Abstract
The insulin-like growth factor I/insulin receptor substrate 1 axis controls, in a nonredundant way, ≈50% of cell and body size in animals from Drosophila to mice and in cells in culture. Although other factors may also intervene, cell size is strongly dependent on ribosome biogenesis, which is under the control of RNA polymerase I activity. We have previously shown that insulin receptor substrate 1 (IRS-1) translocates to the nuclei and nucleoli, where it binds to the upstream binding factor (UBF) 1, a regulator of RNA polymerase I activity. Activation of UBF1 requires its phosphorylation. However, IRS-1 is not a kinase, and we searched for an intermediate kinase that can phosphorylate UBF1. We demonstrate here that IRS-1 binds also to the phosphatidylinositol 3-kinase (PI3-K) subunits in nuclear extracts, and that the p110 subunit of PI3-K directly phosphorylates and activates UBF1, an exclusively nucleolar protein. The interaction of IRS-1, PI3-K, and UBF1 in the nucleoli provides one of the mechanisms for the effects of IRS-1 on cell and body size.
The insulin-like growth factor I/insulin receptor substrate 1 (IGF-I/IRS-1) axis is known to control ≈50% of cell and body size in animals from Drosophila to mice (1, 2) and in cells in culture (3). Cell size is an important component of cell proliferation because the cell must double its size from G1 to G2 before cell division occurs (4, 5). Cell size is regulated by ribosome biogenesis (6). Isolation of size mutants from Saccharomyces cerevisiae has confirmed the importance of ribosome biogenesis in the determination of cell size (7). Ribosome biogenesis is regulated by the activity of RNA polymerase I, which controls the rate of rRNA synthesis (6, 8). The activity of RNA polymerase I at the ribosomal DNA promoter is modulated by a complex of proteins (6, 9), which includes the nucleolar protein upstream binding factor (UBF) 1. UBF1 interacts with the protein complex TIF-1B (SL1 in humans), which consists of the TATA box-binding protein and three associated factors (10). The resulting complex promotes the binding of RNA polymerase I to the ribosomal DNA promoter (11). We have recently found that IRS-1, a docking protein for both the insulin and IGF-I receptors (12), translocates to the nuclei and nucleoli of cells, where it binds UBF1 (13, 14). The significance of this finding lies in the fact that IRS-1 (or its homolog in Drosophila) controls cell and body size in Drosophila, mice, and mammalian cells in culture (see above). Thus, deletion of the Drosophila IRS homologue, called chico, reduces fly weight by ≈50%. The reduction in body and organ size is due to a reduction in both cell number and cell size (1). Chico is the only IRS protein of Drosophila, whereas mammalian cells have four IRS proteins (12). Phosphatidylinositol 3-kinase (PI3-K), Akt, and S6K1 are downstream effectors of IRS-1 (15), and all have Drosophila homologues that regulate body size in Drosophila (16–19). The evidence accumulated in Drosophila can be extended to mice and probably to higher organisms (20). Mice with a targeted disruption of IRS-1 (2) or S6K1 (21) genes are smaller than their WT littermates. The ability of IRS-1 to double cell size has also been observed in 32D myeloid cells (3). The binding of IRS-1 to UBF1 suggests a molecular explanation for the role of IRS-1 in regulating cell size. However, binding to UBF1 does not necessarily mean activation of UBF1. In fact, both the retinoblastoma protein (22) and the IFN-inducible p204 nucleolar protein (23) bind to UBF1, but they inactivate it and repress RNA polymerase I activity.
The activity of UBF1 is regulated, at least in part, by its phosphorylation (6), especially at its C terminus (24). Phosphorylation of UBF1 has been reported in cells stimulated by serum (24–26), but there are no reports on the effect of IGF-I (a strong activator of IRS-1) on UBF1 phosphorylation. Because nuclear localization of IRS-1 caused a sharp increase in rRNA synthesis (13, 14), we have assumed that IRS-1 binding activates UBF1. However, IRS-1 has no detectable kinase activity, and, if it stimulates UBF activity by phosphorylation, it must be doing so through a kinase. IRS-1 is a very strong activator of PI3-K (27); thus, PI3-K itself seems like a good candidate for IGF-I-dependent activation of UBF1. We have therefore asked whether, in cells stimulated to proliferate by IGF-I, PI3-K may serve as the intermediate between the nuclear translocated IRS-1 and the phosphorylation and activation of UBF1 in the nucleolus. This possibility is supported by reports that PI3-K, like other downstream effectors of IRS-1, can increase cell size (19) and can be found in detectable amounts in nuclei (28).
