTR expression across choroid plexi (data not shown). Of note, the directionality of differential gene expression is conserved between the mouse and primate choroid plexi, yet the relative enrichment is variable between species, which may arise from differences in the starting material (use of the entire mouse ChP vs smaller portions of macaque and human ChP). Together, these findings suggest that at the transcriptional level, anterior–posterior positional identity of choroid plexi is evolutionarily conserved, and furthermore, that at least a subset of gene expression differences that define positional identity of tChP versus hChP persist from early development through adulthood.
Figure 4.
Positional identities of telencephalic and hindbrain choroid plexi are conserved in macaque and human brain. A, H&E staining of 13-year-old macaque tChP and hChP. Scale bar, 50 μm. B, qRT-PCR validation of transcription factor expression in six samples of macaque tChP and three samples of hChP, including two paired sets of tChP and hChP from the same animal, confirms that positional identity is evolutionarily conserved. Gene expression in macaque tChP and hChP is represented as mean fold-change (tChP/hChP) ± SEM. EMX2, OTX1, and SIX3 expression were enriched in tChP compared with hChP: EMX2, 69.310 ± 15.467, p = 0.019; OTX1, 509.212 ± 54.897, p = 0.0004; SIX3, 179.401 ± 83.648; not detected in 1 of 3 hChPs tested; p = 0.287), whereas MEIS1, EN2, and HOXA2 expression were enriched in hChP compared with tChP: MEIS1, −64.673 ± 9.168, p < 0.0001; EN2, −666.360 ± 586.365, p < 0.0001; HOXA2, −269.846 ± 269.145, p = 0.171. C, H&E staining of 36-year-old human tChP and hChP. Scale bar, 50 μm. D, qRT-PCR validation of transcription factor expression in human tChP and hChP confirms that positional identity is conserved among primates. Gene expression in human tChP and hChP (n = 5, paired samples) is represented as mean fold-change (tChP/hChP) ± SEM: EMX2, 5.540 ± 2.953, p = 0.0007; OTX1, 308.947 ± 294.251, p < 0.0001; SIX3, 312.103 ± 242.809, p < 0.0001; MEIS1, −9.070 ± 5.420, p = 0.174; EN2, −150.898 ± 95.988, p = 0.157; HOXA2, −113.994 ± 100.118, p = 0.292.
Defining the ChP epithelial cell secretome
Transcriptome analysis of FACS-purified epithelial cells allowed us to characterize the purified ChP epithelial cell secretome. The robust enrichment of vascular genes in whole ChP (e.g., Pecam1, Tie1, and Cd34) and of genes associated with multi-ciliated epithelial cells in the purified ChP epithelial cell samples (e.g., FoxJ1, Ift88, and Lrp2) support successful purification of epithelial cells from vasculature and mesenchyme (Fig. 2B). We performed functional categorization analysis of 18,133 genes detected in the purified ChP epithelium with ≥0.5 normalized read counts and defined the ChP epithelial cell secretome as the compilation of 1064 genes annotated to encode for secreted proteins (Table 3). The ChP transcriptome is significantly enriched for secreted factors, which is the top enriched functional category, followed by extracellular matrix components. Together, these genes contribute to 5.9% of the ChP transcriptome (modified Fisher exact p value, p < 0.05). Ttr and Igf2 are the two most abundantly expressed genes in the ChP epithelial cell secretome (Table 3), and these genes are not differentially expressed between tChP and hChP at this age.
Table 3.
Defining the choroid plexus epithelial cell secretome
| Gene | Mean reads | Mean tChP reads | Mean hChP reads | FDR |
|---|---|---|---|---|
| Ttr | 3847081.62 | 4455127.718 | 3239035.526 | 0.342 |
| Igf2 | 92897.44 | 76313.778 | 109481.117 | 0.360 |
| Enpp2 | 67405.69 | 47432.967 | 87378.416 | 0.054 |
| Sparc | 50425.49 | 56872.126 | 43978.845 | 0.671 |
| Sostdc1 | 29752.30 | 27843.994 | 31660.612 | 1.000 |
| Igfbp2 | 29345.47 | 34796.683 | 23894.247 | 0.105 |
| Clu | 24653.31 | 30431.046 | 18875.581 | 0.350 |
| 1500015O10Rik | 19142.38 | 19654.511 | 18630.256 | 1.000 |
| Fbn1 | 18107.83 | 21813.362 | 14402.295 | 0.180 |
| Sfrp1 | 17118.72 | 17508.331 | 16729.101 | 1.000 |
| Timp3 | 16347.37 | 18360.712 | 14334.026 | 0.725 |
| Hspg2 | 15489.86 | 18985.833 | 11993.877 | 0.049 |
| Lbp | 13679.64 | 15298.781 | 12060.490 | 1.000 |
| Apoe | 12875.60 | 14457.881 | 11293.319 | 0.649 |
| Wfikkn2 | 12229.14 | 17282.475 | 7175.813 | 0.000 |
| Gpc3 | 12095.70 | 14495.916 | 9695.491 | 0.149 |
| Psap | 10913.19 | 12579.000 | 9247.388 | 0.308 |
| Ogn | 10458.96 | 12224.946 | 8692.978 | 0.355 |
| Adamts1 | 10404.69 | 10770.837 | 10038.544 | 1.000 |
| Aebp1 | 10048.43 | 10539.088 | 9557.781 | 1.000 |
| Ace | 10020.11 | 11343.519 | 8696.706 | 0.462 |
| Nid2 | 9347.06 | 10812.944 | 7881.169 | 0.400 |
| Col8a1 | 9244.87 | 6800.335 | 11689.414 | 0.038 |
| Bgn | 8970.90 | 7596.718 | 10345.083 | 0.536 |
| Cst3 | 8481.43 | 9460.944 | 7501.910 | 0.803 |
Functional annotation by DAVID of the overall ChP transcriptome revealed 1064 genes that characterize the ChP epithelial cell-specific secretome. The 25 most abundantly expressed genes annotated to be secreted factors are listed, along with their mean normalized read counts from DESeq and FDR values.
