Abstract
Symbiotic microbes can dramatically impact host health and fitness, and recent research in a diversity of systems suggests that different symbiont community structures may result in distinct outcomes for the host. In amphibians, some symbiotic skin bacteria produce metabolites that inhibit the growth of Batrachochytrium dendrobatidis (Bd), a cutaneous fungal pathogen that has caused many amphibian population declines and extinctions. Treatment with beneficial bacteria (probiotics) prevents Bd infection in some amphibian species and creates optimism for conservation of species that are highly susceptible to chytridiomycosis, the disease caused by Bd. In a laboratory experiment, we used Bd-inhibitory bacteria from Bd-tolerant Panamanian amphibians in a probiotic development trial with Panamanian golden frogs, Atelopus zeteki, a species currently surviving only in captive assurance colonies. Approximately 30% of infected golden frogs survived Bd exposure by either clearing infection or maintaining low Bd loads, but this was not associated with probiotic treatment. Survival was instead related to initial composition of the skin bacterial community and metabolites present on the skin. These results suggest a strong link between the structure of these symbiotic microbial communities and amphibian host health in the face of Bd exposure and also suggest a new approach for developing amphibian probiotics.
Keywords: amphibians, probiotics, skin bacteria, disease, microbiome, chytrid fungus
1. Introduction
Animals host a diversity of symbiotic microorganisms, many of which are vital for host fitness. These microbial communities are made of bacteria, fungi, archaea and viruses that interact with each other and their host along a continuum from mutualistic to parasitic [1–3]. Recent studies suggest that the structure of these symbiotic microbial communities can have direct impacts on their function, and ultimately on host phenotype. For example, in humans, gut communities with high diversity and dominated by Bacteroidetes appear to contribute to lean phenotypes, while gut communities with low diversity and dominated by Actinobacteria appear to contribute to obesity [4]. In marine invertebrates, such as corals and sponges, specific community structures of symbiotic microbes have been linked to healthy and diseased animals [5,6]. Some symbiotic microbes positively impact host physiology and health by preventing pathogen colonization and enhancing immune function (defensive symbiosis) [6,7]. For example, in mice, immunity against the cutaneous parasite Leishmania major is improved through skin microbiota-enhanced effector T-cell responses [8]. Amphibian skin is also inhabited by diverse bacteria that produce a range of antimicrobial metabolites that defend against cutaneous and embryonic pathogens, including the fungal pathogen Batrachochytrium dendrobatidis (Bd) [9–13].
Bd causes the disease chytridiomycosis and is responsible for hundreds of amphibian declines and extinctions, including extirpation of the emblematic Panamanian golden frog, Atelopus zeteki [14,15]. This species was historically present in central-western Panamá until Bd arrived. Fortunately, prior to the arrival of Bd, a few A. zeteki individuals were captured and placed in survival-assurance colonies [14]. Currently, successful breeding colonies exist in both Panama and the USA [16]. However, Bd still remains on amphibian species that are less susceptible to chytridiomycosis in what was the natural habitat of A. zeteki [17], preventing the reintroduction of highly susceptible golden frogs.
One promising reintroduction strategy is the use of probiotics (beneficial microbes) [18]. Probiotic therapies prevent disease, as well as improve growth and survival, in many other systems including agriculture, aquaculture and human medicine [19–21]. For instance, an experimental study demonstrated that the probiotic Lactobacillus sakei was effective at protecting its mouse host against Corynebacterium tuberculostearicum, a bacterium associated with chronic rhinosinusitis in humans [22]. In some amphibian species, bacteria that inhibit Bd in vitro prevent chytridiomycosis when incorporated into their existing skin microbiota as probiotics [9,18], presumably through the production of anti-Bd metabolites. Janthinobacterium lividum is a bacterium commonly found on the skin of North American amphibians [13,23,24] and has been effective as a probiotic treatment to prevent chytridiomycosis in two North American amphibian species [9,11]. Unfortunately, probiotic therapy with J. lividum did not prevent chytridiomycosis in captive A. zeteki, as it failed to persist on the skin of experimental frogs possibly owing to host defences or competition from other resident microbes [25].
The aims of this study were to investigate the relationship between disease outcome and the structure (relative abundance) and function (metabolite profiles) of the cutaneous bacterial community prior to and after pathogen exposure, as well as to develop a probiotic treatment method for A. zeteki. We hypothesized that antifungal bacterial species collected from Panamanian amphibian species would be better able to colonize and persist on the skin of A. zeteki and prevent chytridiomycosis. We isolated four candidate probiotic bacteria from Panamanian amphibians that strongly inhibited Bd growth in vitro and tested them experimentally to determine whether they could prevent chytridiomycosis in captive A. zeteki.
2. Material and methods
(a). Probiotic candidate selection
Probiotic candidates were chosen from 484 bacteria isolated from skin swabs of 11 Panamanian frog species (67 individuals) sampled from January to April 2010 [26]. We tested each isolate's ability to inhibit Bd with a spectrophotometric assay (similar to Bell et al. [27]). We identified each isolate taxonomically by sequencing the 16S rRNA gene [24] and analysing the sequence with the Ribosomal Database Project's Sequence Match tool (http://rdp.cme.msu.edu). We then identified four probiotic candidates that we hypothesized would have the best success at colonizing and persisting on the skin of A. zeteki and preventing chytridiomycosis based on the following criteria: (i) high inhibition against Bd in vitro, (ii) no previous reports of pathogenicity (determined by a literature search), (iii) isolation from a host species that is closely related to A. zeteki and/or a species that persists in the presence of Bd, and (iv) detection on a high proportion of individuals in the host community from which it was collected. The following four bacterial isolates were selected for experimental tests with A. zeteki (electronic supplementary material, table S1): Chryseobacterium sp. (hereafter, Chryseo), Pseudomonas sp. 1 (hereafter, Pseudo1), Pseudomonas sp. 2 (hereafter, Pseudo2) and Stenotrophomonas sp. (hereafter, Steno).
(b). Study species and animal care
For our experiment, we obtained 47 adult captive-bred surplus A. zeteki frogs from the Maryland Zoo (Baltimore, MD, USA), which manages the Association of Zoos and Aquariums Panamanian golden frog Species Survival Program (www.aza.org/species-survival-plan-program). We placed frogs in individual plastic enclosures and followed standard husbandry protocols throughout the experiment as in Becker et al. [25]. Prior to any treatment, frogs underwent a two-week acclimation and observation period.
(c). Experimental design
To initially ensure the probiotic candidates did not have detrimental effects on the frogs, we randomly assigned frogs to five experimental groups; a control group (n = 9) and four probiotic groups (n = 8 each). We then treated all individuals with chlorhexidine (0.05%) every other day for one week to reduce existing cutaneous microbiota that may inhibit colonization of the probiotics. We prepared cultures of each probiotic and treated frogs with 4 × 108 cells of the same probiotic strain (probiotic groups) or sterile water (control group) following procedures in Harris et al. [9]. Afterwards, each individual was returned to its enclosure. Immediately prior to and every 14 days after probiotic treatment, we weighed each individual to assess body condition. We monitored frogs twice a day (morning and afternoon) to assess mortality and morbidity associated with probiotic treatment for 84 days.