In this paper, we demonstrate that IRS-1 binds to PI3-K in nuclear lysates of mouse embryo fibroblasts (MEFs), and that nuclear PI3-K binds to and directly phosphorylates UBF. The results suggest that the IRS-1 regulation of cell and body size in animals and cells in culture is mediated through IRS-1 activation of PI3-K in the nuclei/nucleoli of cells. In turn, the activated PI3-K phosphorylates and activates UBF1, thus regulating rRNA synthesis (6).
Materials and Methods
Cell Cultures. The R+ cell line used in these experiments is derived from R-cells, which are 3T3-like cells originating from mouse embryos with a targeted disruption of the IGF-I genes. The R+ cells express the human IGF-I cDNA under the control of the cytomegalovirus promoter and have substantial levels of IRS-1 (29). In some experiments, we used 32D or 32D-derived myeloid cells, specifically 32D IGF-I and 32D IGF-I/IRS1 cells, which are described in ref. 3.
Western Blot Analysis. All protein samples for Western blot analysis were resolved by 4–15% gradient SDS/PAGE gels. For PI3-K p85 (06-497, Upstate Biotechnology, Lake Placid, NY) Western blot, the membrane was incubated with a 1:1,000 dilution in block buffer overnight at 4°C while rocking. IRS-1 (06-468, Upstate Biotechnology) Western blot was completed with a 1:1,000 dilution in blocking buffer overnight at 4°C while rocking. Cytoplasmic and nuclear markers used were GAPDH (RDI-TRK5G4-6C5, Research Diagnostics, Flanders, NJ) and c-Jun (sc-45, Santa Cruz Biotechnology). Secondary antibodies used were anti-mouse-horseradish peroxidase (The Jackson Laboratory) and anti-rabbit-horseradish peroxidase (Amersham Biosciences), and detection was by chemiluminescence (SuperSignal, Pierce) using Hyperfilm (Amersham Biosciences).
Preparation of Nuclear Extracts. The cells used were R+ cells (see Cell Cultures). The isolation protocol is from a protocol published in ref. 30. Both cytoplasmic and nuclear lysates were centrifuged (Sorvall RC2-B) at 13,000 rpm for 10 min at 4°C and stored at -80°C.
Immunoprecipitations. Nuclear lysates (1 mg) of R+ cells were immunoprecipitated (13, 14) by using 4 μg of p110 Ab (Upstate Biotechnology) followed by rotating at 4°C overnight with protein A/G + (Santa Cruz Biotechnology). IRS-1 (06–248, Upstate Biotechnology) was immunoprecipitated from 500 μgof R+ nuclear lysate with 30 μl of protein A/G+. The subsequent procedure was the same as for p110. 32D cells, which do not express IRS-1, were used as a control.
PI3-K Protein Kinase Assay. Cell extracts were prepared from R+ cells serum-starved for 48 h. Harvested cells were resuspended in 2 ml of radioimmunoprecipitation assay buffer (1× PBS/1% Nonidet P-40/0.5% sodium deoxycholate/0.1% SDS/1× protease inhibitors/1× serine/threonine phosphatase inhibitors/1× tyrosine phosphatase inhibitors) to lyse cells. The cell suspension was left on ice for 30 min and then centrifuged (Sorvall RC2-B) at 13,000 rpm for 10 min at 4°C. Three hundred micrograms of radioimmunoprecipitation assay lysate was immunoprecipitated with 4 μg of UBF antibody (SC-13125, Santa Cruz Biotechnology) rotating for 5 h at 4°C. The lysate was resuspended in 15 μl of kinase buffer and stored at -20°C (37). The kinase assay and buffer conditions were essentially the same as outlined by Stoyanova et al. (31). The immunoprecipitated UBF was phosphorylated by the addition of 20 μl of kinase buffer that included 20 μCi of [γ-32P]ATP (1 Ci = 37 GBq) (UltraTides High Specific Activity 6,000 Ci/mmol, MP Biomedicals), 25 μM ATP (final concentration), and 100 ng of PI3-K protein. The reaction was incubated at 30°C for 20 min and stopped by the addition of 5 μl of 10× SDS/PAGE loading buffer. Samples were loaded onto a 4–15% gradient gel (Bio-Rad Ready Gel) and electrophoresed until bromophenol blue dye was at the bottom. The gel was fixed in 50% MeOH/12% acetic acid three times for 10 min, washed once with distilled water, then exposed to film (Hyperfilm, Amersham Biosciences).
