Abstract
Cell fusion was recently reported to account for the plasticity of adult stem cells in vivo. Adult stem cells, referred to as mesenchymal stem cells or marrow stromal cells, from rat marrow, were infused into 1.5- to 2-day-old chick embryos. After 4 days, the rat cells had expanded 1.3- to 33-fold in one-third of surviving embryos. The cells engrafted into many tissues, and no multinuclear cells were detected. The most common site of engraftment was the heart, apparently because the cells were infused just above the dorsal aorta. Some of the cells in the heart expressed cardiotin, and α-heavy-chain myosin. GFP+ cells reisolated from the embryos had a rat karyotype. Therefore, the cells engrafted and partially differentiated without evidence of cell fusion.
Multipotential cells with many of the characteristics of stem cells have now been isolated from a variety of differentiated tissues, including bone marrow, fat, umbilical blood, synovial membranes, brain, and heart (1–14). To varying degrees, the adult stem cells from different sources have been shown to differentiate into a variety of cell phenotypes in culture, including osteoblasts, adipocytes, chondrocytes, epithelial cells, myoblasts, and early precursors of neural cells. Also, a number of investigators have reported that after infusion into animals, adult stem cells from several sources engraft into multiple tissues and, at least in part, differentiate into the phenotype of the cells found in the tissues. Recently, however, several reports have indicated that some or all of the apparent plasticity observed after adult stem cells are administered in vivo is accounted for by the donor cells fusing with recipient cells (15–19).
Among the most extensively studied adult stem cells (1–4, 6, 14, 20) are the cells from bone marrow referred to as mesenchymal stem cells or marrow stromal cells (MSCs). If plated at very low densities, MSCs generate single-cell-derived colonies, and the colonies can be differentiated into several cell phenotypes in culture (1–4, 14, 20). After infusion into irradiated animals, MSCs home to a variety of tissues, particularly after tissue injury (14). Therefore, MSCs and related cells from bone marrow appear to be part of a natural repair system that supplements the stem-like cells found in multiple tissues that are mobilized by tissue injury. Cell fusion by MSCs was recently observed in a coculture system in which MSCs were added to a monolayer of heat-shocked lung epithelial cells. A few of the fused cells also underwent nuclear fusion (21). However, three-fourths or more of the MSCs that differentiated into epithelial cells underwent the change in phenotype without evidence of cell fusion (21).
In this study, integration and differentiation of rat MSCs were examined in vivo by transplantation into organogenesis-stage embryos. Rat MSCs expressing GFP were grafted in place of epithelial-stage somites of chick embryos that were 1.5–2 days old, i.e., stage 12–13 of development. The grafted cells that survived in the chick host were identified as GFP-expressing cells 4 days postinjection. Real-time PCR assays for rat Y chromosome confirmed that the GFP+ rat cells had integrated into the embryonic tissue. In one embryo, there was a 30-fold increase of rat cells. Histological analysis showed that MSCs had migrated into multiple host tissues, including the heart, liver, and vertebral column. Karyotyping of GFP+ cells isolated from the embryos demonstrated that the cells retained a normal karyotype with 42 rat chromosomes instead of the 78 in chick cells.
Methods
Primary Marrow Stromal Cell Cultures. MSC were collected from femurs and tibias of adult male rats that ubiquitously expressed GFP. The transgenic rats (CZ-004) were kindly provided by M. Okabe, Genome Information Research Center, Osaka University, Osaka (22). Rats were killed with a mixture of 70% CO2 and 30% O2. Tibias and femurs were placed on ice in MEM with α modification (α-MEM; GIBCO/BRL) containing 20% FCS (Atlanta Biologicals, Norcross, GA), 2 mM l-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, and 25 ng/ml amphotericin B (GIBCO/BRL). Epiphyses of femurs and tibias were removed, and the marrow was flushed out by using a syringe filled with medium. Bone marrow was filtered through a 70-μm nylon mesh and plated in 75-cm2 flasks. About 24 h after plating, supernatant containing nonadherent cells was removed, and fresh medium was added. After the cells had grown to near confluency, they were passaged two to five times by being detached with 0.25% trypsin/1 mM EDTA at 37°C for 5 min and replated at a density of ≈5,000 cells per cm2. For injection, MSCs were plated at 100–500 cells per cm2 and incubated for 4–5 days at 37°C. The cells were harvested with trypsin/EDTA and suspended in PBS.