There were no lethal or sublethal (weight loss) effects in response to any initial treatment with any of the probiotic candidates, thus we proceeded to assess the effectiveness of each probiotic for preventing chytridiomycosis. Frogs were reassigned at random to six experimental treatment groups. All individuals in each of the four probiotic groups received the identical probiotic as in the initial trial, but were also exposed to Bd (probiotic + Bd; n = 8 each). The nine frogs from the initial trial control group and an additional six frogs were randomly assigned to a control group (no probiotic, no Bd; n = 7) and a Bd only group (no probiotic + Bd; n = 8). We treated frogs within each probiotic + Bd group with the appropriate probiotic strain as in the initial trial. Three days later, we exposed probiotic + Bd and no probiotic + Bd frogs to 3000 zoospores of Bd strain JEL 310 as in Harris et al. [9]. This strain was isolated from Smilisca phaeota from Fortuna, Panamá and used in a previous A. zeteki probiotic experiment [25]. Control frogs were exposed to sterile water. After these exposures, each frog was returned to its enclosure.
Immediately prior to the second probiotic treatment and approximately every 28 days after Bd exposure, we swabbed each frog with a sterile rayon swab (MW113, Medical Wire & Equipment Co., Corsham, Wiltshire, UK) 10 times on the ventral surface, 10 times on each thigh and five times on each hind foot to assess microbial community dynamics and Bd infection intensity. On the same swabbing days, we weighed each individual to assess body condition. We monitored frogs twice a day (morning and afternoon) to assess mortality and morbidity. After death, frogs were preserved by freezing at −20°C.
The experiment was terminated 241 days after A. zeteki were exposed to Bd, and at that time 23 frogs had died of chytridiomycosis, nine frogs remained alive with Bd infections, five frogs had cleared infection with no Bd detection via qPCR for three consecutive time-points and in histological examination after euthanasia, and all control frogs remained alive (electronic supplementary material, table S2). All surviving frogs were swabbed and then euthanized by subcutaneous injection with 1% tricaine methanesulfonate [25]. We predicted the outcome (clear infection or die) of the nine remaining frogs based on current infection intensity, the change in infection intensity over time and the relationship of those variables to the trajectories in the frogs that actually did die or clear infection during the experiment. Six of the nine remaining individuals were predicted to clear infection because they had a low infection intensity at the end of the experiment (less than 100 zoospore equivalents) and infection intensity was decreasing on those individuals over the last 45 days of the experiment. The other three individuals were predicted to die because they had a high infection intensity at the end of the experiment (more than 100 zoospore equivalents) and infection intensity was increasing on those individuals over the last 45 days of the experiment.
(d). DNA extraction, qPCR and 16S rRNA amplicon sequencing
We extracted DNA from each swab with a Qiagen DNeasy blood and tissue kit (Valencia, CA, USA) following the manufacturer's protocol. To quantify Bd infection intensity, we amplified extracted DNA of all samples in duplicate with TaqMan qPCR following procedures described by Boyle et al. [28]. We used Bd strain JEL 427 to make DNA standards [28]. Samples with greater than 0.1 zoospore genomic equivalents (hereafter, zoospore equivalent) were considered positive for Bd infection. If the number of zoospore equivalents from duplicates was inconsistent, a third qPCR was conducted, and the majority result was retained (e.g. if two of three replicates were positive, we considered the individual Bd positive).
We focused on two experimental time-points for assessing the microbial communities: 3 days prior to Bd exposure and 28 days post-exposure. We chose 28 days after exposure because by then Bd had gone through approximately six generations of zoospore production (based on Bd's rate of development in vitro [29]) and impacts of Bd on the microbial community should be detectable. At those two time-points, we sequenced the microbial communities of frogs that cleared infection (day −3, n = 5; day 28, n = 5), those that died of chytridiomycosis (day −3, n = 10; day 28, n = 9) and individuals in the control group (day −3, n = 4; day 28, n = 4). We prepared extracted DNA for sequencing by amplifying the V4 region of the 16S rRNA gene following Caporaso et al. [30] with the exception that PCR reactions contained 2 µl of template DNA. Controls without template were run for each sample. DNA extracted from a sterile swab was also included as a negative control. We purified PCR products with the Qiagen QIAquik PCR Purification Kit (Qiagen, Valencia, CA, USA) using the manufacturer's protocol. An equimolar mixture of all the samples was then sequenced on an Illumina MiSeq instrument with a 250 bp paired-end strategy at the Dana-Farber Cancer Institute, following methods similar to Caporaso et al. [31]. To compensate for the low base diversity of the amplicon pool, the sample was run with a 10% PhiX control.
16S amplicon sequence data were assembled with Fastq-join (https://code.google.com/p/ea-utils/wiki/FastqJoin) and processed with the Quantitative Insights Into Microbial Ecology pipeline (QIIME v. 1.7.0) [32] according to methods in Becker et al. [33]. Briefly, we clustered quality-filtered sequences into distinct bacterial operational taxonomic units (OTUs, approx. bacterial ‘species’) at a sequence similarity threshold of 97% and assigned taxonomy with RDP classifier and the Greengenes database (v. 13_5). All samples were rarefied to 27 000 sequences to standardize sampling effort. Details of the bioinformatics methods are in the electronic supplementary material.
(e). Extraction of cutaneous metabolites and LCMS
Metabolites produced by amphibian cutaneous bacteria can inhibit Bd in vitro [13,34,35] and in vivo [11] . Therefore, we excised and extracted the skin of each frog to obtain cutaneous metabolites following procedures described by Brucker et al. [13]; briefly, excised skins were extracted three times by shaking in HPLC-grade methanol (5 ml each extraction). The combined crude extracts were filtered (0.45 µm PTFE membrane), concentrated and reconstituted in methanol. Samples were analysed with high performance liquid chromatography-mass spectrometry (LCMS) following Umile et al. [36]. To detect inter-sample contamination, we inserted methanol injections into the LCMS queue after every five samples. Tricaine methanesulfonate was detected in the LCMS results and was removed from data analysis. Only frogs that could be processed within eight hours of death were used for metabolite analysis to minimize the chance of analysing compounds that were produced by microbes after host death. Finally, we removed from all analyses metabolites known to be produced by Bd in culture (T. P. Umile 2014, unpublished data).
(f). Statistical analyses
One frog in Pseudo1 + Bd group and one frog in Pseudo2 + Bd group did not become infected and one frog in the Chryseo + Bd group died from causes other than chytridiomycosis (qPCR analysis and histological examination showed no sign of Bd infection, but mortality occurred). These individuals were removed from all analyses. Unless noted, all data were normally distributed and variances were equal among specific comparisons. Differences in survival among treatment groups were tested with a Mantel–Cox log-rank test. Bd infection intensity (number of zoospore equivalents) data were log-transformed to achieve normality. Differences in infection intensity among frogs infected with Bd were tested with ANOVA.