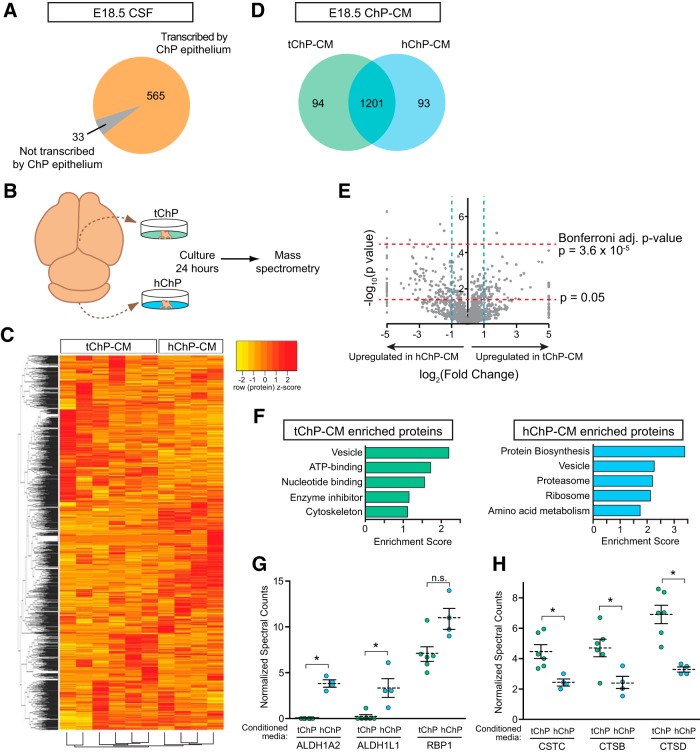
As the choroid plexi are considered the principal source of CSF, we determined the concordance between CSF proteins and ChP-derived factors by comparing MS analyses of native CSF and the predicted secretomes of tChP and hChP. Pooled CSF samples from lateral and fourth ventricles were collected (Zappaterra et al., 2013) from E18.5 mice. We accepted proteins with a false discovery rate of 1%. In total, 598 unique proteins were identified (Fig. 5A; data not shown). Five hundred sixty-five proteins had corresponding genes detected in the ChP transcriptome, and 140 of these proteins were included in our predicted ChP secretome. Furthermore, using this approach, we detected proteins encoded by 6 of the top 10 most abundant genes in the ChP secretome (in order of transcriptomics-defined secretome abundance: TTR, IGF2, ENPP2, SPARC, IGFBP2, CLU; Table 3; data not shown). Of all proteins identified, only 33 proteins did not have corresponding transcripts detected by RNA-seq in the ChP. Our data suggest that the most abundant proteins identified by MS in the embryonic CSF proteome could originate from the ChP. However, in vivo native CSF also receives contributions from the vasculature, ependymal cells, tissues adjacent to the ventricles, as well as the interstitial fluid (Johanson et al., 2008; Damkier et al., 2013). Our data represent an improvement in CSF protein detection by MS over previous analyses (Parada et al., 2006; Zappaterra et al., 2007; Lehtinen et al., 2011). Moreover, our MS analyses captured 53% of the proteins defined by gene ontology as the ChP secretome (Table 3). Of the proteins detected by MS, only 19 of the total 598 proteins were identified by RNA-seq to be differentially expressed between tChP and hChP (Table 1), suggesting that detection of additional region-specific proteins in the native CSF proteome will require an improved analytical depth.
Figure 5.
