Abstract
Farming was established in Central Europe by the Linearbandkeramik culture (LBK), a well-investigated archaeological horizon, which emerged in the Carpathian Basin, in today's Hungary. However, the genetic background of the LBK genesis is yet unclear. Here we present 9 Y chromosomal and 84 mitochondrial DNA profiles from Mesolithic, Neolithic Starčevo and LBK sites (seventh/sixth millennia BC) from the Carpathian Basin and southeastern Europe. We detect genetic continuity of both maternal and paternal elements during the initial spread of agriculture, and confirm the substantial genetic impact of early southeastern European and Carpathian Basin farming cultures on Central European populations of the sixth–fourth millennia BC. Comprehensive Y chromosomal and mitochondrial DNA population genetic analyses demonstrate a clear affinity of the early farmers to the modern Near East and Caucasus, tracing the expansion from that region through southeastern Europe and the Carpathian Basin into Central Europe. However, our results also reveal contrasting patterns for male and female genetic diversity in the European Neolithic, suggesting a system of patrilineal descent and patrilocal residential rules among the early farmers.
Keywords: ancient DNA, mitochondrial DNA, Y chromosomal DNA, Neolithization, Carpathian Basin, Central Europe
1. Introduction
Agriculture was first established in the Near Eastern Fertile Crescent after 10 000 BC; and expanded from the Levant and Anatolia to southeastern Europe [1]. Archaeological research has described the subsequent spread of Neolithic farming into (and throughout) Central and southwestern Europe along two major and largely contemporaneous routes. On the Continental route, the Carpathian Basin connected southeastern Europe to the Central European loess plains, while the Mediterranean route bridged the eastern and western Mediterranean coasts, introducing farming to the Iberian Peninsula in the west [2–6].
The Early Neolithic Starčevo culture (STA) has played a major role in the Neolithization of southeastern Europe. The STA expanded from present-day Serbia to the western part of the Carpathian Basin, encompassing the regions of today's northern Croatia and southwestern Hungary (ca 6000–5400 BC) [7,8] (figure 1). The earliest Linearbandkeramik culture (LBK) emerged in the mid-sixth millennium BC in western Hungary or Transdanubia (hence called ‘LBK in Transdanubia’, or LBKT) [9], marking the beginning of sedentary life in Central Europe. The earliest LBKT coexisted with the STA in Transdanubia for approximately 100–150 years [10]. Archaeological research described an interaction zone between indigenous hunter–gatherer groups and farmers at the northernmost extent of the STA in Transdanubia, which might have given rise to the LBKT [10,11]. After its formative phase in western Hungary, the LBK spread rapidly to Central Europe, reaching central Germany around 5500 BC [2,12]. In the following 500 years, the LBK continually expanded, eventually covering a vast geographical area from the Paris Basin to Ukraine in its latest phase [2,13], while persisting in Transdanubia until approximately 4900 BC (figure 1).
Figure 1.

Geographical distribution of the Starčevo, LBK cultures and the Continental route of the European Neolithization. The shaded areas of the maps show the distribution of the Starčevo culture (STA) and the LBK in Transdanubia (LBKT) and Central Europe (LBK), while the arrows show the directions of the farmers' expansion into Central Europe. The absolute dates refer to the whole dissemination areas of the cultures [2,3,5,7,9]. (Online version in colour.)
Despite the well-established archaeological relations between the STA and LBKT in the Carpathian Basin and the LBK in Central Europe, their genetic relationship is yet unclear. Traditionally, scholars have explained the Neolithic transition either as an expansion of early farmers from the Near East, who brought new ideas as well as new genes (demic diffusion) [14–17], or as an adoption of farming technologies by indigenous hunter–gatherer populations with little or no genetic influence (cultural diffusion) [18–21]. These two contrasting models have been merged into complex integrationist approaches, considering small-scale population movements on regional levels [1,2,10,22].
Inferences drawn from present-day genetic data have yielded contradictory results about the Neolithic impact on the genetic diversity of modern Europeans, showing a disparity between mitochondrial DNA (mtDNA) and Y chromosomal patterns. Several Y chromosome studies supported the Neolithic demic diffusion model [17,23,24], while most mtDNA and some Y chromosomal studies have proposed a continuity of Upper Palaeolithic lineages [20,21,25,26]. The contrasting mtDNA and Y chromosomal evidence has been explained by differences in evolutionary scenarios, such as sex-biased migration [27].
Recent ancient DNA (aDNA) studies have provided direct insights into the mtDNA and nuclear genomic diversity of hunter–gatherers in Europe [28–35] and the Central European LBK [33,36–40], describing genetic discontinuity between local foragers and early farmers [28,31,38,40]. Comparative analyses with present-day populations have revealed Near Eastern affinities of the mitochondrial LBK ancestry, supporting the demic diffusion model and population replacement at the beginning of the Neolithic period [38,39]. Ancient genomic studies have described the early farmers as genetically most similar to extant populations of southern Europe [31,33,40,41]. Y chromosome data from prehistoric Europeans is still scarce but relevant for our study (electronic supplementary material, dataset S20), including hunter–gatherers [33–35,40,42], and early farmers from Germany [38,40], eastern Hungary [41], Austria [43] and southwest Europe [40,44,45].
The postulated Near Eastern origin of LBK farmers has so far only been inferred from modern-day population data, and while the first ancient mtDNA data from early Near Eastern farmers has been reported recently [46], the genetic diversity in the vast territory from the Fertile Crescent to Central Europe remains largely unexplored. Consequently, our aims were to (i) study the genetic diversity of the early farming Carpathian Basin cultures from both the mtDNA and Y chromosome perspectives, (ii) examine whether men and women had different demographic histories, (iii) investigate the contribution of the STA to the genetic variability of the LBKT and LBK, (iv) reveal the potential genetic origins of the first farmers in Eurasia and (v) assess the role of the Continental route in the European Neolithic dispersal.
In this study, we present 84 mtDNA and 9 Y chromosomal DNA data from Mesolithic (6200–6000 BC) and Neolithic specimens of the STA and LBKT from western Hungary and Croatia. Spanning a time transect of the Hungarian Neolithic in Transdanubia over approximately 900 years (ca 5800–4900 BC) allowed us to gain detailed insight into the spread of farming from the Near East.
2. Results
(a) Mitochondrial DNA.
Using well-established aDNA methods (electronic supplementary material, material and methods), we genotyped mtDNA variability by sequencing the hyper-variable segment I and II (HVS-I/II) and 22 single nucleotide polymorphisms (SNPs) in the coding region of the mitochondrial genome [38]. Overall, we investigated 109 skeletons from one Mesolithic, six STA and eight LBKT sites from western Hungary and Croatia (electronic supplementary material, figure S8, datasets S1 and S2). We genotyped endogenous HVS-I sequences of 84 individuals (1 hunter–gatherer, 44 STA and 39 LBKT), yielding a success rate of 76% (electronic supplementary material, dataset S3). We also sequenced parts of HVS-II from 25 individuals with identical HVS-I motifs to increase the phylogenetic resolution and detect potential intra-site maternal kinship. The analysis of haplogroup defining coding region SNPs resulted in reproducible profiles for 96 individuals, with a success rate of 86% (electronic supplementary material, datasets S3 and S4).