Measures of alpha diversity for the bacterial community on each frog (OTU richness, phylogenetic diversity and Shannon diversity index) were computed with QIIME. We analysed alpha diversity measures of microbiota and the number of metabolites on frogs among probiotic treatment groups with ANOVA. To test for significant differences in alpha diversity measures of microbiota and the number of metabolites on frogs that died of chytridiomycosis and those that cleared Bd infection, we used Student's t-tests. Frogs that were predicted to die or clear infection were not included in this and subsequent analyses because we were not positive of their final outcome. To compare the microbial community structure between frogs (beta diversity), a Bray–Curtis distance matrix [37] was generated with square-root transformed data using Primer 6 (v. 6.1.15). We compared metabolite composition between samples with a Sørensen distance matrix [38] using Primer 6. The Bray–Curtis distance metric considers relative abundance of individual OTUs within a community, while the Sørensen distance metric uses only presence/absence information. We focused on metabolite presence/absence because the nature of the metabolite LCMS analysis does not allow us to compare relative abundances of different metabolites on the same frog. From the distance matrices, differences in community structure between frogs that died of chytridiomycosis versus those that cleared Bd infection were statistically analysed with permutational multivariate analysis of variance (PERMANOVA) and visualized with principal coordinates analysis (PCO) using Permanova+ (v. 1.0.5).
The PERMANOVA suggested significant differences in community structure between frogs that died and those that cleared infection. To then identify which OTUs were driving those differences, we used indicator species analysis [39] using the IndVal function in the labdsv package (http://ecology.msu.montana.edu/labdsv/R) in R (v. 3.0.1) [40]. Indicator species analysis calculates an indicator value for each OTU in the dataset by computing the product of the relative abundance and frequency of each OTU in our predefined groups (cleared infection versus died of chytridiomycosis). Statistical significance was calculated for the indicator value of each OTU with Monte Carlo simulations. We considered an OTU as an indicator species if it had a p < 0.05 and an indicator value of more than 0.7 (as in [41,42]). An indicator value of 1 indicates that all the sequences of an OTU were observed in all the samples from one group and completely absent from the other group, while an indicator value of 0 indicates that the OTU was widely distributed across both groups. We corrected all multiple comparisons with the false discovery rate procedure [43].
3. Results
(a). Survival and infection intensity
Treating frogs with anti-Bd bacteria prior to Bd exposure did not alter survival rates when compared with frogs without probiotic treatment exposed to Bd (figure 1; Log-rank test, χ2 = 7.3, p = 0.119). However, five individuals (within the Bd only and Chryseo + Bd treatment groups) became infected with Bd and were able to clear the infection approximately 168 days after exposure (figure 2). By contrast, infection intensities on frogs that died of chytridiomycosis (n = 23) increased throughout the experiment or until death. Infection intensity was similar among all survival/infection outcome groups that contained infected individuals (died, n = 23; predicted to die, n = 3; cleared, n = 5; predicted to clear, n = 6) at day 28 (figure 2; ANOVA, F = 0.980, p = 0.415). This is the only statistical analysis that included frogs with ‘predicted’ outcomes.
Figure 1.
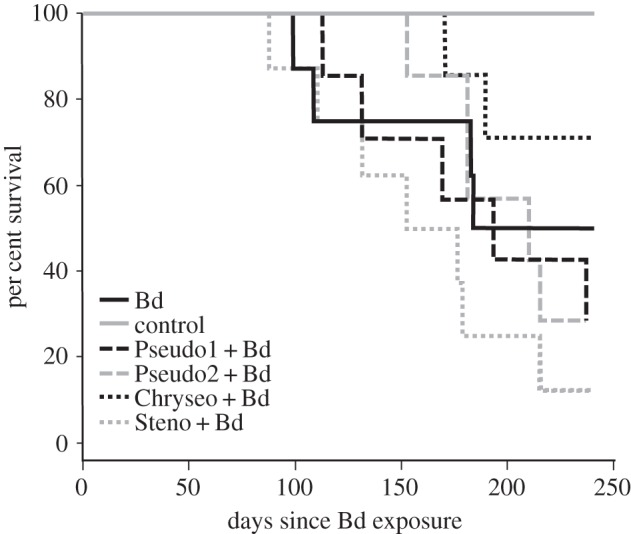
Survival of Atelopus zeteki treated with probiotics and exposed to Bd (‘probiotic’ + Bd), exposed only to Bd (Bd) and exposed to sterile water (control).
Figure 2.
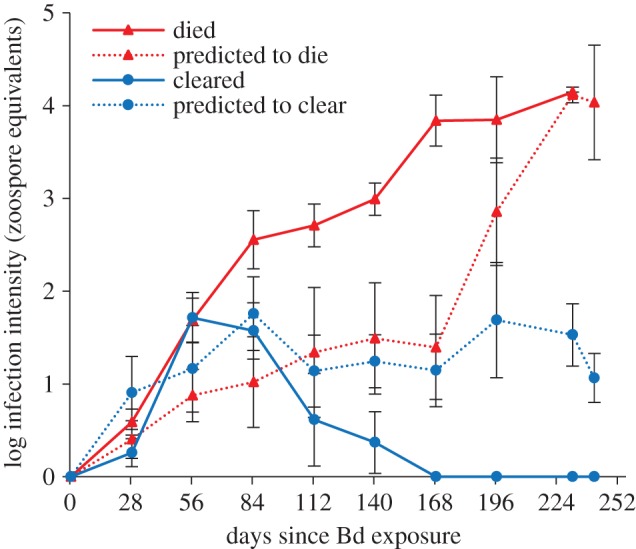
Average infection intensity (zoospore equivalents) of Atelopus zeteki that died of chytridiomycosis, were predicted to die of chytridiomycosis, cleared Bd infections, or were predicted to clear Bd infection. Error bars represent s.e.
(b). Cutaneous symbiotic microbiota
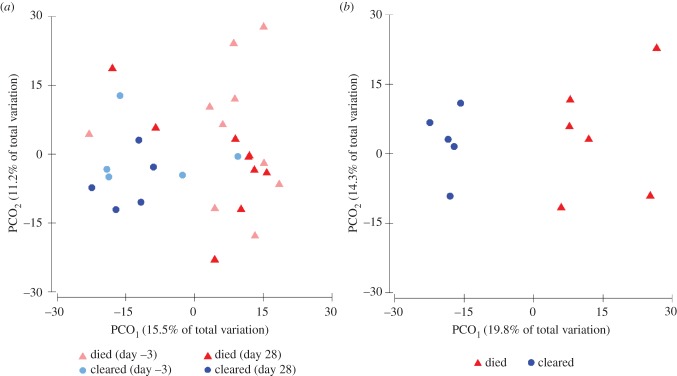
We sequenced the cutaneous bacterial communities of 37 golden frogs and detected an average of 758 OTUs (range = 542–900) on an individual frog. The microbial community structure 3 days prior to Bd exposure was significantly different on individuals that eventually cleared infection compared with those that died (figure 3a; PERMANOVA, Pseudo-F = 1.586, p = 0.042). The community structures also varied among these two groups 28 days after exposure to Bd (figure 3a; PERMANOVA, Pseudo-F = 1.671, p = 0.014). While the ordinations of Bray–Curtis distances (beta diversity) demonstrated differences in the composition of bacterial communities between frogs that cleared infection and those that died, we did not detect differences between these groups in alpha diversity metrics (OTU richness, evenness or phylogenetic diversity) at either day 0 or day 28 (t-tests, p > 0.05). The community structures of infected frogs were not significantly different than those of control frogs prior to exposure to Bd (PERMANOVA, Pseudo-F = 1.317, p = 0.104) or 28 days after exposure (PERMANOVA, Pseudo-F = 1.161, p = 0.177).