Mass spectrometry reveals distinct proteomes between telencephalic and hindbrain choroid plexus-conditioned media. A, Schematic representing mass spectrometry analysis of E18.5 mouse native CSF compared with E18.5 mouse ChP transcriptome. A total of 598 proteins were identified in E18.5 mouse CSF, and 33 of these proteins did not have their corresponding transcript detected in the E18.5 ChP transcriptome. B, Schematic illustrating approach for preparing ChP-conditioned medium for MS analyses. C, Heatmap of normalized spectral counts reveals differential protein availability in replicates of conditioned media obtained from pooled biological samples of conditioned media from tChP (n = 3) and hChP (n = 2). Each conditioned medium sample was run in duplicate, as represented by each lane in the heatmap (tChP, 6 technical runs; hChP, 4 technical runs). Spectral counts were centered and scaled across each row (protein) for display purposes. Red and yellow indicate relatively higher and lower number of spectral counts respectively. D, Schematic representing the numbers of proteins identified as shared and region-specific between conditions analyzed in C. E, Volcano plot showing distribution of proteins obtained by plotting log2(fold-change tChP/hChP) versus −log10(p value from two-tailed t test) of the averages of normalized spectral counts from sample replicates analyzed in C. Blue vertical lines: 2- and 0.5-fold cutoffs. Red horizontal lines: Log2 of Bonferroni-corrected p value cutoff [log2(0.05/1388) = 4.33], log2 of uncorrected p value cutoff [log2(0.05) = 1.3]. F, Functional clustering of differentially expressed proteins identified by MS with fold-change ≥1.5, p = 0.05 (tChP-enriched proteins = 57; hChP-enriched proteins = 137). The top five enriched functional clusters are shown (FDR < 0.05). G, Spectral counts show more abundant availability of ALDH1A2 and ALDH1L1 in hChP-conditioned medium (blue circles) than tChP-conditioned medium (green circles; hChP ALDH1A2 = 3.827 ± 0.346, tChP ALDH1A2 = not detected, t test, p = 0.002; hChP ALDH1L1 = 3.312 ± 1.020, tChP ALDH1L1 = 0.190 ± 0.190, t test, p = 0.046). A similar trend is observed for RBP1 (hChP RBP1 = 10.926 ± 1.096, tChP RBP1 = 7.038 ± 0.797, t test, p = 0.101). Differential expression of these proteins corresponds to similar trends at the transcript level as detected by RNA-seq (mean normalized read counts from DESeq ± SEM; Aldh1a2, hChP = 248.188 ± 125.197, tChP = 13.381 ± 5.824, FDR = 0.260; Aldh1l1, hChP = 236.645 ± 37.892, tChP = 93.027 ± 17.053, FDR = 0.029; Rbp1, hChP = 6811.561 ± 465.403, tChP = 3590.211 ± 151.407, FDR = 0.001). Graphed points represent individual replicates obtained from pooled samples of ChP-conditioned medium. Statistical analyses were performed on pooled samples of ChP-conditioned medium. Dotted line denotes mean. H, Spectral counts show more abundant availability of Cystatin C (CSTC), Cathepsin B (CTSB), and Cathpsin D (CTSD) in tChP-conditioned medium than hChP-conditioned medium (tChP CSTC = 4.466 ± 0.452, hChP CSTC = 2.441 ± 0.208, t test, p = 0.018; tChP CTSB = 4.698 ± 0.576; hChP CTSB = 2.356 ± 0.424, t test, p = 0.006; tChP CTSD = 6.904 ± 0.606, hChP CTSD = 3.286 ± 0.168, t test, p = 0.010). Data graphed and analyzed as described in G.
Molecular heterogeneity contributes to regional choroid plexus protein secretion
Differential gene expression analyses coupled with gene ontology annotation suggest that hChP expresses a greater number of genes encoding secreted proteins than tChP, which may contribute to the production of regionalized CSF proteomes in the fourth versus lateral ventricle. Because obtaining sufficient quantities of pure, ventricle-specific native mouse CSF for MS analyses is not technically feasible, we isolated secreted CSF/ChP-conditioned medium from cultures of whole tChP and hChP (Fig. 5B) and performed quantitative MS on these samples. Differential protein expression analyses showed that conditioned medium from the tChP and hChP clustered in a tissue-specific manner (Fig. 5C). Of the 1388 total proteins detected, 94 were identified only in the tChP-conditioned medium, 93 were identified only in hChP-conditioned medium, and 1201 proteins were found in both (Fig. 5D).
We performed a Volcano plot analysis of the quantitative proteomics data, i.e., log2-transformed tChP/hChP fold-change versus −log10-transformed p values (Fig. 5E). The vertical blue lines indicate a cutoff fold-change of 2 and 0.5, respectively, whereas the horizontal red lines indicate a p value cutoff of 0.05 or 3.6 × 10−5, corresponding to significance after applying the extremely conservative Bonferroni multiple testing correction. The fold-change of all proteins unique to either hChP or tChP were set to 2.0 × 10−5 and 2.0 × 105, respectively (Fig. 5E). The conservative (Bonferroni-corrected) analysis resulted in six proteins that showed significant abundance differences, whereas the looser cutoff resulted in 137 proteins being upregulated in hChP and 57 proteins being upregulated in tChP.
Enrichment analysis by DAVID of the differentially expressed proteins identified in the Volcano plot revealed shared and distinct classes of enriched proteins in the tChP- and hChP-conditioned medium (Fig. 5F). Vesicle and protein biosynthesis categories were among the top enriched classes of proteins identified (Fig. 5F). The differential availability of some proteins was consistent with predictions based on our RNA-seq analyses, including members of the Retinoic acid signaling pathway, which overall were more highly represented in the hChP-conditioned medium than tChP (Fig. 5G). Other groups of proteins, for example members of the Cystatin-Cathepsin signaling pathways, were not predicted to be differentially expressed by our RNA-seq analyses, yet were detected as differentially expressed by our MS analyses (Fig. 5H). These observations reflect how the availability of extracellular and secreted proteins depends on many parameters including but not limited to translation, processing, and transport. We also cannot rule-out protein contributions from ChP resident cells other than epithelial cells in these cultures. Previously suggested ventricular differences in CSF content (Cavanagh et al., 1983; Zappaterra et al., 2007) were not detected at the transcript level based on our RNA-seq analysis. However, 11.5% of the proteins previously identified in rat lateral ventricle CSF, but not rat fourth ventricle CSF (Zappaterra et al., 2007) were confirmed to be differentially expressed by our MS analyses. Differences in CSF protein availability may also vary by CSF-protein source, age, and/or species.