The haplotype of the Mesolithic skeleton from the Croatian Island Korčula could be assigned to mtDNA haplogroup U5b2a5 (electronic supplementary material, dataset S3). Sub-haplogroup U5b has been shown to be common in hunter–gatherer communities across Europe [28–30,32,33,47,48]. Contrary to the low mtDNA diversity observed in Central/North European hunter–gatherers [28–30], we identify a higher variability in early farming communities of the Carpathian Basin including haplogroups N1a, T1, T2, J, K, H, HV, V, W, X, U2, U3, U4 and U5a (electronic supplementary material, table S1). Previous studies described haplogroups N1a, T2, J, K, HV, V, W and X as being characteristic for the Central European LBK and suggested these as the mitochondrial ‘Neolithic package’ that had reached Central Europe in the sixth millennium BC [38,39]. Interestingly, most of these eight haplogroups show comparable frequencies between the STA, LBKT and LBK, and represent the majority of mtDNA variation in each culture (STA = 86.36%, LBKT = 61.54%, LBK = 79.63%) with similar haplotype diversity (STA = 0.97674, LBKT = 0.95277, LBK = 0.95483). By contrast, hunter–gatherer haplogroups are rare in the STA and both LBK groups (electronic supplementary material, table S1). Haplogroup H was not included in the Neolithic package, because it has also been found in pre-agricultural context in Iberia [48]. However, the low resolution of HVS-I does not allow to distinguish between H lineages of Neolithic or pre-Neolithic origins in Transdanubia and would require whole mitochondrial genome analyses.
To evaluate whether the haplogroup and haplotype composition of the STA, LBKT, LBK [36–39] and hunter–gatherers from Central/North Europe [28–30,41] differ significantly from each other, we performed a haplogroup-based Fisher's exact test and a sequence-based genetic distance analysis. We also used a test of population continuity (TPC) [39] to elucidate whether the observed differences can be best explained by genetic drift or by other factors such as migration. These analyses reveal that the mtDNA composition of the Early Neolithic cultures is significantly different from that of the hunter–gatherers, both on the haplogroup (p = 0.0001) and haplotype level (Fst = 0.1764–0.187, p = 0.0000; electronic supplementary material, table S1), indicating genetic discontinuity of maternal lineages at the advent of farming in the Carpathian Basin. The TPC shows that, independent of the tested effective population size, the transition from foraging to farming cannot be explained by genetic drift alone (p < 0.000001; electronic supplementary material, dataset S13). More importantly, non-significant differences between the STA and the LBK groups from Transdanubia and Central Europe (haplogroup composition p = 0.0638–0.5518); haplotype composition (Fst = –0.00521–0.01345, p = 0.46581–0.62012) and TPC (p > 0.177; electronic supplementary material, table S1, dataset S13) support a rather homogeneous mtDNA signature of early farming communities from both regions and population continuity during the Neolithic period.
We combined our Neolithic samples from the Carpathian Basin with 533 published mtDNA sequences from Upper Palaeolithic and Mesolithic [28–30,32,47,48], Neolithic [36–39,41,44,45,47–52] and Early Bronze Age [39] sites across Europe (electronic supplementary material, dataset S6) and analysed principal components (PCA), multidimensional scaling (MDS), molecular variance (AMOVA) and shared haplotypes to compare the mtDNA variability of the STA and LBKT in a broader geographical and chronological context (electronic supplementary material).
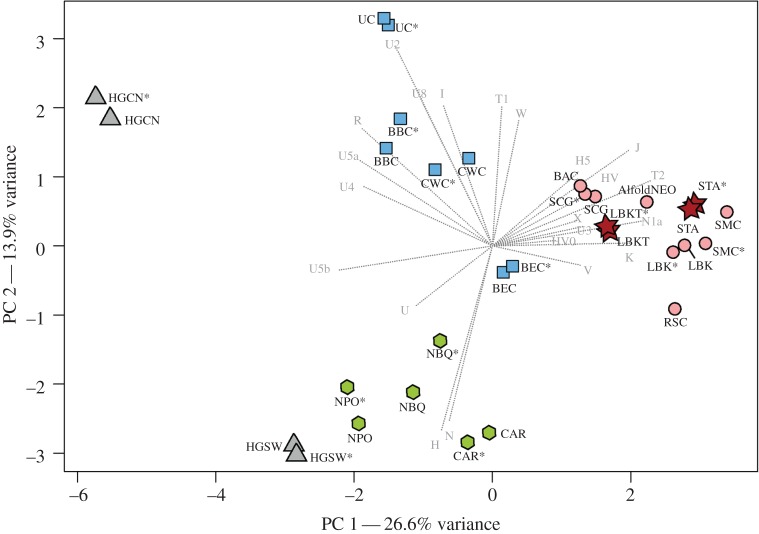
The PCA and MDS show that the mtDNA composition of the STA and LBKT is strikingly similar to the LBK [36–39], eastern Hungarian Neolithic dataset [36,41,52] and to subsequent populations of the fifth/fourth millennia BC in Central Europe [39] (figure 2; electronic supplementary material, figures S1–S3, datasets S7–S10). This is predominately based on a high number of lineages attributed to the ‘Neolithic package’ and low frequencies of hunter–gatherers lineages, which clearly distinguish the cluster of farmers not only from hunter–gatherers of Central/North [28–30,41] and southwestern Europe [32,47,48], but also from Neolithic Iberian populations [44,45,47,48,50] and Central European cultures of the third/second millennia BC [39,49,51]. To exclude biases induced by potential maternal kinship within the prehistoric datasets, we also performed PCA and MDS with reduced datasets, excluding haplotypes with identical HVS-I and II sequences from the same site. The reduced and complete datasets fall closely next to each other, indicating a negligibly small bias of maternal kinship.
Figure 2.