Figure 3.
Principal coordinates plots of (a) Bray–Curtis distances between microbial communities present on Atelopus zeteki 3 days prior to Bd exposure and 28 days after exposure that died of chytridiomycosis or cleared Bd infection, and (b) Sørensen distances between metabolites present on frogs that died of chytridiomycosis or cleared Bd infection.
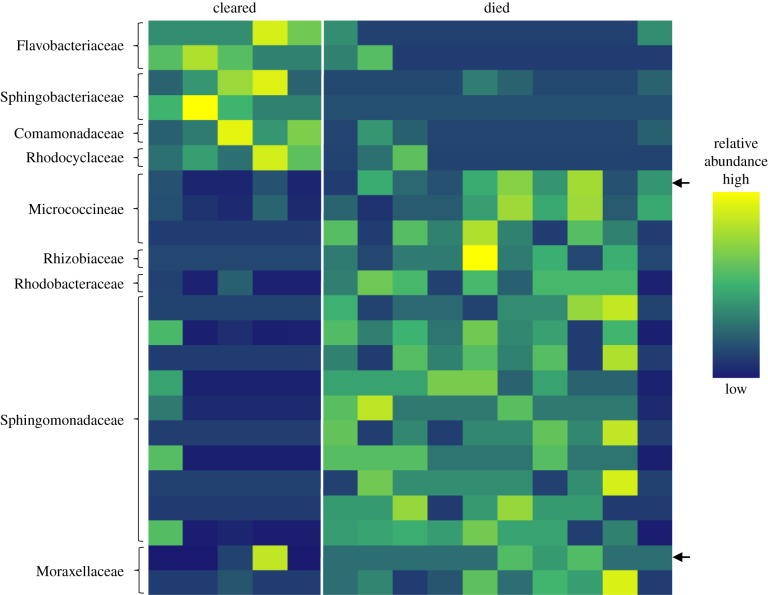
Indicator species analysis revealed a diverse array of OTUs that contributed to the dissimilarity between the microbial community composition on frogs that cleared infection and those that died (figure 4; electronic supplementary material, figure S1). Three days prior to Bd exposure, six OTUs were associated with individuals that eventually cleared infection, and 17 OTUs were associated with frogs that eventually died (figure 4). These indicator OTUs accounted for an average of 2.3% of the total sequences per frog. Three days prior to Bd exposure, indicator OTUs on individuals that eventually cleared infection belonged to the families Flavobacteriaceae, Sphingobacteriaceae, Comamonadaceae and Rhodocyclaceae. OTUs on individuals that died belonged to the families Micrococcineae, Rhizobiaceae, Rhodobacteraceae, Sphingomonadaceae and Moraxellaceae. Twenty-eight days after Bd exposure, 52 OTUs were associated with frogs that eventually cleared infection, and 22 were associated with frogs that died (electronic supplementary material, figure S1). Day 28 indicator OTUs accounted for an average of 7.6% of the total sequences per frog, however, many of the OTUs were rare members of the community (less than 0.1%). Two OTUs, one belonging to the family Micrococcineae and the other to the family Sphingomonadaceae, were significant indicator taxa at both time-points (figure 4; electronic supplementary material, figure S1).
Figure 4.
Heat map of the relative abundances of indicator OTUs from indicator species analysis associated with Atelopus zeteki 3 days prior to Bd exposure that cleared Bd infection or died of chytridiomycosis. Rows indicate unique OTUs and columns indicate individual frogs. Family level taxonomic classification is shown for each OTU. Arrows are indicator OTUs that were detected both 3 days prior to and 28 days after Bd exposure.
Day of sampling had no effect on the microbial community structure of frogs (3 days prior to Bd exposure versus 28 days afterwards; PERMANOVA, Pseudo-F = 0.902, p = 0.668). In addition, treating frogs with probiotics had no effect on the cutaneous microbial community structure 31 days after treatment (PERMANOVA, Pseudo-F = 0.999, p = 0.450). In addition, there were no differences in OTU richness, evenness (Shannon diversity index) and phylogenetic diversity among probiotic treatment groups at this time-point (ANOVAs, p > 0.05). Comparing the probiotic candidate sequences of the Stenotrophomonas sp. and Chryseobacterium sp. isolates to the Illumina sequences with a 99% similarity threshold revealed that neither probiotic could be detected 28 days after exposure to Bd (31 days after probiotic treatment). We were unable to look for the two Pseudomonas sp. probiotic candidates because the 16S rRNA gene is a poor region for distinguishing Pseudomonas species and strains [44].
(c). Cutaneous metabolites
We analysed the skin of 28 golden frogs and detected 554 unique metabolites across all individuals. There was an average of 67 (range = 51–81) metabolites detected on an individual frog. The number of metabolites per frog was similar among all probiotic treatment groups (ANOVA, F = 1.380, p = 0.270) and between frogs that cleared infection and those that died of chytridiomycosis (t-test, t = 1.031, p = 0.327). However, the metabolite profiles were significantly different between frogs that cleared infection and died (figure 3b; PERMANOVA, Pseudo-F = 2.187, p = 0.002).
4. Discussion
We found that treatment of captive A. zeteki with bacteria that are highly inhibitory against Bd in vitro was not successful in preventing the colonization and growth of Bd in vivo. However, although there were no statistically significant differences in survival among probiotic treatments, some A. zeteki frogs acquired a Bd infection and were able to clear this infection. This was unexpected as A. zeteki is highly susceptible to Bd, as demonstrated by three independent studies [25,45,46]. In these prior infection studies, only one of 228 golden frogs infected with Bd was documented as having acquired and then cleared infection. In this study, approximately 30% of golden frogs either cleared infection (14%) or were predicted to clear infection (16%).
Susceptibility to Bd can differ among amphibian species and populations [47], probably due to variation in defence mechanisms of the host [18,48,49], the virulence of Bd [46] and properties of the environment where the host and pathogen interact [50]. The environment and Bd strain in this study were constant across all frogs and were therefore not factors in the variation in Bd susceptibility. Of the known host defence mechanisms (antimicrobial peptides [48], diversity in major histocompatibility complex genes [49], acquired immune response [51] and skin microbial communities [18]), our results suggest that the community composition of skin bacteria probably plays a role in the ability of golden frogs to clear Bd and survive exposure. However, we were only able to examine the latter in this study.