The three genes encoding secreted factors predicted by RNA-seq to be most differentially expressed between hChP and tChP [mean DESeq normalized-read fold-change (FC) ± SEM; Growth differentiation factor 7 (FC = −540.070 ± 244.866, p = 0.0001), Proenkephalin (Penk; FC = −76.546 ± 22.146, p = 0.0009), and Sonic hedgehog (Shh; FC = −44.721 ± 15.414, p = 0.0015)], an important morphogen for hindbrain, cerebellar, and hChP development (Awatramani et al., 2003; X. Huang et al., 2009, 2010; Nielsen and Dymecki, 2010), were not detected by our quantitative MS analyses, potentially due to processing of these proteins into smaller peptides and other physicochemical properties of the peptides (e.g., their size and hydrophobicity). To determine whether these hChP-enriched candidates, along with others such as Superoxide dismutase 3 (Sod3), which encodes the secreted anti-oxidant Extracellular superoxide dismutase (EC-SOD), contribute to producing distinct CSF proteomes, we first confirmed their expression patterns by qRT-PCR (Fig. 6A; Table 2) and immunohistochemistry (Fig. 6B). As predicted by both RNA-seq and qRT-PCR data, PENK and EC-SOD proteins were more abundant in hChP epithelial cells than tChP epithelial cells (Fig. 6B). Next, we again isolated secreted CSF/ChP-conditioned medium from cultures of whole tChP and hChP, this time assaying for specific protein content. Cultured CSF demonstrated that PENK, EC-SOD, and Shh were robustly secreted by the hChP compared with tChP (Figs. 6C–E). Similar trends of differential expression and possibly secretion are predicted by our RNA-seq analyses for various growth factors with known roles in the developing nervous system (e.g., members of the FGF and BMP families; Tables 1, 2). We also observed the equal secretion of proteins indicated to be similarly expressed across tissues by RNA-seq and qRT-PCR including TTR (Figs. 6A–C; Table 2). Immunohistological analyses also revealed a striking regionalization in protein expression domains within the hChP, highly suggestive of hotspots of protein secretion into the CSF in vivo. For example, EC-SOD is more abundant ventrally than dorsally in the hChP (Fig. 6F), akin to observations made for Shh, whose regionalized expression pattern within the hChP, and descendants of these Shh-expressing cells, exhibit regionalized character (Awatramani et al., 2003).
Finally, we observed that the regionalized expression and secretion of ChP proteins is regulated in an age-dependent manner. We first confirmed that many genes exhibiting positional heterogeneity at E18.5 also exhibited similar patterns of positional heterogeneity earlier in development at E14.5 by qRT-PCR (data not shown). Because overall CSF protein content (Fig. 6G,H; Lehtinen et al., 2011), as well as specific proteins including Shh (X. Huang et al., 2009; Nielsen and Dymecki, 2010), IGF2 (Lehtinen et al., 2011), and transporters in the tChP (Liddelow et al., 2012) show age-dependent expression, we next assayed gene expression in adult ChP. Overall, ChP gene expression decreased in adulthood (Fig. 6A; data not shown). However unexpectedly, many of the gene expression patterns observed embryonically also changed postnatally. For example, Penk expression decreased in adult hChP compared with tChP, leading to less PENK secretion by hChP than tChP into conditioned medium (Fig. 6C). Ttr, whose expression and protein secretion were similar between E18.5 tChP and hChP (Fig. 6A–C; Table 2), was found to be differentially expressed and secreted by adult ChP (Fig. 6A,C). Finally, the differential expression pattern of Sod3 was maintained in the adult ChP (Fig. 6A,C), and the decrease in overall adult ChP Sod3 expression was accompanied by reduced SOD activity in native adult CSF (Fig. 6I). Collectively, these findings demonstrate regional, age-dependent regulation of ChP gene expression from development into adulthood.
Consistent with our findings across rodent choroid plexi, we detected SHH and IGF2 in juvenile and adult primate choroid plexi. By qRT-PCR, we observed SHH expression in macaque hChP compared with tChP [mean fold-change (tChP/hChP) ± SEM, −16.085 ± 13.642, p = 0.3314; not detected in 3 of 6 tChPs tested]. Likewise, we detected SHH in two of five human hChP samples, whereas it was not detected in any of five tChP samples tested (data not shown). We also detected by qRT-PCR IGF2 expression in human tChP and hChP [mean fold-change (tChP/hChP) ± SEM, 1.879 ± 0.900, n = 5, p = 0.394]. Together, our findings suggest that the regionalized distribution of ChP-CSF factors regulating neurogenesis is conserved from rodent to primate brain.
Discussion
We show that ChP gene expression is spatially heterogeneous; a characteristic that may enable ventricle-specific instruction of brain development and health. The four key findings of our study are as follows: (1) the identification of positional heterogeneity in gene expression between the telencephalic and hindbrain mouse choroid plexi which parallels gene expression in axial tissues of origin, (2) the discovery that this positional identity of tChP and hChP is conserved across macaque and human choroid plexi, (3) the definition of secretomes from purified tChP and hChP epithelial cells (compared with the whole ChP), and (4) the identification of gene expression domains that contribute to functional differences in ChP-specific protein secretion in an age-dependent manner. Collectively, our findings suggest that specialized domains of ChP secretory cells may contribute to region-specific CSF-signaling niches in the rodent and primate brain.