Microsurgery. Chick embryos incubated at 39°C for 40–48 h at stages 12–14 were used to inject the MSCs. To lower the embryo, 2 ml of albumin was removed from the tapered end of the egg by using an 18-gauge needle and a syringe. An oval opening ≈1 inch long was made in the shell to view the embryo. Filtered 5% India Ink (Pelikan) in PBS was injected under the embryo to increase the contrast. The two or three most recently formed somites of 12–20 somites present in the chick embryos were crushed or removed by using a titanium needle (McCrone Microscopes, Westmount, IL). Rat GFP+ MSCs were suspended at 5,000–20,000 cells per microliter in PBS at 4°C, and 1–5 μl of cells suspension was injected into the space created by removing the somites. An equal volume of PBS was injected into control embryos. The cell suspension was kept on ice until injection to prevent cell aggregation. The embryos were harvested 4 days postinjection and used for immunofluorescence or real-time analysis.
Immunocytochemistry. Embryos were harvested, rinsed with PBS, fixed in 4% paraformaldehyde in PBS fixative overnight at 4°C transferred to 30% sucrose solution, and flash frozen, and 20-μm sections were cut in a cryostat. Every 9th and 10th section was mounted on a slide for further analysis, and the remaining sections were collected into a tube for real-time PCR analysis. Antibodies to cardiotin and α-heavy-chain myosin (Chemicon Temecula, CA, and Abcam, San Antonio, TX) were incubated at 1:100 dilution for 36 h at 4°C. The slides were washed three times for 5 min with PBS. They were incubated with Alexa-594-tagged (Molecular Probes) secondary anti-mouse antibody at 1:1,000 dilution for 1 h. Controls included omitting the primary antibody. Slides were evaluated by using epifluorescence (Eclipse 800; Nikon) and 3D deconvolution microscopy [Leica (Deerfield, IL) DM RXA2 and SlideBook from Intelligent Imaging Innovations, Denver].
Real-Time PCR Analysis for the Rat Y Chromosome. Both frozen sections and whole embryos were used to extract DNA. The tissues were incubated with 1 ml of proteinase K solution (0.4 mg/ml proteinase K/10 mg/ml SDS in Tris buffer at pH 7.4; Sigma) at 45°C for 24 h. The DNA was extracted by using phenol/chloroform extraction protocol (Sigma). The precipitated DNA was purified by using DNeasy tissue kit (Qiagen, Chatsworth, CA). Approximately 300 ng of chicken embryo DNA was used to analyze the engraftment by using a universal PCR mix (Applied Biosystems). The primers and probe set used were caagtcaagcgccccatgaa and ttgagccaacttgtgcctctctc and FAM-tgcatttatggtgtggtcccgcg-TAMRA. The conditions were initial denaturation at 94°C for 10 min, then 45 cycles with denaturation at 94°C for 1 min and annealing at 60°C (or -5° Tm) for 1 min (23). Standards were run simultaneously by using pure uninjected chicken embryo DNA and purified DNA from rat MSCs.
Fluorescence-Activated Cell Sorting (FACS) and Karyotyping. Embryos injected with rat GFP+ cells were dispersed with 1 mg/ml collagenase and dispase in PBS (Roche Molecular Biochemicals) for 1 h at 37°C. GFP+ cells were sorted from the suspension by using a flow cytometer (FACS Vantage SE; Becton Dickinson). The sorted cells were cultured for 3 days at 10–15 cells/cm2 and harvested by using trypsin/EDTA. Harvested cells were treated with hypotonic solution of 0.075 M potassium chloride and fixed in 3:1 methanol/glacial acetic acid. G bands were stained with a standard G band procedure by using trypsin and Wright's stain (24).
Results
Rat MSCs expressing GFP were infused into stage 12–13 chick embryos prepared by warming fertilized eggs to 37°C for 2 days. Two of the most recently formed somites on the right side were surgically removed (25), and 5,000–50,000 male rat MSCs expressing GFP were infused into the space created (Fig. 1). About half the embryos infused with either rat MSCs (101/235) or PBS (50/101) survived the manipulations. The embryos were harvested 4 days after the infusion, at which time there was considerable variation in size but most had developed to stages 24–26. The average initial wet weight content of the embryos was ≈3 mg; 4 days later, the wet weights varied between 300 and 500 mg, indicating a 100- to 160-fold increase in size. A preliminary screen for engraftment was carried out by searching for GFP+ cells in whole mounts of the embryos. Fifteen of 65 live embryos tested were shown to contain the donor cells by real-time PCR assays for the rat Y chromosome (Table 1). The extent of chimerism varied widely, but 5 of the 15 embryos showed 0.03–0.8% chimerism and 1.5- to 33-fold increase in male rat chromosomes.