IGF-I-Dependent UBF Phosphorylation and LY294002 Inhibition. A six-well plate was seeded with R+ cells, and the cells were allowed to grow for 72 h. After 48 h in serum-free medium, two of the wells had LY294002 added (final concentration, 30 μM). After 1.5 h, IGF-I was added (final concentration, 50 ng/ml) to selected wells, and the cells were stimulated for 13 h. Then 1 mCi of [32P]orthophosphate was added to each well, and they were incubated for 5 h. The cells were washed three times with cold PBS and lysed in 0.15 mM NaCl/0.05 mM Tris·HCl, pH 7.2/1% Triton X-100/1% sodium deoxycholate/0.1% SDS buffer. Three hundred micrograms of cell lysate was immunoprecipitated with 4 μg of UBF antibody and rotated overnight at 4°C. The next day 30 μl of protein A/G+ was added and rotated for 1 h at 4°C. The pellet was washed three times with 1 ml of HNTG (20 mM Hepes, pH 7.5/150 mM NaCl/10% gylcerin/0.1% Triton X-100), then 30 μl of 2× SDS/PAGE sample buffer was added. The samples were separated on a 4–15% gradient gel, transferred to nitrocellulose, and autoradiographed.
Results
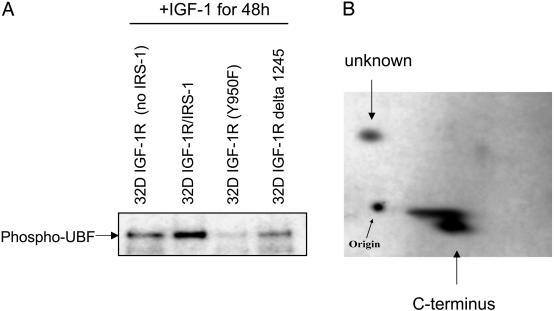
Phosphorylation of UBF by IGF-I. We first wished to determine whether IGF-I alone could induce phosphorylation of UBF1, and whether this phosphorylation was dependent on IRS-1. This experiment was performed in 32D myeloid cells, because parental 32D cells do not express IRS-1 (or IRS-2) (3, 15). Therefore, we used two 32D-derived cell lines, specifically 32D IGF-I cells (that have the IGF-I but do not express IRS1) and 32 IGF-I/IRS-1 cells (3). The cells were shifted from IL-3 to IGF-I (50 ng/ml) for 48 h, at which time both cell lines were growing exponentially (3). The cells were labeled with [32P]orthophosphate (final concentration, 1 mCi/ml) for 4 h, and lysates were made and immunoprecipitated with an antibody to UBF (Santa Cruz Biotechnology). The results are shown in Fig. 1A. The presence of IRS-1 in these cells markedly increases phosphorylation of UBF in response to IGF-I stimulation. To confirm that this phosphorylation was not due simply to differences between cell lines, we repeated the experiment in two other 32D-derived cells lines, where the IGF-IR has a mutation at Y950 or a truncation at residue 1245. These receptors still respond to IGF-I with mitogenesis (32). These two cell lines do not express IRS-1, and UBF phosphorylation is even lower than in 32D cells with the WT IGF-IR only. A control without IGF-IR is shown (see Fig. 5A) in another cell type. Although we do not have an explanation at this point, the decreased phosphorylation of UBF1 in the cell lines with defective IGF-IRs suggests that signals originating from Y950 and the C terminus of the IGF-I may contribute to the phosphorylation of UBF1. These receptors are mitogenic but fail to transform MEFs (32).
Fig. 1.
Effect of IRS-1 on the phosphorylation of UBF1. (A) The cells used were 32D-derived cells as follows: lane 1, 32D IGF-I cells (no IRS-1); lane 2, 32D IGF-I/IRS1 cells; lane 3, 32D cells with IGF-I mutation at Y950 (no IRS-1); lane 4, 32D cells with IGF-I truncated at residue 1245 (no IRS-1). All cell lines were shifted from IL-3 to IGF-I, then, after 24 h, they were labeled with [32P]orthophosphate for 8 h. The lysates were immunoprecipitated with an antibody to UBF, and the blot was autoradiographed. (B) Phospho-peptide map after IGF-I stimulation in MEFs. R+ cells stimulated with IGF-I were labeled with [32P]orthophosphate (1 mCi/ml) for 16 h, the lysates were immunoprecipitated with an antibody to UBF, and the precipitate was digested with trypsin. Digestion and 2D gel analysis were carried out exactly as described by Voit et al. (24). The large spot in the lower part of the map is known to be the C terminus of UBF, which contains >20 serines (29).
Fig. 5.