Many cells are known to secrete signals that instruct development, and it is becoming clear that different subtypes of cells, such as astrocytes (Molofsky et al., 2014), demonstrate local heterogeneity in what signals are generated and secreted. During development, neural stem cells are initially in direct contact with the CSF, and the ChP secretes signals that play an active role in instructing neural stem cell proliferation and survival (X. Huang et al., 2010; Lehtinen et al., 2011). However, the question of whether regional heterogeneity in ChP-secreted signals exists has not been examined in detail. The possibility that CSF composition may be regionally specialized is intriguing, as it would provide a powerful means for region-specific regulation of stem cells. At early stages of development, ventricles arise within the newly closed neural tube (Lowery and Sive, 2009; Zappaterra and Lehtinen, 2012), and substantial mixing of CSF between ventricles is likely to occur. For example, the cerebral aqueduct first emerges as a wide channel that projects caudally from the third ventricle over the superior aspect of the pontine flexure and then connects to the fourth ventricle. Over the course of embryonic brain development, the aqueduct narrows, thus anatomically restricting flow from the lateral and third ventricles to the fourth ventricle (Fig. 1A). According to the traditional model of CSF flow, lateral and third ventricle CSF and factors therein will eventually arrive at the fourth ventricle if not bound to receptors on cells lining the ventricles (Brinker et al., 2014). Because factors in fourth ventricle CSF are less likely to flow upstream to the lateral ventricle, we reasoned that: (1) certain factors may be present only in fourth ventricle CSF, and (2) such factors might arise from a selective production and secretion by fourth ventricle choroid plexus (hChP) but not lateral ventricle choroid plexus (tChP).
Our results lend support to both of these hypotheses. First, although previous studies have shown that signals such as CSF-Shh act specifically on periventricular cells in the fourth ventricle at late stages of development (X. Huang et al., 2010), it was not known whether factors, such as Shh are produced locally, or whether they originate, at least in part, from upstream lateral ventricle ChP. We confirmed the abundant availability of Shh and several other factors in hChP-conditioned medium compared with tChP-conditioned medium (Figs. 5, 6), suggesting local production of these factors by hChP. These data were confirmed by our finding that Shh expression was greater in hChP than tChP (Fig. 6A; Tables 1, 2). However, additional investigation is needed to determine whether the hChP may be more receptive to Shh signaling as some classic downstream signaling factors (e.g., Gli1) are upregulated, whereas others (e.g., Ptch1 and Smo) are more similarly expressed between tChP and hChP. More generally, there are greater numbers of differentially expressed genes encoding for secreted proteins found in the hChP than in the tChP (Fig. 2). Our analyses are unique and highly sensitive to detect such regional differences, in part because of our approach to FACS-purify ChP epithelial cells. Validation of differential protein secretion by the ChP was performed on isolated ChP cultures and their conditioned medium because obtaining sufficient quantities of pure, regionalized in vivo embryonic mouse CSF for protein assays is rate limiting. We cannot rule out protein contributions from non-ChP epithelial cells in these cultures. Future studies will examine the molecular mechanisms that selectively regulate age-dependent changes in ChP protein expression and secretion, which should further inform therapeutic approaches of neurologic disease (Bateman et al., 2006; Potter et al., 2013; Xie et al., 2013).
Differentially expressed genes encoding secreted proteins originating from either the hChP or the tChP and positional transcription factor expression between choroid plexi may contribute to the production of regionalized CSF. It is possible that the retention of HoxA gene function in the hChP may be analogous to the developing limb bud, where the Hox clusters exhibit temporal and spatial collinearity and coordinate gene expression including Shh (Kmita et al., 2005; Tarchini et al., 2006; Philippidou and Dasen, 2013). However, abundance of expression and secretion may not correlate with principal sites of biological action. Igf2 is the second most abundantly expressed secreted gene by ChP epithelial cells (Table 3). By RNA-seq analyses, Igf2 expression was not more abundantly expressed by the hChP than tChP, yet its differential expression was suggested by qRT-PCR (Table 2). Yet, at the protein level, IGF2 was not observed to be differentially available in tChP- versus hChP-conditioned medium, as assayed by quantitative MS. IGF2 has confirmed sites of action along the ventricular zone of the developing cerebral cortex (Lehtinen et al., 2011), and Igf2-deficient mice appear to have a smaller cerebellum than controls (Lehtinen et al., 2011). Thus, CSF-IGF2-based signaling may play additional, as yet uncharacterized roles in development and maintenance of hindbrain structures as well.
Previous transcriptome to secretome analyses have revealed a decoupling between transcription and release of protein (Meissner et al., 2013). Comparisons of our RNA-seq data with proteomic analyses of ChP-conditioned medium suggest that differences between gene expression, protein expression, and protein secretion exist in the ChP as well. Unconventional means of protein secretion by the ChP, and other cells in proximity to the ventricles, may also contribute to the CSF proteome in unexpected ways (Nickel and Rabouille, 2009). Nevertheless, our data show that positional heterogeneity in gene expression between the tChP and hChP contributes to the production of regionalized CSF proteomes, which may provide region-specific CSF cues to individual neural stem cell populations located along the brain's ventricles.
Regionalization of the hindbrain ChP epithelium has been demonstrated by previous fate-mapping studies (Awatramani et al., 2003). Our findings build on these studies, showing that positional identities of both the tChP and hChP are instructed by patterned cues with origins in the axial nervous system. However, in vivo CSF is home to signals derived from sources other than the ChP, including the ventricular lining, vasculature, interstitial fluid, and axons innervating the ventricular walls (Johanson et al., 2008; Damkier et al., 2013). Many factors provided by non-ChP cell types have known roles in patterning the forebrain (e.g., BMPs, WNTs, FGFs; Grove and Monuki, 2013), and the choroid plexi express receptors to receive such cues (Table 1). Thus, the choroid plexi may well be patterned by signals distributed in the fluid that bathes them as well. Finally, the choroid plexi are highly vascularized tissues, and complementary patterning cues may derive from the vasculature or possibly from anterior versus posterior circulation.