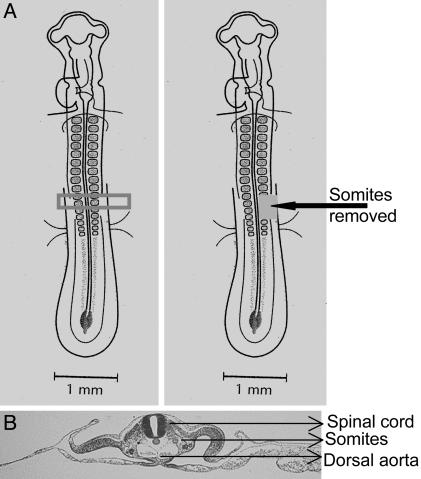
Fig. 1.
Schematic representation of the surgery performed on chick embryos. (A) The two or three most recently formed somites at 48 h were crushed or removed. From 5,000 to 50,000 male rat GFP+ MSCs were injected into the space generated with a glass capillary pipet. (B) Transverse section of stage 12 embryo. [Diagrams reprinted with permission from ref. 28 (Copyright 1997, Elsevier).]
Table 1. Engraftment of rat GFP cells in chick embryos.
| Embryo number | Fold increase of rat MSCs | Percentage chimerism, % |
|---|---|---|
| 8 | 33.3 | 0.8 |
| 9 | 13.3 | 0.3 |
| 5 | 4.0 | 0.167 |
| 6 | 2.0 | 0.066 |
| 4 | 1.5 | 0.033 |
Rat MSCs engraft and multiply in chick embryos. Real-time PCR analysis for rat Y chromosome was performed 4 days postinjection. Fifteen of 65 embryos tested were positive for the Y chromosome. Five of the 15 embryos positive for the Y chromosome showed an increase in the number of rat Y chromosomes relative to the number injected.
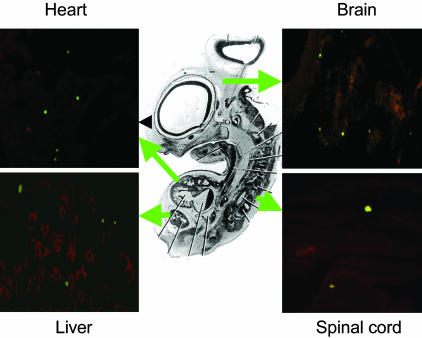
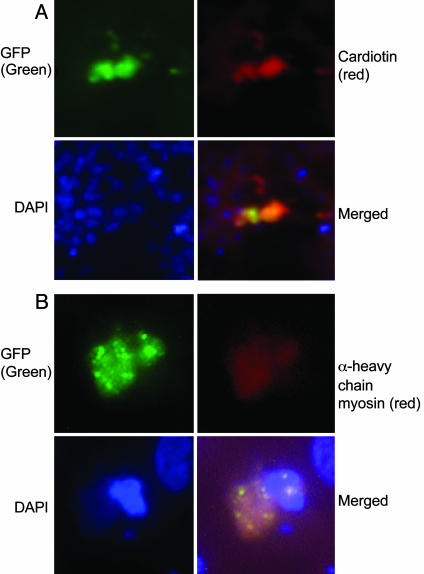
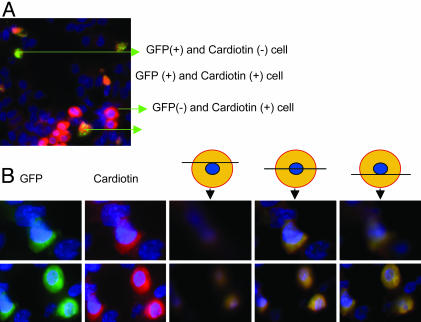
In histological sections of the embryos, GFP+ cells were detected in multiple developing organs of the embryos, including heart, liver, brain, and spinal cord (Fig. 2). The most common site of appearance of the GFP+ cells was the heart, apparently because the site of infusion was just above the dorsal aorta (Fig. 1). The cells were found in several regions of the developing heart (Fig. 3). Immunostaining of sections of heart demonstrated that some of the GFP+ cells expressed cardiotin, a protein found in the longitudinal sarcoplasmic reticulum of mature cardiomyocytes (Figs. 4A and 5). In addition, some stained for α-heavy-chain myosin (Fig. 4B). Because of the early stage of development (26), the hearts also contained both GFP+ cells and endogenous GFP- cells that did not express cardiotin or α-heavy-chain myosin but had the morphology of the proliferating immature cardiomyocytes that form the trabecular myocardium in stage 21–29 chick embryos (26). Examination of the sections by 3D deconvolution microscopy demonstrated that both the GFP+ cells positive for cardiotin and negative for cardiotin were mononuclear (Fig. 5 A and B).