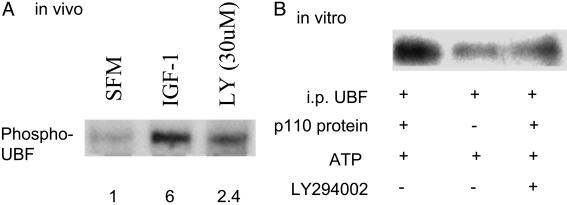
A PI3-K inhibitor inhibits UBF phosphorylation in vivo and in vitro. (A) R+ cells in serum-free medium or stimulated with IGF-I were labeled with [32P]orthophosphate as described in Fig. 1. In one set of cells stimulated with IGF-I, the PI3-K inhibitor LY294002 was added 1 h before stimulation. Autoradiograph of the labeled and immunoprecipitated UBF is shown. The numbers below the lanes are the densitometric ratios in arbitrary units. (B) The experiment was repeated in vitro by using the same conditions as in Fig. 4.
Mass spectrometry analysis of the immunoprecipitates indicated that most of the UBF immunoprecipitated in these cells was UBF1, although there was a modest amount of UBF2 (data not shown). Thus, at least in these cells, UBF1 is the predominant form, a finding that is compatible with the reports in the literature that UBF2 is inactive in the up-regulation of the ribosomal DNA promoter and, therefore, of the control of cell size (6).
The same results (increase in UBF phosphorylation) were obtained when R+ cells (MEFs with high levels of IGF-I and IRS-1) (32), were stimulated with IGF-I (data not shown, but see Fig. 5A). All subsequent experiments were done in MEFs, where isolation of pure nuclei is easier than in 32D-derived cells. We used R+ cells to obtain a map of the UBF peptides phosphorylated by IGF-I. In previous reports, phosphorylation of UBF by serum was reported to occur predominantly in the highly acidic C terminus (25, 26). The phosphorylation of the C terminus is on serines (the C terminus contains >20 serines), and it is necessary for the activation of UBF. A C-terminally truncated UBF is inactive (25). UBF was immunoprecipitated after IGF-I stimulated R+ cells had been labeled with [32P]orthophosphate (1 mCi/ml) for 8 h, and tryptic peptides were analyzed as described in Materials and Methods. The results of a typical experiment are shown in Fig. 1B. Most of the phosphorylation is in a large spot, known to be the C terminus (25, 26). There is a second spot in Fig. 1B that was reported in ref. 26 but has not been identified yet. The phosphorylation of the C terminus is not detectable in IGF-I stimulated R-cells that do not express the IGF-I and where IRS-1 is cytoplasmic (13, 14) (data not shown). Taken together, these experiments establish UBF phosphorylation by IGF-I, the importance of IRS-1 in this process, and the predominant phosphorylation of the C terminus.
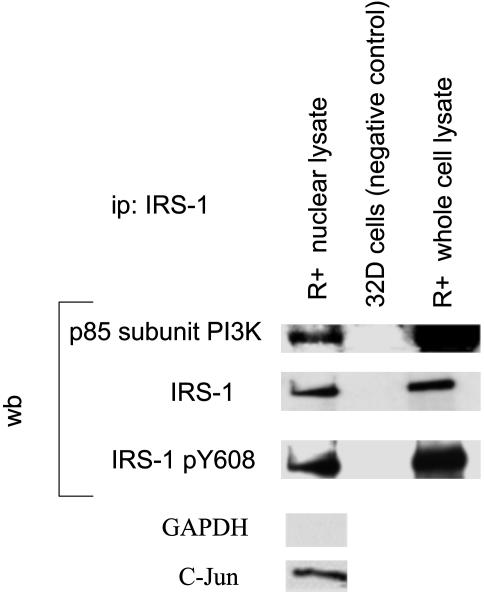
Nuclear Interactions. We next asked whether a nuclear PI3-K would be a reasonable candidate for the serine phosphorylation of UBF. Pure nuclei were prepared by the method of Zhou et al. (30) from R+ cells, which were first serum-starved and then stimulated with IGF-I. Nuclear lysates were prepared, and IRS-1 was immunoprecipitated by using an anti-IRS-1 antibody (Upstate Biotechnology). The blots were developed with an antibody to the p85 regulatory subunit of PI3-K (Upstate Biotechnology). Fig. 2 shows that p85 is immunoprecipitated by an anti-IRS-1 antibody from both nuclear extracts and whole-cell lysates. 32D cells, which do not express IRS-1 (15), were used as control to show that an antibody to IRS-1 will not immunoprecipitate p85 from IRS-1- cells. Stripping and reprobing of the membrane was performed with an antibody to IRS-1. A blot was developed with an antibody specific for the phosphorylated tyrosine at 612 of IRS-1 (Y608 in mouse IRS-1). This tyrosine is one of the IRS-1 binding sites for PI3-K (33). Both IRS-1 from the nuclear fraction and whole-cell lysate are phosphorylated at residue 608, a finding compatible with the binding of nuclear IRS-1 to the p85 subunit of PI3-K. Unstimulated R+ cells gave negative results (data not shown, but see also below) because in unstimulated MEFs IRS-1 is localized to the cytoplasm (13).