Both 3 days prior to Bd exposure and 28 days after exposure, frogs that eventually cleared Bd had a significantly different bacterial skin community structure than those that died. This was also true for the metabolite profiles of frogs that died and cleared infection. These results suggest that bacteria and metabolites present on the skin of golden frogs may be responsible for the ability of some frogs to clear infection. Additionally, the fact that the bacterial community structure on frogs that eventually cleared infection and those that died of chytridiomycosis was significantly different prior to Bd exposure suggests the community was not responding to Bd and therefore is responsible for the clearance of Bd. This conclusion is consistent with results from other studies demonstrating the importance of cutaneous symbiotic bacteria and their antifungal metabolites in protecting amphibians from fungal pathogens, including Bd [10–12]. Predicting disease susceptibility with microbial community data has also been done for human intestinal pathogens [52,53].
In our study, the resident microbiota appeared to influence the interactions between Bd and A. zeteki. Inhibition of Bd could occur via direct interaction between particular resident microbes and Bd through competition and the production of antifungal metabolites [11,13]. In addition, the resident microbiota may indirectly prevent Bd growth by stimulating the host immune system (immunomodulation) [21]. These interactions are commonly seen in others systems [54]. For example, commensal bacteria that reside in the human gut competitively exclude pathogenic bacteria by producing antibiotics, preventing attachment to epithelial cells, competing for resources and stimulating the host's immune system [21,55]. In this study, many indicator OTUs which were associated with frogs that cleared infection belong to bacterial families that inhibit Bd in vitro and have been isolated from non-susceptible Panamanian amphibians (Flavobacteriaceae, Comamonadaceae, Pseudomonadaceae; [26]). For example, a Comamonadaceae indicator OTU (figure 4) had a 98% sequence similarity (16S rRNA gene) to four cultured isolates that inhibit Bd growth [26]. Bacteria belonging to these families have also been detected from skin swabs collected from a captive population of A. zeteki housed at the National Zoo and from a wild population of A. zeteki in Panama [33]. However, the ability of bacteria to inhibit Bd in vitro is dependent on environmental conditions [56] and varies greatly at the genus and even the strain level [26].
Indicator species analysis also revealed OTUs 3 days prior to Bd exposure and 28 days after exposure that were associated with frogs that died. These OTUs may have facilitated the colonization and growth of Bd or may have been opportunistic pathogens. In addition, the relative abundance of these OTUs may have increased because Bd was able to negatively affect the abundance of other OTUs [57]. Facilitation of Bd growth and colonization could occur either when symbiotic bacteria shift their metabolite profiles in a way that favours Bd [35] or by reducing the capability of other microbes to inhibit Bd. In a recent study, bacteria isolated from cyclamen and tomato plants, including a Novosphingobium sp. and a Sphingomonas sp., suppressed production of antimicrobial compounds produced by symbiotic bacteria that inhibit phytopathogens [58]. Interestingly, in our study, several indicator OTUs in the genera Novosphingobium or Sphingomonas were associated with dying frogs (3 days prior to Bd, 8 out of 17 OTUs; day 28, 2 out of 10 OTUs), suggesting that the same mechanism could operate in this system. Indicator taxa that were associated with dying frogs at day 28 may also be opportunistic pathogens that caused secondary infections. Opportunistic secondary infections involving bacteria are common in amphibians [59]. In particular, secondary or co-infections of the bacteria Aeromonas hydrophila and the fungus Saprolegnia sp. have been documented in amphibians with chytridiomycosis [60,61] and these may occur through microbial invasion of the damaged epidermis [59].
The finding that the cutaneous metabolite profiles of frogs that died were significantly different from those that cleared infection further supports the hypothesis that either microbial or host-produced metabolites influenced infection dynamics. Our methods for chemical analysis (reverse-phase LCMS) have been optimized for observation of bacterial metabolites [36]. Many known metabolites of amphibian origin that inhibit pathogens and provide defence against predators, such as antimicrobial peptides [62] and zetekitoxin [63], respectively, are too polar to be observed in our conditions. Antimicrobial peptides have not been detected in captive A. zeteki (B. Sheafor 2014, personal communication). Thus, while select metabolites of amphibian origin may be present in our dataset, it is highly likely that the bulk of the features assessed are of bacterial origin. Bacterially produced metabolites are capable of inhibiting Bd in vitro [13,34] and their concentrations on the skin are correlated with survival in Bd-infected salamanders (Plethodon cinereus) [11].
Although the bacterial isolates used as probiotics in our experiment were highly inhibitory against Bd in in vitro assays, they were not successful at preventing infection and subsequent death when applied to the skin of A. zeteki. Thirty-one days after treating the frogs with probiotic candidates, we were unable to detect the presence of two probiotic isolates, Chryseobacterium sp. and Stenotrophomonas sp., that we looked for in the 16S rRNA sequence data. This suggests that the isolates simply may not have been able to colonize and grow on A. zeteki skin. Even if they were there at low abundance, the probiotic isolates may not have been able to produce anti-Bd compounds on A. zeteki skin. The lack of an in vivo probiotic effect, despite compelling in vitro results, has been documented in fish [64]. The probiotic candidates could have been inhibited by compounds produced by either resident microbiota or the host [65], or a constant environmental inoculum may be needed for the isolates to persist on the skin [66]. In many cases, probiotics designed to prevent and treat human intestinal diseases rely on continual administration to be effective [67]. Furthermore, Bd exposure and probiotic treatment had no effect on the cutaneous bacterial community 28 and 31 days post treatment, respectively. This suggests that A. zeteki microbial communities are at least somewhat resistant and/or resilient to invasion. This is contrary to a recent study that found the cutaneous bacterial community structure of Rana sierrae was affected by the presence of Bd and its density on the frog [57].
Defensive symbioses are commonly found in many systems including mammals, insects, marine invertebrates, algae and plants [7,54,68]. For example, some species of fungus-growing ants have a symbiotic relationship with Actinomycetales bacteria that prevent parasite overgrowth in their fungal gardens [69], and some seaweed species are protected from fouling microorganisms through association with epibiotic bacteria [70]. Our results suggest that members of the amphibian skin microbial community are also important defensive symbionts, potentially through production of antimicrobial metabolites that prevent pathogen colonization or growth. For the critically endangered A. zeteki, these skin microbes may be an important determinant for survival in Bd-infected individuals, and isolating the bacteria that were correlated with clearance of Bd in this study may be the best way forward for probiotic development for Atelopus species. Additionally, the ability to detect microbial community composition differences among survival/infection outcome groups prior to infection is important and, in combination with ecological factors [71], may allow conservationists to predict susceptibility in free-living amphibian populations threatened by Bd.
Supplementary Material
Acknowledgements
We thank the Maryland Zoo in Baltimore and the Association of Zoos and Aquariums Panamanian golden frog Species Survival Program for providing animals used in this study. We are thankful for the insightful comments and careful review of the manuscript provided by J. Becker, R. Brucker, D. Hawley, S. Hopkins, A. Pruden and T. P. Umile as well as technical assistance provided by Z. Herbert of the Dana-Farber Cancer Institute Molecular Biology Core Facilities. We also wish to thank W. Lynch, G. Reynolds, M. Evans, K. Hope and M. Valitutto of the Smithsonian Conservation Biology Institute.