Fig. 2.
Rat MSCs expressing GFP detected in organs of 6-day-old chicken embryo by epifluorescence microscopy (×10).
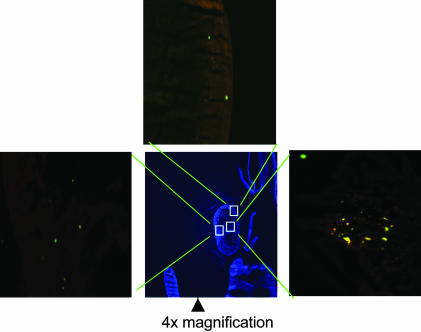
Fig. 3.
Rat GFP+ MSCs engrafted and partially differentiated in heart 4 days postinfusion. Five of six embryos assayed had GFP+ cells in heart; section of 20 μm shown at ×4 and ×10.
Fig. 4.
Epifluorescent immunohistology of sections from chick embryos infused with rat GFP+ MSCs. (A) Cells positive for both GFP (green) and cardiotin (red) or (B) cells positive for both GFP (green) and α heavy-chain myosin (red). 4′,6-Diamidino-2-phenylindole stain was used to identify nuclei (×63).
Fig. 5.
Photomicrograph by 3D deconvolution microscopy of immunohistology sections from chick embryos infused with rat GFP+ MSCs. (A) 3D deconvolution microscopy of section with GFP+ (green) cells and stained for cardiotin (red). The GFP+ cells were mononuclear. (B) 3D deconvolution microscopy to demonstrate that cells that were GFP+ and cardiotin+ were mononuclear (×63).
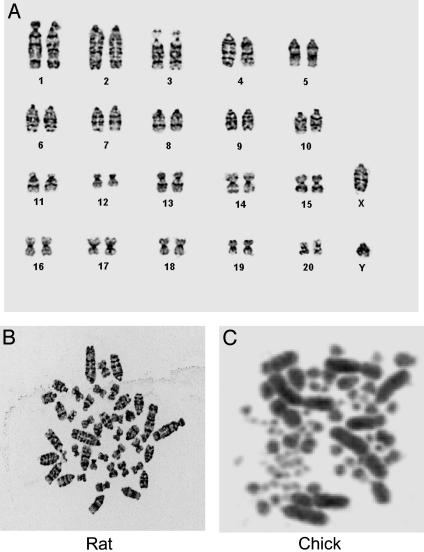
Embryos that contained GFP+ cells were dissociated and cells isolated by fluorescence-activated cell sorting. Metaphase spreads of 35 GFP+ cells pooled from five embryos were then karyotyped. All 35 cells contained the normal complement of 42 rat chromosomes with the expected low-resolution G banding pattern (Fig. 6). The cells were therefore distinctly different from chick cells that contain 78 chromosomes.
Fig. 6.
Karyotyping of cells expressing GFP. (A) Ideogram of GFP-expressing cells isolated by fluorescence-activated cell sorting from embryos 4 days postinjection. Thirty-five metaphase spreads of GFP-expressing cells were analyzed, and all of the cells contained 42 chromosomes characteristic of rat cells. (B) Metaphase spread of rat chromosomes. (C) Metaphase spread of chick chromosomes. The image was kindly provided by M. E. Delany, University of California, Davis.