PCA plot comparing mtDNA data of 17 prehistoric populations. PCA is based on the frequencies of 22 mtDNA haplogroups in the STA, LBKT and 15 prehistoric populations. Colour shadings and symbols denote populations of different periods or European regions: hunter–gatherers (triangles), sixth millennium BC Transdanubian populations (stars), sixth/fourth millennia BC populations in Central Europe and eastern Hungary (circles), Central European third/second millennia BC populations (rectangles), sixth/fifth millennia BC populations of the Iberian Peninsula (hexagons). The reduced version of each dataset is marked by an asterisk. Detailed information about the comparative data and haplogroup frequencies are listed in electronic supplementary material, dataset S7. Population/culture abbreviations: hunter–gatherers in Central and North Europe (HGCN), hunter–gatherers in southwestern Europe (HGSW), Starčevo (STA), LBK in Transdanubia (LBKT), LBK in Central Europe (LBK), Rössen (RSC), Schöningen (SCG), Baalberge (BAC), Salzmünde (SMC), Bernburg (BEC), Corded Ware (CWC), Bell Beaker (BBC), Únětice (UC), Cardial and Epicardial (CAR), Neolithic population in Basque Country and Navarre (NBQ), Neolithic population in Portugal (NPO). (Online version in colour.)
We used AMOVA to evaluate whether the observed affinities of STA and LBKT with the LBK and fifth/fourth millennia BC cultures from Central Europe are the result of a shared population structure. We pooled HVS-I sequences from the STA and LBKT and nine sixth–second millennia BC populations from Central Europe [36–39,49,51] into different groups, and tested 82 different constellations to identify the one with the highest among-group variance and simultaneously with low variation within the groups (electronic supplementary material, dataset S11). The highest among-group variance was observed when STA and LBKT was grouped with the Central European LBK and with all fifth/fourth millennia BC cultures, while the third/second millennia BC cultures were grouped separately (among-group variation = 3.50%, Fst = 0.03501, p = 0.00396; within-group variation = 0.20%, Fst = 0.00203, p = 0.31139). These results suggest a common genetic structure of the sixth–fourth millennia BC populations.
We used shared haplotype analysis [53] and modified this approach by accounting for the temporal succession of cultures (termed ancestral shared haplotype analysis or ASHA; figure 3; electronic supplementary material, dataset S12). The results show that ancestral hunter–gatherer lineages were rare in the STA (2.27%), LBKT (0%) and LBK (1.85%), as well as in fifth/fourth millennia BC cultures (0%), and became more common in Central Europe during the third/second millennia BC (2.86–11.76%) [39]. By contrast, we identified a high proportion of ancestral STA lineages in all subsequent periods (LBKT = 61.54%, LBK = 55.56%, fifth/fourth millennia BC = 36.84–63.64%, third/second millennia BC = 36.17–43.18%). The subsequent LBKT reveals a smaller distinctive influence on its successors, as only 12.96% of the LBK, 0–10.53% of the fifth/fourth millennia BC and 0–3.19% of the third/second millennia BC cultures can be traced back to ancestral lineages first observed in the LBKT. The number of new ‘ancestral’ lineages is even lower in the LBK of Central Europe, with no effect on the third/second millennia BC populations. This highlights the importance of the STA in the formation of the mtDNA diversity in early farmers of Central Europe.
Figure 3.
Results of the ancestral shared haplotype analysis. The bar plot shows the proportions of ancestral mtDNA lineages associated to hunter–gatherer, STA, LBKT, LBK and other subsequent populations in the Central/North European hunter–gatherer dataset, the two Carpathian Basin and nine Central European populations ranging from the LBK to the Early Bronze Age. Details of the ASHA are provided in electronic supplementary material, dataset S12. Culture/population abbreviations are according to figure 2. (Online version in colour.)
To identify affinities of our Neolithic datasets with present-day populations, we further collated 67 996 published HVS-I sequences from Eurasian populations, analysed principal components and mapped genetic distances (electronic supplementary material).
The PCA shows that the frequencies of N1a, T1, T2, K, J and HV, and the absence of Asian and African lineages in the Carpathian Basin populations result in a clustering of the STA with populations of the Near East and the Caucasus, while the LBKT falls between the latter and populations from South and southeast Europe (Greeks, Bulgarians and Italians), which is explained by a higher frequency of haplogroup H in the LBKT (electronic supplementary material, figure S4, dataset S14). However, the dominant frequencies of haplogroup N1a, T2, and K in the STA and LBKT result in a differentiation from all present-day populations along the third component.
Sequence-based genetic distance maps are largely consistent with PCA and reveal the greatest similarities of the STA to populations of the Near East (Iraq, Syria) and the Caucasus (Azerbaijan, Georgia, Armenia) as well as some European populations, such as Italy, Austria, Romania and Macedonia (electronic supplementary material, figure S5a, dataset S15). The LBKT displays affinities that are overall similar to the STA, which includes populations from Azerbaijan, Syria and Iraq (electronic supplementary material, figure S5b, dataset S15). We also observe similarities to present-day Europeans, such as the populations of Great Britain, Portugal, Romania, Crete and Russia. These similarity peaks are probably explained by elevated frequencies of shared lineages due to shared genetic drift in modern-day populations.
(b). Y chromosomal DNA
We analysed 33 Y-haplogroup defining SNPs located on the non-recombining part of the Y chromosome (NRY), using multiplex [38] and singleplex PCR. We successfully generated unambiguous NRY SNP profiles for nine male individuals (STA = 7, LBKT = 2; electronic supplementary material, datasets S3 and S5). Three STA individuals belong to the NRY haplogroup F* (M89) and two specimens can be assigned to the haplogroup G2a2b (S126), and one each to G2a (P15) and I2a1 (P37.2). The two investigated LBKT samples carry haplogroups G2a2b (S126) and I1 (M253). Furthermore, incomplete SNP profiles of eight specimens potentially belong to the same haplogroups—STA: three G2a2b (S126), two G2a (P15) and one I (M170); LBKT: one G2a2b (S126) and one F* (M89).
G2a2b and F* are rare in present-day Europe. Frequencies of haplogroup G and its subgroups increase slightly towards the Near East and reach the highest values (70%) in populations of the west Caucasus [54–56], while G2a2b and its sub-clusters have been detected in modern-day Sardinia and Caucasus [56,57]. Haplogroup F* shows a diffuse dissemination pattern in Eurasia, which is based on its rarity and insufficient sub-haplogroup resolution in most population genetic studies. One STA F* haplogroup has been further analysed, and could be assigned to haplogroup H2 (L281) [40], which has been found in the extant Sardinian population [57]. Haplogroups I1 and I2a1 are most frequent in present-day populations of Europe, with the highest frequencies in Scandinavian [58–60] and southeast European populations, respectively [58].
We used PCA and genetic distance maps to identify affinities of the Carpathian Basin samples with 49 516 NRY SNP profiles from present-day Eurasian and African populations (electronic supplementary material). Owing to the small sample size and similarities in Y chromosome composition, we pooled ancient STA, Alföld LBK [41], LBKT and LBK data [38,40].
The elevated haplogroup G frequency in populations of the west Caucasus results in a clustering with the STA–LBK group on the second principal component. However, the predominant frequencies of haplogroups G and F* lead to a clear separation of the STA–LBK group from all present-day populations along the third principal component (electronic supplementary material, figure S6, dataset S16).