Fig. 2.
Interaction of the p85 regulatory subunit of PI3-K with nuclear IRS-1. Lysates were immunoprecipitated with an antibody to IRS-1, and the blots developed successively with antibodies (from top to bottom) to p85, IRS-1, and pY608 (the last one is an antibody specific for tyrosine 612 of the human IRS-1). Lane 1 is nuclear lysate of R+ cells, lane 2 is parental 32D cells, and lane 3 is whole-cell lysate of R+ cells. The R+ cells were stimulated with IGF-I. Monitoring for the purity of nuclei was carried out only on nuclear lysates by using antibodies to c-Jun for the nuclei and to GAPDH for the presence of cytoplasmic residues.
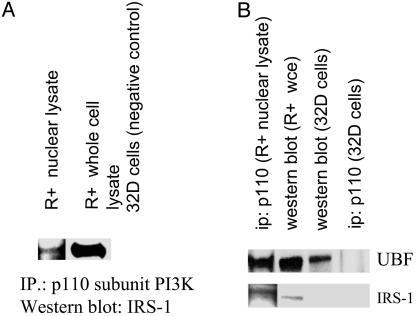
UBF1 is in a complex with other proteins (see Introduction), and the next question was whether the p110 catalytic subunit of PI3-K is also part of this complex. p110β was immunoprecipitated from R+ nuclear lysates by using an antibody to the p110β subunit of PI3-K, and the membrane was probed with an anti-IRS-1 antibody. Fig. 3A shows that nuclear IRS-1 is immunoprecipitated by a p110β antibody (lane 1). R+ whole-cell extract is used as a positive control for IRS-1. Although there is less IRS-1 immunoprecipitated by the p110β antibody in the nuclear extracts than in the cytosolic fraction, the interaction in the nucleus is still evident. Lysates from 32D cells were again used as the negative controls (IRS-1-; Fig. 3A, lane 3). The lack of good anti-p110 antibodies for Western blots made reprobing unsuitable. The purity of the nuclei was monitored as in Fig. 2 (data not shown).
Fig. 3.
The p110 subunit of PI3-K interacts in the nuclei with IRS-1 and UBF. (A) Lysates were made from R+ cells stimulated with IGF-I (lanes 1 and 2). Immunoprecipitation was performed with an antibody to p110, and the blot was developed with IRS-1. Lane 1, nuclear lysate; lane 2, whole-cell lysate; lane 3, lysate from 32D cells (IRS-1 negative) used as the negative control. (B) Lane 1, immunoprecipitation of nuclear lysate with an antibody to p110, the blot was developed with antibodies to UBF and IRS-1. Lanes 2 and 3, Western blots (no immunoprecipitation) of whole-cell lysates from R+ and 32D cells. Notice that UBF, but not IRS-1, is detectable in 32D cells. Lane 4, immunoprecipitation of lysates from parental 32D cells with an antibody to the p110 subunit of PI3-K.
The antibody to the p110β subunit of PI3-K also immunoprecipitates UBF from lysates of R+ cells serum-starved then stimulated with IGF-I (Fig. 3B, lane 1). UBF is detectable in Western blots of R+ and 32D whole cell lysates (lanes 2 and 3, respectively), but not in 32D cells lysate immunoprecipitated with IRS-1 (lane 4). Taken together, these results suggest that IRS-1, the two subunits of PI3-K and UBF, form a complex in the nuclei or nucleoli of R+ cells stimulated by IGF-I. Under these conditions, IRS-1 can be detected in the nucleoli of R+ cells, where it stimulates rRNA synthesis (13, 14). We have shown that the activation of rRNA synthesis is roughly proportional to the amount of UBF immunoprecipitated by an antibody to IRS-1 (14).
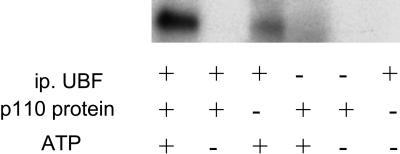
In Vitro Phosphorylation of UBF1 by PI3-K. We next asked whether purified p110 can phosphorylate UBF in vitro. R+ cells were serum-starved for 48 h, then a whole-cell lysate was made. UBF was immunoprecipitated by using anti-UBF antibodies and when used as substrate. Highly purified p110 (a kind gift from Steven Stirdivant, Merck Research Laboratories, West Point, PA) was added, along with [γ-32P]ATP, to immunoprecipitated UBF, and the reaction was carried out exactly as described by Stoyanova et al. (31). Fig. 4 shows that UBF is phosphorylated when purified p110 is added (lane 1). There is slight UBF phosphorylation even when p110 is not added, but it is much less than with p110 (lane 3). As a negative control, p110 was incubated without immunoprecipitated UBF, and no phosphorylation was detected (lane 4).