Discussion
The data presented here demonstrate that after rat MSCs expressing GFP were infused into early stage chick embryos, about one-third of the surviving embryos harvested 4 days later contained rat cells in multiple tissues. The number of rat GFP+ cells in a few of the embryos increased 1.5- to 33-fold as the size of the embryos increased 100- to 160-fold; therefore, the rat cells expanded, but not as rapidly as the endogenous chick cells. GFP+ cells harvested from the embryos had the normal rat karyotype of 42 chromosomes instead of the distinctive 78 chromosomes found in chick cells. Therefore, the rat cells had engrafted and expanded under conditions that did not produce any major genomic alterations. In the heart, most of the cells assumed the expected morphology of mononuclear proliferating cardiomyocytes, and few of the cells had partially differentiated, as demonstrated by expression of cardiotin and α-heavy chain myosin. The simplest explanation for the data therefore is that the rat cells both engrafted and partially differentiated without cell fusion. The data, however, do not rule out the possibility that some of the rat cells fused with the chick cells, and that some of the fused cells underwent reductive division. However, to account for the normal rat karyotype of a large fraction of the reisolated GFP+ cells and the failure to detect multinuclear cells, it would be necessary to assume that the fused cells are transient intermediates, and that the reductive division to generate cells with a rat karyotype is highly efficient.
All of the observations on differentiation or fusion of adult stem cells can probably be resolved within the context of information now available. One consideration is that cell fusion is a normal event during the development of several tissues. Fusion has long been known to be essential for formation of skeletal and cardiac muscle cells (8). Multinuclear and polyploid cells have long been observed in liver, and recent observations demonstrated that they are primarily caused by cell fusion and not by an interruption of mitosis (17, 18). Also, recent observations have demonstrated that Purkinje cells continue to fuse with cells of hematopoietic origin during adult life (27). A second consideration is that recent observations suggest several types of stem cells are particularly prone to fuse with differentiated cells both in culture and in vivo (19, 21). A third consideration is that the experimental strategies used to follow differentiation or fusion are generally biased to detect either one process or the other. Also, most of the experimental strategies are still not fully tested, and the biological systems for in vivo experiments are complex. Therefore, negative data in any one system are generally not conclusive. With these considerations, all of the data can be reconciled by concluding that adult stem cells can undergo both transdifferentiation and fusion in vivo, and that the relative degree of detection of each process depends highly on the analytical techniques used, the tissue examined, and other factors such as the developmental stage of the organism and the presence of tissue injury (14).
Acknowledgments
This work was supported in part by National Institutes of Health Grants AR 47796, HL073755, and AR 48323; the Oberkotter Foundation; HCA the Healthcare Company; the Louisiana Gene Therapy Research Consortium (to D.J.P.); and the National Institutes of Health/National Institute of Arthritis and Musculoskeletal and Skin Diseases National Research Service Award (to R.R.P). The metaphase spread of chick chromosomes image was kindly provided by Dr. Mary E. Delany, Department of Animal Science, One Shields Avenue, University of California, Davis.
Abbreviation: MSC, marrow stromal cells.
References
- 1.Friedenstein, A. J., Petrakova, K. V., Kurolesova, A. I. & Frolova, G. P. (1968) Transplantation 6, 230-247. [PubMed] [Google Scholar]
- 2.Owen, M. & Friedenstein, A. J. (1988) Ciba Found. Symp. 136, 42-60. [DOI] [PubMed] [Google Scholar]