The conservation of ChP positional identity from mouse to primate brain suggests that our findings may inform future therapies for patients with a range of CNS disorders. For example, our data may provide clues to putative susceptibility loci for tumors of the ChP, which arise predominantly in the tChP in children and in the hChP in adults (Safaee et al., 2013). Our analyses show that Twist, a bHLH transcription factor associated with neoplastic ChP epithelial cells (Hasselblatt et al., 2009), is typically enriched in the hindbrain ChP. In addition, common surgical treatments for patients with hydrocephalus include cauterization of the ChP and/or shunting CSF. Following these procedures, particularly in cases of overshunting, patients may lack important regional CSF signals, raising the possibility that a region-specific, synthetic CSF supplement could be engineered as an adjuvant therapy. With the advent of nervous system therapies using induced pluripotent stem cells to derive and graft specific cell types, including ChP epithelial cells (Watanabe et al., 2012; Lehtinen et al., 2013), it will be essential to engineer cells and fluids with appropriate regional identity and age specificity.
Footnotes
This work was supported by an ABTA Medical Student Summer Fellowship and Glenn/AFAR Scholarship for Research in the Biology of Aging (M.P.L.); Nancy Lurie Marks Family Foundation Postdoctoral Fellowship (M.B.J.); California Institute for Regenerative Medicine (CIRM) Training Grant TF2-01152 (M.W.); NSF Graduate Research Fellowship (K.F.C.); São Paulo Research Foundation FAPESP 2013/24501-9 (T.A.); NIH U19 AI070489 (M.J.H.); NIH award MH106934 (N.S.); CIRM RN2-00915-1 and UCI Institute for Clinical and Translational Science Pilot Project Award (E.S.M.); Pediatric Hydrocephalus Foundation, Eleanor and Miles Shore Fellowship Program for Scholars in Medicine/Boston Children's Hospital Career Development Award, NIH K99/R00 NS072192 and R01 NS088566 (M.K.L.); and BCH IDDRC P30 HD18655. M.K.L. is a fellow of the Alfred P. Sloan Foundation. We thank C. A. Walsh, M. W. Zappaterra, and members of the Lehtinen laboratory for helpful discussions; and C. Caldwell, V. Ho, and A. Stough for technical assistance.
The authors declare no competing financial interests.
References
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, McCarthy DJ, Chen Y, Okoniewski M, Smyth GK, Huber W, Robinson MD. Count-based differential expression analysis of RNA sequencing data using R and bioconductor. Nat Protoc. 2013;8:1765–1786. doi: 10.1038/nprot.2013.099. [DOI] [PubMed] [Google Scholar]
- Awatramani R, Soriano P, Rodriguez C, Mai JJ, Dymecki SM. Cryptic boundaries in roof plate and choroid plexus identified by intersectional gene activation. Nat Genet. 2003;35:70–75. doi: 10.1038/ng1228. [DOI] [PubMed] [Google Scholar]
- Bateman RJ, Munsell LY, Morris JC, Swarm R, Yarasheski KE, Holtzman DM. Human amyloid-beta synthesis and clearance rates as measured in cerebrospinal fluid in vivo. Nat Med. 2006;12:856–861. doi: 10.1038/nm1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinker T, Stopa E, Morrison J, Klinge P. A new look at cerebrospinal fluid circulation. Fluids Barriers CNS. 2014;11:10. doi: 10.1186/2045-8118-11-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh ME, Cornelis ME, Dziegielewska KM, Evans CA, Lorscheider FL, Møllgård K, Reynolds ML, Saunders NR. Comparison of proteins in CSF of lateral and IVth ventricles during early development of fetal sheep. Brain Res. 1983;313:159–167. doi: 10.1016/0165-3806(83)90213-4. [DOI] [PubMed] [Google Scholar]
- Currle DS, Cheng X, Hsu CM, Monuki ES. Direct and indirect roles of CNS dorsal midline cells in choroid plexus epithelia formation. Development. 2005;132:3549–3559. doi: 10.1242/dev.01915. [DOI] [PubMed] [Google Scholar]
- Damkier HH, Brown PD, Praetorius J. Cerebrospinal fluid secretion by the choroid plexus. Physiol Rev. 2013;93:1847–1892. doi: 10.1152/physrev.00004.2013. [DOI] [PubMed] [Google Scholar]
- Delgado AC, Ferrón SR, Vicente D, Porlan E, Perez-Villalba A, Trujillo CM, D'Ocón P, Fariñas I. Endothelial NT-3 delivered by vasculature and CSF promotes quiescence of subependymal neural stem cells through nitric oxide induction. Neuron. 2014;83:572–585. doi: 10.1016/j.neuron.2014.06.015. [DOI] [PubMed] [Google Scholar]
- Dziegielewska KM, Ek J, Habgood MD, Saunders NR. Development of the choroid plexus. Microsc Res Tech. 2001;52:5–20. doi: 10.1002/1097-0029(20010101)52:1<5::AID-JEMT3>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Emerich DF, Skinner SJ, Borlongan CV, Vasconcellos AV, Thanos CG. The choroid plexus in the rise, fall and repair of the brain. Bioessays. 2005;27:262–274. doi: 10.1002/bies.20193. [DOI] [PubMed] [Google Scholar]
- Grove EA, Monuki ES. Morphogens, patterning centers, and their mechanisms of action. In: Rubenstein JL, Rakic P, editors. Comprehensive developmental neuroscience: patterning and cell type specification in the developing CNS and PNS. San Diego: Academic; 2013. pp. 25–44. [Google Scholar]