Fig. 4.
In vitro phosphorylation of UBF by the p110 catalytic subunit of PI3-K. Immunoprecipitated UBF was phosphorylated in vitro with a purified p110 subunit of PI3-K (see Materials and Methods). Cell extracts were prepared from R+ cells serum-starved for 48 h. Under these conditions, UBF should be very little phosphorylated. UBF was immunoprecipitated with an antibody to UBF (Santa Cruz Biotechnology), and the kinase assay was carried out exactly as described by Stoyanova et al. (31).
If the p110 catalytic subunit can directly phosphorylate UBF1, its effect should be decreased by inhibitors of PI3-K. R+ cells were serum-starved for 48 h. The PI3-K inhibitor LY294002 (30 μM) was added to cells 1 h before stimulation with IGF-I. The cells were stimulated for 13 h, then [32P]orthophosphate (1 mCi/ml) was added, and the cells were labeled for 5 h. UBF was immunoprecipitated with an antibody, and the gel transferred to a membrane was autoradiographed (Fig. 5A). IGF-I again sharply increases UBF phosphorylation (lane 2) as compared with serum-starved cells. LY294002 inhibits IGF-I mediated phosphorylation (lane 3) although it is greater than in the serum-starved cells. This result was not surprising, because UBF can also be phosphorylated by extracellular signal-regulated protein kinases (34), a signaling pathway that can be independent from IRS-1.
The experiment was repeated in vitro by using the same cell-culture conditions given in Fig. 4. Again, UBF is highly phosphorylated in vitro by p110 (Fig. 5B, lane 1), and the reaction is almost completely abolished by the addition of LY294002 (lane 3).
Discussion
The data presented here can be summarized as follows: (i) IGF-I induces phosphorylation of UBF1, especially of the C terminus; (ii) UBF phosphorylation by IGF-I is markedly increased by the presence in cells of IRS-1; (iii) in the nucleus, IRS-1 coprecipitates the p85 regulatory subunit of PI3-K, whereas the p110 catalytic subunit coprecipitates with both IRS-1 and UBF1, suggesting the formation of a complex; (iv) the p110 subunit of PI3-K directly phosphorylates in vitro UBF1; and (v) phosphorylation of UBF by p110 is inhibited in vivo and in vitro by inhibitors of PI3-K.
The significance of these findings is linked to previous reports that the IGF-I/IRS-1 axis controls ≈50% of cell and body size in Drosophila and mice (see Introduction). Deletion of the IGF-I in mice also causes an ≈50% decrease in the size of mouse embryos (35), emphasizing that, at least in some multicellular organisms (and some cells in culture), the IGF-I/IRS-1 axis controls, in a nonredundant way, about half of cell and body size. Our present data indicate that one of the mechanisms of IGF-I-mediated determination of cell size is the activation of nuclear PI3-K by IRS-1, and the phosphorylation of UBF1 by the p110 catalytic subunit of PI3-K. We have shown in previous papers that nuclear translocation of IRS-1 and binding to UBF1 result in increased rRNA synthesis and activation of the ribosomal DNA promoter (13, 14).
Activation of UBF1 requires its phosphorylation, especially of its C terminus (6, 25). This C terminus has a peculiar sequence, because it is very acidic (numerous glutamic and aspartic acid residues) and contains at least 20 serines. Removal of the C terminus markedly reduces the activity of UBF1 (25). The phosphorylation of the C terminus of UBF1 by IGF-I stimulation therefore makes sense, because PI3-K is a serine kinase (36). I. Grummt and coworkers (24, 25) have repeatedly shown the serine phosphorylation of the UBF1 C terminus, but their cells were stimulated by serum, which contains a variety of growth factors. Lesser phosphorylation at serine residues 484 and 388 (26) has also been reported, whereas EGF has been shown to induce phosphorylation at threonines 117 and 201 through the activation of extracellular signal-regulated protein kinases (34). Perhaps phosphorylation of these additional sites increases further UBF1 activation and is therefore responsible for the other 50% of cell size regulation.
UBF1 is an exclusively nucleolar protein (24), and it completely disappears when the nucleolus involutes during differentiation (37). By confocal microscopy, in R+ cells stimulated with IGF-I, IRS-1 colocalizes with nucleolin to the nucleoli (13, 14). Further support comes from the fact that the interactions among IRS-1, PI3-K, and UBF1 in this paper were all studied in nuclear extracts. Interestingly, in differentiated cells, IRS-1 is cytoplasmic (37), UBF1 is not activated (22, 37, 38), and rRNA synthesis is markedly decreased (38). Our findings are compatible with the recent report that ribosomal gene transcription requires S6K1 and is mediated by the phosphorylation of the C-terminal domain of UBF (39). S6K1 is activated by IRS-1 (27).