- 3.Caplan, A. I. (1990) Biomaterials 11, 44-46. [PubMed] [Google Scholar]
- 4.Prockop, D. J. (1997) Science 276, 71-74. [DOI] [PubMed] [Google Scholar]
- 5.Eglitis, M. A. & Mezey, E. (1997) Proc. Natl. Acad. Sci. USA 94, 4080-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liechty, K. W., MacKenzie, T. C., Shaaban, A. F., Radu, A., Moseley, A. M., Deans, R., Marshak, D. R. & Flake, A. W. (2000) Nat. Med. 6, 1282-1286. [DOI] [PubMed] [Google Scholar]
- 7.Krause, D. S., Theise, N. D., Collector, M. I., Henegariu, O., Hwang, S., Gardner, R., Neutzel, S. & Sharkis, S. J. (2001) Cell 105, 369-377. [DOI] [PubMed] [Google Scholar]
- 8.LaBarge, M. A. & Blau, H. M. (2002) Cell 111, 589-601. [DOI] [PubMed] [Google Scholar]
- 9.Jiang, Y., Henderson, D., Blackstad, M., Chen, A., Miller, R. F. & Verfaillie, C. M. (2003) Proc. Natl. Acad. Sci. USA 100 Suppl 1, 11854-11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McKinney-Freeman, S. L., Majka, S. M., Jackson, K. A., Norwood, K., Hirschi, K. K. & Goodell, M. A. (2003) Exp. Hematol. 31, 806-814. [DOI] [PubMed] [Google Scholar]
- 11.D'Amour, K. A. & Gage, F. H. (2003) Proc. Natl. Acad. Sci. USA 100 Suppl 1, 11866-11872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Ugarte, D. A., Morizono, K., Elbarbary, A., Alfonso, Z., Zuk, P. A., Zhu, M., Dragoo, J. L., Ashjian, P., Thomas, B., Benhaim, P., et al. (2003) Cells Tissues Organs 174, 101-109. [DOI] [PubMed] [Google Scholar]
- 13.De Bari, C., Dell'Accio, F., Vandenabeele, F., Vermeesch, J. R., Raymackers, J. M. & Luyten, F. P. (2003) J. Cell Biol. 160, 909-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prockop, D. J., Gregory, C. A. & Spees, J. L. (2003) Proc. Natl. Acad. Sci. USA 100 Suppl 1, 11917-11923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ying, Q. L., Nichols, J., Evans, E. P. & Smith, A. G. (2002) Nature 416, 545-548. [DOI] [PubMed] [Google Scholar]
- 16.Terada, N., Hamazaki, T., Oka, M., Hoki, M., Mastalerz, D. M., Nakano, Y., Meyer, E. M., Morel, L., Petersen, B. E. & Scott, E. W. (2002) Nature 416, 542-545. [DOI] [PubMed] [Google Scholar]
- 17.Wang, X., Willenbring, H., Akkari, Y., Torimaru, Y., Foster, M., Al Dhalimy, M., Lagasse, E., Finegold, M., Olson, S. & Grompe, M. (2003) Nature 422, 897-901. [DOI] [PubMed] [Google Scholar]
- 18.Vassilopoulos, G., Wang, P. R. & Russell, D. W. (2003) Nature 422, 901-904. [DOI] [PubMed] [Google Scholar]
- 19.Alvarez-Dolado, M., Pardal, R., Garcia-Verdugo, J. M., Fike, J. R., Lee, H. O., Pfeffer, K., Lois, C., Morrison, S. J. & Alvarez-Buylla, A. (2003) Nature 425, 968-973. [DOI] [PubMed] [Google Scholar]
- 20.Sekiya, I., Larson, B. L., Smith, J. R., Pochampally, R., Cui, J. G. & Prockop, D. J. (2002) Stem Cells 20, 530-541. [DOI] [PubMed] [Google Scholar]
- 21.Spees, J. L., Olson, S. D., Ylostalo, J., Lynch, P. J., Smith, J., Perry, A., Peister, A., Wang, M. Y. & Prockop, D. J. (2003) Proc. Natl. Acad. Sci. USA 100, 2397-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okabe, M., Ikawa, M., Kominami, K., Nakanishi, T. & Nishimune, Y. (1997) FEBS Lett. 407, 313-319. [DOI] [PubMed] [Google Scholar]
- 23.Religa, P., Bojakowski, K., Maksymowicz, M., Bojakowska, M., Sirsjo, A., Gaciong, Z., Olszewski, W., Hedin, U. & Thyberg, J. (2002) Transplantation 74, 1310-1315. [DOI] [PubMed] [Google Scholar]
- 24.Gustashaw, K. M. (1991) Cytogenetics Laboratory Manual, ed. M. J. Barch (Raven, New York), pp. 205-298.
- 25.Goldstein, R. S., Drukker, M., Reubinoff, B. E. & Benvenisty, N. (2002) Dev. Dyn. 225, 80-86. [DOI] [PubMed] [Google Scholar]
- 26.Sedmera, D., Reckova, M., deAlmeida, A., Coppen, S. R., Kubalak, S. W., Gourdie, R. G. & Thompson, R. P. (2003) Anat. Rec. 274A, 773-777. [DOI] [PubMed] [Google Scholar]
- 27.Weimann, J. M., Johansson, C. B., Trejo, A. & Blau, H. M. (2003) Nat. Cell Biol. 5, 959-966. [DOI] [PubMed] [Google Scholar]
- 28.Bellairs, R. & Osmond, M. (1997) The Atlas of Chick Development (Academic, London).