Similarly, the Y chromosome distance map shows the greatest similarities to populations of the west and south Caucasus, and Sardinians (electronic supplementary material, figure S7, dataset S17), which can be explained by the high frequency of haplogroup G/G2a [55,61] in these populations. This might reflect genetic drift, caused by isolation and small effective population size after a direct gene flow from the Near East, which led to a fixation of this haplogroup [54]. Intriguingly, populations of the northeast Caucasus show greater distances to the STA–LBK samples due to lower abundance of haplogroup G/G2a [55]. Genomic data from early farmers have been shown to be the most similar to modern-day Sardinians [33,40,41], which could be explained by the geographical isolation of Sardinia, having led to a conservation of the Neolithic genetic signature. Interestingly, our mtDNA population genetic analyses did not highlight the Neolithic–Sardinian affinity, but was observed on our genetic distance map. We caution that the haplogroup resolution of most modern comparative population datasets does not allow further differentiation between the sub-clusters of haplogroups C, F or G, and therefore does not provide sufficient Y-subgroup information at a smaller geographical scale.
3. Discussion
This study provides the first in-depth population survey of early farming cultures from the western Carpathian Basin and demonstrates its essential role in the genesis of the first farming communities of Central Europe. Population genetic analyses (Fisher's exact test, PCA, MDS, AMOVA, TPC) reveal similar mtDNA haplogroup composition and haplotype diversity in early farming populations from the Carpathian Basin and the populations of the Central European LBK, indicating a shared maternal genetic ancestry (figure 2; electronic supplementary material, table S1, figures S1–S3, dataset S11, S13).
The ASHA shows that about 55% of the LBK lineages ascribed to characteristic ‘Neolithic package’ haplogroups could be traced back to the STA and LBK in Transdanubia (figure 3; electronic supplementary material, dataset S12). It is therefore likely that this mtDNA signature was also present in ancient populations preceding the STA, in accordance with the archaeological record, which suggests cultural links to seventh/sixth millennia BC farming groups of the Aegean and the southern Balkans [5]. Interestingly, the STA mtDNA signature was still dominant in Neolithic cultures of the fifth/fourth millennia BC in Central Europe (figure 3; electronic supplementary material, dataset S12), attesting a direct and enduring genetic legacy of the STA/LBKT farmers in the Central European Neolithic, with minimal or no additional maternal genetic influence from outside for the subsequent 2500 years. Congruently with our results, recent genomic data also support the genetic homogeneity of early European farmers across large geographical distances [40].
Importantly, our comparative analyses with modern population data (PCA and genetic distance maps) emphasizes that both the mtDNA and NRY variability observed in the Carpathian Basin are most likely to have originated in the Near East with connections to the Caucasus (electronic supplementary material, figures S4–S7), in accordance with previous mtDNA studies of the Central European LBK [38,39]. The continuation of lineages through space and time suggests a scenario in which the genetic makeup of early farmers originated in the Near Eastern Fertile Crescent, from where it spread to Central Europe via the western Carpathian Basin, a region which acted as a natural corridor and a transient zone of adaptation to cooler climatic conditions along the Continental route. The shared Near Eastern affinities of the STA, LBKT and LBK, and the genetic continuity in the maternal and paternal gene pools, are consistent with the archaeological record, which describes the genesis of the early LBK (LBKT) from STA communities, followed by a rapid dispersal of the early LBK culture from Transdanubia towards the northwestern part of Central Europe [3,9,13]. We caution that use of modern-day data from the Near East could potentially lead to a biased interpretation. However, we still consider it the best available proxy of the prehistoric Near Eastern mtDNA variability, until aDNA datasets become available.
Recent studies using ancient genomic data of Central and North European early farmers have shown an affinity to South European rather than Near Eastern populations [33,40,41]. Of note, the proportion of western hunter–gatherer ancestry is higher in all modern-day Europeans and the early farmer ancestry being comparably best preserved in, for example, Sardinians [33,40]. These results are not necessarily contradictory but show that uniparental markers are more conservative, preserving a Near Eastern signature more consistently. Ancient genomic data from the Pre-Neolithic and Early Neolithic Near East will be needed to confirm these assumptions.
The very low frequencies of hunter–gatherer lineages (0–2.27%), in the STA, LBKT and LBK sample sets (figure 3) indicate that the arrival of agriculture in the Carpathian Basin and Central Europe was accompanied by a strong reduction of the currently known Mesolithic mtDNA substratum, resulting in a distinct and contrasting mtDNA haplogroup composition, and significant differences between hunter–gatherers and early farmers (figures 2 and 3; electronic supplementary material, figures S1–S3, table S1, datasets S7–S10, S12 and S13). This scenario is consistent with coalescent-based simulations that have revealed genetic discontinuity between Central European hunter–gatherers and LBK communities [28,38]. The detection of haplogroup U5b in the Mesolithic individual from Croatia matches previous observations, which describe sub-haplogroups of U as characteristic mtDNA substratum in Mesolithic Europe [28,39]. Residual Neolithic hunter–gatherer isolates, as reported from Central Europe by Bollongino et al. [30], probably also existed during the Early Neolithic of the Carpathian Basin, as shown by the hunter–gatherer ancestry of the sixth millennium BC human remains from an Early Neolithic Körös settlement pit in eastern Hungary [41]. According to the low proportion of hunter–gatherer mtDNA lineages in the LBK gene pool, we assume that admixture between hunter–gatherers and colonizing LBK farmers was also low in Central Europe.
Considering the relative size and speed of the LBK expansion, we have to presume a substantial population growth during the earliest LBKT, which might have resulted in a population pressure and led to emigration from Transdanubia [62]. Although such a radical population size increase was not palpable from the Early Neolithic archaeological records [7], recent extensive archaeological excavations have revealed large-scale LBKT settlements in western Hungary [9,63,64], suggesting larger source communities for expansion than previously assumed.