- Hasselblatt M, Mertsch S, Koos B, Riesmeier B, Stegemann H, Jeibmann A, Tomm M, Schmitz N, Wrede B, Wolff JE, Zheng W, Paulus W. TWIST-1 is overexpressed in neoplastic choroid plexus epithelial cells and promotes proliferation and invasion. Cancer Res. 2009;69:2219–2223. doi: 10.1158/0008-5472.CAN-08-3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009a;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009b;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Ketova T, Fleming JT, Wang H, Dey SK, Litingtung Y, Chiang C. Sonic hedgehog signaling regulates a novel epithelial progenitor domain of the hindbrain choroid plexus. Development. 2009;136:2535–2543. doi: 10.1242/dev.033795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Liu J, Ketova T, Fleming JT, Grover VK, Cooper MK, Litingtung Y, Chiang C. Transventricular delivery of Sonic hedgehog is essential to cerebellar ventricular zone development. Proc Natl Acad Sci U S A. 2010;107:8422–8427. doi: 10.1073/pnas.0911838107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter NL, Dymecki SM. Molecularly and temporally separable lineages form the hindbrain roof plate and contribute differentially to the choroid plexus. Development. 2007;134:3449–3460. doi: 10.1242/dev.003095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson CE, Duncan JA, 3rd, Klinge PM, Brinker T, Stopa EG, Silverberg GD. Multiplicity of cerebrospinal fluid functions: new challenges in health and disease. Cerebrospinal Fluid Res. 2008;5:10. doi: 10.1186/1743-8454-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappers JA. The development of the paraphysis cerebri in man with comments on its relationship to the intercolumnar tubercle and its significance for the origin of cystic tumors in the third ventricle. J Comp Neurol. 1955;102:425–509. doi: 10.1002/cne.901020205. [DOI] [PubMed] [Google Scholar]
- Keep RF, Jones HC. A morphometric study on the development of the lateral ventricle choroid plexus, choroid plexus capillaries and ventricular ependyma in the rat. Brain Res Dev Brain Res. 1990;56:47–53. doi: 10.1016/0165-3806(90)90163-S. [DOI] [PubMed] [Google Scholar]
- Kmita M, Tarchini B, Zàkàny J, Logan M, Tabin CJ, Duboule D. Early developmental arrest of mammalian limbs lacking HoxA/HoxD gene function. Nature. 2005;435:1113–1116. doi: 10.1038/nature03648. [DOI] [PubMed] [Google Scholar]
- Kokovay E, Goderie S, Wang Y, Lotz S, Lin G, Sun Y, Roysam B, Shen Q, Temple S. Adult SVZ lineage cells home to and leave the vascular niche via differential responses to SDF1/CXCR4 signaling. Cell Stem Cell. 2010;7:163–173. doi: 10.1016/j.stem.2010.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon GS, Hadjantonakis AK. Transthyretin mouse transgenes direct RFP expression or Cre-mediated recombination throughout the visceral endoderm. Genesis. 2009;47:447–455. doi: 10.1002/dvg.20522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtinen MK, Walsh CA. Neurogenesis at the brain-cerebrospinal fluid interface. Annu Rev Cell Dev Biol. 2011;27:653–679. doi: 10.1146/annurev-cellbio-092910-154026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtinen MK, Zappaterra MW, Chen X, Yang YJ, Hill AD, Lun M, Maynard T, Gonzalez D, Kim S, Ye P, D'Ercole AJ, Wong ET, LaMantia AS, Walsh CA. The cerebrospinal fluid provides a proliferative niche for neural progenitor cells. Neuron. 2011;69:893–905. doi: 10.1016/j.neuron.2011.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtinen MK, Bjornsson CS, Dymecki SM, Gilbertson RJ, Holtzman DM, Monuki ES. The choroid plexus and cerebrospinal fluid: emerging roles in development, disease, and therapy. J Neurosci. 2013;33:17553–17559. doi: 10.1523/JNEUROSCI.3258-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddelow SA, Temple S, Møllgård K, Gehwolf R, Wagner A, Bauer H, Bauer HC, Phoenix TN, Dziegielewska KM, Saunders NR. Molecular characterisation of transport mechanisms at the developing mouse blood-CSF interface: a transcriptome approach. PLoS One. 2012;7:e33554. doi: 10.1371/journal.pone.0033554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddelow SA, Dziegielewska KM, Ek CJ, Habgood MD, Bauer H, Bauer HC, Lindsay H, Wakefield MJ, Strazielle N, Kratzer I, Møllgård K, Ghersi-Egea JF, Saunders NR. Mechanisms that determine the internal environment of the developing brain: a transcriptomic, functional and ultrastructural approach. PLoS One. 2013;8:e65629. doi: 10.1371/journal.pone.0065629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery LA, Sive H. Totally tubular: the mystery behind function and origin of the brain ventricular system. Bioessays. 2009;31:446–458. doi: 10.1002/bies.200800207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallamaci A, Muzio L, Chan CH, Parnavelas J, Boncinelli E. Area identity shifts in the early cerebral cortex of Emx2−/− mutant mice. Nat Neurosci. 2000;3:679–686. doi: 10.1038/76630. [DOI] [PubMed] [Google Scholar]