In conclusion, we propose that the effect of IRS-1 on cell and body size is mediated (at least in part) by the nuclear translocation of IRS-1 and PI3-K and by the activation of UBF1 by PI3-K through serine phosphorylation of UBF1. This interpretation should not be construed as excluding other kinases as activators of UBF1. As mentioned above, UBF1 can be phosphorylated also by extracellular signal-regulated protein kinases (34) and CK2 (25). PI3-K is simply the most reasonable candidate for an IGF-I-mediated activation of UBF1. We propose that these experiments suggest that nuclear IRS-1 forms complexes in the nuclei and nucleoli with other signaling proteins, such as PI3-K and possibly Akt, which can also be translocated to the nuclei (40). In this location, IRS-1, PI3-K, and Akt may have distinct and different actions from those of the same molecules in the cytoplasm, because proteins like UBF are not found in the cytoplasm.
Acknowledgments
This work was supported by National Institutes of Health Grants CA56309 and AG20956.
Abbreviations: IGF-I, insulin-like growth factor I; IRS-1, insulin receptor substrate 1; MEF, mouse embryo fibroblast; PI3-K, phosphatidylinositol 3-kinase; UBF, upstream binding factor.
References
- 1.Bohni, R., Riesco-Escovar, J., Oldham, S., Brogiolo, W., Stocker, H., Andruss, B. F., Beckingham, K. & Hafen, E. (1999) Cell 97, 865-875. [DOI] [PubMed] [Google Scholar]
- 2.Pete, G., Fuller, G. R., Oldham, J. M., Smith, D. R., D'Ercole, A. J., Kahn, C. R. & Lund, P. K. (1999) Endocrinology 140, 5478-5487. [DOI] [PubMed] [Google Scholar]
- 3.Valentinis, B., Navarro, M., Zanocco-Marani, T., Edmonds, P., McCormick, J., Morrione, A., Sacchi, A., Romano, G., Reiss, K. & Baserga, R. (2000) J. Biol. Chem. 275, 25451-25459. [DOI] [PubMed] [Google Scholar]
- 4.Fraser, R. S. S. & Nurse, P. (1978) Nature 27, 726-730. [DOI] [PubMed] [Google Scholar]
- 5.Hartwell, L. H. (1978) J. Cell Biol. 77, 627-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grummt, I. (1999) Mol. Biol. 62, 109-153. [DOI] [PubMed] [Google Scholar]
- 7.Jorgensen, P., Nishikawa, J. L., Breitkreutz, B. J. & Tyers, M. (2002) Science 297, 395-400. [DOI] [PubMed] [Google Scholar]
- 8.Moss, T. & Stefanovsky, V. Y. (1995) Prog. Nucleic Acid Res. Mol. Biol. 50, 25-66. [DOI] [PubMed] [Google Scholar]
- 9.Reeder, R. H. (1999) Prog. Nucleic Acid Res. Mol. Biol. 62, 293-327. [DOI] [PubMed] [Google Scholar]
- 10.Kihm, A. J., Hershey, J. C., Haystead, T. A. J., Madsen, C. S. & Owens, G. K. (1998) Proc. Natl. Acad. Sci. USA 95, 14816-14820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beckmann, H., Chen, J. L., O'Brien, T. & Tjian, R. (1995) Science 270, 1506-1519. [DOI] [PubMed] [Google Scholar]
- 12.White, M. F. (1998) Mol. Cell. Biochem. 182, 3-11. [PubMed] [Google Scholar]
- 13.Tu, X., Batta, P., Innocent, N., Prisco, M., Casaburi, I., Belletti, B. & Baserga, R. (2002) J. Biol. Chem. 277, 44357-44365. [DOI] [PubMed] [Google Scholar]
- 14.Sun, H., Tu, X., Prisco, M., Wu, A., Casiburi, I. & Baserga, R. (2003) Mol. Endocrinol. 17, 472-486. [DOI] [PubMed] [Google Scholar]
- 15.Wang, L. M., Myers, M.G., Jr., Sun, X. J., Aaronson, S. A., White, M. & Pierce, J. H. (1993) Science 261, 1591-1594. [DOI] [PubMed] [Google Scholar]
- 16.Leevers, S. J., Weinkove, D., MacDougall, L. K., Hafen, E. & Waterfield, M. D. (1996) EMBO J. 15, 6584-6594. [PMC free article] [PubMed] [Google Scholar]
- 17.Montagne, J., Stewart, M. J., Stocker, H., Hafen, E., Kozma, S. C. & Thomas, G. (1999) Science 285, 2126-2129. [DOI] [PubMed] [Google Scholar]
- 18.Verdu, J., Buratovich, M. A., Wilder, E. L. & Birnbaum, M. J. (1999) Nat. Cell Biol. 1, 500-506. [DOI] [PubMed] [Google Scholar]
- 19.Kozma, S. C. & Thomas, G. (2002) BioEssays 24, 65-71. [DOI] [PubMed] [Google Scholar]
- 20.Baserga, R., Prisco, M. & Yuan, T. (2003) in Insulin-Like Growth Factor Receptor Signaling, Molecular Biology Intelligence Unit, eds. LeRoith, D., Zumkeller, W. & Baxter, R. C. (Plenum, New York; Landes Biosciences, Georgetown, TX).