The discontinuity between hunter–gatherer and farmer ancestry is also visible in our Y chromosome results. Y chromosome study of modern-day Europeans has suggested a post-LGM expansion from a Franco-Cantabrian refugium for clade I1, and southeast European refugium for I2a1 based on high divergence time estimates [58]. I2a has indeed been found in Mesolithic and Neolithic Central and North European hunter–gatherers [33,34,40,41], as well as in Neolithic remains of southwestern Europe [44,45]. Haplogroup I2a (and possibly I1) might represent a pre-farming legacy of the NRY variation in Europe, alongside the recently described pre-Neolithic C (M130) haplogroups in Russia and Spain [35,42]. Y chromosome haplogroups from STA and LBKT samples, such as haplogroups G2a2b and F*, have also been reported from the Central European LBK [38], and support a homogeneity of paternal lineages among early farmers. Genetic studies on modern-day populations have discussed haplogroup G [25,65] and its subgroup G2a as potential representatives of the spread of farming from the Near East to Europe [26]. This scenario has recently been supported by Neolithic data from northern Spain [44] and southern France [45], which attested a pivotal role for G2a in the Neolithic expansion on the Mediterranean route. Furthermore, G2a has also been reported from four LBK individuals (G2a2a) [40] and the Tyrolean Iceman (G2a2a1b (L91)) [43]. Taken together, these findings suggest that sub-haplogroups of G2a were frequent in Neolithic populations of the sixth–fourth millennia BC across Europe. Thus, if we take Y chromosomal haplogroup I2a (and possibly I1) as proxy for a Mesolithic paternal genetic substratum in Europe, we observe a similar pattern to the changeover in the mitochondrial DNA variability, in which NRY G lineages were predominant among Neolithic farmers across Europe and I lineages became rare [38,40,43–45,49,51]. While the newly discovered NRY C haplogroup suggests additional Y lineages in eastern Hungary [41], the eastern Hungarian NRY data are still too few to estimate, whether there was paternal genetic difference between the populations of western and eastern Carpathian Basin.
The most characteristic mtDNA haplogroup of early farmers from the Carpathian Basin and Central Europe is N1a. N1a has previously been discussed as a potential marker of the spread of farming [36]. The presence of N1a in early farmers from the Carpathian Basin (6.82–10.26%) and Central Europe (12.04%; electronic supplementary material, table S1) lends further support to its pivotal role as a marker for the Continental route of the Neolithic expansion. On the other hand, mtDNA N1a and NRY G2a haplogroups are rare in present-day European populations, which is also reflected in the separation of the sixth millennium BC cultures from all present-day populations along the third principal component on the PCA plots (electronic supplementary material, figures S4 and S6). Intriguingly, R1a and R1b, which represent the most frequent European Y chromosome haplogroups today, are absent in early farmers of Central Europe (electronic supplementary material, dataset S20), but have been reported from cultures arriving in Europe during the third/second millennia BC [40,49,51]. These findings nonetheless indicate further demographic events after the Early/Middle Neolithic period that shaped modern-day mtDNA and NRY variability. However, we caution that the Y chromosome record is still very limited. Recent evidence from ancient mtDNA and genome-wide SNP analyses has stressed the role of further migrations from the eastern steppes reaching Central Europe during the third millennium BC [40], while three Bronze and Iron Age individuals from eastern Hungary suggest that both eastern and western influences reached the Carpathian Basin in post-Neolithic times [41].
Interestingly, recent model-based statistical analyses of contemporary NRY and mtDNA data, testing a series of population scenarios for the Neolithic transition, have revealed a shared admixture history for men and women, but not the same demographic history [66]. This study has shown that females had a larger effective population size, probably based on differential effects of social and cultural practices including increasing sedentism alongside a shift to monogamy and patrilocality in early farmer communities. It is therefore important to interpret our new uniparental genetic data in the light of those findings. Considering the entire set of published ancient NRY records available for Europe thus far, the low paternal diversity at the onset of the Neolithic is quite remarkable: G2a is the prevailing haplogroup (65.5%) in the sixth–fourth millennia BC Neolithic dataset (electronic supplementary material, datasets S5 and S20) [38,40,44,45]. The limited variation in NRY haplogroups in contrast to the high mtDNA haplogroup diversity suggests a smaller effective population size for males than females. One plausible explanation for this phenomenon is patrilocality (where women move to their husband's birth place after marriage), while other possibilities include polygyny or male-biased adult mortality. A patrilocal residential rule was probably linked to a system of descent along the father's line (patrilineality) in early farming communities. Ethnographic studies have suggested a change of residential rules at the advent of Neolithization, showing different trends in residential rules among modern foragers and non-foragers [67]. Increasing sedentism promotes territorial defence and control of resources, favouring men in the inheritance of land and property, which consequently leads to patrilocal residence [67]. However, residence patterns have to be temporarily flexible in expanding populations, allowing some of the sons to settle in new territories following population pressure and/or natural limitation of resources (e.g. after the carrying capacity of a particular region has been reached) [66]. Patrilocality has also been suggested in recent bioarchaeological studies, for example by investigating aDNA evidence from Neolithic site Treilles [45], or stable isotopes from LBK communities in Central Europe [68].
We note that patrilocality does not contradict the demic diffusion model, and it appears that both phenomena have left a discernible mark on the European Neolithic genetic diversity. While patrilocality and lineality might have caused high mtDNA and low NRY within population diversity, the demic diffusion model best explains the mtDNA and NRY affinity of the early farmers to the modern-day Near East, and the observed genetic homogeneity within southeastern and Central Europe. Importantly, local processes of sex-biased migration are unlikely to have an effect on genetic variation at broader spatial scales. Observations from many sites in Europe therefore argue for a common set of cultural and social practices across larger distances for early farming cultures in Europe. We caution that the observed differences in genetic diversity between males and females could also be influenced by resolution biases, resulting from the different sets of studied mtDNA and NRY markers. However, examining sex-specific dynamics of prehistoric societies emerges as an important area of ancient genetic research to be able to address underlying parameters such as migration rate, level of exogamy and distances of marriage-related dispersals.
The novel ancient data from early farming Neolithic populations of the Carpathian Basin help to fill the geographical gap on the Continental route of the Neolithic expansion from the Near Eastern Fertile Crescent to Central Europe. The joint analyses of mitochondrial and Y chromosomal DNA data support a demic diffusion of early farmer men and women through western Hungary, and demonstrate the paramount importance of this region as a prehistoric corridor of the migration. We point out that archaeological cultures of the Carpathian Basin provided the genetic basis of the first Central European farmers that affected subsequent prehistoric cultures for a long period of time. Considering our results in the light of newly available ancient genomic data, we observe a remarkable consistency of the different marker systems. Importantly, the new NRY data complement the sporadic European Y chromosomal dataset, and lend further support to patrilocal residential rules and patrilineal social system of the first farmers, underlining the role of demographic factors, which, depending strongly on cultural practices, notably shaped prehistoric and extant genetic diversity.
4. Material and methods
We sampled one Mesolithic, 47 Starčevo and 61 LBKT individuals, excavated in Croatia and western Hungary. The ancient DNA work was carried out at the Institute of Anthropology at the Johannes Gutenberg University of Mainz, following well-established protocols [36,38,39,49]. For minor modifications in the procedure of HVS-I, II, and coding region SNP typing of the mitochondrial genome and SNP typing of the Y chromosome, see electronic supplementary material, datasets S3–S5 and S18.