- Marques F, Sousa JC, Coppola G, Gao F, Puga R, Brentani H, Geschwind DH, Sousa N, Correia-Neves M, Palha JA. Transcriptome signature of the adult mouse choroid plexus. Fluids Barriers CNS. 2011;8:10. doi: 10.1186/2045-8118-8-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner F, Scheltema RA, Mollenkopf HJ, Mann M. Direct proteomic quantification of the secretome of activated immune cells. Science. 2013;340:475–478. doi: 10.1126/science.1232578. [DOI] [PubMed] [Google Scholar]
- Molofsky AV, Kelley KW, Tsai HH, Redmond SA, Chang SM, Madireddy L, Chan JR, Baranzini SE, Ullian EM, Rowitch DH. Astrocyte-encoded positional cues maintain sensorimotor circuit integrity. Nature. 2014;509:189–194. doi: 10.1038/nature13161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Nickel W, Rabouille C. Mechanisms of regulated unconventional protein secretion. Nat Rev Mol Cell Biol. 2009;10:148–155. doi: 10.1038/nrm2617. [DOI] [PubMed] [Google Scholar]
- Nielsen CM, Dymecki SM. Sonic hedgehog is required for vascular outgrowth in the hindbrain choroid plexus. Dev Biol. 2010;340:430–437. doi: 10.1016/j.ydbio.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer TD, Willhoite AR, Gage FH. Vascular niche for adult hippocampal neurogenesis. J Comp Neurol. 2000;425:479–494. doi: 10.1002/1096-9861(20001002)425:4<479::AID-CNE2>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Parada C, Gato A, Aparicio M, Bueno D. Proteome analysis of chick embryonic cerebrospinal fluid. Proteomics. 2006;6:312–320. doi: 10.1002/pmic.200500085. [DOI] [PubMed] [Google Scholar]
- Philippidou P, Dasen JS. Hox genes: choreographers in neural development, architects of circuit organization. Neuron. 2013;80:12–34. doi: 10.1016/j.neuron.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter R, Patterson BW, Elbert DL, Ovod V, Kasten T, Sigurdson W, Mawuenyega K, Blazey T, Goate A, Chott R, Yarasheski KE, Holtzman DM, Morris JC, Benzinger TL, Bateman RJ. Increased in vivo amyloid-beta42 production, exchange, and loss in presenilin mutation carriers. Sci Transl Med. 2013;5:189ra77. doi: 10.1126/scitranslmed.3005615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puelles L, Rubenstein JL. Expression patterns of homeobox and other putative regulatory genes in the embryonic mouse forebrain suggest a neuromeric organization. Trends Neurosci. 1993;16:472–479. doi: 10.1016/0166-2236(93)90080-6. [DOI] [PubMed] [Google Scholar]
- Safaee M, Oh MC, Bloch O, Sun MZ, Kaur G, Auguste KI, Tihan T, Parsa AT. Choroid plexus papillomas: advances in molecular biology and understanding of tumorigenesis. Neuro Oncol. 2013;15:255–267. doi: 10.1093/neuonc/nos289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schambra U. Prenatal mouse brain atlas. New York: Springer; 2008. [Google Scholar]
- Sturrock RR. A morphological study of the development of the mouse choroid plexus. J Anat. 1979;129:777–793. [PMC free article] [PubMed] [Google Scholar]
- Tarchini B, Duboule D, Kmita M. Regulatory constraints in the evolution of the tetrapod limb anterior–posterior polarity. Nature. 2006;443:985–988. doi: 10.1038/nature05247. [DOI] [PubMed] [Google Scholar]
- Tavazoie M, Van der Veken L, Silva-Vargas V, Louissaint M, Colonna L, Zaidi B, Garcia-Verdugo JM, Doetsch F. A specialized vascular niche for adult neural stem cells. Cell Stem Cell. 2008;3:279–288. doi: 10.1016/j.stem.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Kang YJ, Davies LM, Meghpara S, Lau K, Chung CY, Kathiriya J, Hadjantonakis AK, Monuki ES. BMP4 sufficiency to induce choroid plexus epithelial fate from embryonic stem cell-derived neuroepithelial progenitors. J Neurosci. 2012;32:15934–15945. doi: 10.1523/JNEUROSCI.3227-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilting J, Christ B. An experimental and ultrastructural study on the development of the avian choroid plexus. Cell Tissue Res. 1989;255:487–494. doi: 10.1007/BF00218783. [DOI] [PubMed] [Google Scholar]
- Xie L, Kang H, Xu Q, Chen MJ, Liao Y, Thiyagarajan M, O'Donnell J, Christensen DJ, Nicholson C, Iliff JJ, Takano T, Deane R, Nedergaard M. Sleep drives metabolite clearance from the adult brain. Science. 2013;342:373–377. doi: 10.1126/science.1241224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappaterra MD, Lisgo SN, Lindsay S, Gygi SP, Walsh CA, Ballif BA. A comparative proteomic analysis of human and rat embryonic cerebrospinal fluid. J Proteome Res. 2007;6:3537–3548. doi: 10.1021/pr070247w. [DOI] [PubMed] [Google Scholar]
- Zappaterra MW, Lehtinen MK. The cerebrospinal fluid: regulator of neurogenesis, behavior, and beyond. Cell Mol Life Sci. 2012;69:2863–2878. doi: 10.1007/s00018-012-0957-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappaterra MW, LaMantia AS, Walsh CA, Lehtinen MK. Isolation of cerebrospinal fluid from rodent embryos for use with dissected cerebral cortical explants. J Vis Exp. 2013;73:e50333. doi: 10.3791/50333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Huang G, Shornick LP, Roswit WT, Shipley JM, Brody SL, Holtzman MJ. A transgenic FOXJ1-Cre system for gene inactivation in ciliated epithelial cells. Am J Respir Cell Mol Biol. 2007;36:515–519. doi: 10.1165/rcmb.2006-0475RC. [DOI] [PMC free article] [PubMed] [Google Scholar]