- 21.Shima, H., Pende, M., Chen, Y., Fumagalli, S., Thomas, G. & Kozma, S. C. (1998) EMBO J. 17, 6649-6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cavanaugh, A. H., Hempel, W. M., Taylor, L. J., Rogalsky, V., Todorov, G. & Rothblum, L. I. (1995) Nature 374, 177-180. [DOI] [PubMed] [Google Scholar]
- 23.Liu, C., Wang, H. & Lengyel, P. (1999) EMBO J. 18, 2845-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Voit, R., Kuhn, A., Sander, E. E. & Grummt. I. (1995) Nucleic Acids Res. 23, 2593-2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Voit, R., Hoffmann, M. & Grummt, I. (1999) EMBO J. 18, 1891-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voit, R. & Grummt, I. (2001) Proc. Natl. Acad. Sci. USA 98, 13631-13636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Myers, M. G., Jr., Grammer, T. C., Wang, L. M., Sun, X. J., Pierce, J. H., Blenis, J. & White, M. F. (1994) J. Biol. Chem. 269, 28783-28789. [PubMed] [Google Scholar]
- 28.Neri, L. M., Borgatti, P., Capitani, S. & Martelli, A. M. (2002) Biochim. Biophys. Acta 1584, 73-80. [DOI] [PubMed] [Google Scholar]
- 29.Sell, C., Rubini, M., Rubin, R., Liu, J., Efstratiadis, A. & Baserga, R. (1993) Proc. Natl. Acad. Sci. USA 90, 11217-11221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou, B. P., Liao, Y., Xia, W., Spohn, B., Lee, M. H. & Hung, M. C. (2001) Nat. Cell Biol. 3, 245-252. [DOI] [PubMed] [Google Scholar]
- 31.Stoyanova, S., Bulgarelli-Leva, G., Kirsch, C., Hanck, T., Klinger, R. & Wetzker, R. (1997) Biochem. J. 324, 489-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Romano, G., Prisco, M., Zanocco-Marani, T., Peruzzi, F., Valentinis, B. & Baserga, R. (1999) J. Cell Biochem. 72, 294-310. [DOI] [PubMed] [Google Scholar]
- 33.Sun, X. J., Crimmins, D. L., Myers, M. G., Jr., Miralpeix, M. & White, M. F. (1993) Mol. Cell. Biol. 13, 7418-7428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stefanovsky, V. Y., Pelletier, G., Hannan, R., Gagnon-Kugler, T., Rothblum, L. I. & Moss, T. (2001) Mol. Cell 8, 1063-1073. [DOI] [PubMed] [Google Scholar]
- 35.Efstratiadis, A. (1998) Int. J. Dev. Biol. 42, 955-976. [PubMed] [Google Scholar]
- 36.Cantley, L. (2002) Science 206, 1655-1677. [Google Scholar]
- 37.Tu, X., Baffa, R., Luke, S., Prisco, M. & Baserga, R. (2003) Exp. Cell Res. 288, 119-130. [DOI] [PubMed] [Google Scholar]
- 38.Comai, L., Song, Y., Tan, C. & Bui, T. (2000) Cell Growth Differ. 11, 63-70. [PubMed] [Google Scholar]
- 39.Hannan, K. M., Brandenburger, Y., Jenkins, A., Sharkey, K., Cavanaugh, A., Rothblum, L., Moss, T., Poortinga, G., McArthur, G. A., Pearson, R. B. & Hannan, R. D. (2003) Mol. Cell. Biol. 23, 8862-8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pekarsky, Y., Koval, A., Hallas, C., Bichi, R., Tresini, M., Malstrom, S., Russo, G., Tsichils, P. & Croce, C. M. (2000) Proc. Natl. Acad. Sci. USA 97, 3028-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]