The genetic results were evaluated by population genetic analyses, using ancient and modern DNA datasets for comparison (electronic supplementary material, datasets S6 and S14–17). We performed Fst and AMOVA analyses in Arlequin 3.5.1 [53]. Furthermore, we calculated Fisher's exact test and ran PCA and MDS in R v. 3.0.2 [69]. PCAs were based on mtDNA and Y chromosome haplogroup frequencies (electronic supplementary material, datasets S7–S8, S14 and S16). MDSs were based on Slatkin linearized Fst values, calculated from mitochondrial HVS-I sequences (electronic supplementary material, datasets S9 and S10). Haplotype diversity was computed in DnaSP v. 5.10.01 software [70].
MtDNA HVS-I sequence data and Y chromosomal haplogroup frequencies were applied for the genetic distance calculation, comparing the STA dataset, LBKT mitochondrial DNA dataset and a combined STA–LBK Y chromosomal dataset with 130 and 100 modern populations, respectively (electronic supplementary material, datasets S15 and S17). Genetic distance maps from the Fst values were generated in ArcGis v. 10.2.
In our modified approach of shared haplotype analysis [53], we traced each HVS-I lineage of a given population back to its earliest appearance within a defined chronology. Each haplotype was regarded either as ancestral or as a new lineage, receiving its name after the culture/population where it was detected first (electronic supplementary material, dataset S12).
The adjusted parameters of the TPC [39] (electronic supplementary material, dataset S13) and further points of each analysis are detailed in the electronic supplementary material.
Supplementary Material
Supplementary Material
Acknowledgements
We thank Rozália Kustár, Olga Vajda-Kiss, Gábor Ilon and Zsuzsanna K. Zoffmann for providing archaeological, anthropological information and samples, and Gabor Krizsma for informatics support.
Data accessibility
The datasets supporting this article are available as electronic supplementary material, datasets 1–20. The sequence data are deposited in GenBank under the accession numbers KP828071—KP828154.
Author contribution
K.W.A., E.B., G.B., A.S.N. and J.J. designed the study; A.S.N. performed the palaeogenetic analyses; A.S.N., J.J., V.Ke., M.B.G. and M.F. collected the samples; G.B. and A.S.N. collected reference data; S.M.R. and A.S.N. cloned the PCR products; A.S.N., G.B. and W.H. performed the biostatistics analyses; K.K., M.B.G., B.Ő., G.T., E.M., G.P., M.Š., M.N. and N.P.Š. accomplished the anthropological analyses including individualization; K.O., T.M., V.Ki., A.O., K.S., A.C., V.V. and T.P. provided the samples and the archaeological information; K.O., T.M., J.J. and A.S.N. summarized and re-evaluated the archaeological data; B.K. made the radiocarbon dating; A.S.N., G.B., W.H., V.Ke., E.B. and K.W.A. wrote the paper; all authors discussed the results and commented on the manuscript.
Funding statement
This study is part of a 4-year research project funded by the German Research Foundation (AL 287-10-1).
References
- 1.Perlès C. 2005. From the Near East to Greece: let's reverse the focus—cultural elements that did not transfer. In BYZAS 2. How did farming reach Europe? Anatolian-European relations from the second half of the 7th through the first half of the 6th Millennium cal BC. Veröffentlichungen des Proceedings of the International Workshop Istanbul, 20–22 May 2004 (ed. Lichter C.), pp. 275–290. Istanbul, Turkey: Veröffentlichungen des Deutschen Archäologischen Instituts Istanbul. Ege Yayinlari. [Google Scholar]
- 2.Gronenborn D. 1999. A variation on a basic theme: the transition to farming in southern Central Europe. J. World Prehistory 13, 123–210. ( 10.1023/A:1022374312372) [DOI] [Google Scholar]
- 3.Gronenborn D. 2007. Beyond the models: ‘Neolithisation’ in Central Europe. In Going over: the Mesolithic-Neolithic transition in North-West Europe (eds Whittle A, Cummings V.), pp. 73–98. Oxford, UK: Oxford University Press/British Academy. [Google Scholar]
- 4.Rowley-Conwy P. 2011. Westward ho! The spread of agriculture from Central Europe to the Atlantic. Curr. Anthropol. 52, S431–S451. ( 10.1086/658368) [DOI] [Google Scholar]
- 5.Price TD. (ed). 2000. Europe's first farmers. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 6.Zilhão J. 2001. Radiocarbon evidence for maritime pioneer colonization at the origins of farming in west Mediterranean Europe. Proc. Natl Acad. Sci. USA 98, 14 180–14 185. ( 10.1073/pnas.241522898) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalicz N. 2010. An Grenze ‘zweier Welten’ -Transdanubien (Ungarn) im Frühneolithikum. In Die Neolithisierung Mitteleuropas. The spread of neolithic to Central Europe. International Conference, Mainz, 24–26th June 2005. (eds Gronenborn D, Petrasch J.), pp. 235–254. Mainz: RGZM. [Google Scholar]
- 8.Minichreiter K, Bronic IK. 2006. New radiocarbon dates for the Early Starčevo Culture in Croatia. Pril. Inst. Arheol. Zagrebu 23, 5–16. [Google Scholar]
- 9.Oross K, Bánffy E. 2009. Three successive waves of Neolithisation: LBK development in Transdanubia. Doc. Praehistorica 36, 175–189. ( 10.4312/dp.36.11) [DOI] [Google Scholar]
- 10.Bánffy E. 2004. The 6th millennium BC boundary in western Transdanubia and its role in the Central European Neolithic transition (the Szentgyörgyvölgy-Pityerdomb settlement). Budapest, Hungary: Archaeological Institute of the Hungarian Academy of Science. [Google Scholar]
- 11.Bánffy E, Eichmann WJ, Marton T. 2007. Mesolithic foragers and the spread of agriculture in western Hungary. In Proc. XV UISPP World Congress vol. 6, Lisbon, 4–9 September 2006, BAR IS 1726 (eds Kozlowski JK, Nowak M.), pp. 53–62. Oxford, UK: Archaeopress. [Google Scholar]
- 12.Dolukhanov P, Shukurov A, Gronenborn D, Sokoloff D, Timofeev V, Zaitseva G. 2005. The chronology of Neolithic dispersal in Central and Eastern Europe. J. Archaeol. Sci. 32, 1441–1458. ( 10.1016/j.jas.2005.03.021) [DOI] [Google Scholar]
- 13.Bogucki P, Grygiel R. 1993. The first farmers of Central Europe: a survey article. J. Field Archaeol. 20, 399–426. ( 10.1179/jfa.1993.20.4.399) [DOI] [Google Scholar]
- 14.Ammerman AJ, Cavalli-Sforza LL. 1984. The Neolithic transition and the genetics of populations in Europe. Princeton, NJ: Princeton University Press. [Google Scholar]