Ethics statement
The United States Fish and Wildlife Service provided permits to conduct the experiment and the Animal Care and Use Committees at the Smithsonian National Zoological Park and the Maryland Zoo in Baltimore approved the animal care protocol.
Data accessibility
The 16S rRNA sequences for the probiotic isolates have been deposited in GenBank (accession numbers KM006931–KM006934). Raw 16S rRNA read sequences from Illumina sequencing have been deposited in the European Nucleotide Archive (accession number PRJEB6565). Illumina data will be publicly available 1 year after publication.
Author contributions
M.H.B., J.B.W., R.N.H., L.K.B. and B.G. contributed to the design of the study. M.H.B., J.B.W., S.C., A.E.S., N.M., C.N.S., K.P.C.M. and B.G. performed research. M.H.B. analysed data and wrote the manuscript. All authors contributed to revisions and approved the final version of the manuscript.
Funding statement
This research was financially supported by the United States Fish and Wildlife Service Amphibians in Decline Fund, the National Science Foundation (DEB-1136640 and DEB-1136662), the Shared Earth Foundation and an anonymous frog-friendly foundation.
Competing interests
We have no competing interests.
References
- 1.Schommer NN, Gallo RL. 2013. Structure and function of the human skin microbiome. Trends Microbiol. 21, 660–668. ( 10.1016/j.tim.2013.10.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McKenzie VJ, Bowers RM, Fierer N, Knight R, Lauber CL. 2012. Co-habiting amphibian species harbor unique skin bacterial communities in wild populations. ISME J. 6, 588–596. ( 10.1038/ismej.2011.129) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taylor MW, Radax R, Steger D, Wagner M. 2007. Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol. Mol. Biol. Rev. 71, 295–347. ( 10.1128/MMBR.00040-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turnbaugh PJ, et al. 2009. A core gut microbiome in obese and lean twins. Nature 457, 480–484. ( 10.1038/nature07540) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Webster NS, Taylor MW. 2012. Marine sponges and their microbial symbionts: love and other relationships. Environ. Microbiol. 14, 335–346. ( 10.1111/j.1462-2920.2011.02460.x) [DOI] [PubMed] [Google Scholar]
- 6.Krediet CJ, Ritchie KB, Paul VJ, Teplitski M. 2013. Coral-associated micro-organisms and their roles in promoting coral health and thwarting diseases. Proc. R. Soc. B 280, 20122328 ( 10.1098/rspb.2012.2328) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clay K. 2014. Defensive symbiosis: a microbial perspective. Funct. Ecol. 28, 293–298. ( 10.1111/1365-2435.12258) [DOI] [Google Scholar]
- 8.Naik S, et al. 2012. Compartmentalized control of skin immunity by resident commensals. Science 337, 1115–1119. ( 10.1126/science.1225152) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris RN, et al. 2009. Skin microbes on frogs prevent morbidity and mortality caused by a lethal skin fungus. ISME J. 3, 818–824. ( 10.1038/ismej.2009.27) [DOI] [PubMed] [Google Scholar]
- 10.Banning JL, Weddle AL, Wahl GW, Simon MA, Lauer A, Walters RL, Harris RN. 2008. Antifungal skin bacteria, embryonic survival, and communal nesting in four-toed salamanders, Hemidactylium scutatum. Oecologia 156, 423–429. ( 10.1007/s00442-008-1002-5) [DOI] [PubMed] [Google Scholar]
- 11.Becker MH, Brucker RM, Schwantes CR, Harris RN, Minbiole KPC. 2009. The bacterially produced metabolite violacein is associated with survival of amphibians infected with a lethal fungus. Appl. Environ. Microbiol. 75, 6635–6638. ( 10.1128/AEM.01294-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Becker MH, Harris RN. 2010. Cutaneous bacteria of the redback salamander prevent morbidity associated with a lethal disease. PLoS ONE 5, e10957 ( 10.1371/journal.pone.0010957) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brucker RM, Harris RN, Schwantes CR, Gallaher TN, Flaherty DC, Lam BA, Minbiole KPC. 2008. Amphibian chemical defense: antifungal metabolites of the microsymbiont Janthinobacterium lividum on the salamander Plethodon cinereus. J. Chem. Ecol. 34, 1422–1429. ( 10.1007/s10886-008-9555-7) [DOI] [PubMed] [Google Scholar]
- 14.Gagliardo R, Crump P, Griffith E, Mendelson J, Ross H, Zippel K. 2008. The principles of rapid response for amphibian conservation, using the programmes in Panama as an example. Int. Zoo Yearb. 42, 125–135. ( 10.1111/j.1748-1090.2008.00043.x) [DOI] [Google Scholar]
- 15.Skerratt LF, Berger L, Speare R, Cashins S, McDonald KR, Phillott AD, Hines HB, Kenyon N. 2007. Spread of chytridiomycosis has caused the rapid global decline and extinction of frogs. Ecohealth 4, 125–134. ( 10.1007/s10393-007-0093-5) [DOI] [Google Scholar]
- 16.Poole V. 2008. Project golden frog. Endanger. Species Bull. 33, 7–10. [Google Scholar]
- 17.Perez R, Richards-Zawacki CL, Krohn AR, Robak M, Griffith EJ, Ross H, Gratwicke B, Ibáñez R, Voyles J. 2014. Field surveys in Western Panama indicate populations of Atelopus varius frogs are persisting in regions where Batrachochytrium dendrobatidis is now enzootic. Amphib. Reptile Conserv. 8, 30–35(e85). [Google Scholar]
- 18.Bletz MC, Loudon AH, Becker MH, Bell SC, Woodhams DC, Minbiole KPC, Harris RN. 2013. Mitigating amphibian chytridiomycosis with bioaugmentation: characteristics of effective probiotics and strategies for their selection and use. Ecol. Lett. 16, 807–820. ( 10.1111/ele.12099) [DOI] [PubMed] [Google Scholar]
- 19.Mohapatra S, Chakraborty T, Kumar V, DeBoeck G, Mohanta KN. 2013. Aquaculture and stress management: a review of probiotic intervention. J. Anim. Physiol. Anim. Nutr. 97, 405–430. ( 10.1111/j.1439-0396.2012.01301.x) [DOI] [PubMed] [Google Scholar]
- 20.Berg G. 2009. Plant-microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture. Appl. Microbiol. Biotechnol. 84, 11–18. ( 10.1007/s00253-009-2092-7) [DOI] [PubMed] [Google Scholar]
- 21.Reid G, Younes JA, Van der Mei HC, Gloor GB, Knight R, Busscher HJ. 2011. Microbiota restoration: natural and supplemented recovery of human microbial communities. Nat. Rev. Microbiol. 9, 27–38. ( 10.1038/nrmicro2473) [DOI] [PubMed] [Google Scholar]