- 15.Renfrew C. 1987. Archaeology and language: the puzzle of Indo-European origins. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 16.Pinhasi R, von Cramon-Taubadel N. 2009. Craniometric data supports demic diffusion model for the spread of agriculture into Europe. PLoS ONE 4, e6747 ( 10.1371/journal.pone.0006747) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chikhi L, Nichols RA, Barbujani G, Beaumont MA. 2002. Y genetic data support the Neolithic demic diffusion model. Proc. Natl Acad. Sci. USA 99, 11 008–11 013. ( 10.1073/pnas.162158799) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barker G. 1985. Prehistoric farming in Europe. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 19.Whittle A. 1996. Europe in the Neolithic. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 20.Richards M, et al. 1996. Paleolithic and neolithic lineages in the European mitochondrial gene pool. Am. J. Hum. Genet. 59, 185–203. [PMC free article] [PubMed] [Google Scholar]
- 21.Richards M, et al. 2000. Tracing European founder lineages in the Near Eastern mtDNA pool. Am. J. Hum. Genet. 67, 1251–1276. ( 10.1016/S0002-9297(07)62954-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zvelebil M. 1989. On the transition to farming in Europe, or what was spreading with the Neolithic: a reply to Ammerman. Antiquity 63, 379–383. ( 10.1017/S0003598X00076110) [DOI] [Google Scholar]
- 23.Semino O, et al. 2004. Origin, diffusion, and differentiation of Y-chromosome haplogroups E and J: inferences on the Neolithization of Europe and later migratory events in the Mediterranean area. Am. J. Hum. Genet. 74, 1023–1034. ( 10.1086/386295) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balaresque P, et al. 2010. A predominantly neolithic origin for European paternal lineages. PLoS Biol. 8, e1000285 ( 10.1371/journal.pbio.1000285) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Semino O, et al. 2000. The genetic legacy of paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective. Science 290, 1155–1159. ( 10.1126/science.290.5494.1155) [DOI] [PubMed] [Google Scholar]
- 26.Battaglia V, et al. 2009. Y-chromosomal evidence of the cultural diffusion of agriculture in Southeast Europe. Eur. J. Hum. Genet. 17, 820–830. ( 10.1038/ejhg.2008.249) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rasteiro R, Bouttier P-A, Sousa VC, Chikhi L. 2012. Investigating sex-biased migration during the Neolithic transition in Europe, using an explicit spatial simulation framework. Proc. R. Soc. B 279, 2409–2416. ( 10.1098/rspb.2011.2323) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bramanti B, et al. 2009. Genetic discontinuity between local hunter-gatherers and central Europe's first farmers. Science 326, 137–140. ( 10.1126/science.1176869) [DOI] [PubMed] [Google Scholar]
- 29.Fu Q, et al. 2013. A revised timescale for human evolution based on ancient mitochondrial genomes. Curr. Biol. 23, 553–559. ( 10.1016/j.cub.2013.02.044) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bollongino R, Nehlich O, Richards MP, Orschiedt J, Thomas MG, Sell C, Fajkosová Z, Powell A, Burger J. 2013. 2000 years of parallel societies in Stone Age Central Europe. Science 342, 479–481. ( 10.1126/science.1245049) [DOI] [PubMed] [Google Scholar]
- 31.Skoglund P, Malmström H, Raghavan M, Storå J, Hall P, Willerslev E, Gilbert MTP, Götherström A, Jakobsson M. 2012. Origins and genetic legacy of Neolithic farmers and hunter-gatherers in Europe. Science 336, 466–469. ( 10.1126/science.1216304) [DOI] [PubMed] [Google Scholar]
- 32.Sánchez-Quinto F, et al. 2012. Genomic affinities of two 7,000-year-old Iberian hunter-gatherers. Curr. Biol. 22, 1494–1499. ( 10.1016/j.cub.2012.06.005) [DOI] [PubMed] [Google Scholar]
- 33.Lazaridis I, et al. 2014. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature 513, 409–413. ( 10.1038/nature13673) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skoglund P, et al. 2014. Genomic diversity and admixture differs for Stone-age Scandinavian foragers and farmers. Science 344, 747–750. ( 10.1126/science.1253448) [DOI] [PubMed] [Google Scholar]
- 35.Seguin-Orlando A, et al. 2014. Genomic structure in Europeans dating back at least 36,200 years. Science 346, 1113–1118. ( 10.1126/science.aaa0114) [DOI] [PubMed] [Google Scholar]
- 36.Haak W, et al. 2005. Ancient DNA from the first European farmers in 7500-year-old Neolithic sites. Science 310, 1016–1018. ( 10.1126/science.1118725) [DOI] [PubMed] [Google Scholar]
- 37.Bramanti B. 2008. Ancient DNA: genetic analysis of aDNA from sixteen skeletons of the Vedrovice. Anthropologie (Brno) 46, 153–160. [Google Scholar]
- 38.Haak W, et al. 2010. Ancient DNA from European early Neolithic farmers reveals their Near Eastern affinities. PLoS Biol. 8, e1000536 ( 10.1371/journal.pbio.1000536) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brandt G, et al. 2013. Ancient DNA reveals key stages in the formation of Central European mitochondrial genetic diversity. Science 342, 257–261. ( 10.1126/science.1241844) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haak W, et al. In press. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature. ( 10.1038/nature14317) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gamba C, et al. 2014. Genome flux and stasis in a five millennium transect of European prehistory. Nat. Commun. 5, 5257 ( 10.1038/ncomms6257) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olalde I, et al. 2014. Derived immune and ancestral pigmentation alleles in a 7,000-year-old Mesolithic European. Nature 507, 225–228. ( 10.1038/nature12960) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keller A, et al. 2012. New insights into the Tyrolean Iceman's origin and phenotype as inferred by whole-genome sequencing. Nat. Commun. 3, 698 ( 10.1038/ncomms1701) [DOI] [PubMed] [Google Scholar]