- 22.Abreu NA, Nagalingam NA, Song Y, Roediger FC, Pletcher SD, Goldberg AN, Lynch SV. 2012. Sinus microbiome diversity depletion and Corynebacterium tuberculostearicum enrichment mediates rhinosinusitis. Sci. Transl. Med. 4, 151ra124 ( 10.1126/scitranslmed.3003783) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lauer A, Simon MA, Banning JL, André E, Duncan K, Harris RN. 2007. Common cutaneous bacteria from the eastern red-backed salamander can inhibit pathogenic fungi. Copeia 2007, 630–640. ( 10.1643/0045-8511(2007)2007[630:CCBFTE]2.0.CO;2) [DOI] [Google Scholar]
- 24.Lauer A, Simon MA, Banning JL, Lam BA, Harris RN. 2008. Diversity of cutaneous bacteria with antifungal activity isolated from female four-toed salamanders. ISME J. 2, 145–157. ( 10.1038/ismej.2007.110) [DOI] [PubMed] [Google Scholar]
- 25.Becker MH, Harris RN, Minbiole KPC, Schwantes CR, Rollins-Smith LA, Reinert LK, Brucker RM, Domangue RJ, Gratwicke B. 2012. Towards a better understanding of the use of probiotics for preventing chytridiomycosis in Panamanian golden frogs. Ecohealth 8, 501–506. ( 10.1007/s10393-012-0743-0) [DOI] [PubMed] [Google Scholar]
- 26.Becker MH, et al. In press Phylogenetic distribution of symbiotic bacteria from Panamanian amphibians that inhibit growth of the lethal fungal pathogen Batrachochytrium dendrobatidis. Mol. Ecol. ( 10.1111/mec.13135) [DOI] [PubMed] [Google Scholar]
- 27.Bell SC, Alford RA, Garland S, Padilla G, Thomas AD. 2013. Screening bacterial metabolites for inhibitory effects against Batrachochytrium dendrobatidis using a spectrophotometric assay. Dis. Aquat. Organ. 103, 77–85. ( 10.3354/dao02560) [DOI] [PubMed] [Google Scholar]
- 28.Boyle DG, Boyle DB, Olsen V, Morgan JAT, Hyatt AD. 2004. Rapid quantitative detection of chytridiomycosis (Batrachochytrium dendrobatidis) in amphibian samples using real-time Taqman PCR assay. Dis. Aquat. Organ. 60, 141–148. ( 10.3354/dao060141) [DOI] [PubMed] [Google Scholar]
- 29.Longcore JE, Pessier AP, Nichols DK, Longcorel JE. 1999. Batrachochytrium dendrobatidis gen. et sp. nov., a chytrid pathogenic to amphibians. Mycologia 91, 219–227. ( 10.2307/3761366) [DOI] [Google Scholar]
- 30.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. 2010. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl Acad. Sci. USA 108(Suppl. 1), 4516–4522. ( 10.1073/pnas.1000080107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caporaso JG, et al. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624. ( 10.1038/ismej.2012.8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caporaso JG, et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. ( 10.1038/nmeth.f.303) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Becker MH, Richards-Zawacki CL, Gratwicke B, Belden LK. 2014. The effect of captivity on the cutaneous bacterial community of the critically endangered Panamanian golden frog (Atelopus zeteki). Biol. Conserv. 176, 199–206. ( 10.1016/j.biocon.2014.05.029) [DOI] [Google Scholar]
- 34.Brucker RM, Baylor CM, Walters RL, Lauer A, Harris RN, Minbiole KPC. 2008. The identification of 2,4-diacetylphloroglucinol as an antifungal metabolite produced by cutaneous bacteria of the salamander Plethodon cinereus. J. Chem. Ecol. 34, 39–43. ( 10.1007/s10886-007-9352-8) [DOI] [PubMed] [Google Scholar]
- 35.Loudon AH, Holland JA, Umile TP, Burzynski EA, Minbiole KPC, Harris RN. 2014. Interactions between amphibians’ symbiotic bacteria cause the production of emergent anti-fungal metabolites. Front. Microbiol. 5, 441 ( 10.3389/fmicb.2014.00441) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Umile TP, et al. 2014. Nonlethal amphibian skin swabbing of cutaneous natural products for HPLC fingerprinting. Anal. Methods 6, 3277–3284. ( 10.1039/c4ay00566j) [DOI] [Google Scholar]
- 37.Bray JR, Curtis JT. 1957. An ordination of the upland forest communities of southern Wisconsin. Ecol. Monogr. 27, 325–349. ( 10.2307/1942268) [DOI] [Google Scholar]
- 38.Sørensen T. 1948. A method of establishing groups of equal amplitude in plant sociology based on similarity of species and its application to analyses of the vegetation on Danish commons. Biol. Skr. 5, 1–34. [Google Scholar]
- 39.Dufrene M, Legendre P. 1997. Species assemblages and indicator species: the need for a flexible asymmetrical approach. Ecol. Monogr. 67, 345–366. ( 10.1890/0012-9615(1997)067[0345:SAAIST]2.0.CO;2) [DOI] [Google Scholar]
- 40.R Core Team 2014. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; (http://www.r-project.org/) [Google Scholar]
- 41.Castro-luna AAA, Sosa VJ, Castillo-Campos G. 2007. Quantifying phyllostomid bats at different taxonomic levels as ecological indicators in a disturbed tropical forest. Acta Chiropterologica 9, 219–228. ( 10.3161/1733-5329(2007)9[219:QPBADT]2.0.CO;2) [DOI] [Google Scholar]
- 42.Van Rensburg BJ, McGeoch MA, Chown SL, Van Jaarsveld AS. 1999. Conservation of heterogeneity among dung beetles in the Maputaland Centre of Endemism, South Africa. Biol. Conserv. 88, 145–153. ( 10.1016/S0006-3207(98)00109-8) [DOI] [Google Scholar]
- 43.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300. [Google Scholar]
- 44.Hilario E, Buckley TR, Young JM. 2004. Improved resolution on the phylogenetic relationships among Pseudomonas by the combined analysis of atpD, carA, recA and 16S rDNA. Antonie Van Leeuwenhoek 86, 51–64. ( 10.1023/B:ANTO.0000024910.57117.16) [DOI] [PubMed] [Google Scholar]
- 45.Bustamante HM, Livo LJ, Carey C. 2010. Effects of temperature and hydric environment on survival of the Panamanian golden frog infected with a pathogenic chytrid fungus. Integr. Zool. 5, 143–153. ( 10.1111/j.1749-4877.2010.00197.x) [DOI] [PubMed] [Google Scholar]
- 46.Langhammer PF, Lips KR, Burrowes PA, Tunstall T, Palmer CM, Collins JP. 2013. A fungal pathogen of amphibians, Batrachochytrium dendrobatidis, attenuates in pathogenicity with in vitro passages. PLoS ONE 8, e77630 ( 10.1371/journal.pone.0077630) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blaustein AR, Romansic JM, Scheessele EA, Han BA, Pessier AP, Longcore JE. 2005. Interspecific variation in susceptibility of frog tadpoles to the pathogenic fungus Batrachochytrium dendrobatidis. Conserv. Biol. 19, 1460–1468. ( 10.1111/j.1523-1739.2005.00195.x) [DOI] [Google Scholar]
- 48.Rollins-Smith LA. 2009. The role of amphibian antimicrobial peptides in protection of amphibians from pathogens linked to global amphibian declines. Biochim. Biophys. Acta 1788, 1593–1599. ( 10.1016/j.bbamem.2009.03.008) [DOI] [PubMed] [Google Scholar]
- 49.Savage AE, Zamudio KR. 2011. MHC genotypes associate with resistance to a frog-killing fungus. Proc. Natl Acad. Sci. USA 108, 16 705–16 710. ( 10.1073/pnas.1106893108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murphy PJ, St-Hilaire S, Corn PS. 2011. Temperature, hydric environment, and prior pathogen exposure alter the experimental severity of chytridiomycosis in boreal toads. Dis. Aquat. Organ. 95, 31–42. ( 10.3354/dao02336) [DOI] [PubMed] [Google Scholar]
- 51.Voyles J, Rosenblum EB, Berger L. 2011. Interactions between Batrachochytrium dendrobatidis and its amphibian hosts: a review of pathogenesis and immunity. Microbes Infect. 13, 25–32. ( 10.1016/j.micinf.2010.09.015) [DOI] [PubMed] [Google Scholar]
- 52.Stecher B, et al. 2010. Like will to like: abundances of closely related species can predict susceptibility to intestinal colonization by pathogenic and commensal bacteria. PLoS Pathog. 6, e1000711 ( 10.1371/journal.ppat.1000711) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morrow AL, et al. 2013. Early microbial and metabolomic signatures predict later onset of necrotizing enterocolitis in preterm infants. Microbiome 1, 13 ( 10.1186/2049-2618-1-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.White J, Jr, Torres F, Mónica S. (eds). 2010. Defensive mutualism in microbial symbiosis. Boca Raton, FL: CRC Press. [Google Scholar]
- 55.Guarner F, Malagelada J-R. 2003. Gut flora in health and disease. Lancet 361, 512–519. ( 10.1016/S0140-6736(03)12489-0) [DOI] [PubMed] [Google Scholar]
- 56.Woodhams DC, et al. 2014. Interacting symbionts and immunity in the amphibian skin mucosome predict disease risk and probiotic effectiveness. PLoS ONE 9, e96375 ( 10.1371/journal.pone.0096375) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jani AJ, Briggs CJ. 2014. The pathogen Batrachochytrium dendrobatidis disturbs the frog skin microbiome during a natural epidemic and experimental infection. Proc. Natl Acad. Sci. USA 111, E5049–E5058. ( 10.1073/pnas.1412752111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Someya N, Akutsu K. 2009. Indigenous bacteria may interfere with the biocontrol of plant diseases. Naturwissenschaften 96, 743–747. ( 10.1007/s00114-009-0524-y) [DOI] [PubMed] [Google Scholar]
- 59.Densmore CL, Green DE. 2007. Diseases of amphibians. ILAR J. 48, 235–254. ( 10.1093/ilar.48.3.235) [DOI] [PubMed] [Google Scholar]
- 60.Taylor SK, Williams ES, Thorne ET, Mills KW, Withers DI, Pier AC. 1999. Causes of mortality of the Wyoming toad. J. Wildl. Dis. 35, 49–57. ( 10.7589/0090-3558-35.1.49) [DOI] [PubMed] [Google Scholar]
- 61.Bodinof CM. 2010. Translocation and conservation of hellbenders (Cryptobranchus alleganiensis) in Missouri. MS Thesis, University of Missouri, Columbia, pp. 1–25. [Google Scholar]
- 62.Conlon JM. 2011. The contribution of skin antimicrobial peptides to the system of innate immuity in anurans. Cell Tissue Res. 343, 201–212. ( 10.1007/s00441-010-1014-4) [DOI] [PubMed] [Google Scholar]
- 63.Yotsu-Yamashita M, Kim YH, Dudley SC, Choudhary G, Pfahnl A, Oshima Y, Daly JW. 2004. The structure of zetekitoxin AB, a saxitoxin analog from the Panamanian golden frog Atelopus zeteki: a potent sodium-chanel blocker. Proc. Natl Acad. Sci. USA 101, 4346–4351. ( 10.1073/pnas.0400368101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gram L, Løvold T, Nielsen J, Melchiorsen J, Spanggaard B. 2001. In vitro antagonism of the probiont Pseudomonas fluorescens strain AH2 against Aeromonas salmonicida does not confer protection of salmon against furunculosis. Aquaculture 199, 1–11. ( 10.1016/S0044-8486(01)00565-8) [DOI] [Google Scholar]
- 65.Rollins-Smith LA, Ramsey JP, Pask JD, Reinert LK, Woodhams DC. 2011. Amphibian immune defenses against chytridiomycosis: impacts of changing environments. Integr. Comp. Biol. 51, 552–562. ( 10.1093/icb/icr095) [DOI] [PubMed] [Google Scholar]
- 66.Loudon AH, Woodhams DC, Parfrey LW, Archer H, Knight R, McKenzie V, Harris RN. 2013. Microbial community dynamics and effect of environmental microbial reservoirs on red-backed salamanders (Plethodon cinereus). ISME J. ( 10.1038/ismej.2013.200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bezkorovainy A. 2001. Probiotics: determinants of survival and growth in the gut. Am. J. Clin. Nutr. 73, 399–405. [DOI] [PubMed] [Google Scholar]
- 68.Gallo RL, Nakatsuji T. 2011. Microbial symbiosis with the innate immune defense system of the skin. J. Invest. Dermatol. 131, 1974–1980. ( 10.1038/jid.2011.182) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fernández-Marín H, Bruner G, Gomez EB, Nash DR, Boomsma JJ, Wcislo WT. 2013. Dynamic disease management in Trachymyrmex fungus-growing ants (Attini: Formicidae). Am. Nat. 181, 571–582. ( 10.1086/669664) [DOI] [PubMed] [Google Scholar]
- 70.Egan S, Harder T, Burke C, Steinberg P, Kjelleberg S, Thomas T. 2013. The seaweed holobiont: understanding seaweed-bacteria interactions. FEMS Microbiol. Rev. 37, 462–476. ( 10.1111/1574-6976.12011) [DOI] [PubMed] [Google Scholar]
- 71.Bielby J, Cooper N, Cunningham AA, Garner TWJ, Purvis A. 2008. Predicting susceptibility to future declines in the world's frogs. Conserv. Lett. 1, 82–90. ( 10.1111/j.1755-263X.2008.00015.x) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 16S rRNA sequences for the probiotic isolates have been deposited in GenBank (accession numbers KM006931–KM006934). Raw 16S rRNA read sequences from Illumina sequencing have been deposited in the European Nucleotide Archive (accession number PRJEB6565). Illumina data will be publicly available 1 year after publication.