- 44.Lacan M, Keyser C, Ricaut F-X, Brucato N, Tarrús J, Bosch A, Guilaine J, Crubézy E, Ludes B. 2011. Ancient DNA suggests the leading role played by men in the Neolithic dissemination. Proc. Natl Acad. Sci. USA 108, 18 255–18 259. ( 10.1073/pnas.1113061108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lacan M, Keyser C, Ricaut F-X, Brucato N, Duranthon F, Guilaine J, Crubézy E, Ludes B. 2011. Ancient DNA reveals male diffusion through the Neolithic Mediterranean route. Proc. Natl Acad. Sci. USA 108, 9788–9791. ( 10.1073/pnas.1100723108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fernández E, Pérez-Pérez A, Gamba C, Prats E, Cuesta P, Anfruns J, Molist M, Arroyo-Pardo E, Turbón D. 2014. Ancient DNA analysis of 8000 BC near eastern farmers supports an Early Neolithic pioneer maritime colonization of Mainland Europe through Cyprus and the Aegean Islands. PLoS Genet. 10, e1004401 ( 10.1371/journal.pgen.1004401) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chandler H, Sykes B, Zilhão J. 2005. Using ancient DNA to examine genetic continuity at the Mesolithic-Neolithic transition in Portugal. In Actas dell III Congreso del Neolítico en la Península Ibérica, Santander, Monografías del Instituto internacional de Investigaciones Prehistóricas de Cantabria 1, (eds Arias P, Ontañón R, García-Moncó C.), pp. 781–786. Santander, Spain: Servicio de Publicaciones Universidad de Cantabria. [Google Scholar]
- 48.Hervella M, Izagirre N, Alonso S, Fregel R, Alonso A, Cabrera VM, de la Rúa C. 2012. Ancient DNA from hunter-gatherer and farmer groups from Northern Spain supports a random dispersion model for the Neolithic expansion into Europe. PLoS ONE 7, e34417 ( 10.1371/journal.pone.0034417) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haak W, et al. 2008. Ancient DNA, Strontium isotopes, and osteological analyses shed light on social and kinship organization of the Later Stone Age. Proc. Natl Acad. Sci. USA 105, 18 226–18 231. ( 10.1073/pnas.0807592105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gamba C, et al. 2012. Ancient DNA from an Early Neolithic Iberian population supports a pioneer colonization by first farmers. Mol. Ecol. 21, 45–56. ( 10.1111/j.1365-294X.2011.05361.x) [DOI] [PubMed] [Google Scholar]
- 51.Lee EJ, et al. 2012. Emerging genetic patterns of the European Neolithic: perspectives from a late Neolithic Bell Beaker burial site in Germany. Am. J. Phys. Anthropol. 148, 571–579. ( 10.1002/ajpa.22074) [DOI] [PubMed] [Google Scholar]
- 52.Szécsényi-Nagy A, Keerl V, Jakucs J, Brandt G, Bánffy E, Alt KW. 2014. Ancient DNA evidence for a homogeneous maternal gene pool in sixth millennium cal BC Hungary and the Central European LBK. In Early farmers: the View from Archaeology and Science Proceedings of the British Academy 198. (eds Whittle A, Bickle P.), pp. 71–93. Oxford, UK: Oxford University Press/British Academy. [Google Scholar]
- 53.Excoffier L, Lischer HEL. 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. ( 10.1111/j.1755-0998.2010.02847.x) [DOI] [PubMed] [Google Scholar]
- 54.Balanovsky O, et al. 2011. Parallel evolution of genes and languages in the Caucasus region. Mol. Biol. Evol. 28, 2905–2920. ( 10.1093/molbev/msr126) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yunusbayev B, et al. 2012. The Caucasus as an asymmetric semipermeable barrier to ancient human migrations. Mol. Biol. Evol. 29, 359–365. ( 10.1093/molbev/msr221) [DOI] [PubMed] [Google Scholar]
- 56.Rootsi S, et al. 2012. Distinguishing the co-ancestries of haplogroup G Y-chromosomes in the populations of Europe and the Caucasus. Eur. J. Hum. Genet. 20, 1275–1282. ( 10.1038/ejhg.2012.86) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Francalacci P, et al. 2013. Low-pass DNA sequencing of 1200 Sardinians reconstructs European Y-chromosome phylogeny. Science 341, 565–570. ( 10.1126/science.1237947) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rootsi S, et al. 2004. Phylogeography of Y-chromosome haplogroup I reveals distinct domains of prehistoric gene flow in Europe. Am. J. Hum. Genet. 75, 128–137. ( 10.1086/422196) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Karlsson AO, Wallerström T, Götherström A, Holmlund G. 2006. Y-chromosome diversity in Sweden—a long-time perspective. Eur. J. Hum. Genet. 14, 963–970. ( 10.1038/sj.ejhg.5201651) [DOI] [PubMed] [Google Scholar]
- 60.Lappalainen T, Laitinen V, Salmela E, Andersen P, Huoponen K, Savontaus M, Lahermo P. 2008. Migration waves to the Baltic Sea region. Ann. Hum. Genet. 72, 337–348. ( 10.1111/j.1469-1809.2007.00429.x) [DOI] [PubMed] [Google Scholar]
- 61.Morelli L, Contu D, Santoni F, Whalen MB, Francalacci P, Cucca F. 2010. A comparison of Y-chromosome variation in Sardinia and Anatolia is more consistent with cultural rather than demic diffusion of agriculture. PLoS ONE 5, e10419 ( 10.1371/journal.pone.0010419) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Petrasch J. 2001. Seid fruchtbar und mehret euch und füllet die Erde und machet sie euch untertan. Archäologisches Korrespondenzblatt 31, 13–25. [Google Scholar]
- 63.Marton T, Oross K. 2010. Siedlungsforschung in linienbandkeramischen Fundorten in Zentral- und Südtransdanubien- Wiege, Peripherie oder beides? In Siedlungsstruktur und Kulturwandel in der Bandkeramik. Beiträge der internationalen Tagung ‘Neue Fragen zur Bandkeramik oder alles beim Alten?!’ Leipzig, 23. bis 24. September 2010. Arbeits- und Forschungsberichte zur sächsischen Bodendenkmalpflege, Beihe (eds Kreienbrink F, Cladders M, Stäuble H, Tischendorf T, Wolfram S.), pp. 220–239. Dresden, Germany: Druckhaus Dresden GmbH. [Google Scholar]
- 64.Oross K, Marton T. 2012. Neolithic burials of the Linearbandkeramik settlement at Balatonszárszó and their European context. Acta Archaeol. Acad. Sci. Hung. 63, 257–299. ( 10.1556/AArch.63.2012.2.1) [DOI] [Google Scholar]
- 65.Behar DM, et al. 2004. Contrasting patterns of Y chromosome variation in Ashkenazi Jewish and host non-Jewish European populations. Hum. Genet. 114, 354–365. ( 10.1007/s00439-003-1073-7) [DOI] [PubMed] [Google Scholar]
- 66.Rasteiro R, Chikhi L. 2013. Female and male perspectives on the Neolithic transition in Europe: clues from ancient and modern genetic data. PLoS ONE 8, e60944 ( 10.1371/journal.pone.0060944) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marlowe FW. 2004. Marital residence among foragers. Curr. Anthropol. 45, 277–284. ( 10.1086/382256) [DOI] [Google Scholar]
- 68.Bentley RA, et al. 2012. Community differentiation and kinship among Europe's first farmers. Proc. Natl Acad. Sci. USA 109, 9326–9330. ( 10.1073/pnas.1113710109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.R Core Team 2012. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; (http://www.R-project.org/) [Google Scholar]
- 70.Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. ( 10.1093/bioinformatics/btp187) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting this article are available as electronic supplementary material, datasets 1–20. The sequence data are deposited in GenBank under the accession numbers KP828071—KP828154.